Background: Peptide RJPXD33 binds to and inhibits both LpxA and LpxD acyltransferases.
Results: The crystal structure of the antibacterial peptide RJPXD33 complexed to E. coli LpxA was determined.
Conclusion: RJPXD33 binds to E. coli LpxA in a unique modality that mimics the (R)-β-hydroxyacyl pantetheine moiety of substrate acyl-ACP.
Significance: Bioactive, dual binding LpxA/LpxD peptides raise the possibility of designing less resistance-prone peptidomimetics and/or small molecule antibacterials.
Keywords: Acyl Carrier Protein (ACP), Antimicrobial Peptide (AMP), Enzyme Inhibitor, Lipopolysaccharide (LPS), Peptides, X-ray Crystallography, Acyltransferase, Lipid A, LpxA
Abstract
UDP-N-acetylglucosamine acyltransferase (LpxA) and UDP-3-O-(acyl)-glucosamine acyltransferase (LpxD) constitute the essential, early acyltransferases of lipid A biosynthesis. Recently, an antimicrobial peptide inhibitor, RJPXD33, was identified with dual affinity for LpxA and LpxD. To gain a fundamental understanding of the molecular basis of inhibitor binding, we determined the crystal structure of LpxA from Escherichia coli in complex with RJPXD33 at 1.9 Å resolutions. Our results suggest that the peptide binds in a unique modality that mimics (R)-β-hydroxyacyl pantetheine binding to LpxA and displays how the peptide binds exclusive of the native substrate, acyl-acyl carrier protein. Acyltransferase binding studies with photo-labile RJPXD33 probes and truncations of RJPXD33 validated the structure and provided fundamental insights for future design of small molecule inhibitors. Overlay of the LpxA-RJPXD33 structure with E. coli LpxD identified a complementary peptide binding pocket within LpxD and serves as a model for further biochemical characterization of RJPXD33 binding to LpxD.
Introduction
Gram-negative microbes are encapsulated by an asymmetric lipid bilayer composed mainly of lipopolysaccharide (LPS) on the environmental face and phospholipids on the periplasmic face (1, 2). This protective sheath imparts a physiochemical barrier that helps defend these pathogens from hydrophobic antibacterial compounds (3, 4). The glycolipid, lipid A, which is the minimal motif required for the survival of most Gram-negative organisms (5) and is responsible for the endotoxic effects associated with Gram-negative bacterial sepsis, anchors LPS to the outer cell membrane (2, 5). Furthermore, although lipid A has proven to be a viable target for antimicrobial drug discovery (6, 7), there are currently no clinically approved antimicrobial agents targeting lipid A.
The first and third enzymes of the Raetz lipid A pathway (UDP-N-acetylglucosamine acyltransferase, LpxA; UDP-3-O-(R-3-hydroxyacyl)-glucosamine acyltransferase, LpxD) are type II acyl-carrier protein (ACP)3-dependent acyltransferases. Both enzymes utilize R-3-hydroxymyristoyl-ACP and UDP-glucosamine core substrates (Fig. 1) in Escherichia coli (8, 9) and display a unique left-handed β-helix motif that was first identified in LpxA (10–12). Such structural and functional similarities provide the basis for the possibility that a single inhibitor that targets both enzymes could be identified.
FIGURE 1.

Lipopolysaccharide biosynthesis in E. coli. Acyltransferases LpxA and LpxD catalyze functionally similar acylations of their respective UDP-glucosamine-based substrates using R-3-hydroxymyristoyl-ACP as the acyl donor.
Recently, through a phage display screen against LpxD, an inhibitor (RJPXD33; TNLYMLPKWDIP-NH2) was identified that demonstrated affinity and inhibitory activity for both LpxA and LpxD (13). Although peptides display susceptibility to proteases and poor bioavailability, they have become a coveted tool in chemists' and biologists' repertoires because of the ease of synthesis and functional utility. Thus, much attention has been given to designing “peptide mimics,” or peptidomimetics, which display greater bioavailability, enhanced three-dimensional structural characteristics, and proteolytic stability compared with their natural counterparts (14). However, a structural and biochemical understanding of how the peptide interacts with its host complex is crucial to the development of peptidomimetics.
The molecular mechanism by which RJPXD33 interacts with either LpxA or LpxD is currently unknown. Herein, we have utilized x-ray crystallography and peptide photoaffinity probes to provide insights into the binding interactions between the LpxA acyltransferase and RJPXD33 (13). Furthermore, acyltransferase binding studies using truncations of RJPXD33 were undertaken to provide biochemical verification of the structural data obtained.
EXPERIMENTAL PROCEDURES
Materials
Bio-Rad Protein assay and BioGel P2 size exclusion gel were purchased from Bio-Rad. LB (Lennox) broth and agar were purchased from Difco. Fluorescein isothiocyanate (FITC) was purchased from Acros Organics. Natural amino acids, Rink amide resin, and Fmoc-OSu were purchased from Anaspec. Photo-leucine (PhotoLeu) was purchased from Thermo Fisher/Pierce. Benzonase was purchased from Novagen. All buffers and antibiotics were purchased in the highest grade from Sigma or Fisher.
