Background: NF-κB post-translational modifications contribute to gene-specific transcriptional activation.
Results: Symmetrical dimethylation of NF-κB p65 by PRMT5 enhances CXCL10 induction in response to TNF-α.
Conclusion: Arginine methylation of p65 is a novel mechanism regulating inflammatory chemokine induction.
Significance: Methylation of specific NF-κB arginine residues contributes to promoter targeting and transcriptional activation.
Keywords: Endothelial Cell, Gene Expression, NF-Kappa B (NF-KB), Post-Translational Modification (PTM), Protein Methylation, Tumor Necrosis Factor (TNF)
Abstract
The chemokine CXCL10/IP-10 facilitates recruitment of Th1-type leukocytes to inflammatory sites. In this study, we show that the arginine methyltransferase PRMT5 is critical for CXCL10 transcription in TNF-α-activated human endothelial cells (EC). We found that depletion of PRMT5 results in significantly reduced levels of CXCL10 mRNA, demonstrating a positive role for PRMT5 in CXCL10 induction. Chromatin immunoprecipitation experiments revealed the presence of the symmetrical dimethylarginine modification catalyzed by PRMT5 associated with the CXCL10 promoter in response to TNF-α. However, symmetrical dimethylarginine-modified proteins were not detected at the promoter in the absence of PRMT5, indicating that PRMT5 is essential for methylation to occur. Furthermore, NF-κB p65, a critical driver of TNF-α-mediated CXCL10 induction, was determined to be methylated at arginine residues. Crucially, RNAi-mediated PRMT5 depletion abrogated p65 methylation and CXCL10 promoter binding. Mass spectrometric analysis in EC identified five dimethylated arginine residues in p65, four of which are uncharacterized in the literature. Expression of Arg-to-Lys point mutants of p65 demonstrated that both Arg-30 and Arg-35 must be dimethylated to achieve full CXCL10 expression. In conclusion, we have identified previously uncharacterized p65 post-translational modifications critical for CXCL10 induction.
Introduction
Activation of endothelial cells (EC)2 in response to inflammatory agonists lead to the production of growth, regulatory, and vasoactive substances that are not produced by unstimulated EC (1). Studies from our laboratory and others identify arginine methylation as a contributing factor in induction of the inflammatory genes E-selectin, VCAM-1, IL-2, CXCL8/IL-8, and IκBα (2–5). Arginine residues in mammalian proteins can carry zero, one, or two methyl groups on their terminal (ω) nitrogen atoms. Methylation eliminates a potential hydrogen bond, adds bulk, and increases hydrophobicity but does not alter arginine's positive charge. Methylarginine residues may impede or promote protein-substrate interactions (6–9). Protein arginine methyltransferase 5 (PRMT5) is the major enzyme that catalyzes SDMA formation in mammalian cells and also produce monomethylated products. PRMT5 regulates transcription through multiple mechanisms, including alterations in chromatin structure and methylation of proteins involved in transcription, elongation, and splicing (7).
To complement our previous studies of arginine methylation in the expression of leukocyte adhesion molecules (2), we chose to examine roles of PRMT5 in expression of chemoattractive cytokines (chemokines). Most proinflammatory chemokines are secreted proteins that promote recruitment, adherence, and extravasation of circulating leukocytes (10, 11). We determined that the chemokine CXCL10 requires PRMT5 to achieve full induction in response to TNF-α. CXCL10 has been extensively studied since its discovery in 1985 by Luster et al. (12) due to its contributions to numerous pathologies involving Th1-type inflammation, including atherosclerosis, coronary artery disease, multiple sclerosis, rheumatoid arthritis, psoriasis, asthma, and immune responses to solid organ transplant and infections (10). Despite its prominence in such a wide variety of pathologies, only one published study to our knowledge has examined CXCL10 expression in EC (13). CXCL10 is rapidly induced in response to TNF-α, IFN-α/β/γ, IL-1β, or LPS (10). Secreted CXCL10 recruits and retains Th1 (type-1 helper) CD4+ T cells, CD8+ cytotoxic effector cells, natural killer, natural killer T cells, plasmocytoid dendritic cells, and some B-cell subsets at inflammatory lesions (10, 11). CXCL10 also has non-immune cell effects as a potent smooth muscle cell mitogen and chemotactic agent and as a vascular angiostatic factor (14, 15).
CXCL10 induction is driven in part by NF-κB transcription factors known as master regulators of inflammation and immunity (16). In canonical NF-κB signaling, the latent cytosolic transcription factor NF-κB is activated by kinases that phosphorylate p65 and trigger the degradation of inhibitory IκB subunits. These events free NF-κB to translocate into the nucleus where it associates with κB sites in target promoters (16).
Multiple types of post-translation modifications of p65 are established, including roles for phosphorylation, acetylation, and ubiquitination. Roles of many of these modifications have been identified, including regulators of protein localization, DNA-binding affinity, interactions with other proteins, and the duration and strength of transcription (16, 17). However, a major unresolved question in the study of NF-κB remains how the various post-translational modifications of NF-κB enable specific gene activation, differential kinetics, transcription magnitude, and inducer-specific responses (18). Many of these processes are governed by post-translational modifications of NF-κB. Here, we report that the arginine methyltransferase PRMT5 post-translationally modifies the p65 subunit of the NF-κB Rel-homology domain, a step imperative to the CXCL10 induction by TNF-α.
EXPERIMENTAL PROCEDURES
Reagents
Primary human EC were isolated from discarded patient samples as described previously (19). Fetal bovine serum was from Atlas Biologicals (Fort Collins, CO). Targefect F-2 and peptide enhancer transfection reagents were purchased from Targeting Systems (El Cajon, CA). siRNAs complementary to the coding sequence of PRMT5 (sense, 5′-GAGGGAGUUCAUUCAGGAAUU-3′) and the NF-κB p65 3′-UTR (sense, 5′-GGAUUCAUUACAGCUUAAUUU-3′) were designed using the Whitehead Institute siRNA design tool. siRNAs were synthesized by Ambion and contain the Silencer Select modifications. Nontargeting control Silencer Select siRNA (4390843) was purchased from Ambion. ChIP was performed using a kit from Millipore (Billerica, MA; catalog no. 17-295). Antibodies used for ChIP, IP, and Western blots included anti-PRMT5 (Millipore, catalog no. 07-405), anti-p65 (Millipore, catalog no. 06-418), anti-dimethyl-arginine, symmetric (SYM10, Millipore, catalog no. 07-412), anti-HOXA9 (Millipore, catalog no. 07-178), anti-FLAG M2 (Sigma Aldrich), anti-mouse IgG1 (Cell Signaling), and anti-normal rabbit IgG (Millipore, catalog no. 12-370). Antibodies used for Western blot included anti-PRMT5 (sc-22132; Santa Cruz Biotechnology), anti-GAPDH (sc-20357; Santa Cruz Biotechnology), and anti-α-tubulin (T5168; Sigma). Recombinant TNF-α from R&D Systems was used at 2 ng/ml for all experiments. Human p65 cDNA was acquired from Addgene (construct 21966). PCR was performed to insert an N-terminal KpnI site followed by a FLAG sequence (amino acid sequence: DYKDDDDK) in-frame with the start codon. The resulting FLAG-tagged cDNA was subcloned into the KpnI and HindIII sites of pCDNA3. The FLAG-tagged fusion protein is expressed under control of the CMV promoter.
Cell Culture
Early passage (P2–4) EC pooled from multiple individual biological samples were used for all experiments. Cells were maintained in MCDB 107 medium augmented with 15% FBS, 150 μg/ml endothelial cell growth supplement, and 90 μg/ml heparin.
