Background: Factors regulating NAD+ metabolism and homeostasis remain unclear because of the dynamic nature of NAD+ synthesis pathways.
Results: Pof1 is a novel NMN-specific NMNAT that mediates NAD+ production.
Conclusion: Enzymes with redundant function may provide flexibility to maintain NAD+ homeostasis.
Significance: Novel NAD+ metabolic factors are identified, aiding our understanding of the complex NAD+ pathways.
Keywords: Cell Metabolism, Metabolic Regulation, NAD Biosynthesis, Yeast Genetics, Yeast Metabolism, Nicotinamide Mononucleotide Adenylyltransferase, Nicotinamide Riboside Salvage
Abstract
NAD+ is an essential metabolic cofactor involved in various cellular biochemical processes. Nicotinamide riboside (NR) is an endogenously produced key pyridine metabolite that plays important roles in the maintenance of NAD+ pool. Using a NR-specific cell-based screen, we identified mutants that exhibit altered NR release phenotype. Yeast cells lacking the ORF YCL047C/POF1 release considerably more NR compared with wild type, suggesting that POF1 plays an important role in NR/NAD+ metabolism. The amino acid sequence of Pof1 indicates that it is a putative nicotinamide mononucleotide adenylyltransferase (NMNAT). Unlike other yeast NMNATs, Pof1 exhibits NMN-specific adenylyltransferase activity. Deletion of POF1 significantly lowers NAD+ levels and decreases the efficiency of NR utilization, resistance to oxidative stress, and NR-induced life span extension. We also show that NR is constantly produced by multiple nucleotidases and that the intracellular NR pools are likely to be compartmentalized, which contributes to the regulation of NAD+ homeostasis. Our findings may contribute to the understanding of the molecular basis and regulation of NAD+ metabolism in higher eukaryotes.
Introduction
Pyridine nucleotides NAD+(H) and NADP+(H) are essential coenzymes participating in many cellular redox reactions in all living systems. NAD+ and its derivatives also function as substrates and signaling molecules in key cellular processes such as regulation of Ca2+ signaling, chromatin structure, DNA repair, and life span (1–5). Many of these processes consume NAD+; therefore cells have developed complex interconnecting biosynthetic and signaling pathways to monitor and replenish intracellular NAD+ levels.
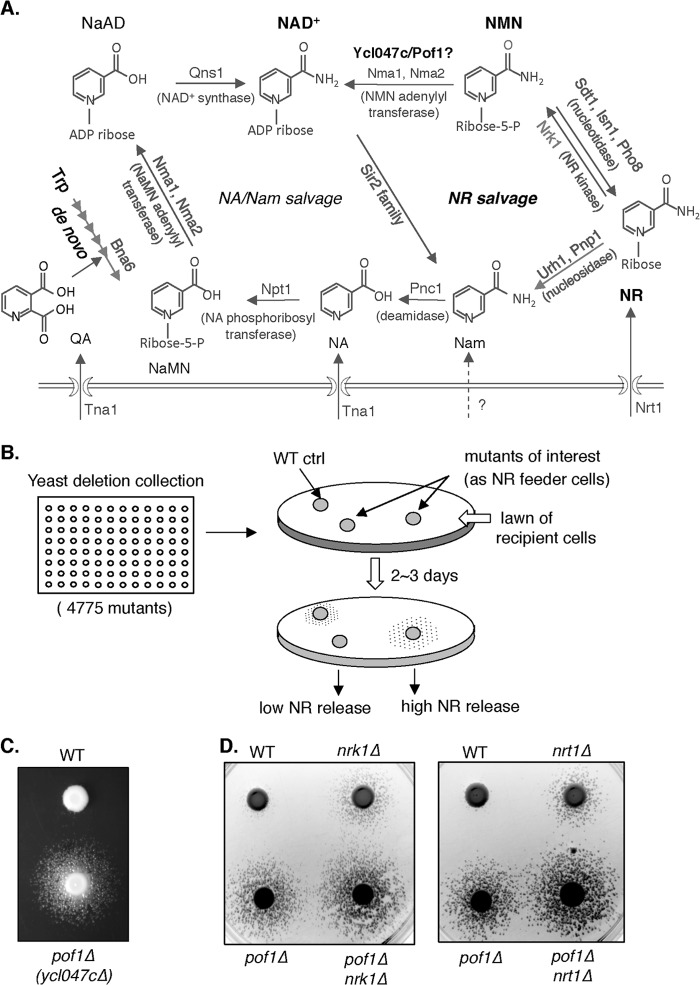
NAD+ is synthesized from multiple precursors. In yeast, the cellular pool of NAD+ is maintained by biosynthesis from nicotinic acid mononucleotide (NaMN)2 or nicotinamide mononucleotide (NMN) (see Fig. 1A). NaMN is produced by transferring the phosphoribose moiety of phosphoribosyl pyrophosphate to nicotinic acid (NA) or to tryptophan-derived quinolinic acid (QA), catalyzed by phosphoribosyltransferases Npt1 and Bna6, respectively (6–8) (see Fig. 1A). QA and NA are intermediate metabolites generated by de novo synthesis or by salvaging reactions that utilize exogenous pyridines or internal pyridines derived from NAD+ recycling. NMN is generated by phosphorylating nicotinamide riboside (NR), which is catalyzed by the kinase Nrk1 at the expense of ATP (9, 10). It has been shown that yeast cells produce NAD+ predominantly via the NA/Nam salvage pathway during exponential growth (11). More recently, NR has been shown to be an efficient NAD+ precursor that contributes to the NAD+ pool and supports NAD+-dependent reactions (9, 10). Intact NR salvaging pathway is essential for maintaining NAD+ homeostasis and life span (12, 13). Because yeast cells constantly release and reassimilate NR, it has been suggested that this NR pool might confer metabolic flexibility for prompt adjustment of cellular NAD+ levels (12, 13). How NAD+ synthesis routes are regulated in response to different growth conditions remains to be elucidated.
FIGURE 1.
Deletion of YCL047C/POF1 causes altered NR metabolism. A, a simplified model of Saccharomyces cerevisiae NAD+ synthesis pathway. In yeast, NAD+ is synthesized de novo from tryptophan (Trp) and by salvaging NA, Nam, QA, and NR. Cells can also salvage nicotinic acid riboside by converting it to NA or NaMN. NaAD, deamido NAD+. B, overview of the genetic screen to identify mutants that exhibit altered NR metabolism. Haploid single deletion mutants (feeder cells) are spotted onto a lawn of pyridine auxotrophic mutant npt1Δbna6Δpho5Δ (recipient cells), whose growth is dependent on NR released from the feeder cells in a dose-dependent manner. We have previously reported that Pho5 can convert extracellular NMN to NR. To rule out NMN unitization as a possible confounding factor in our assay, PHO5 was deleted in the recipient cell strain. C, deletion of YCL047C/POF1 confers increased NR release. Various feeder strains are spotted onto a lawn of pyridine auxotrophic npt1Δbna6Δpho5Δ recipient cells. The number and size of the satellite colonies surrounding the feeder spots positively correlate with the amount of NR released from the feeder cells. D, deleting POF1 in nrk1Δ (left panel) or nrt1Δ (right panel) mutant further enhances NR release. For clarity, inverse images are shown. ctrl, control.