Protein Crystallography
Protein purification was carried out as previously described (15). Before crystallization, purified LpxA (10 mg/ml) was incubated for 1 h at 4 °C in the presence of 600 μm RJPXD33 (final concentration of DMSO was 2% v/v). The mixture was centrifuged at 10,000 × g for 10 min at 4 °C to pellet any insoluble material. Crystals were grown by vapor diffusion in a sitting drop tray. Droplets contained 2 μl of the LpxA-RJPXD33 solution and 2 μl of a well solution containing 0.8–1.8 m phosphate buffer (pH 6.3–6.8) and 31–34% DMSO. Crystals formed within 24 h. Crystals were cryoprotected in well solution containing 20% glycerol and flash-frozen in liquid nitrogen. X-ray data were collected at LS-CAT 21-ID-F and -G lines at the Advanced Photon Source at Argonne National Laboratory and processed using HKL2000 software (16). The complex was solved by molecular replacement with Phaser (17) using a previously solved structure of LpxA (containing no ligands; PDB code 1LXA) as the starting model (12). Iterative rounds of refinement and model building were completed using Buster (18) and Coot (19). MolProbity (20, 21) and Parvarti (22) were used to validate the structures. Figures were generated in PyMOL 1.7.0.3 (Schrödinger LLC) in which pairwise structural alignments were generated using align or cealign.
Fluorescence Polarization Assay
Peptides were synthesized as previously reported for solid phase peptide synthesis (23). The fluorescence polarization assay was performed as previously described (13). In 384-well black Costar plates, LpxA or His6-LpxD were serially diluted while holding the fluorescent peptide at 20 nm in a final volume of 50 μl in 20 mm HEPES pH 8.0 (0.01% DMSO). The wells were gently mixed and incubated at 30 °C in the dark for 15 min. Polarization was measured on a SpectraMax M5 plate reader in triplicate with readings taken at λex = 485 nm and λem = 525 nm. The binding data were first fit to a standard binding isotherm (Equation 1),
 |
where mP is the experimentally determined polarization, mPf is the polarization of free FITC-RJPXD33, [P] is the total acyltransferase concentration, Kd is the dissociation constant of peptide-protein complex, and mPb is the polarization value of fully bound fluorescent peptide. The calculated mPb was used to normalize the experimental data to fit the binding curves to the Hill equation (Equation 2), where α is the fraction of FITC-peptide bound, h is the Hill coefficient, [P] is the total concentration of acyltransferase, and Kd is the dissociation constant of the peptide-protein complex.
 |
For competition binding assays, 220–660 nm acyltransferase was incubated in the presence of varying concentrations of unlabeled peptides for 10 min at 30 °C in the dark. The [I]50 was determined from the competition binding curve, and the dissociation constant of unlabeled ligand was calculated as previously described (24) using Equation 3,
 |
where [I]50 is the unlabeled peptide concentration (inhibitor) at 50% inhibition, [L]50 is the free ligand (fluorescent tracer) concentration at 50% inhibition, [P]0 is the free protein concentration at 0% inhibition, Kd is the dissociation constant of fluorescent peptide (tracer), and Ki is the calculated dissociation constant for unlabeled peptide.
Fmoc-Photoleucine
Fmoc-PhotoLeu was synthesized as previously described (25). In an aluminum foil-covered round-bottom flask, PhotoLeu (l-2-amino-4,4′-azi-pentanoic acid; 1.4 mmol, 200 mg) and sodium bicarbonate (2.8 mmol, 236 mg) were dissolved in 20 ml of H2O and chilled to 0 °C on ice. Fmoc-OSu (2.1 mmol, 708 mg) was dissolved in 10 ml of THF and dropwise added over 15 min to the covered round-bottom flask. The resulting slurry was stirred on ice for 10 min and then allowed to warm to room temperature for 8 h. The flask was placed on ice, and the reaction was stopped by the addition of 25 ml of H2O and 50 ml of ethyl acetate (EtOAc) followed by acidification to pH 2 with concentrated HCl. The product was extracted (3×) with 50 ml EtOAc and concentrated by rotary evaporation. The resulting white solid was purified by flash chromatography with 1% methanol (v/v) and 1% acetic acid (v/v) in dichloromethane. The product was obtained as a white solid (464 mg, 91%). 1H NMR (400 MHz, DMSO) d 12.70 (s, 1H), 7.87 (d, J = 7.5, 2H), 7.71 (dd, J = 7.6, 4.4, 3H), 7.39 (t, J = 7.4, 2H), 7.30 (t, J = 7.4, 2H), 4.38–4.16 (m, 3H), 3.89–3.76 (m, 1H), 1.93–1.81 (m, 1H), 1.63 (dd, J = 14.8, 10.8, 1H), 0.99 (s, 3H). High resolution MS (ESI+): calculated for [M+Na]+ was 388.1278; observed was 388.1276.
Covalent Cross-linking of RJPXD33 Photo-affinity Probes to LpxA
Photo-activatable peptides (24 μm) were incubated with LpxA (10 μm) in the presence or absence of unlabeled peptide (200 μm) in 96-well half-area clear plates on ice in a final volume of 30 μl in 20 mm HEPES, pH 8.0 (DMSO 1% v/v). Plates, remaining on ice, were irradiated using a UV lamp (UVP model UVGL-58) at λ = 365 nm for 10 min at a distance of ∼2 cm. Samples were subsequently loaded and run on a 12% Tris-glycine SDS-PAGE gel. Gels were washed 3 times for 20 min at room temperature with deionized water to remove SDS. For fluorescein-labeled peptides, gels were analyzed for in-gel fluorescence using a Typhoon 9400 imaging system set to fluorescein wavelength (λex = 485 nm and λem = 525 nm), photomultiplier tube sensitivity at 500, and pixel size at 50 μm. The resulting data were visualized using ImageQuant 5.2 software. Following in-gel fluorescence analysis, gels were stained using SimplyBlue SafeStain (Invitrogen).