Transient Transfection
Cells were transfected within 24 h of passage. Both siRNA and cDNA were transfected in serum-free DMEM using targefect F-2 (5 μl/ml) and peptide enhancer (5 μl/ml) from Targeting Systems (El Cajon, CA). siRNA was transfected at a final concentration of 50 nm. cDNA was transfected at 250 ng/ml. All transfections were incubated at 37 °C for 4 h, followed by replacement of the DMEM-transfection solution with complete medium containing 15% FBS. Cells were cultured for 24–48 h to allow for cDNA expression or protein knockdown.
Preparation of Nuclear and Cytosolic Extracts
Cells were grown to confluence on 150-mm plates. Cells were separated into nuclear and cytosolic fractions using the CellLytic NuCLEAR Extraction Kit (Sigma) according to the manufacturer's instructions. Briefly, cells were harvested with a cell scraper, swelled in hypotonic lysis buffer, and lysed with IGEPAL-630. Following centrifugation, the supernatant was saved as the cytosolic fraction. The pellet was washed three times with hypotonic lysis buffer and lysed in 420 mm NaCl high salt extraction buffer to collect the crude nuclear fraction.
Immunoprecipitation
Cells were lysed in 1× radioimmune precipitation assay buffer (1% Nonidet P-40, 0.1% SDS, 50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 150 mm NaCl) containing protease inhibitors. Endogenous proteins were immunoprecipitated using anti-PRMT5 or anti-p65 antibody from Millipore and protein A/G-agarose beads (Pierce). Overexpressed N-terminal tagged FLAG-p65 was pulled down using anti-FLAG-agarose magnetic beads (Sigma).
Western Blotting
Lysates were resolved on 10% bis-tris acrylamide gels (2). Proteins were transferred to PVDF membranes and blocked with 5% nonfat milk or protein-free blocking buffer (Pierce; catalog no. 37570). Blots were washed with 1× TBST or milli-Q water and incubated with the appropriate HRP-conjugated secondary antibody.
RNA Purification and Real-time Quantitative PCR
After transfection, cells were allowed to recover for 48 h. Following TNF-α treatment, cells were lysed with 350 μl of buffer RLT containing 1% 2-mercaptoethanol and processed with the RNeasy kit (Qiagen) to collect RNA. cDNA was synthesized from RNA using SuperScript First-Strand Synthesis reagents (Invitrogen). cDNA was diluted 6-fold, and real-time PCR was performed with Fast SYBR Green PCR Reagents (PE Applied Biosystems) using an ABI StepOnePlus quantitative real-time PCR machine. Primers are separated by an intron and amplified 100–150 bp of spliced products. Results were normalized to GAPDH expression following the −ΔΔCt method. Primer sequences are CXCL10/IP-10, 5′-GCAGAGGAACCTCCAGTCTCAGCA-3′ (forward) and 5′-GCTGATGCAGGTACAGCGTACAGT-3′ (reverse); CCL2/MCP-1, 5′-TCTCTTCCTCCACCACTAATGCA-3′ (forward) and 5′-GGCTGAGACAGCACGTGGAT-3′ (reverse); CX3CL1/fractalkine, 5′-CATCATGCGGCAAACGCGCA-3′ (forward) and 5′-AGCAGCCTGGCGGTCCAGAT-3′ (reverse); CXCL8/IL-8, 5′-GGCCGTGGCTCTCTTGGCAG-3′ (forward) and 5′-TGTGTTGGCGCAGTGTGGTCC-3′ (reverse); GAPDH, 5′-TCAACAGCGACACCCACTCC-3′ (forward) and 5′-TGAGGTCCACCACCCTGTTG-3′ (reverse); 18 S rRNA, 5′-GCTTAATTTGACTCAACACGGGA-3′ (forward) and 5′-AGCTATCAATCTGTCAATCCTGTC-3′ (reverse); and CXCL10 pre-mRNA, 5′-CCACGTGTTGAGATGTGAGTGA-3′ (forward) and 5′-TGGCATACGCAGTTCTGAAGTCAG-3′ (reverse). For the mRNA stability experiment, actinomycin D (Sigma) was used at 1 μg/ml. Amplification, melt curves, and sequenced products indicate a single product identical to the reference sequence.
CXCL10 ELISA
Cells were plated in six-well dishes and transfected. After 24 h, 1 ml of fresh was applied. At 40 h, cells were stimulated with TNF-α for 8 h. At the end of treatment, conditioned medium was collected and assayed for the presence of CXCL10 using a sandwich ELISA kit (R&D Systems; DIP-100) and read with an iMark plate reader (Bio-Rad).
ChIP
Cells were cultured in 10-cm dishes, treated with TNF-α, and cross-linked with 1% formaldehyde (Sigma) in MCDB medium. Immunoprecipitated DNA was purified using the PrepEase DNA Cleanup kit (Affymetrix, Santa Clara, CA). Primers for the CXCL10 proximal promoter are 5′-AGGAGCAGAGGGAAATTCCGTAAC-3′ (forward) and 5′-AACGTGGGGCTAGTGTGCCA-3′ (reverse). They amplify a 182-bp fragment between −186/−5 containing the κB2, κB1 (distal and proximal κB sites, respectively), AP-1, CAAT, and TATA elements. The proximal promoter of CX3CL1 containing the κB site was amplified by 5′-GGCATGTTCCCAGCTTGTGGCAGG-3′ (forward) and 5′-GTTGCCAAGGAACCAAGCCGGC-3′ (reverse). The MCP-1 promoter fragment containing the κB site was amplified by 5′-CCCATTTGCTCATTTGGTCTCAGC-3′ (forward) and 5′-GCTGCTGTCTCTGCCTCTTATTGA-3′ (reverse). The ChIP assay was standardized using E-selectin promoter primers (20) and NF-κB p65 antibody. DNA was quantified in 15 μl quantitative real-time PCR reactions using Power SYBR green (Applied Biosystems) with 40 cycles of amplification.
CXCL10 Promoter Activity
Cells were plated in triplicate in 24-well plates and transfected with a plasmid encoding −875 to +97 bp of the human CXCL10 promoter (GL-IP10) upstream of firefly luciferase in promoterless pGL3-basic. The GL-IP10 construct was a gift of Richard Ransohoff (21). Cells were cultured for 24 h and then treated with TNF-α for 6 h, washed twice with MCDB medium, and lysed in 60 μl/well passive lysis buffer (Promega). 20 μl of lysate was pipetted in triplicate into 96-well luciferase plates, activated with luciferase substrate (Promega), and measured with a Dynex luminometer.
Mass Spectrometry
Coomassie-stained bands of overexpressed p65 were excised (∼65 kDa) and digested in-gel with trypsin, chymotrypsin, or GluC, and analyzed by capillary column LC-tandem mass spectrometry to identify methylated peptides. Enzymatic digestion was carried out at room temperature overnight. We used a Finnigan LTQ (linear trap quadrapole) ion trap mass spectrometer (ThermoFinnigan, San Jose, CA) and a self-packed 9 cm × 75 μm inner diameter Phenomenex Jupiter C18 reversed phase capillary chromatography column. Peptides were analyzed using collision-induced dissociation spectra to search the NCBI non-redundant protein database and manually interpreted with reference to the Sequest and BLAST programs as needed.
In Vitro Methyltransferase Assay
Active, recombinant human PRMT5 expressed in HEK293 (Sigma-Aldrich, SRP0145), full-length recombinant human p65 produced in HEK293 (OriGene, TA301086), and the methyl donor S-adenosylmethionine (New England Biolabs, B9003S) were incubated for 90 min at 37 °C in a 30-μl volume according to published methods (2). In each reaction, 0.3 μg of PRMT5, 1.0 μg of p65, and S-adenosylmethionine was added to a final concentration of 80 μm. Buffer conditions were 25 mm HEPES (pH 7.6), 5 mm MgCl2, 100 mm NaCl, and 2 mm dithiothreitol (DTT). Reactions were terminated with the addition of 10 μl of 4× Laemmli buffer. Arginine methylation was detected by immunoblotting for SDMA.