To date, factors regulating NAD+ metabolism and homeostasis remain unclear because of the dynamic nature and redundancy of NAD+ synthesis pathways. One major challenge has been the lack of a specific and sensitive genetic screen system. Employing the property of yeast cells that constantly release and retrieve NR, we developed a unique NR reporter-based assay (14). A genetic screen utilizing this reporter system led to the identification of a novel NAD+ biosynthesis pathway component. In this study, we describe the role of POF1/YCL047C in NR and NAD+ homeostasis. We show evidence supporting the function of Pof1 as the third nicotinamide adenylyltransferase (NMNAT) in yeast.
EXPERIMENTAL PROCEDURES
Yeast Strains, Growth Media, and Plasmids
Haploid yeast strain BY4742 MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 acquired from Open Biosystems was used for this study (15). Yeast extract-peptone-dextrose (YPD) medium (2% Bacto peptone, 1% yeast extract, 1.5% agar supplemented with sterilized glucose at a final concentration of 2%) was made as described (16). All gene deletions were generated by replacing wild type genes with a reusable loxP-Kanr-loxP cassette as described (16). Multiple deletions were carried out by removing the Kanr marker using a galactose-inducible Cre-recombinase. The Nrk1 overexpression construct pADH1-Nrk1 was made in the integrative pPP81 (LEU2) vector as described (16). The resulting construct was verified by DNA sequencing. The POF1-URA3 episomal plasmid (pRS316-POF1) was derived from pADH1-POF1, which was constructed in the integrative pPP81 (LEU2) vector as described (16). pADH1-POF1 was digested with SacI and XhoI, and the excised fragment was ligated into pRS316 (URA3). The NRK1-URA3 episomal plasmid (pPS1527-NRK1) was constructed by PCR amplification of the NRK1 coding region with 1 kb upstream and downstream sequence. The PCR fragment was digested with NotI and XhoI and then ligated into pPS1527 (URA3).
NR Cross-feeding Spot Assays
To study mutants with altered NR release, 3 × 104 cells of each strain were spotted onto YPD plates spread with NAD+ auxotrophic recipient cells (the npt1Δbna6Δpho5Δ mutant or the qns1Δpho5Δ mutant) (14) at a density of ∼9,000 cells/cm2. Growth of the recipient cells relies on NR released from the spotted NAD+ prototrophic strains, and the extent of the recipient cell growth indicates the levels of NR release. After incubation at 30 °C for 3 days, we scored the cross-feeding activity of each strain by comparing the diameter of the cross-feeding zones to that of the wild type. Mutants with increased cross-feeding activity showed larger cross-feeding zones, whereas mutants with decreased cross-feeding activity showed smaller cross-feeding zones.
Determinations of NR and NAD+
Total intracellular levels of NAD+ were determined using an enzymatic cycling reaction as described (16, 17). Relative NR levels were determined by a liquid-based cross-feeding bioassay (12). To prepare cell extracts for intracellular NR determination, ∼1.5 × 109 (∼150 A600 unit cells) of cells (donors of interest) grown to late log-phase (∼12 h growth from an A600 of 0.1) were lysed by bead beating in 800 μl of ice-cold 50 mm ammonium acetate solution. After filter sterilization, 16 μl of cleared extract was used to supplement 8 ml of culture of recipient cells (the npt1Δbna6Δpho5Δ mutant) (12, 14) with starting A600 of 0.05 in YPD. To determine NR release levels, supernatant of donor cell culture was collected and filter-sterilized, and then 1 ml was added to 7 ml of recipient cell culture with starting A600 of 0.05 in YPD. A control culture of recipient cells in YPD without supplementation was included in all experiments. After incubation at 30 °C for 24 h, growth of the recipient cells (A600) was measured and normalized to the cell number of each donor strain. A600 readings were then converted to NR concentrations using the NR standard curve as previously described (12).
Fluorescence Microscopy
Strain expressing Pof1-GFP fusion protein was made by integrating GFP gene directly into the yeast genome at a site immediately upstream of WT POF1 using pFA6a-kanMX6-PGAL1-GFP as described (18). The resultant strain carries only the modified GFP-POF1 gene on its chromosome because haploid strains are used for our studies. GAL80 gene was deleted to allow Pof1-GFP expression in glucose-containing medium. Proper integration was verified by PCR, and the function of POF1 fused to the GFP tag was confirmed by a cell-based cross-feeding assay (see Fig. 3A, bottom panel). Localization analysis of Pof1-GFP was performed by fluorescence microscopy with a Nikon Eclipse 80i microscope at ×1,000 magnification.
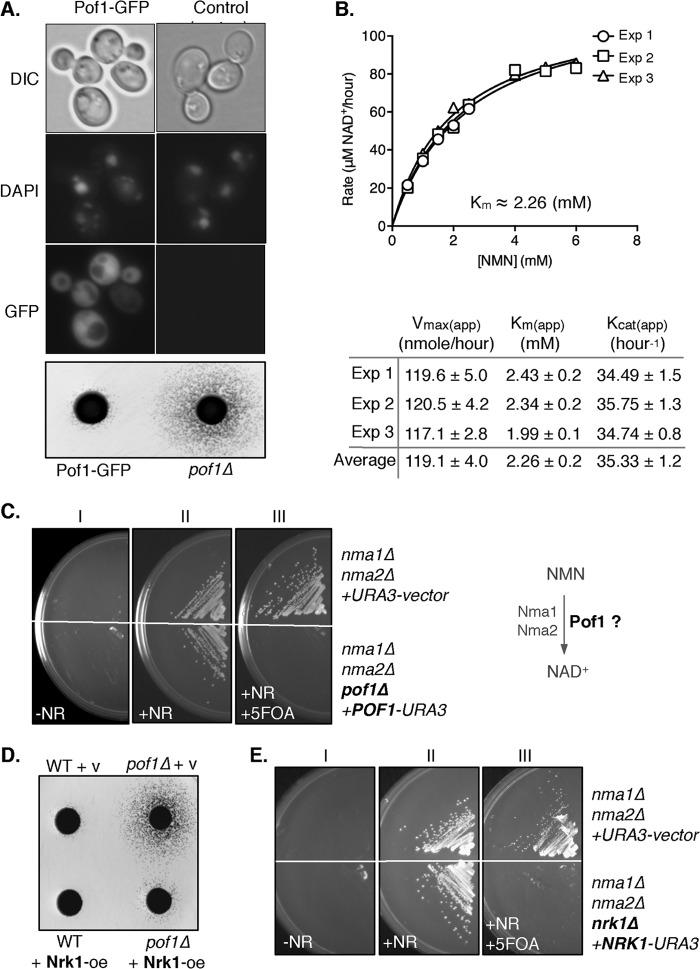
FIGURE 3.