RESULTS AND DISCUSSION
Recognition of RJPXD33 by LpxA
RJPXD33 is an antimicrobial peptide that inhibits both E. coli LpxA (Kd = 22 μm) and LpxD (Kd = 6 μm) acyltransferases in early lipid A biosynthesis. The peptide binds to acyltransferases in a mutually exclusive manner with the acyl-ACP substrate. To design potent peptidomimetics and/or small molecule antimicrobials with the same unique dual binding properties of the parent peptide, a fundamental understanding of the molecular interactions between peptide and the target protein is vital. In this present study the EcLpxA-RJPXD33 crystal structure has assisted in elucidating the molecular mechanism of this peptide-protein interaction.
The co-crystal structure of EcLpxA-RJPXD33 (TNLYMLPKWDIP-NH2) diffracted to a resolution of 1.9 Å. The structure was solved by molecular replacement using a previously solved structure of LpxA (PDB code 1LXA), containing no bound ligands, as the model structure (12). The structure was refined to an Rfactor and Rfree of 0.179 and 0.188, respectively, while maintaining a good MolProbity score and low clash score (21). The complete crystallography and refinement statistics can be found in Table 1.
TABLE 1.
Data collection and refinement statistics
| Data set | EcLpxA + RJPXD33 |
|---|---|
| Data collection | |
| Space group | P213 |
| Unit cell a, b, c (Å) | 96.40, 96.40, 96.40 |
| Resolution (Å) | 1.9 (1.93-1.90) |
| Rsym (%) | 5.7 (29.7) |
| I/σI | >20 (5) |
| Completeness | 99.9 |
| Redundancy | 10.6 (10.1) |
| Refinement | |
| Resolution | 1.90 |
| Unique reflections | 23,751 |
| R-Factor (%) | 0.179 |
| Rfree (%) | 0.188 |
| Protein atoms | 2004 |
| Solvent molecules | 213 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.06 |
| MolProbity | 1.21 |
| Protein Data Bank code | 4J09 |
One molecule of LpxA and RJPXD33 occupied the asymmetric unit. However, three asymmetric units form around a 3-fold crystallographic axis, which is in agreement with previous structures and data showing that LpxA is a homotrimer (9, 26, 27). All of the 262 amino acids of LpxA, with the exception of the N-terminal methionine, were visible in the electron density map. The N-terminal half of RJPXD33 binds within the region between two monomeric subunits, constituting the LpxA active site, with a portion of the Met-5 side chain missing (28). RJPXD33 binds to LpxA in an unforeseen binding mode (Fig. 2). The six N-terminal amino acids of RJPXD33, TNLYML, run vertically up the bifurcated active site with Thr-1 positioned at coil 6 of LpxA left-handed β-helix motif and Leu-6 at coil 10. The side chains of Thr-1, Leu-3, and Met-5 all point inward toward the hydrophobic cleft between the parallel β strand 3 (PB3) face of the LpxA subunit within the asymmetric unit and the parallel β strand 2 (PB2) face of the adjacent subunit (12). The side chains of Tyr-4 and Leu-6 occupy separate hydrophobic pockets along parallel β strand 2 of the adjacent subunit, whereas the Asn-2 side chain makes a favorable amide-π center stacked interaction with the Tyr-4 side chain (29).
FIGURE 2.
Crystal structure of RJPXD33 bound to LpxA. A, frontal view of RJPXD33 (cyan; spheres) in complex with LpxA trimer (gray; ribbon). RJPXD33 is positioned vertically between coils 6 and 10 (shaded in green) of the left-handed β-helix motif. Only the first six residues of RJPXD33 are shown in the figure, as the rest of the peptide is disordered in the electron density maps. RJPXD33 binds between two monomeric subunits of the trimer; only one molecule of RJPXD33 is shown. B, top down view of the RJPXD33-bound LpxA with each of the trimer subunits labeled (S1-S3) along with each face of the parallel β sheets of each subunit depicted (PB1-PB3). C, surface structure of RJPXD33 (cyan; spheres) in complex with LpxA trimer (gray, surface map). D, close-up view of the LpxA active site with RJPXD33 (cyan; spheres) bound. LpxA monomers and associated amino acids are differentiated by shades of gray.
Located at the top of the LpxA binding cleft, 2.8 Å from Leu-6 of RJPXD33 is His-191. His-191 serves as a “hydrocarbon ruler” capping the size of acyl groups allowed to bind to the enzyme (26). The density of the C-terminal six amino acids of RJPXD33, PKWDIP, could not be visualized beyond this point in the structure (Fig. 3) most likely due to the protrusion of this portion of the peptide away from the LpxA active site and into solvent-exposed space.
FIGURE 3.

Positioning of key active site residues in the EcLpxA-RJPXD33 complex. A, the 2Fo − Fc electron density map of the RJPXD33-LpxA complex contoured to 1σ around RJPXD33 (cyan; stick) with amino acids from LpxA (gray; stick) that surround the bound ligand. Although the full density of the methionine side chain from RJPXD33 could not be visualized, His-160 of LpxA kinks away from the hydrophobic cleft, allowing space for the methionine side chain to occupy the binding pocket. B, close-up overlay stereo view of the EcLpxA-RJPXD33 complex (PDB code 4J09; enzyme (gray); RJPXD33 (cyan)) and EcLpxA (PDB code 1LXA; green) with no bound ligands. PyMOL alignment r.m.s.d. = 0.35 over 256 residues.