Site-directed Mutagenesis
A FLAG tag was added to the N terminus of the RelA-pCMV4 construct (22) (Addgene construct 21966) via PCR and digestion with Kpn1 and Bsm1. Arginine residues identified as methylation targets were mutated to lysine via the GeneArt Mutagenesis System (Invitrogen).
Arginine-to-lysine NF-κB Substitution Assay
Arginine-to-lysine mutant constructs were co-transfected along with CMV-lacZ reporter plasmid and 3′-UTR targeting sip65, introduced to knockdown endogenous p65. Cells were maintained for 45 h and then stimulated with TNF-α for 3 h. RNA was collected, reverse-transcribed, and analyzed for chemokine expression.
Statistical Analysis
Results are presented as means ± S.E. Comparisons between groups were evaluated by one- or two-way analysis of variance followed by Bonferroni post hoc tests to evaluate pairwise comparisons. Multiple comparisons were performed by Student's t test. A p value ≤ 0.05 was selected as the cutoff for significance. p values are expressed in the figures as follows: *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.005. For all experiments, n = 3–4, with the exception of the mass spectrometric analysis (n = 2).
RESULTS
PRMT5 Positively Regulates TNF-α-induced CXCL10 Gene Transcription in EC
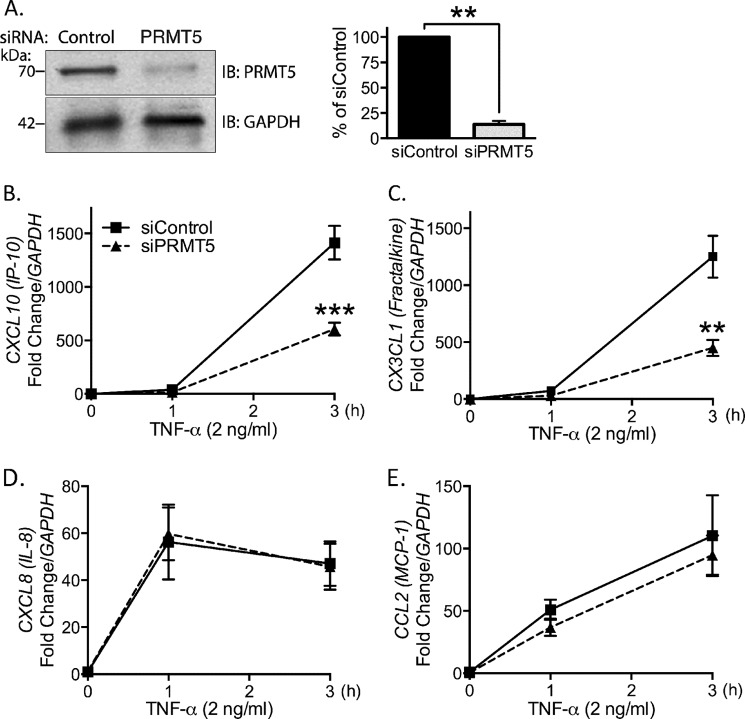
Our group has reported previously that PRMT5 positively regulates expression of leukocyte adhesion molecules VCAM-1 and E-selectin in TNF-α-stimulated EC (2). Here, we extended our study and explored the regulatory role of PRMT5 in the TNF-α induction of chemokines that are involved in vascular disease pathogenesis. To identify the role of PRMT5 in inflammatory chemokine gene expression, we compared TNF-α-induced mRNA levels of chemokines in PRMT5 intact versus PRMT5-depleted EC. Transfection of PRMT5 siRNA yielded >80% knockdown of PRMT5 protein (Fig. 1A). Depletion of PRMT5 in EC led to substantial decrease (>60%) in induction of both CXCL10 (Fig. 1B) and CX3CL1/fractalkine (Fig. 1C) message levels. Thus, PRMT5 is necessary to achieve full induction of these chemokines in stimulated EC. In contrast, expression of both CXCL8/IL-8 (Fig. 1D) and CCL2/MCP-1 (Fig. 1E) were unaffected by the absence of PRMT5. Thus, we conclude that PRMT5 is required for the expression of specific proinflammatory chemokines, including CXCL10 (Fig. 1B) and CX3CL1 (Fig. 1C) in EC. In this study, we focused on elucidating the mechanism of PRMT5's contribution to CXCL10 expression.
FIGURE 1.
PRMT5 is necessary for the induction of CXCL10 and CX3CL1, but not IL-8 or MCP-1 in TNF-α-stimulated EC. A, Western blot analysis of PRMT5 levels from control and siPRMT5 transfected cells are shown in A, and quantified at right (p < 0.01). B–E, real-time quantitative PCR was performed to measure mRNA of the following chemokines from control and siPRMT5 transfected EC. B, CXCL10 (p < 0.0001); C, CX3CL1/fractalkine (p < 0.001); D, CXCL8/IL-8 (p > 0.05); and E, CCL2/MCP1 (p > 0.05). IB, immunoblot; **, p < 0.01; ***, p < 0.005. Means are presented as S.E. from 3–4 independent experiments.
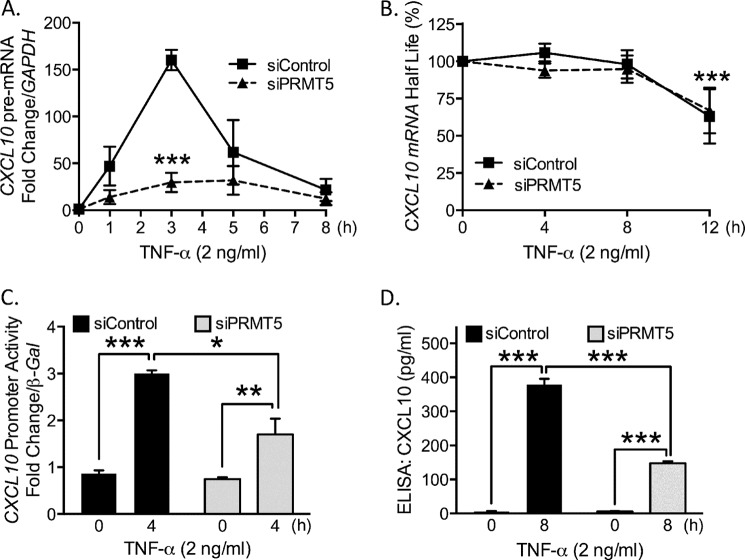
To differentiate between potential roles for PRMT5 in transcriptional versus post-transcriptional processes, we measured CXCL10 pre-mRNA in TNF-α-stimulated EC with quantitative real-time PCR. The levels of CXCL10 pre-mRNA in PRMT5-depleted cells were significantly lower compared with control cells (Fig. 2A), indicating that regulation occurs at the level of transcription. Under some circumstances, chemokine production is modulated by regulating mRNA half-life (23). To assess a possible role for PRMT5 in CXCL10 mRNA stability, we blocked de novo transcription with actinomycin D and observed no significant difference in message stability with and without PRMT5 (Fig. 2B). To provide further evidence for the role of PRMT5 in CXCL10 expression, we transfected into EC a promoter-reporter construct consisting of a 1-kb fragment of the CXCL10 promoter proximal to the transcription start site cloned upstream of the luciferase reporter. Results from this assay showed significantly diminished promoter activity from PRMT5-depleted cells compared with control (Fig. 2C). We also analyzed CXCL10 protein levels via ELISA assay in EC conditioned medium following 8 h of TNF-α stimulation. EC lacking PRMT5 showed significant reduction as compared with control siRNA-transfected cells (Fig. 2D). This finding is consistent with the mRNA result presented in Fig. 1B. Taken together, we conclude that PRMT5 regulates TNF-α-stimulated induction of CXCL10 at the transcriptional level.
FIGURE 2.