Characterization of the Pof1 protein. A, cellular localization of Pof1 in yeast cells. The fluorescence signal of Pof1-GFP along with the differential interference contrast (DIC) and DAPI (which marks the nuclear and mitochondrial DNA) images indicate that Pof1 localizes both in the cytoplasm and the nucleus. Direct genomic integration of a GFP tag (upstream of the POF1 ORF) into haploid WT yeast genome does not interfere with the normal function of Pof1, because unlike the pof1Δ mutant, the GFP-tagged strain releases WT level of NR, determined by cross-feeding plate assay (bottom panel). B, rPof1 protein catalyzes the conversion of NMN to NAD+. The Michaelis-Menten plot of rPof1 is shown. The apparent kinetic parameters of rPof1 protein are shown in the table. C, Pof1 exhibits NMNAT activity in vivo. The results show that POF1 is required for the growth of nma1Δnma2Δ mutants on NR-supplemented medium. Cells lacking both NMA1 and NMA2 require NR for viability (panels I and II). 5-FOA is toxic to cells carrying the URA3 gene (panel III), which therefore forces cells to lose the URA3 plasmid for viability. Loss of the POF1-expressing plasmid (POF1-URA3) in the triple deletion mutant nma1Δnma2Δpof1Δ results in lethality even in the presence of NR (panel III, bottom panel), suggesting that Pof1 is indispensable in the absence of other NMNATs. D, Nrk1 overexpression rescues the high NR release phenotype of the pof1Δ mutant. E, Pof1 does not have NR kinase function. The results show that NRK1 is required for the growth of nma1Δnma2Δ on NR-supplemented medium. The loss of the NRK1-expressing plasmid (NRK1-URA3) in the triple deletion mutant nma1Δnma2Δnrk1Δ results in lethality even in the presence of NR (and POF1) (panel III, bottom panel), suggesting that Nrk1 is the only NR kinase in budding yeast. Exp, experiment.
Cloning, Expression, Purification, and Kinetic Analysis of Recombinant Pof1 (rPof1)
The POF1 ORF was amplified from yeast genomic DNA using primers 5′-ATTAGGATCCTATGAAGAAGACGTTCGAGCAGTTTCGAAA-3′ and 5′-AGGCCTCGAGATCAAATATTGTCTTATTATTGATCAAATAAT-3′. The amplified DNA was cloned into BamHI and XhoI sites of plasmid pET28a (Novagen). The resulting plasmid pET28-POF1 was transformed into Escherichia coli Rosetta (DE3) cells (Novagen), and cells were grown in M9 medium supplemented with 50 μg/ml of kanamycin and 25 μg/ml of chloramphenicol. Overexpression of Pof1 was induced with 0.5 mm isopropyl β-d-thiogalactopyranoside at 37 °C for 16–18 h. Harvested cells were frozen, resuspended in 10 volumes of lysis buffer (50 mm Tris-HCl, pH 8.0, 500 mm NaCl, 5 mm imidazole, 10% glycerol, 1 mm PMSF, 1 μg/ml leupeptin, 1 mg/ml lysozyme, 1 mg/ml DNase I, 0.1% Triton X-100), and lysed by multiple freeze and thaw cycles. Purification of Pof1 with N-terminal His6 tag was carried out using nickel-nitrilotriacetic acid-agarose resin (MCLab). The column was washed with buffer (50 mm Tris-HCl, pH 8.0, 500 mm NaCl) containing 30 mm imidazole, and the recombinant protein was eluted with buffer containing 250 mm imidazole.
Recombinant Pof1 activity was measured based on the NMNAT discontinuous assay described (19) with modifications. Briefly, 100 μg of rPof1 was added to a reaction mix containing 20 mm AMP-HCl, pH 10, 1 mm ATP, 5 mm MgCl2 and various concentrations of NMN and incubated for 1 h at 30 °C. NAD+ produced in this reaction was subsequently amplified by enzymatic cycling and measured in the same manner described for NAD+ measurement. The kinetic parameters for NMN were estimated from the results of three independent measurements (each carried out in triplicate) using GraphPad Prism 6. Fresh rPof1 was purified for each experiment shown in this study.
Replicative Life Span
All replicative life span (RLS) analyses were carried out on YPD plates supplemented with glucose at a final concentration of 2% with 50 cells/strain for each experiment (17) using a micromanipulator. Statistical analysis was carried out using the JMP statistics software (SAS), and the Wilcoxon rank sum test p values were calculated for each pair of life spans.
Quantitative PCR Analysis of Gene Expression Levels
Cells were grown to log phase or late log phase in YPD (∼6 or 12 h growth from A600 of 0.1). Total RNA was isolated using RNeasy mini kit, and cDNA was synthesized using Quantitect reverse transcription kit (Qiagen) according to the manufacturer's instructions. For each quantitative PCR, 50 ng of cDNA and 500 nm of each primer were used. Quantitative PCR was run on Roche LightCycler 480 using LightCycler 480 SYBR green I Master Mix (Roche) with the following cycle conditions: preincubation (95 °C for 5 min) and 43 cycles of amplification (denaturation at 95 °C for 10 s, annealing at 60 °C for 10 s, and extension at 72 °C for 10 s) followed by melting curve generation (65–97 °C, 0.11 °C per s). Average size of the amplicon for each gene was ∼150 bp. The target mRNA transcript levels were normalized to ACT1 transcript levels.
RESULTS
YCL047C/POF1 Affects NR Metabolism and Encodes a Putative NMNAT
To identify novel players in the NR/NAD+ metabolic pathway, we exploited the NR release property of yeast cells (12) and carried out a genetic screen to identify mutants that showed altered NR release activity, using the nonessential haploid single gene deletion mutants (20) (Fig. 1B). We found that cells lacking the ORF YCL047C/POF1 displayed strong cross-feeding activity (NR release), indicating altered NR metabolism in this mutant (Fig. 1C). When compared with previously characterized NR utilization mutants, nrk1Δ (defective in NR assimilation) and nrt1Δ (defective in NR transport), the pof1Δ mutant showed the strongest NR release phenotype, supporting a role for Pof1 in NR metabolism. Interestingly, pof1Δ appeared to further enhance NR release in these mutants (Fig. 1D).