Conformational Change of His-160 Side Chain
Mutation of His-160 to Ala has been shown to reduce the activity of EcLpxA to ∼5% of the wild-type enzyme (28). A noticeable difference between our current structure and that of unbound EcLpxA is the conformation of the His-160 side chain (Fig. 3). In unbound EcLpxA this side chain faces inward toward the hydrophobic cleft. Upon binding of RJPXD33, the His-160 side chain rotates outward toward the solvent front. This conformational change is what is commonly observed among all LpxA structures containing a bound acylated ligand (26). This is also seen in Leptospira interrogans LpxA, where the analogous histidine residue, His-155, is kinked outward upon binding of its acylated substrate (30). The opposite is true for the structures of the complexes bound with only UDP-GlcNAc (PDB code 2JF3), where His-160 faces inward toward the hydrophobic cleft, as it does in the unbound form of LpxA (12, 27, 31). These results demonstrated that His-160 of LpxA must kink outward to allow the fatty acyl chain to access the hydrophobic cleft and that RJPXD33 is able to mimic this binding motif. Although the full electron density of the RJPXD33 Met-5 side chain cannot be accounted for, these results would suggest that Met-5 would occupy the binding region formed when His-160 is kinked outward (Fig. 3). This conformational change may allow His-160 to form polar contacts with acidic residues of ACP and thus would be important in substrate binding and release.
Comparison of RJPXD33 Binding with Other E. coli LpxA Ligands
The hydroxyl moiety of the Thr-1 side chain of RJPXD33 forms polar contacts with His-122, Gln-73, and an active site water of LpxA (Fig. 3). These moieties have been implicated as binding contacts to the R-3-hydroxymyristoyl moiety of UDP-3-O-(R-3-hydroxymryistoyl)-GlcNAc, the LpxA product (26). When the structures of RJPXD33-bound EcLpxA and the product-bound EcLpxA (PDB code 2QIA) were superimposed, the Thr-1 β-hydroxyl group was shown to occupy the same space as the R-3-hydroxy functional group of the myristate chain (Fig. 4). In addition to the Thr-1 residue, the Leu-3 and Met-5 side chains of RJPXD33 also occupy the fatty acyl binding region.
FIGURE 4.

Position of RJPXD33 in EcLpxA in comparison to acylated LpxA ligands. A, overlay of EcLpxA bound RJPXD33 (cyan) and EcLpxA-bound UDP-3-O-(R-3-hydroxymyrisoyl)-GlcNAc (PDB code 2QIA; wheat). PyMOL alignment r.m.s.d. = 0.18 over 256 residues. B, overlay of EcLpxA-bound RJPXD33 (cyan) and conformation 1 of bound (R)-β-hydroxylauroyl-methylphosphopantetheine (PDB code 3I3A; gray) in LiLpxA. PyMOL alignment r.m.s.d. = 1.62 over 256 residues. Each of the above bound ligands is associated with chain A of their respective asymmetric units.
RJPXD33 binds to LpxA in a unique fashion when compared with the LpxA-specific peptide P920 (WMLDPIAGKWSR; Fig. 5) despite a similar primary sequence motif (YMLP versus WMLDP) between the two (32). P920 forms a β-hairpin fold oriented perpendicular to the LpxA symmetry axis, where the head of the loop, isoleucine, occupies a small portion of the fatty acid binding cleft of LpxA similar to Leu-3 of RJPXD33 (27). The N and C termini of P920 face out of the pocket toward the solvent face, overlapping the nucleotide binding region identified from several UDP ligand-bound structures (26, 31). In contrast, RJPXD33 is oriented parallel to the LpxA symmetry axis, occupying most of the hydrophobic fatty acid binding cleft whereas demonstrating only a minimal overlap between its Asn-2/Tyr-4 side chains and the UDP binding site. Unlike RJPXD33, P920 does not extend into the lower or upper R-β-hydroxy acyl binding pocket occupied by Thr-1 and Met-5 of RJPXD33, respectively. This is highlighted by the lack in conformational change of His-160 in the P920-bound structure compared with unbound LpxA.
FIGURE 5.

Comparison of the binding of RJPXD33 and P920 to LpxA. A, top down view of RJPXD33 (cyan; sticks) and P920 (pink; sticks) complexed with LpxA trimer. Subunit A of each LpxA peptide complex is shown as ribbon (PDB code 4J09, gray; PDB code 2AQ9, magenta), whereas the surface of the other two trimer subunits is shown in gray, and only one molecule of RJPXD33/P920 is shown. B, frontal, close-up, stereo view of the same complexes above. PyMOL alignment r.m.s.d. = 0.18 over 237 atoms.