Knockdown of PRMT5 results in reduced levels of CXCL10 at the transcriptional level. A, CXCL10 pre-mRNA levels were measured following TNF-α-stimulation (p < 0.005). B, CXCL10 mRNA stability was quantified following stimulation (3 h) and application of actinomycin D (1 μg/ml). C, CXCL10 promoter activity was measured by luciferase luminescence intensity (p < 0.005). D, ELISA was used to measure levels of CXCL10 in conditioned medium (p < 0.0005; *, p < 0.05; **, p < 0.01; ***, p < 0.005). Error bars represent S.E. (n = 3).
PRMT5-catalyzed Symmetrically Dimethylated Proteins Associate with the CXCL10 Promoter
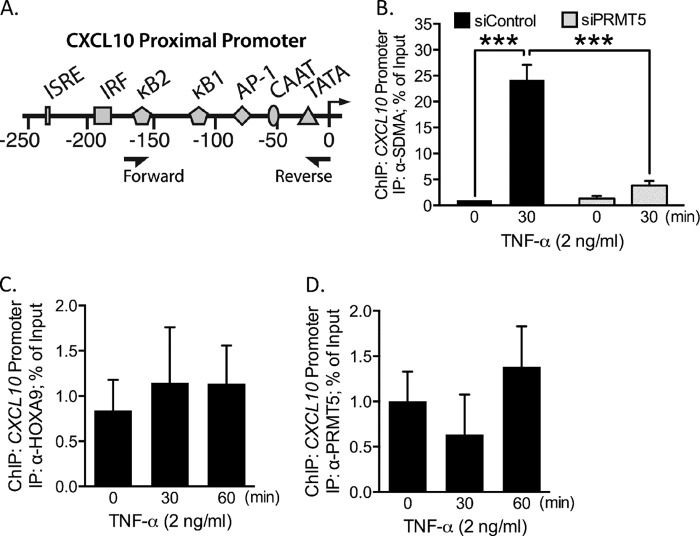
Considering the positive contribution of PRMT5 to CXCL10 transcription, we next asked whether the SDMA modification could be detected at the CXCL10 promoter. To address this question, we employed ChIP assays using an antibody specific for SDMA, and PCR primers that amplify a region of the proximal promoter critical for CXCL10 induction (Fig. 3A). As shown in Fig. 3B, our ChIP results demonstrated the presence of SDMA-containing proteins on the CXCL10 proximal promoter in TNF-α-stimulated EC. Importantly, when PRMT5 is knocked down in stimulated cells, promoter amplification was at background level (Fig. 3B). These results therefore suggest that PRMT5 symmetrically dimethylates at least one protein associated with the CXCL10 promoter in response to TNF-α-stimulation in EC.
FIGURE 3.
PRMT5 activity leads to the association of SDMA-containing proteins with the CXCL10 promoter following TNF-α stimulation. A, illustration depicting the CXCL10 promoter 250 kb upstream of the transcription start site. Major transcription factor binding sites are noted, including the two functional κB sites, which are numbered according to their position relative to the transcription start site. Arrowheads under the promoter denote the ChIP primer binding sites. B, the CXCL10 promoter was immunoprecipitated in a ChIP assay using anti-SDMA modification antibody. PRMT5 depletion blunted TNF-α-induced association of SDMA-containing proteins with the CXCL10 promoter. CXCL10 promoter amplification was also performed in ChIP assays with anti-PRMT5 (C) (p > 0.05), and anti-HOXA9 (D) (p > 0.05) IP antibodies. ***, p < 0.005. Error bars represent S.E. of three independent experiments.
In a previous investigation, we showed that PRMT5 and HOXA9 associate on the promoters of E-selectin and VCAM-1 upon TNF-α exposure in EC. We investigated whether CXCL10 induction occurs though the same mechanism (2). We performed ChIP experiments to assess whether HOXA9 could be identified together with PRMT5 at the CXCL10 promoter. We did not detect CXCL10 promoter enrichment of either HOXA9 (Fig. 3C) or PRMT5 (Fig. 3D) in unstimulated or stimulated EC. Together, the ChIP results indicated that PRMT5 catalyzes methylation of at least one DNA-binding protein that associates with the CXCL10 promoter following TNF-α activation of EC. Methylation appears to occur prior to association with the CXCL10 promoter.
NF-κB p65 Is Symmetrically Dimethylated by PRMT5
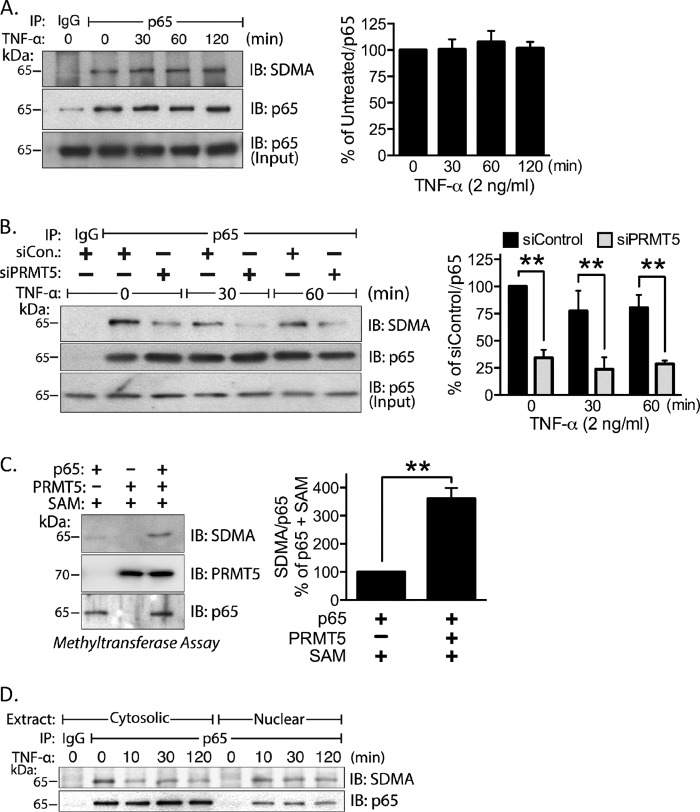
To identify SDMA-modified proteins that associate with the CXCL10 promoter following TNF-α activation, we immunoprecipitated transcription factors known to associate with the CXCL10 proximal promoter (Fig. 3A) such as NF-κB p65, p300, and STAT1 (21, 24). Of these factors, only NF-κB p65 was symmetrically dimethylated in both the stimulated and unstimulated states as detected by the anti-symmetrical dimethylarginine antibody (Fig. 4A, and data not shown). To test whether the arginine methylation detected on p65 is catalyzed by PRMT5, we depleted PRMT5 with siRNA, immunoprecipitated p65, and assessed the SDMA levels by Western blot. PRMT5 knockdown resulted in a substantial decrease (>80%) in arginine methylation levels of both untreated and TNF-α-stimulated lysates (Fig. 4B). We performed an in vitro methyltransferase assay using human recombinant PRMT5 and p65 to test whether PRMT5 is capable of directly methylating p65. We observed an increase in p65 methylation as detected by the anti-SDMA antibody following the methyltransferase reaction (Fig. 4C). To assess whether arginine methylated forms of p65 translocated to the nucleus, cytosolic and nuclear fractions from control and TNF-α-treated cells were prepared. Endogenous p65 was immunoprecipitated from each fraction and analyzed by Western blot. Results show that the SDMA-containing forms of p65 are located in the cytosol in the absence of TNF-α. After stimulation, they migrate to the nucleus (Fig. 4D).
FIGURE 4.