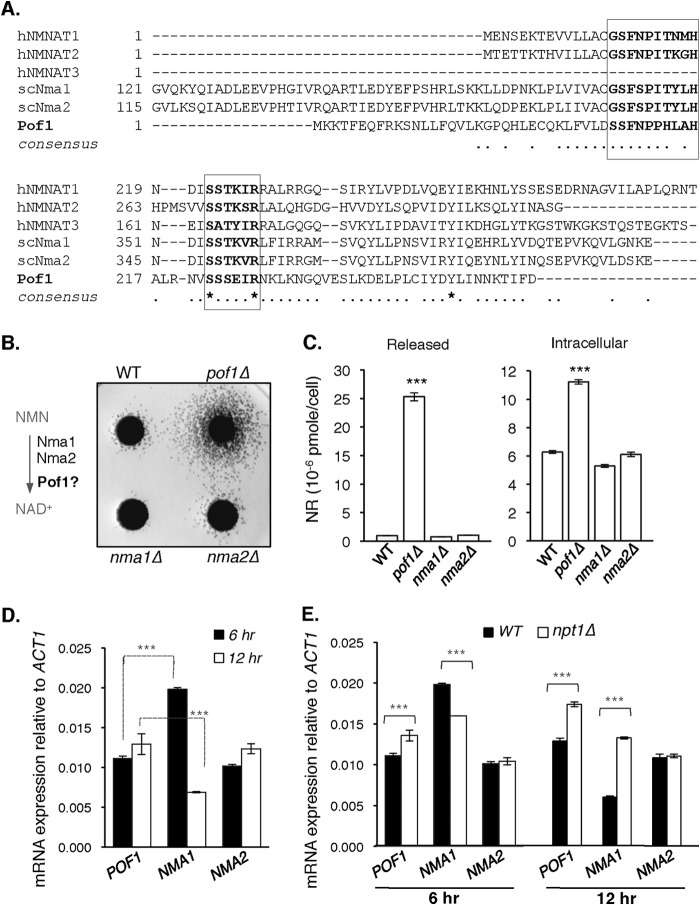
Pof1 was previously reported to harbor ATPase activity, which might be involved in protein quality control (21). Although the amino acid sequence of Pof1 does not show significant overall homology with any characterized protein, Pof1 possesses the characteristics of NMNAT. As shown in Fig. 2A, comparison of the amino acid sequence of Pof1 to known human NMNATs (hNMNAT1–3) and yeast NMNAT (Nma1 and Nma2) revealed that Pof1 possesses two ATP recognition motifs GXXXPX(T/H)XXH and SXTXXR, signature of all NMNATs characterized to date (22–24). To further understand the role of Pof1 in NR metabolism, we first examined whether pof1Δ and other yeast NMNAT mutants displayed similar phenotypes. Interestingly, only the pof1Δ mutant showed increased NR release determined by cross-feeding plate assays (Fig. 2B). This result was further confirmed using a quantitative liquid-based assay (12). As shown in Fig. 2C, pof1Δ mutant cells showed a ∼10-fold increase in NR release (Fig. 2C, left panel), which was accompanied by increased intracellular NR level (Fig. 2C, right panel). This unique phenotype of pof1Δ suggested that Pof1, Nma1, and Nma2 may play different roles in NR metabolism and therefore may be regulated differently. In line with this hypothesis, NMA1 expression is higher than NMA2 and POF1 during the early log phase (6 h), which significantly dropped as the cells entered the late log phase (12 h) (Fig. 2D). On the contrary, the expression of NMA2 and POF1 was slightly induced in the late log phase (Fig. 2D). Next, we determined whether their expression was affected by low intracellular NAD+. As shown in Fig. 2E, the expression of POF1 was significantly induced in a low NAD+ npt1Δ mutant (both 6 and 12 h), whereas NMA2 expression remained constant. Although NMA1 expression was not increased in the npt1Δ mutant during the early log phase (6 h), it remained stable and did not significantly drop (as in the WT cells) as cells entered late log-phase (12 h). These results suggested that NMA1 is the dominant NMNAT during early phases of growth when NA, the most abundant NAD+ precursor in standard growth media, is still abundant. As NA becomes depleted during the late log phase, NMA1 expression is decreased accordingly. In the npt1Δ mutant, the NA/Nam salvage route is blocked; therefore cells rely more on the NR salvage pathway for NAD+ synthesis (Fig. 1A). Dual function of NMA1 in both NA/Nam and NR salvage pathways may contribute to its complex expression patterns. Induction of POF1 expression in the npt1Δ mutant suggested that Pof1 might be a dominant NMNAT in the NR salvage pathway in conditions where NR is the main NAD+ precursor.
FIGURE 2.
POF1 encodes a putative NMNAT. A, amino acid sequence comparison between Pof1 and human NMNATs (hNMNAT1–3) and yeast NMNATs (scNma1 and scNma2). Pof1 possesses characterized ATP recognition motifs GXXXPX(T/H)XXH and SXTXXR. B, comparison of NR release in the pof1Δ mutant and other NMNAT deletion mutants, nma1Δ and nma2Δ. C, measurements of released (left panel) and intracellular (right panel) levels of NR in nma1Δ, nma2Δ, and pof1Δ mutants. NR is determined in both growth media (released) and cell extracts (intracellular). The pof1Δ mutant shows significant increases in both released and intracellular NR levels. D, comparison of gene expression of POF1 and other NMNATs (NMA1 and NMA2). E, comparison of NMNAT gene expression in wild type and the low NAD+ npt1Δ mutant. For C–E, the data shown are representative of three independent experiments each conducted in triplicate. The error bars denote standard deviations. The p values are calculated using Student's t test. ***, p < 0.005.
Pof1 Functions as an NMNAT, Which Catalyzes the Conversion of NMN to NAD+
To further characterize the function of Pof1, we first determined its cellular localization. Adding a GFP tag to Pof1 did not compromise its function, as determined by cross-feeding assays (Fig. 3A, bottom panel). Similar to Nma1, Nma2, and other salvage enzymes (25), Pof1 is localized both in the cytoplasm and the nucleus (Fig. 3A, top three panels). Next we examined whether Pof1 possessed NMNAT activity using His6-tagged rPof1. For comparison and as a validation of our assay system, we tested the activity of recombinant Nma1, which exhibited robust activity toward NMN (23, 26) (data not shown). A steady-state kinetic study was conducted on the conversion of NMN to NAD+ by rPof1. Using 100 μg of rPof1, Km (app) and Vmax for NMN were ∼2.26 mm and 119 (nmol/h), respectively. Km and Vmax for ATP were not determined because a higher ATP concentration (>1 mm) was inhibitory (data not shown) and steady-state approximation was not applicable (Fig. 3B). This pattern of inhibition indicates that Pof1 may exhibit an ordered sequential mechanism, which was also observed with human NMNAT1 (27, 28). Interestingly, Pof1 displayed an alkaline optimum pH (pH 10). Some NMNATs have been reported to be more active under alkaline conditions (29, 30). It is possible that alkaline pH might affect Pof1 structure to mimic its in vivo state, thereby influencing the catalytic activity.