Comparison of RJPXD33 Binding with (R)-β-Hydroxylauroyl-methylphosphopantetheine
Although derivatives of phosphopantetheine (PPan), the post-translational modification necessary for acyl chain assembly on ACP, are not substrates for EcLpxA, R-3-hydroxylauryl methyl-phosphopantetheine has been shown to be a substrate for L. interrogans LpxA (30, 33). The co-crystal structure of EcLpxA-RJPXD33 was superimposed onto the C-chain of the LiLpxA-(R)-3-hydroxylauroyl-methyl-PPan co-crystal structure (Fig. 4; PDB code 3I3A) (30). As with the EcLpxA product-bound structure, the Thr-1, Leu-3, and Met-5 side chains of RJPXD33 overlay with the laurate chain of the LiLpxA-bound (R)-β-hydroxylauroyl-methyl-PPan structure. The PPan portion of the LiLpxA substrate was modeled in two conformations due to the electron density obtained for the phosphopantoyl moiety. The peptide backbone of RJPXD33 occupied the same space as the acyl-PPan substrate (conformation 1) of LiLpxA. This is highlighted at the amide bond between Thr-1 and Asn-2 of RJPXD33, which superimposes with the thioester bond between the acyl chain and the methyl-PPan cysteamine moiety (Fig. 4).
These findings suggest that RJPXD33 mimics the (R)-β-hydroxyacyl pantetheine moiety of acyl-ACP. This similarity to the acyl-PPan accounts for the dual targeting nature of RJPXD33 to both LpxA and LpxD, due to the fact that both enzymes utilize (R)-β-hydroxyacyl-ACP as a substrate.
Photoaffinity Labeling of EcLpxA with Photoleucine-containing FITC-RJPXD33
To determine the effects of incorporating a photo-affinity label on LpxA binding, derivatives of FITC-RJPXD33 containing a leucine-based diazirine moiety (PhotoLeu) were synthesized. PhotoLeu differs from leucine in that the side chain penultimate carbon carries a diazirine moiety, making PhotoLeu more hindered and sterically cumbersome (34). PhotoLeu was substituted for Leu-3 (FITC-Photo 1), Leu-6 (FITC-Photo 3), or Ile-11 (FITC-Photo 4) of FITC-RJPXD33. Fluorescence polarization binding experiments were performed with FITC-photopeptides against LpxA, and each of the photopeptides bound with Hill coefficients between 0.8 and 0.9 and Kd values = 22–50 μm (Table 2). Incorporation of the PhotoLeu in place of Ile-11 was least disruptive of binding to the acyltransferase, based on dissociation constant, consistent with this portion of the molecule contributing little to the overall binding of RJPXD33 to LpxA.
TABLE 2.
Binding constants of RJPXD33 truncations to EcLpxA
FITC, fluorescein; βa, β-alanine; X, l-photoleucine.
| Peptide (sequence) | Kd LpxA | Kd LpxD |
|---|---|---|
| μm | μm | |
| FITC-RJPXD33 (FITC-(βa)TNLYMLPKWDIP-CONH2) | 17 ± 1.6 | 0.6 ± 0.04 |
| FITC-Photo 1 (FITC-(βa)TNXYMLPKWDIP-CONH2) | 50 ± 1.8 | 2.1 ± 0.3 |
| FITC-Photo 3 (FITC-(βa)TNLYMXPKWDIP-CONH2) | 32 ± 1.1 | 1.1 ± 0.1 |
| FITC-Photo 4 (FITC-(βa)TNLYMLPKWDXP-CONH2) | 22 ± 1.7 | 0.8 ± 0.1 |
| FITC-RJPXD33Δ6 (FITC-(βa)TNLYML-CONH2) | 12 ± 1 | 22 ± 1 |
| FITC-RJPXD33Δ6-COOH (FITC-(βa)TNLYML-COOH) | 4.4 ± 0.2 | 36 ± 3.5 |
| RJPXD33Δ6 (TNLYML-CONH2) | 23 ± 2.0 | |
| RJPXD33Δ6-COOH (TNLYML-COOH) | 2.1 ± 0.2 |
When bound to LpxA and irradiated with UV light, PhotoLeu generates a reactive carbene group that can react with groups on LpxA in close proximity and covalently cross-link the peptide to the bound protein. Photopeptides 1, 3, and 4 (24 μm) were subjected to UV irradiation (10 min) in the presence of 10 μm LpxA with and without 200 μm unlabeled RJPXD33 (Fig. 6). When UV light was omitted no cross-linking occurred. However, when irradiated with UV light each of the peptides was able to covalently cross-link to LpxA as visualized through in-gel fluorescence. The addition of unlabeled RJPXD33 significantly decreased the fluorescent signal, indicating that the covalent cross-linking is due to specific peptide-protein interactions and that the photopeptides bind to LpxA in a similar manner as the parent peptide. As a control, reactions containing LpxA without the photopeptides were subjected to UV irradiation, and no observable fluorescence was detected.
FIGURE 6.

In-gel fluorescence of PhotoLeu-containing peptides. Affinity labeling was accomplished by UV irradiation (λ = 365 nm) while holding LpxA and FITC-photopeptides constant at 10 and 24 μm, respectively, in the presence or absence of unlabeled RJPXD33 (200 μm). Under these conditions photo-labeling of LpxA was linear up to 10 min, which is the time used in the above gel samples. Gels were analyzed for in-gel fluorescence using a Typhoon 9400 imaging system set to fluorescein wavelength (λex = 485 nm and λem = 525 nm), photomultiplier tube sensitivity at 500, and pixel size at 50 μm. LpxA was stained with Coomassie Blue to demonstrate consistency of protein loading.