NF-κB p65 is methylated by PRMT5. A, immunoprecipitated endogenous p65 was probed by Western blot for the SDMA modification following TNF-α stimulation. Densitometry of the SDMA signal is shown at right (p > 0.05). B, endogenous p65 was immunoprecipitated from control and siPRMT5-transfected cells and probed for SDMA by Western blot (left) and quantified by densitometry (right; p < 0.01). C, in vitro methyltransferase assay was analyzed by Western blot with anti-SDMA antibody (left) and quantified in the right panel (p < 0.01). D, endogenous p65 was immunoprecipitated from cytosolic and nuclear extracts and probed with the anti-SDMA antibody. SAM, S-adenosylmethionine; IB, immunoblot; **, p < 0.01. Error bars represent S.E. of three independent experiments.
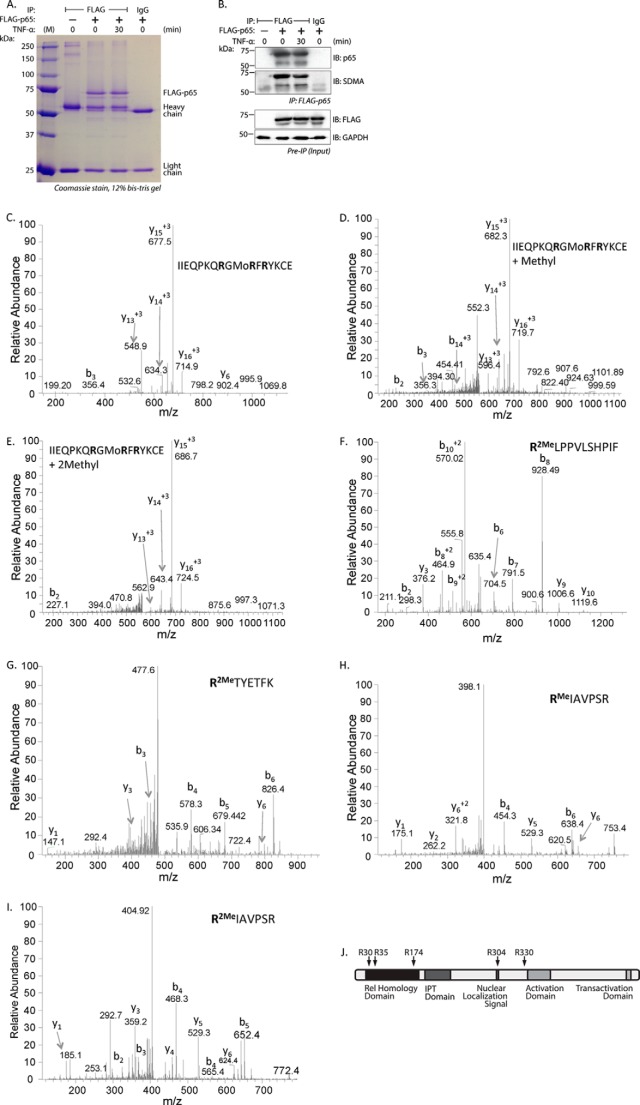
NF-κB p65 Is Methylated at Five Arginine Residues
We utilized mass spectrometric analysis to identify the specific dimethylarginine residues in p65. FLAG-tagged p65 was overexpressed in EC, immunoprecipitated, and subsequently visualized on a Coomassie-stained gel (Fig. 5A). Simultaneously, we probed a small sample of these immunoprecipitates for SDMA by Western blot to detect the modification levels in these samples (Fig. 5B). p65-containing bands in the Coomassie-stained gel were digested with trypsin, chymotrypsin, and GluC and analyzed by mass spectrometry to detect mono- and dimethylarginine. Comparison between the mass spectra of the 23IIEQPKQRGMoRFRYKCE39 unmodified peptide (Fig. 5C) versus its monomethylated product-containing (Fig. 5D) and SDMA-containing forms (Fig. 5E) suggested methylarginine residues at Arg-30 or Arg-35. Mass shift consistent with dimethylation was detected at Arg-174 in the 174RLPPVLSHPIF184 peptide (Fig. 5F). Dimethylation of Arg-304 is present in the 304RTYETFK310 peptide (Fig. 5G). Both monomethylation (Fig. 5H) and dimethylation (Fig. 5I) were found in the 330RIAVPSR336 peptide at Arg-330. In all, we identified four or five different arginine loci containing either monomethylated products or dimethylarginine (Table 1). These residues are Arg-30 or Arg-35, and Arg-174 present in the Rel homology domain, Arg-304 in the nuclear localization signal, and Arg-330, present in an interdomain region (Fig. 5J). We did not observe any significant change in methylation of any of these residues by TNF-α.
FIGURE 5.
Five arginine residues are dimethylated in EC transfected with wild type p65. A, lysates from wild type p65-transfected EC were immunoprecipitated with anti-FLAG antibody, resolved by SDS-PAGE, and stained with Coomassie Blue. B, top panel, a fraction of the immunoprecipitate in A was probed separately for anti-p65 by immunoblot. The membrane was then stripped and reprobed with anti-SDMA. Bottom panel, preimmunoprecipitated input material was probed with anti-FLAG antibody to show FLAG-p65 expression. The lower molecular mass band in A and B appears to be related to FLAG-p65. C–E, full-length FLAG-p65 bands were analyzed by mass spectrometry for the presence of methylarginine residues. Shown are MS/MS spectra of the unmethylated (C), monomethylated (D), and dimethylated (E) peptide 23IIEQPKQRGMoRFRYKCE39 containing Arg-30 and Arg-35. F, MS/MS spectra of the 174RLPPVLSHPIF184 peptide containing Arg-174. G, MS/MS spectra of the 304RTYETFK310, including Arg-304. H, MS/MS spectra of the 330RIAVPSR336 containing Arg-330. I, MS/MS spectra of the 330RIAVPSR336 peptide. J, the location of methylated arginine residues are shown schematically on human p65. IB, immunoblot; IPT, Ig-like, plexin, transcription factor domain.
TABLE 1.
Methylated p65 peptides obtained from MS/MS experiments
Presented here are the proteases used and corresponding methylarginine-containing peptides of p65 detected in the MS/MS experiments.
| Peptide | Enzyme | Residue | Type |
|---|---|---|---|
| IIEQPKKQRGMoRFRYKCE | GluC | Arg-30, Arg-35 | Mono, dimethyl |
| PLRLPPVLSHPIFDNR | Trypsin | Arg-174 | Dimethyl |
| RLPPVLSHPIF | Chymotrypsin | Arg-174 | Dimethyl |
| RLPPVLSHPIFDNRAPN | Chymotrypsin | Arg-174 | Dimethyl |
| RTYETFK | Trypsin | Arg-304 | Dimethyl |
| RIAVPSR | Trypsin | Arg-330 | Mono, dimethyl |
Knockdown of PRMT5 Reduces p65 Association with the CXCL10 Promoter
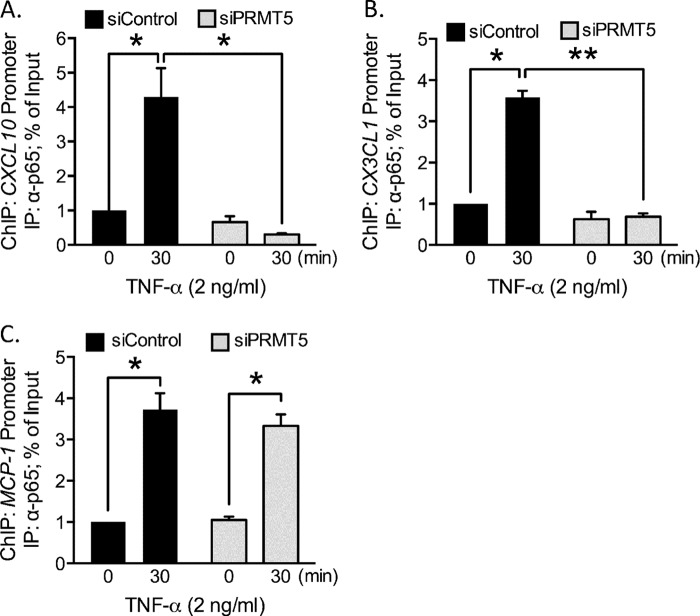
We performed a ChIP assay in the absence of PRMT5 and assessed the association of p65 with the CXCL10 promoter. Under control transfection conditions, we observed an ∼4-fold increase in p65 promoter association upon addition of TNF-α. In contrast, in the absence of PRMT5, promoter amplification was not altered significantly between the TNF-α treated versus unstimulated conditions (Fig. 6A). A similar pattern was observed with p65 association with the CX3CL1 promoter (Fig. 6B). In contrast, p65 association with the MCP-1 promoter upon TNF-α stimulation was similar in both the control and PRMT5 knockdown conditions (Fig. 6C).