Next, we examined whether Pof1 could function as an NMNAT in vivo. Because the adenylylation of NaMN or NMN is a required step in NAD+ synthesis (Fig. 1A), deleting both NMA1 and NMA2 is expected to result in synthetic lethality. However, we found that nma1Δnma2Δ cells were viable in media supplemented with NR (Fig. 3C, panels I and II). We therefore reasoned that a third NMNAT exists, which could function in the absence of NMA1 and NMA2 to support cell growth on NR. To test whether Pof1 fulfills this function, we examined whether the nma1Δnma2Δ mutant could grow when POF1 is lost. We constructed a triple deletion mutant nma1Δnma2Δpof1Δ carrying wild type POF1 on an episomal URA3 plasmid for viability in NR-supplemented media. When these cells were grown in media containing 5-FOA (which is toxic to URA3+ cells), the only way to remain viable was to lose the URA3 plasmids. Under this 5-FOA selection condition, nma1Δnma2Δpof1Δ mutant was unable to grow, even in the presence of NR, which indicated that Pof1 functions as the third NMNAT (Fig. 3C, panel III).
Furthermore, Pof1 may play other in vivo roles. Some eubacterial NMNATs are known to possess multifunctional roles (30–32). For example, NadR in Salmonella typhimurium is known as a trifunctional protein, which serves both regulatory and synthetic roles that contribute to pyridine nucleotide metabolism (32). NadR mainly functions as the transcriptional repressor of the de novo NAD+ pathway. However, it also functions as a NR kinase and possesses weak NMNAT activity (32). The uniquely strong NR release phenotype of the pof1Δ mutant suggests that Pof1 may also possess NR kinase activity. To test this, we first examined whether pof1Δ-induced NR release could be rescued by overexpressing Nrk1. Nrk1 overexpression complemented pof1Δ and lowered NR release back to the wild type level, suggesting that Pof1 and Nrk1 might have overlapping function (Fig. 3D). We next asked whether Pof1 could function as a NR kinase by testing whether Nrk1 is the only NR kinase in yeast. In the nma1Δnma2Δ mutant, Pof1 is essential for growth on NR (Fig. 3C). If Pof1 indeed has NR kinase activity, the nma1Δnma2Δ mutant should remain viable upon loss of NRK1 because they still have POF1. To test this, we constructed the nma1Δnma2Δnrk1Δ mutant carrying the wild type NRK1 on an episomal URA3 plasmid. Fig. 3E showed that these cells were unable to grow in NR-supplemented medium when forced to lose the NRK1-URA3 plasmid (via simultaneous selection on 5-FOA) (Fig. 3E). These results indicated that Nrk1 is the only NR kinase for the utilization of NR and that Pof1 does not possess NR kinase activity.
Pof1 Contributes to NAD+ Pool by Functioning in the NR Utilization Pathway
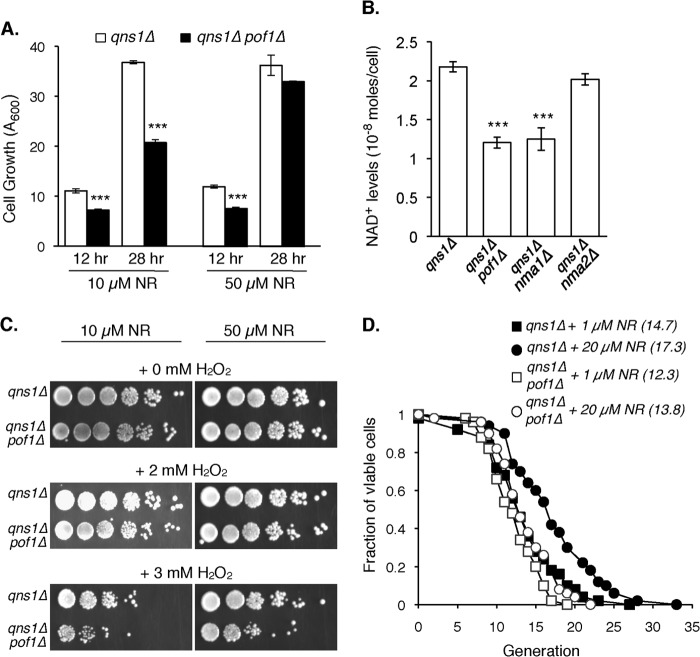
Next, we examined whether Pof1 plays a role in NAD+ production. Because of the complex functional redundancies in the NAD+ synthetic pathways (Fig. 1A), we employed the qns1Δ mutant to study the role of Pof1 specifically in the NR salvage pathway. In this NR-specific background, cells cannot synthesize NAD+ de novo or via the NA/Nam salvage pathway (Fig. 1A) and therefore depend on NR supplementation for viability. We took advantage of this NR-dependent property of qns1Δ cells and examined whether deleting POF1 would affect NR utilization. As shown in Fig. 4A (left panel), the qns1Δpof1Δ mutant exhibited a growth defect in standard NR-supplemented (10 μm) YPD medium. This pof1Δ-induced growth defect was specific to NR utilization deficiency because higher concentration of NR (50 μm) rescued the growth defect (Fig. 4A, right panel). Next, we determined intracellular NAD+ levels in qns1Δ mutant lacking POF1, NMA1, or NMA2. Deletion of POF1 or NMA1 significantly impaired NAD+ production, suggesting that POF1 and NMA1 both played important roles in the conversion of NMN to NAD+ (Fig. 4B). We also examined how POF1 deletion might affect cellular fitness. In line with the observation by Costa et al. (21), deleting POF1 in qns1Δ cells conferred sensitivity to H2O2, which was partially rescued by supplementing a higher concentration of NR (Fig. 4C). Functional NR salvage has been shown to play important roles in maintaining the replication potential of yeast cells (10, 12), and NR can restore the RLS of NAD+ biosynthetic mutants (10, 12). We therefore examined whether deleting POF1 would affect NR mediated RLS extension. As shown in Fig. 5D, RLS of the NR-dependent qns1Δ mutant was extended by higher concentrations of NR, and POF1 deletion blocked NR-induced RLS extension.
FIGURE 4.
Characterization of the pof1Δ mutants. A, deletion of POF1 significantly impairs the growth of NR-dependent cells. To enhance the sensitivity and specificity toward NR, we employ the pyridine auxotrophic qns1Δ mutant for NR-dependent growth assay. The results show the cell growth of qns1Δpof1Δ mutant in YPD supplemented with 10 μm or 50 μm NR. The growth defect of the qns1Δpof1Δ mutant in 10 μm NR (normal condition) (left) can be rescued with higher concentration of NR (50 μm) (right panel), suggesting that the observed growth defect in the qns1Δpof1Δ mutant (left panel) is mainly due to decreased NR utilization. B, comparisons of intracellular NAD+ levels in nma1Δ, nma2Δ, and pof1Δ deletion mutants in NR-dependent cells (qns1Δ). Cells are grown in YPD supplemented with 10 μm NR. Deletion of NMA1 or POF1 significantly decreases NAD+ levels. For A and B, the data shown are representative of three independent experiments, each conducted in triplicate. The error bars denote standard deviations. The p values are calculated using Student's t test. ***, p < 0.005. C, deletion of POF1 renders sensitivity toward H2O2. D, deletion of POF1 abolishes the RLS extension by NR. The results show the RLS of the qns1Δ and qns1Δpof1Δ cells grown in YPD containing different concentrations of NR. The average RLS of qns1Δ + 20 μm NR versus qns1Δpof1Δ + 20 μm NR are significantly different (p = 0.0013) as determined by the Wilcoxon rank sum test.