Binding of RJPXD33 Truncation to LpxA
To corroborate our crystal structure and evaluate the potential for possible RJPXD33 truncations, FITC-RJPXD33Δ6 was synthesized. FITC-RJPXD33Δ6 lacks the six terminal amino acids of full-length FITC-RJPXD33. Fluorescence polarization binding experiments using wild-type LpxA were performed to quantitate the binding affinity of the truncated RJPXD33 peptide. When the assay was performed with EcLpxA, the observed Kd for FITC-RJPXD33Δ6 was 12 ± 1 μm (Table 2). This value was in close proximity to the previously published value (Kd = 17 ± 1.6 μm) for the full-length FITC-RJPXD33 (13). This suggested that the six terminal amino acids are not making extensive binding contacts to LpxA, in agreement with the crystal structure.
In addition to its role as the hydrocarbon ruler, His-191 may be important in binding of the phosphopantetheine moiety of ACP substrates. His-191 occupies a space in EcLpxA similar to Lys-171 of LiLpxA, which is postulated to play a similar role as hydrocarbon ruler and is also in close proximity to the phosphate group of the R-3-hydroxylauryl methyl-phosphopantetheine substrate (30). In each of the above peptides the C terminus is modified as an amide. FITC-RJPXD33Δ6-COOH was synthesized to increase the affinity of the Δ6 peptide by exposing the acidic C terminus to interact with the nearby basic His-191. FITC-RJPXD33Δ6-COOH binds with approximately 3 times higher affinity to EcLpxA than its amide capped homolog. This trend remains true with the removal of the fluorescein moiety. In competition binding assays RJPXD33Δ6-COOH binds 10 times greater than the amide-capped RJPXD33Δ6.
Binding of RJPXD33 to LpxD
It remains unclear as to how exactly RJPXD33 interacts with E. coli LpxD due to unsuccessful attempts at co-crystallization. RJPXD33 was first identified by phage display against immobilized LpxD (13). Although no EcLpxD-RJPXD33 structure is available, this current work suggests that RJPXD33 would overlap with the acyl-PPan arm of acyl-ACP, thereby inhibiting acyl-ACP from binding to LpxD. This is in agreement with our previous results that demonstrated RJPXD33 bound exclusive of acyl-ACP to LpxD.
Fluorescence polarization binding experiments using wild-type LpxD were performed to quantitate the binding affinity of the FITC-photopeptides and truncated RJPXD33 peptides. Similar to their binding to LpxA, each of the photopeptides bound with Hill coefficients between 0.8 and 0.9. The Kd values for the photopeptides (0.8–2.1 μm; Table 2) were in close proximity to the value for FITC-RJPXD33 for LpxD (Kd = 0.6 ± 0.04 μm) (13) and showed the same binding trend as seen in LpxA, with photopeptide 4 being least disruptive of binding. When the assay was performed with truncated peptide, the observed Kd for FITC-RJPXD33Δ6 was 22 ± 1 μm, similar to its binding to LpxA (12 ± 1 μm). However, the binding of FITC-RJPXD33Δ6-COOH (36 ± 3.5 μm) did not show increased affinity to LpxD as it did to LpxA (Table 2) and reflects the fact that the role of hydrocarbon ruler in EcLpxD is fulfilled by Met-290 instead of a basic histidine residue as with LpxA (Fig. 7D) (10, 35).
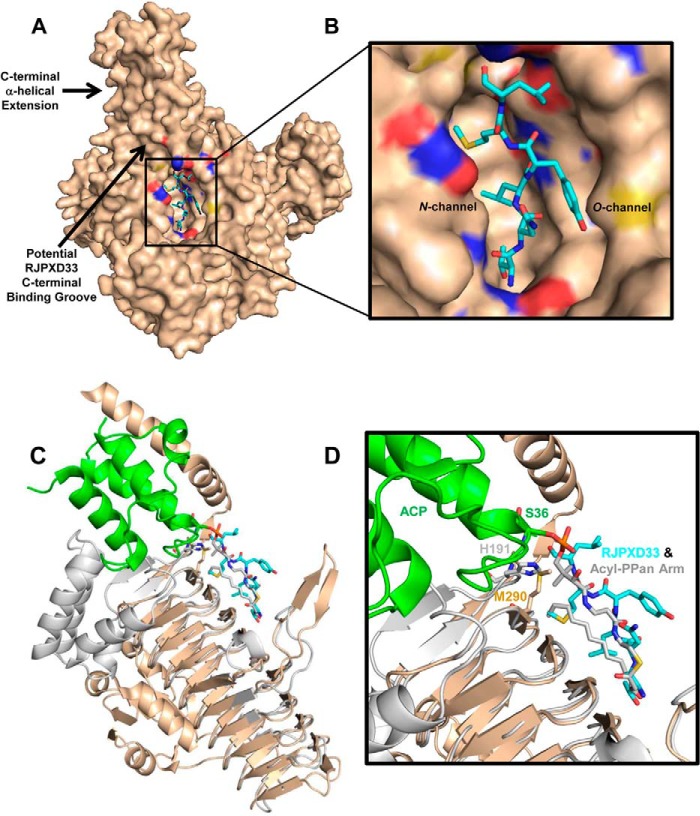
FIGURE 7.