FIGURE 6.
PRMT5 is necessary for p65 association with the CXCL10 promoter. A–C, in a ChIP assay, protein-DNA complexes were immunoprecipitated with anti-p65 antibody from control and PRMT5 siRNA-transfected, TNF-α-stimulated EC. A, enrichment of the CXCL10 promoter was enhanced upon TNF-α stimulation in control-transfected cells (p < 0.05), but not in PRMT5-depleted conditions. B, p65 association with the CX3CL1 promoter was also increased upon application of TNF-α in control transfected cells (p < 0.05). C, association of p65 with the MCP-1 promoter was not altered by PRMT5 depletion. *, p < 0.05; **, p < 0.01. Error bars represent S.E. of three independent experiments.
Methylation of p65 Rel Homology Domain Is Necessary for CXCL10 Induction
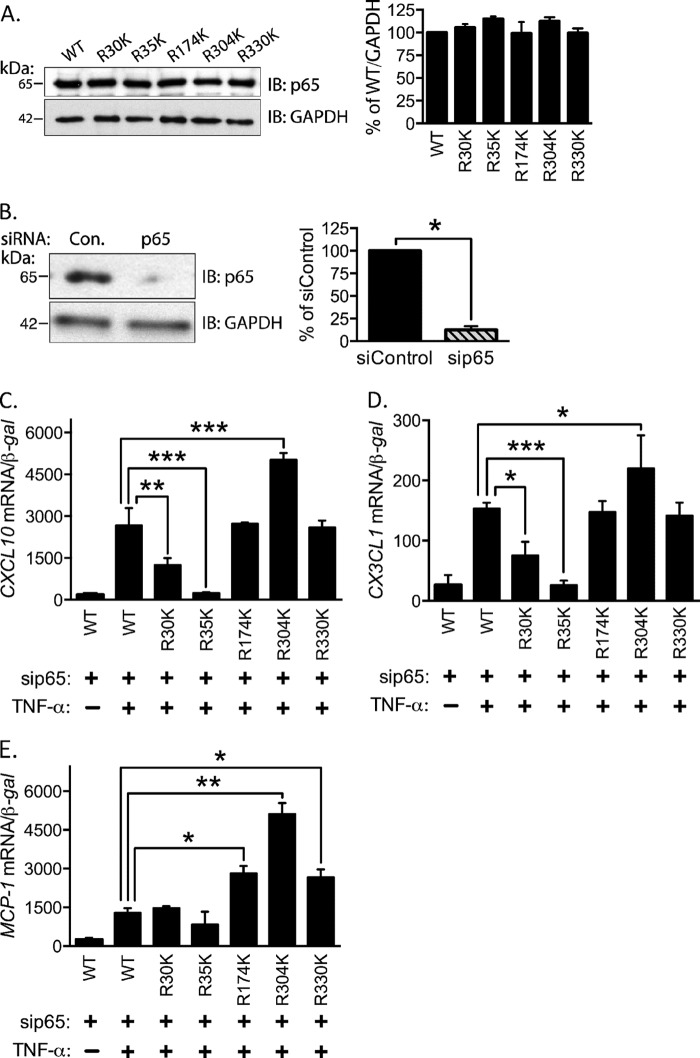
We next tested the hypothesis that at least one arginine residue must be methylated to induce CXCL10 expression by TNF-α using site-directed arginine to lysine point mutants. This conservative mutation preserves the positive charge at the position but prevents methylation by arginine methyltransferases. Following transfection in EC, the mutant constructs express p65 at levels comparable with the wild type (Fig. 7A). Additionally, we used siRNA targeted against the 3′-UTR of p65 to knockdown endogenous protein levels without affecting expression of the transfected p65 cDNA (Fig. 7B). To identify specific dimethylarginine residues required for CXCL10 induction, EC were transfected with either wild type or mutant p65 cDNA along with the above mentioned p65 siRNA and treated with TNF-α. RNA was isolated and analyzed for specific chemokine mRNA expression. R30K and R35K mutants were ineffective in recapitulating wild type levels of CXCL10 expression (Fig. 7C). Expression of R304K significantly increased CXCL10 expression, and all other Arg-to-Lys mutants led to levels of CXCL10 not statistically different from the wild type reconstituted levels (Fig. 7C). Similar results were obtained for expression of CX3CL1 (Fig. 7D). In contrast, expression of MCP1 by R30K and R35K mutants was not different from wild type, indicating these mutants are functional (Fig. 7E). R174K, R304K, and R330K enhanced MCP1 expression relative to wild type (Fig. 7E).
FIGURE 7.
SDMA-p65 is critical for CXCL10 induction. A, p65 arginine-to-lysine mutants at Arg-30, Arg-35, Arg-174, Arg-304, and Arg-330 were expressed and immunoblotted for p65. Quantification of the immunoblot is shown at the right (p > 0.05). B, lysates from p65 3′-UTR targeting siRNA and control transfection are presented (left) and quantified (right; p < 0.05). C–E, knockdown of endogenous p65 and reconstitution with R30K, R35K, R174K, R304K, and R330K was performed in EC. TNF-α-mediated induction of CXCL10 (C), CX3CL1 (D), and MCP-1 (E) in EC reconstituted with p65 mutants is shown. *, p < 0.05; **, p < 0.01; ***, p < 0.005. Error bars represent S.E. of 3–4 independent experiments.
DISCUSSION
We previously demonstrated the requirement of PRMT5 for the induction of the EC-leukocyte adhesion molecules E-selectin and VCAM-1 (2). Here, we report a critical role of PRMT5 in specific chemokine gene induction in EC. The mechanism we report here differs from our previous report of E-selectin and VCAM-1 induction in that HOXA9 is not involved and that PRMT5 is not detected by ChIP on the promoter itself. We report that PRMT5 is capable of methylating NF-κB p65 both in vitro and in cultured EC. Mass spectroscopy approaches identify that NF-κB p65 is methylated at multiple residues in EC. We show that p65 recruited to the CXCL10 and CX3CL1 promoters in response to TNF-α was methylated by PRMT5 at arginine residues. Furthermore, we show that arginine methylation of Arg-30 and Arg-35 of the p65 Rel homology domain is critical for TNF-α-stimulated induction of the proinflammatory chemokine CXCL10. Together, we propose that methylation of p65 at Arg-30 and Arg-35 by PRMT5 enhances p65 association and transcription of a subset of TNF-responsive proinflammatory genes in EC.