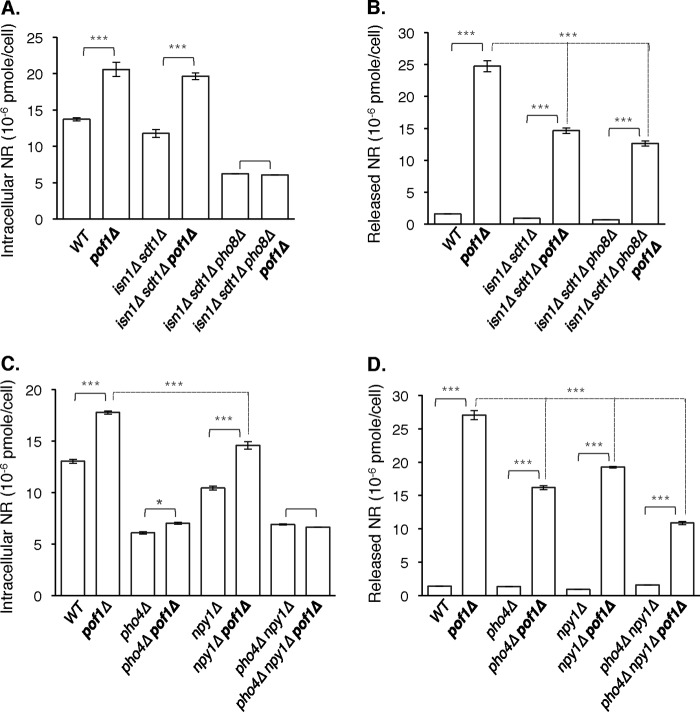
FIGURE 5.
Determining the nucleotidases that contribute to NR production in the pof1Δ mutant. A and B, deletions of ISN1, SDT1, and PHO8 completely abolish intracellular NR (A) increase but only partially reduce the NR release (B) in the pof1Δ mutant. C and D, deletion of NPY1 also significantly reduces increased intracellular NR (C) and released NR (D) in the pof1Δ mutant. Deletion of NPY1 further decreases the NR level in the pho4Δ mutant. The data shown are representative of three independent experiments, each conducted in triplicate. The error bars denote standard deviations. The p values are calculated using Student's t test. *, p < 0.05; ***, p < 0.005.
Multiple Phosphatases and Nucleotidases Contribute to NR Production in the pof1Δ Mutant
Based on our results thus far, we have concluded that Pof1 is the third NMNAT in Saccharomyces cerevisiae. Cells lacking POF1 release and accumulate high levels of NR, and it is possible that blocked NMN utilization shifts/increases the flux toward NR production. Cytosolic nucleotidases Isn1 and Sdt1 and the vacuolar phosphatase Pho8 have previously been shown to hydrolyze NMN to NR (13, 14). We first tested whether these enzymes contribute to increased NR production in pof1Δ mutant. Consistent with previous reports, deletion of ISN1 and SDT1 significantly decreased NR release in pof1Δ cells, and PHO8 deletion did not further decrease the level of release (Fig. 5B). Conversely, the intracellular NR level remained high in pof1Δisn1Δsdt1Δ cells but significantly decreased when combined with pho8Δ (Fig. 5A). This phenomenon could be due to compartmentalization of intracellular NR pools; NR is produced both in the cytosol and the vacuole is and stored mainly in the vacuole. NR produced by cytosolic nucleotidases (Isn1 and Sdt1) is more likely to be assimilated or excreted; therefore measured intracellular NR is predominantly the vacuolar fraction (mainly affected by Pho8).
Because these mutants did not lower released NR to wild type level, we sought to identify other nucleotidases that may contribute to NR production. A link between NAD+ metabolism and the inorganic phosphate-sensing PHO pathway has been established in our previous studies (14). Also, Pho8 (33) and Sdt1 (34) have been either shown or suggested to be under PHO regulation. Therefore, we reasoned that additional nucleotidases/phosphatases under PHO regulation might be involved in NR production. To test this, we examined intracellular and released NR levels in the pho4Δ mutant, in which PHO signaling is blocked (35). PHO4 deletion significantly decreased both intracellular (Fig. 5C) and released NR (Fig. 5D) in pof1Δ cells. Interestingly, deletion of PHO4 also reduced intracellular NR level in the wild type background (Fig. 5C). To identify nucleotidases that are not under PHO regulation, we screened a set of genes based on previously characterized property of hydrolyzing nucleotides and pyridine nucleotides. Deletion mutants of these genes were tested for NR release (data not shown). We found that deleting NPY1 was sufficient to moderately reduce intracellular NR and NR release in pof1Δ cells, and the NR level was further decreased when combined with pho4Δ (Fig. 5D). NPY1 was characterized as a peroxisomal NAD+(H) pyrophosphatase (or Nudix hydrolase) which could produce NMN(H) from NAD+(H) (36). These results suggested that cells constantly convert NAD+(H) to NMN by Npy1-like enzymes. NMN is subsequently converted to NR by nucleotidases/phosphatases for release, storage, or reassimilation into NAD+. In the pof1Δ mutant, NMN assimilation into NAD+ is decreased; therefore, there is more NMN flow to the NR production route. Overall, our studies demonstrated the dynamic nature of interchangeable NAD+ intermediate pools and identified several key players.
DISCUSSION
In this study, we characterized Pof1 as a novel component of yeast NAD+ synthesis pathway. Our genetic screen revealed that deleting POF1 significantly increased NR levels, which suggested a role for Pof1 in NR and NAD+ metabolism. We showed that Pof1 is endowed with NMNAT activity that is specific for NMN in vitro and in vivo. Although rPof1 appeared to be a less efficient NMNAT in vitro when compared with rNma1 and rNma2, which exhibit low Km toward NMN (∼ 100 μm) (23), Pof1 activity may be robust in its physiological context. Precedents exist for efficient enzymes that exhibit high Km for their substrates. For example, Nma1 has high Km for NaMN (∼5 mm) but is a key enzyme in the de novo and NA salvage pathways for NAD+ synthesis (27), and deletion of Nma1 significantly decreases NAD+ levels (12). Our study also suggested that Pof1 activity is specific for NMN unlike Nma1 and Nma2, which exhibit dual substrate specificity toward NMN and NaMN (Fig. 1A) (23, 24, 26). Although we did not directly determine the catalytic activity of Pof1 toward NaMN, Km for NMN was not affected under high concentration of NaMN (4 mm), indicating that there is no competition or inhibition by NaMN (data not shown). In addition, if Pof1 could adenylylate NaMN, the nma1Δnma2Δ mutant would not be lethal without NR supplement (Fig. 3C). Collectively, these studies indicated that Pof1 is the only other NMNAT, and it contributes to NAD+ biosynthesis by converting NMN to NAD+.