Binding model of the RJPXD33-LpxD complex. A, the EcLpxA-RJPXD33 complex was superimposed onto EcLpxD (PDB code 3EH0; wheat). PyMOL alignment r.m.s.d. = 0.70 over 141 atoms. RJPXD33 (cyan) occupies the fatty acyl binding cleft between two monomers in EcLpxD. The six C-terminal amino acids of RJPXD33 would potentially extend into a binding groove near the C-terminal α-helical extension of EcLpxD. B, close-up view of the putative EcLpxD RJPXD33 binding pocket. C, comparison of RJPXD33-LpxA complex with acyl-ACP-LpxD complex (PDB code 4IHF). Overlay of subunit A of EcLpxA (gray; ribbon) bound RJPXD33 (cyan; sticks) and EcLpxD (wheat; ribbon) bound acyl-ACP (green; ribbon). RJPXD33 occupies the same region as the acyl-PPan prosthetic group (gray; sticks). D, close-up view of the LpxD active site of the EcLpxA-RJPXD33, EcLpxD-acyl-ACP alignment. PyMOL alignment r.m.s.d. = 0.71 over 141 atoms.
Like acyl-ACP, RJPXD33 binds to LpxD without the prior binding of other ligands. To shed light on this interaction, structural overlays between our RJPXD33-EcLpxA complex, EcLpxD (PDB code 3EH0), and the recently published EcLpxD-acyl-ACP (PDB code 4IHF) were performed (Fig. 7). From these overlays, it is clear that RJPXD33 would occupy a similar fatty acyl binding groove in EcLpxD when compared with LpxA. The overlays align RJPXD33 with the R-3-hydroxymyristoyl phosphopantetheine prosthetic group of acyl-ACP. The Thr-1, Leu-3, and Met-5 side chains of RJPXD33 occupy the N-channel of LpxD and overlay with the R-3-hydroxymyristoyl chain, whereas the peptide backbone of RJPXD33 aligns well with the PPan moiety. The amide bond between Thr-1 and Asn-2 of RJPXD33 superimposes with the thioester bond between the acyl chain and the PPan cysteamine moiety. The Tyr-4 and Leu-6 side chains of RJPXD33 occupy the O-channel, which is postulated to be the binding site for the ester-linked acyl group of substrate UDP-3-O-acyl-GlcN. The six C-terminal residues of RJPXD33 would potentially extend into a binding groove near the C-terminal α-helical domain of EcLpxD, which is not present in EcLpxA and has been identified as the ACP binding domain (35). This would explain the role of the six C-terminal residues with respect to the increased affinity of RJPXD33 for LpxD when compared with LpxA as well as the greater loss in affinity between full-length peptide and the C-terminal Δ6 peptide for LpxD over LpxA.
Our present work unravels the molecular mechanism underlying RJPXD33 binding to acyltransferases in early lipid A biosynthesis, showing that it mimics the (R)-β-hydroxyacyl pantetheine moiety of substrate acyl-ACP both in terms of its overall chemical space occupancy and its ability to induce similar conformational changes to the acyltransferase upon binding. Considering the diverse array of protein-protein interactions between ACP and its client proteins, other acyl-PPan mimics may find use as antimicrobial inhibitors of essential ACP-protein interactions. The co-crystal structure of RJPXD33-LpxA and the fluorescence polarization experiments associated with FITC-RJPXD33Δ6 demonstrate that certain RJPXD33 truncations can bind with similar affinity to both LpxA and LpxD. Such truncations would be important in developing smaller peptidomimetics capable of crossing the cell membrane.
Acknowledgments
Use of the Advanced Photon Source, an Office of Science User Facility operated for the United States Department of Energy Office of Science by Argonne National Laboratory, was supported by the United States Department of Energy Contract DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). We thank Dr. David Smith of LS-CAT for help with remote data collection.
This work was supported, in whole or in part, by National Institutes of Health Grant 5T32GM008597-14 (University of Michigan Chemistry-Biology Interface training program; to R. J. J.). This work was also supported in part by a Valteich Research Award administered by the College of Pharmacy, University of Michigan and by a Pre-doctoral Research Grant administered by the University of Michigan Rackham Graduate School.
The atomic coordinates and structure factors (code 8J09) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ACP
- acyl-carrier protein
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- PPan
- phosphopantetheine
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Raetz C. R., Whitfield C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Raetz C. R., Reynolds C. M., Trent M. S., Bishop R. E. (2007) Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vaara M. (1993) Outer membrane permeability barrier to azithromycin, clarithromycin, and roxithromycin in gram-negative enteric bacteria. Antimicrob. Agents Chemother. 37, 354–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vuorio R., Vaara M. (1992) The lipid A biosynthesis mutation lpxA2 of Escherichia coli results in drastic antibiotic supersusceptibility. Antimicrob. Agents Chemother. 36, 826–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meredith T. C., Aggarwal P., Mamat U., Lindner B., Woodard R. W. (2006) Redefining the requisite lipopolysaccharide structure in Escherichia coli. ACS Chem. Biol. 1, 33–42 [DOI] [PubMed] [Google Scholar]
- 6. Vaara M. (1996) Lipid A: target for antibacterial drugs. Science 274, 939–940 [DOI] [PubMed] [Google Scholar]
- 7. Onishi H. R., Pelak B. A., Gerckens L. S., Silver L. L., Kahan F. M., Chen M. H., Patchett A. A., Galloway S. M., Hyland S. A., Anderson M. S., Raetz C. R. (1996) Antibacterial agents that inhibit lipid A biosynthesis. Science 274, 980–982 [DOI] [PubMed] [Google Scholar]