Most reports of the role of PRMT5 in transcription describe PRMT5 as a transcriptional repressor that operates through SDMA modification of histones H2A, H3, and H4 (7, 25). In contrast, methylation of non-histone DNA-binding proteins such as HOXA9 (2) and p53 (26) results in transcriptional enhancement. In CXCL10 expression, we show that SDMA-containing proteins associate with the promoter only when PRMT5 is present and following TNF-α simulation. Through immunoprecipitation, mass spectrometry, and siRNA-based approaches, we demonstrate that PRMT5 methylates NF-κB p65 at multiple arginine residues. Methylation arginine residues 35, 174, 304, and 330 have not been reported in the literature. The fifth, Arg-30, was recently reported by Wei et al. (27) and is also catalyzed by PRMT5. Arginine methylated p65 is present at low abundance. However, small amounts of post-translationally modified species are sufficient to initiate physiological responses, particularly in the context of transcription. Through replacement of endogenous p65 with Arg-to-Lys mutants, we show that dimethylation of Arg-30 and Arg-35 is critical for TNF-α-induced CXCL10 expression in EC. Of the two residues, methylation of Arg-35 appears to be of greater significance in CXCL10 induction. Both R30K and R35K mutants remain competent to induce MCP1 expression, however, which shows that these mutants are still transcriptionally functional. We also find that reconstitution with the R304K mutant resulted in significantly increased CXCL10, CX3CL1, and MCP-1 expression in EC as compared with the wild type. We suggest that Arg-304 methylation may be an inhibitory modification that may interfere with cofactor interactions. Alternatively, the presence of lysine instead of arginine at that location alone may be enough to change p65 conformation and enhance expression of these genes.
Arg-30 and Arg-35 are present in the Rel homology domain of p65, a motif necessary for dimerization, DNA binding, and cytosolic localization of p65 (16). Arg-35 directly contacts the 3′-subsite of κB sites (28). However, the 3′-subsite for p65 is not well conserved, requiring conformational flexibility of κB dimers to allow for base-specific interactions with the DNA (28). As reported by Wei et al. (27), Arg-30 dimethylation probably enhances DNA affinity. Methylation of Arg-35 may function though a similar mechanism, suggesting that one or both of the CXCL10 κB elements may require methylarginine, in contrast to the κB element(s) present in the promoters of other chemokines.
We show that p65 is methylated under non-stimulated conditions in EC. We did not detect changes in methylation levels by anti-SDMA Western blot following TNF-α exposure. However, the anti-methylarginine antibody may be interacting with several p65 methylarginine sites and changes at individual sites may be masked or not be detected by the antibody. Conditions may exist whereby methylarginine at individual sites are modulated.
The permanence of arginine methylation is an open question. Historically, arginine methylation has been seen as a permanent modification and may provide a means for long-lived transcriptional effects. Roles for arginine demethylases are yet to be established. In this case, p65 methylation does not appear to be enhanced by TNF-α. Termination of the methylarginine-containing p65 could be achieved through canonical NF-κB down-regulatory mechanisms, including cytosolic resequestration by IκB upon nuclear export or degradation through proteolytic mechanisms (29).
A major active area of NF-κB research focuses on determining the underlying molecular mechanisms of specific gene induction. A primary driver of context-dependent gene expression is the post-translational modifications of p65, including phosphorylation, acetylation, lysine methylation, and ubiquitination (16, 30, 31). Individual modifications can also influence the presence or absence of subsequent modifications through post-translational modification cross-talk. Comprehensive study of histone post-translational modification in mouse brain reveals that post-translational modifications are present in defined combinations with different ratios (32). Studies have noted that methylation of H3 at Arg-2 enhances the probability of Lys-4 acetylation, and together, these modifications control interaction with other proteins (33, 34). Post-translational modification cross-talk has been demonstrated in non-histone substrates as well. Arginine methylation of the nuclease FEN1 inhibits nearby phosphorylation and enhances binding to the DNA repair enzyme proliferating cell nuclear antigen (35). In another example, arginine methylation of EGF receptor by PRMT5 at Arg-1175 enhances trans-autophosphorylation of Tyr-1173, leading to increased ERK activation (36). Arginine methylation of p65 is likely to be a factor in other combinatorial mechanisms regulating p65 functionality and promoter targeting.
Development of strategies to limit CXCL10 production by inhibiting PRMT5 or the PRMT5-p65 interaction may ultimately be beneficial in the management of inflammatory and immune pathologies. Inhibitor utilization will have to be weighed against a potential increase in susceptibility to other diseases. For example, although CXCL10 is atherogenic and promotes development of coronary artery disease (37–40), it is protective against the formation and rupture of abdominal aortic aneurysms in a murine model (41).
This study is the first to demonstrate that PRMT5 enhances expression of CXCL10, a chemokine regulator of Th1 effector populations at sites of infection and inflammation. This occurs through PRMT5-mediated methylation of NF-κB at Arg-30 and Arg-35. Methylarginine-containing p65 subsequently interacts with the CXCL10 promoter, where it may enhance promoter association. Future studies are warranted to address the underlying molecular mechanism of p65 Rel homology domain methylation in gene specific transcriptional activation.
Acknowledgments
We thank Lisa Dechert, Chad Braley, and Emily Tillmaand for cell culture assistance. We thank Unni Chandrasekharan, Matthew Waitkus, and Joel Boerckel for helpful comments during manuscript preparation. Human umbilical vein endothelial cells were harvested from human umbilical cords collected from the Birthing Services Department at the Cleveland Clinic (Hillcrest Hospital) and the Perinatal Clinical Research Center (PCRC) at the Cleveland MetroHealth Hospital. The PCRC has been supported by a grant (UL1TR000439) awarded to the Clinical and Translational Science Collaborative of Cleveland by the National Center for Advancing Translational Sciences component of the NIH and the NIH Roadmap for Medical Research. The Orbitrap Elite instrument was purchased via a National Institutes of Health shared instrument grant (1S10RR031537-01).
This work was supported by National Institutes of Health Grant HL29582 (to P. E. D.).
- EC
- endothelial cell(s)
- PRMT5
- protein arginine methyltransferase 5.