Although it is the third NMNAT in yeast, one striking phenotype associated with POF1 deletion (but not with deletions of the other two NMNAT) is the significantly increased NR production. Given the NMNAT activity of Pof1 and its substrate specificity for NMN, this mutant phenotype is likely due to decreased utilization of NMN. One interesting question is why pof1Δ mutant cells release more NR. Our study revealed that multiple nucleotidases are responsible for increased NR production in pof1Δ (Fig. 5, A and B). However, expression of these nucleotidase genes did not appear to be significantly up-regulated in pof1Δ mutant (data not shown). Therefore, these nucleotidases may constantly convert NMN to NR to maintain the flow of NAD+ synthesis and salvage. This flow is altered in the pof1Δ mutant, leading to increased NR accumulation because NMN, a substrate for the nucleotidases, becomes more available when Pof1 is absent. Interestingly, one novel factor contributing to increased NR level in pof1Δ mutant is Npy1, a peroxisomal Nudix hydrolase that produces NMN (36). It remains unclear how this peroxisomal enzyme contributes to NAD+ homeostasis. In addition, increased NR production has also been associated with activated PHO signaling (14). It would be interesting to determine the interaction between Pof1 and these NMN/NR producing factors.
Although Pof1 appears to share redundant function with Nma1 and Nma2, the significance of Pof1 in NAD+ synthesis in the NR salvage pathway is evident. These NMNATs may have distinct roles because their expression varies under different growth conditions (Fig. 2, D and E). It appears that POF1 expression is stimulated when NAD+ level decreases such as in late growth phase or in low NAD+ mutants. NMA1 expression is more complicated in that it can be stimulated under both high NAD+ and low NAD+ conditions. NMA1 expression may also be influenced by certain NAD+ precursors such as NA. Supporting this possibility, we showed that NMA1 expression remained high in the low NAD+ npt1Δ mutant in late growth stage (Fig. 2E). In npt1Δ mutant, NA salvaging is blocked, and depletion of NA is expected to be slower. Additional evidence supports that NA also regulates other NAD+ salvaging factors. Bogan et al. (13) reported that Isn1 protein expression responds positively to extracellular NA and glucose availability. This suggests that cytosolic degradation of NMN is regulated in response to available extracellular resources. It is possible that NAD+ synthesis is mainly mediated via the NA/Nam route when NA is abundant, and the key NMNAT involved is Nma1, whose expression is induced by NA. In saturated culture, depletion of NA might signal the cells to switch the route of NAD+ synthesis from NA/Nam salvage to de novo synthesis and NR salvage.
NMNAT family proteins have been reported to affect cellular functions independent of their enzymatic function. For example, Salmonella NadR is a multifunctional protein with a role in transcriptional regulation (32). Some eukaryotic NMNATs have been reported to possess cytoprotective functions. For example, chaperone activity that contributes to axonal protection was described for Drosophila NMNAT (37). In yeast, Nma1 and Nma2 have been shown to alleviate proteotoxicity in yeast models of proteinopathies (38). Studies by Costa et al. (21) suggested that Pof1 has a role in protein quality control. Their study also showed that pof1Δ exhibited compromised resistance to oxidative stress, heat shock, and chemical-induced ER stress. In our study, cells lacking POF1 also showed increased sensitivity to oxidative stress in NR-dependent mutant background (Fig. 4C). Apparent low catalytic activity of Pof1, together with the strong NR production phenotype (Fig. 2E), suggests that Pof1 may also maintain NMN/NAD+ homeostasis by sequestering NMN. For example, the majority of NADH was found to be protein-bound, and only a small fraction exists as free molecules (2, 39, 40). Therefore, it is possible that a significant fraction of NMN is bound to Pof1, and increased NR accumulation in pof1Δ mutant may be a result of the degradation of increased free NMN. Supporting this, pof1Δ further enhanced the NR release phenotypes of the nrk1Δ and nrt1Δ mutants (Fig. 1D) (Nrk1 and Nrt1 are two major players in NR salvage pathway) (Fig. 1A), suggesting that Pof1 does not simply function to convert NMN to NAD+. In addition, low levels of free NMN can still be efficiently assimilated into NAD+, because Nma1 and Nma2 have low Km for NMN (23). Further studies are required to understand the interaction among these NMNATs and to elucidate whether they possess additional roles in other cellular processes.
Our results suggest that intracellular NR pools are compartmentalized into two major pools: the stored pool and the cytosolic pool. The stored NR pool is maintained mainly in the vacuole, where Pho8 is a major NR-producing enzyme. The cytosolic NR pool is more dynamic and mainly generated from NMN by nucleotidases such as Isn1 and Sdt1. NR in this pool can be excreted or transported to the vacuole if not assimilated into NAD+. For each individual cell, the level of released NR reflects the size of its dynamic cytosolic NR pool, and the steady-state NR level determined in total cell extract reflects the size of the stored pool. Supporting this idea, deletions of genes encoding cytosolic nucleotidases Isn1 and Sdt1 significantly decreased NR release (cytosolic pool) (Fig. 5B) but only slightly affected intracellular NR level (stored pool) (Fig. 5A) in pof1Δ mutant. Likewise, deleting PHO8 largely decreased intracellular NR level (Fig. 5A) but only slightly affected the level of NR release (Fig. 5B) in pof1Δ mutant. Deleting these genes in wild type cells caused similar effects, but at a smaller scale (Fig. 5, A and B) (14), because wild type cells have lower levels of intracellular NR and NR release compared with pof1Δ cells. This model predicts that perturbing the vacuolar NR storage will increase cytosolic NR, and therefore more NR is released. Indeed, cells lacking a putative NR transporter Fun26 showed increased NR release (14). In higher eukaryotes, stored NR pool is likely to reside in the lysosome. Human equilibrative nucleoside transporters are Fun26 homologs that facilitate the transport of variety of purine and pyrimidine nucleosides (41), which may also have a role in intracellular NR homeostasis. Vacuolar NR production and uptake of extracellular NR may be stimulated concurrently to replenish the cytosolic NR pool for NAD+ synthesis when cells are in need for alternative NAD+ precursors.