- 8. Kelly T. M., Stachula S. A., Raetz C. R., Anderson M. S. (1993) The firA gene of Escherichia coli encodes UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase. The third step of endotoxin biosynthesis. J. Biol. Chem. 268, 19866–19874 [PubMed] [Google Scholar]
- 9. Anderson M. S., Bull H. G., Galloway S. M., Kelly T. M., Mohan S., Radika K., Raetz C. R. (1993) UDP-N-acetylglucosamine acyltransferase of Escherichia coli. The first step of endotoxin biosynthesis is thermodynamically unfavorable. J. Biol. Chem. 268, 19858–19865 [PubMed] [Google Scholar]
- 10. Bartling C. M., Raetz C. R. (2009) Crystal structure and acyl chain selectivity of Escherichia coli LpxD, the N-acyltransferase of lipid A biosynthesis. Biochemistry 48, 8672–8683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buetow L., Smith T. K., Dawson A., Fyffe S., Hunter W. N. (2007) Structure and reactivity of LpxD, the N-acyltransferase of lipid A biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 4321–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raetz C. R., Roderick S. L. (1995) A left-handed parallel β helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science 270, 997–1000 [DOI] [PubMed] [Google Scholar]
- 13. Jenkins R. J., Dotson G. D. (2012) Dual targeting antibacterial peptide inhibitor of early lipid a biosynthesis. ACS Chem. Biol. 7, 1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yin H., Hamilton A. D. (2005) Strategies for targeting protein-protein interactions with synthetic agents. Angew. Chem. Int. Ed. Engl. 44, 4130–4163 [DOI] [PubMed] [Google Scholar]
- 15. Jenkins R. J., Dotson G. D. (2012) A continuous fluorescent enzyme assay for early steps of lipid A biosynthesis. Anal. Biochem. 425, 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. In Methods in Enzymology: Macromolecular Crystallography, part A (Carter C. W., Jr., Sweet R. M., ed.) pp. 307–326, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 17. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O., Vonrhein C., Womack T. (2011) Buster. 2.10.0 Ed., Global Phasing Ltd., Cambridge, United Kingdom [Google Scholar]
- 19. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 20. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zucker F., Champ P. C., Merritt E. A. (2010) Validation of crystallographic models containing TLS or other descriptions of anisotropy. Acta Crystallogr. D. Biol. Crystallogr. 66, 889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chan W. C., White P. D. (2000) Fmoc Solid Phase Peptide Synthesis, Oxford University Press, New York [Google Scholar]
- 24. Nikolovska-Coleska Z., Wang R., Fang X., Pan H., Tomita Y., Li P., Roller P. P., Krajewski K., Saito N. G., Stuckey J. A., Wang S. (2004) Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal. Biochem. 332, 261–273 [DOI] [PubMed] [Google Scholar]
- 25. Janz J. M., Ren Y., Looby R., Kazmi M. A., Sachdev P., Grunbeck A., Haggis L., Chinnapen D., Lin A. Y., Seibert C., McMurry T., Carlson K. E., Muir T. W., Hunt S., 3rd, Sakmar T. P. (2011) Direct interaction between an allosteric agonist pepducin and the chemokine receptor CXCR4. J. Am. Chem. Soc. 133, 15878–15881 [DOI] [PubMed] [Google Scholar]
- 26. Williams A. H., Raetz C. R. (2007) Structural basis for the acyl chain selectivity and mechanism of UDP-N-acetylglucosamine acyltransferase. Proc. Natl. Acad. Sci. U.S.A. 104, 13543–13550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Williams A. H., Immormino R. M., Gewirth D. T., Raetz C. R. (2006) Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide. Proc. Natl. Acad. Sci. U.S.A. 103, 10877–10882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wyckoff T. J., Raetz C. R. (1999) The active site of Escherichia coli UDP-N-acetylglucosamine acyltransferase. Chemical modification and site-directed mutagenesis. J. Biol. Chem. 274, 27047–27055 [DOI] [PubMed] [Google Scholar]
- 29. Hughes R. M., Waters M. L. (2006) Effects of lysine acetylation in a β-hairpin peptide: comparison of an amide-π and a cation-π interaction. J. Am. Chem. Soc. 128, 13586–13591 [DOI] [PubMed] [Google Scholar]
- 30. Robins L. I., Williams A. H., Raetz C. R. (2009) Structural basis for the sugar nucleotide and acyl-chain selectivity of Leptospira interrogans LpxA. Biochemistry 48, 6191–6201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ulaganathan V., Buetow L., Hunter W. N. (2007) Nucleotide substrate recognition by UDP-N-acetylglucosamine acyltransferase (LpxA) in the first step of lipid A biosynthesis. J. Mol. Biol. 369, 305–312 [DOI] [PubMed] [Google Scholar]
- 32. Benson R. E., Gottlin E. B., Christensen D. J., Hamilton P. T. (2003) Intracellular expression of peptide fusions for demonstration of protein essentiality in bacteria. Antimicrob. Agents Chemother. 47, 2875–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Majerus P. W., Alberts A. W., Vagelos P. R. (1965) Acyl carrier protein: Iv. the identification of 4′-phosphopantetheine as the prosthetic group of the acyl carrier protein. Proc. Natl. Acad. Sci. U.S.A. 53, 410–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suchanek M., Radzikowska A., Thiele C. (2005) Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat. Methods 2, 261–267 [DOI] [PubMed] [Google Scholar]
- 35. Masoudi A., Raetz C. R., Zhou P., Pemble C. W. (2014) Chasing acyl carrier protein through a catalytic cycle of lipid A production. Nature 505, 422–426 [DOI] [PMC free article] [PubMed] [Google Scholar]