REFERENCES
- 1. Medzhitov R., Horng T. (2009) Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 9, 692–703 [DOI] [PubMed] [Google Scholar]
- 2. Bandyopadhyay S., Harris D. P., Adams G. N., Lause G. E., McHugh A., Tillmaand E. G., Money A., Willard B., Fox P. L., Dicorleto P. E. (2012) HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Mol. Cell. Biol. 32, 1202–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boisvert F. M., Richard S. (2004) Arginine methylation regulates the cytokine response. Mol. Cell 15, 492–494 [DOI] [PubMed] [Google Scholar]
- 4. Hassa P. O., Covic M., Bedford M. T., Hottiger M. O. (2008) Protein arginine methyltransferase 1 coactivates NF-kappaB-dependent gene expression synergistically with CARM1 and PARP1. J. Mol. Biol. 377, 668–678 [DOI] [PubMed] [Google Scholar]
- 5. Richard S., Morel M., Cléroux P. (2005) Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5). Biochem. J. 388, 379–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang Y., Bedford M. T. (2013) Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50 [DOI] [PubMed] [Google Scholar]
- 7. Krause C. D., Yang Z. H., Kim Y. S., Lee J. H., Cook J. R., Pestka S. (2007) Protein arginine methyltransferases: Evolution and assessment of their pharmacological and therapeutic potential. Pharmacol. Ther. 113, 50–87 [DOI] [PubMed] [Google Scholar]
- 8. Arkov A. L., Wang J. Y., Ramos A., Lehmann R. (2006) The role of Tudor domains in germline development and polar granule architecture. Development 133, 4053–4062 [DOI] [PubMed] [Google Scholar]
- 9. Boisvert F. M., Chénard C. A., Richard S. (2005) Protein interfaces in signaling regulated by arginine methylation. Sci. STKE 2005, re2. [DOI] [PubMed] [Google Scholar]
- 10. Groom J. R., Luster A. D. (2011) CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol. Cell Biol. 89, 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zernecke A., Shagdarsuren E., Weber C. (2008) Chemokines in atherosclerosis: an update. Arterioscler. Thromb. Vasc. Biol. 28, 1897–1908 [DOI] [PubMed] [Google Scholar]
- 12. Luster A. D., Unkeless J. C., Ravetch J. V. (1985) γ-Interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature 315, 672–676 [DOI] [PubMed] [Google Scholar]
- 13. Marx N., Mach F., Sauty A., Leung J. H., Sarafi M. N., Ransohoff R. M., Libby P., Plutzky J., Luster A. D. (2000) Peroxisome proliferator-activated receptor-γ activators inhibit IFN-γ-induced expression of the T cell-active CXC chemokines IP-10, Mig, and I-TAC in human endothelial cells. J. Immunol. 164, 6503–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang X., Yue T. L., Ohlstein E. H., Sung C. P., Feuerstein G. Z. (1996) Interferon-inducible protein-10 involves vascular smooth muscle cell migration, proliferation, and inflammatory response. J. Biol. Chem. 271, 24286–24293 [DOI] [PubMed] [Google Scholar]
- 15. Angiolillo A. L., Sgadari C., Taub D. D., Liao F., Farber J. M., Maheshwari S., Kleinman H. K., Reaman G. H., Tosato G. (1995) Human interferon-inducible protein 10 is a potent inhibitor of angiogenesis in vivo. J. Exp. Med. 182, 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang B., Yang X. D., Lamb A., Chen L. F. (2010) Posttranslational modifications of NF-κB: another layer of regulation for NF-κB signaling pathway. Cell. Signal. 22, 1282–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leung T. H., Hoffmann A., Baltimore D. (2004) One nucleotide in a κB site can determine cofactor specificity for NF-κB dimers. Cell 118, 453–464 [DOI] [PubMed] [Google Scholar]
- 18. Baltimore D. (2011) NF-κB is 25. Nat. Immunol. 12, 683–685 [DOI] [PubMed] [Google Scholar]
- 19. Scarpati E. M., DiCorleto P. E. (1996) Identification of a thrombin response element in the human platelet-derived growth factor B-chain (c-sis) promoter. J. Biol. Chem. 271, 3025–3032 [DOI] [PubMed] [Google Scholar]
- 20. Bandyopadhyay S., Ashraf M. Z., Daher P., Howe P. H., DiCorleto P. E. (2007) HOXA9 participates in the transcriptional activation of E-selectin in endothelial cells. Mol. Cell. Biol. 27, 4207–4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Majumder S., Zhou L. Z., Chaturvedi P., Babcock G., Aras S., Ransohoff R. M. (1998) p48/STAT-1α-containing complexes play a predominant role in induction of IFN-γ-inducible protein, 10 kDa (IP-10) by IFN-γ alone or in synergy with TNF-α. J. Immunol. 161, 4736–4744 [PubMed] [Google Scholar]
- 22. Ballard D. W., Dixon E. P., Peffer N. J., Bogerd H., Doerre S., Stein B., Greene W. C. (1992) The 65-kDa subunit of human NF-κB functions as a potent transcriptional activator and a target for v-Rel-mediated repression. Proc. Natl. Acad. Sci. U.S.A. 89, 1875–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hamilton T. A., Novotny M., Datta S., Mandal P., Hartupee J., Tebo J., Li X. (2007) Chemokine and chemoattractant receptor expression: post-transcriptional regulation. J. Leukoc. Biol. 82, 213–219 [DOI] [PubMed] [Google Scholar]
- 24. Ohmori Y., Hamilton T. A. (1993) Cooperative interaction between interferon (IFN) stimulus response element and κ B sequence motifs controls IFN γ- and lipopolysaccharide-stimulated transcription from the murine IP-10 promoter. J. Biol. Chem. 268, 6677–6688 [PubMed] [Google Scholar]
- 25. Fabbrizio E., El Messaoudi S., Polanowska J., Paul C., Cook J. R., Lee J. H., Negre V., Rousset M., Pestka S., Le Cam A., Sardet C. (2002) Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 3, 641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jansson M., Durant S. T., Cho E. C., Sheahan S., Edelmann M., Kessler B., La Thangue N. B. (2008) Arginine methylation regulates the p53 response. Nat. Cell Biol. 10, 1431–1439 [DOI] [PubMed] [Google Scholar]
- 27. Wei H., Wang B., Miyagi M., She Y., Gopalan B., Huang D. B., Ghosh G., Stark G. R., Lu T. (2013) PRMT5 dimethylates R30 of the p65 subunit to activate NF-κB. Proc. Natl. Acad. Sci. U.S.A. 110, 13516–13521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen F. E., Huang D. B., Chen Y. Q., Ghosh G. (1998) Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature 391, 410–413 [DOI] [PubMed] [Google Scholar]
- 29. Ruland J. (2011) Return to homeostasis: downregulation of NF-κB responses. Nat. Immunol. 12, 709–714 [DOI] [PubMed] [Google Scholar]
- 30. Oeckinghaus A., Hayden M. S., Ghosh S. (2011) Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12, 695–708 [DOI] [PubMed] [Google Scholar]
- 31. Smale S. T. (2011) Hierarchies of NF-κB target-gene regulation. Nat. Immunol. 12, 689–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tweedie-Cullen R. Y., Brunner A. M., Grossmann J., Mohanna S., Sichau D., Nanni P., Panse C., Mansuy I. M. (2012) Identification of combinatorial patterns of post-translational modifications on individual histones in the mouse brain. PLoS One 7, e36980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feng Y., Wang J., Asher S., Hoang L., Guardiani C., Ivanov I., Zheng Y. G. (2011) Histone H4 acetylation differentially modulates arginine methylation by an in Cis mechanism. J. Biol. Chem. 286, 20323–20334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Molina-Serrano D., Schiza V., Kirmizis A. (2013) Cross-talk among epigenetic modifications: lessons from histone arginine methylation. Biochem. Soc. Trans. 41, 751–759 [DOI] [PubMed] [Google Scholar]
- 35. Guo Z., Zheng L., Xu H., Dai H., Zhou M., Pascua M. R., Chen Q. M., Shen B. (2010) Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nat. Chem. Biol. 6, 766–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hung C. M., Li C. (2004) Identification and phylogenetic analyses of the protein arginine methyltransferase gene family in fish and ascidians. Gene 340, 179–187 [DOI] [PubMed] [Google Scholar]
- 37. Fernandes J. L., Mamoni R. L., Orford J. L., Garcia C., Selwyn A. P., Coelho O. R., Blotta M. H. (2004) Increased Th1 activity in patients with coronary artery disease. Cytokine 26, 131–137 [DOI] [PubMed] [Google Scholar]
- 38. Heller E. A., Liu E., Tager A. M., Yuan Q., Lin A. Y., Ahluwalia N., Jones K., Koehn S. L., Lok V. M., Aikawa E., Moore K. J., Luster A. D., Gerszten R. E. (2006) Chemokine CXCL10 promotes atherogenesis by modulating the local balance of effector and regulatory T cells. Circulation 113, 2301–2312 [DOI] [PubMed] [Google Scholar]
- 39. Kawamura A., Miura S., Fujino M., Nishikawa H., Matsuo Y., Tanigawa H., Tomita S., Tsuchiya Y., Matsuo K., Saku K. (2003) CXCR3 chemokine receptor-plasma IP10 interaction in patients with coronary artery disease. Circ J. 67, 851–854 [DOI] [PubMed] [Google Scholar]
- 40. Mach F., Sauty A., Iarossi A. S., Sukhova G. K., Neote K., Libby P., Luster A. D. (1999) Differential expression of three T lymphocyte-activating CXC chemokines by human atheroma-associated cells. J. Clin. Invest. 104, 1041–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. King V. L., Lin A. Y., Kristo F., Anderson T. J., Ahluwalia N., Hardy G. J., Owens A. P., 3rd, Howatt D. A., Shen D., Tager A. M., Luster A. D., Daugherty A., Gerszten R. E. (2009) Interferon-γ and the interferon-inducible chemokine CXCL10 protect against aneurysm formation and rupture. Circulation 119, 426–435 [DOI] [PMC free article] [PubMed] [Google Scholar]