Intracellular concentrations of many of the NAD+ intermediates are maintained at low levels (42), which is characteristic of signaling molecules. NR and NMN, similar to other NAD+ intermediates, may function as signaling molecules to regulate NAD+ homeostasis or other cellular processes. We have previously discovered that low NaMN level is associated with activation of the PHO pathway (14). Moreover, nicotinic acid adenine dinucleotide phosphate has been shown to function as a signaling molecule to regulate calcium homeostasis in variety of organisms (43, 44). Supporting this possibility, our results showed that most NR is stored in the vacuole and that Pof1 may also function to sequester NMN. In addition to functioning as signaling molecules, high concentrations of intracellular NAD+ intermediates may be unfavorable for certain cellular processes. For example, NAD+-dependent DNA ligase in bacteria is inhibited by NMN, and it is suggested that NMN deamidase contributes to maintaining a small intracellular NMN pool (45). In addition, Nam is known as an inhibitor of Sir2, and clearance of Nam is critical for maintaining Sir2 activity and life span (46). Nam clearance is facilitated by a Nam deamidase Pnc1 in yeast or Nampt, a Nam phosphoribosyltransferase in mammals (47, 48). A recent report described that Nam is also methylated, and this modification induces a hormetic response to protect cells from oxidative damage (49). In this study, we showed that yeast cells constantly convert NMN to NR, which is more mobile and can be readily excreted, stored, or reassimilated. A recent report showed that QA is also produced and excreted like NR (50). Thus, it is possible that pyridine nucleotides and their metabolites are involved in a variety of cellular processes, and balancing their concentrations would be critical for the regulation of these processes. Overall, our studies have contributed to the understanding of the complex NAD+ homeostasis pathways and may also provide insights into the underlying mechanisms of diseases related to defects in NAD+ metabolism.
Acknowledgments
We thank members of the Lin laboratory for discussions and suggestions and Dr. T. Powers for providing the pFA6a-kanMX6-PGAL1-GFP plasmid. We also thank Dr. J. Roth, Dr. D. Wilson, and Dr. C. Brenner for suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM102297.
- NaMN
- nicotinic acid mononucleotide
- NR
- nicotinamide riboside
- NMNAT
- nicotinamide mononucleotide adenylyltransferase
- NMN
- nicotinamide mononucleotide
- NA
- nicotinic acid
- QA
- quinolinic acid
- Nam
- nicotinamide
- RLS
- replicative life span
- rPof1
- recombinant Pof1
- 5-FOA
- 5-fluoroorotic acid.
REFERENCES
- 1. Lin S. J., Defossez P. A., Guarente L. (2000) Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289, 2126–2128 [DOI] [PubMed] [Google Scholar]
- 2. Lin S. J., Guarente L. (2003) Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr. Opin. Cell. Biol. 15, 241–246 [DOI] [PubMed] [Google Scholar]
- 3. Rusche L. N., Kirchmaier A. L., Rine J. (2003) The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72, 481–516 [DOI] [PubMed] [Google Scholar]
- 4. Bürkle A. (2005) Poly(ADP-ribose). The most elaborate metabolite of NAD+. FEBS J. 272, 4576–4589 [DOI] [PubMed] [Google Scholar]
- 5. Chini E. N. (2009) CD38 as a regulator of cellular NAD: a novel potential pharmacological target for metabolic conditions. Curr. Pharm. Des. 15, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Panozzo C., Nawara M., Suski C., Kucharczyka R., Skoneczny M., Bécam A. M., Rytka J., Herbert C. J. (2002) Aerobic and anaerobic NAD+ metabolism in Saccharomyces cerevisiae. FEBS Lett. 517, 97–102 [DOI] [PubMed] [Google Scholar]
- 7. Preiss J., Handler P. (1958) Biosynthesis of diphosphopyridine nucleotide: II. enzymatic aspects. J. Biol. Chem. 233, 493–500 [PubMed] [Google Scholar]
- 8. Preiss J., Handler P. (1958) Biosynthesis of diphosphopyridine nucleotide: I. identification of intermediates. J. Biol. Chem. 233, 488–492 [PubMed] [Google Scholar]
- 9. Bieganowski P., Brenner C. (2004) Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 117, 495–502 [DOI] [PubMed] [Google Scholar]
- 10. Belenky P., Racette F. G., Bogan K. L., McClure J. M., Smith J. S., Brenner C. (2007) Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 129, 473–484 [DOI] [PubMed] [Google Scholar]
- 11. Sporty J., Lin S. J., Kato M., Ognibene T., Stewart B., Turteltaub K., Bench G. (2009) Quantitation of NAD+ biosynthesis from the salvage pathway in Saccharomyces cerevisiae. Yeast 26, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu S. P., Kato M., Lin S. J. (2009) Assimilation of endogenous nicotinamide riboside is essential for calorie restriction-mediated life span extension in Saccharomyces cerevisiae. J. Biol. Chem. 284, 17110–17119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bogan K. L., Evans C., Belenky P., Song P., Burant C. F., Kennedy R., Brenner C. (2009) Identification of Isn1 and Sdt1 as glucose- and vitamin-regulated nicotinamide mononucleotide and nicotinic acid mononucleotide [corrected] 5′-nucleotidases responsible for production of nicotinamide riboside and nicotinic acid riboside. J. Biol. Chem. 284, 34861–34869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu S. P., Lin S. J. (2011) Phosphate-responsive signaling pathway is a novel component of NAD+ metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 286, 14271–14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 16. Easlon E., Tsang F., Skinner C., Wang C., Lin S. J. (2008) The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev. 22, 931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Easlon E., Tsang F., Dilova I., Wang C., Lu S. P., Skinner C., Lin S. J. (2007) The dihydrolipoamide acetyltransferase is a novel metabolic longevity factor and is required for calorie restriction-mediated life span extension. J. Biol. Chem. 282, 6161–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 19. Balducci E., Emanuelli M., Raffaelli N., Ruggieri S., Amici A., Magni G., Orsomando G., Polzonetti V., Natalini P. (1995) Assay methods for nicotinamide mononucleotide adenylyltransferase of wide applicability. Anal. Biochem. 228, 64–68 [DOI] [PubMed] [Google Scholar]
- 20. Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., Chu A. M., Connelly C., Davis K., Dietrich F., Dow S. W., El Bakkoury M., Foury F., Friend S. H., Gentalen E., Giaever G., Hegemann J. H., Jones T., Laub M., Liao H., Liebundguth N., Lockhart D. J., Lucau-Danila A., Lussier M., M'Rabet N., Menard P., Mittmann M., Pai C., Rebischung C., Revuelta J. L., Riles L., Roberts C. J., Ross-MacDonald P., Scherens B., Snyder M., Sookhai-Mahadeo S., Storms R. K., Véronneau S., Voet M., Volckaert G., Ward T. R., Wysocki R., Yen G. S., Yu K., Zimmermann K., Philippsen P., Johnston M., Davis R. W. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 [DOI] [PubMed] [Google Scholar]
- 21. Costa I. M., Nasser T. H., Demasi M., Nascimento R. M., Netto L. E., Miyamoto S., Prado F. M., Monteiro G. (2011) The promoter of filamentation (POF1) protein from Saccharomyces cerevisiae is an ATPase involved in the protein quality control process. BMC Microbiol. 11, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garavaglia S., D'Angelo I., Emanuelli M., Carnevali F., Pierella F., Magni G., Rizzi M. (2002) Structure of human NMN adenylyltransferase. A key nuclear enzyme for NAD homeostasis. J. Biol. Chem. 277, 8524–8530 [DOI] [PubMed] [Google Scholar]
- 23. Emanuelli M., Amici A., Carnevali F., Pierella F., Raffaelli N., Magni G. (2003) Identification and characterization of a second NMN adenylyltransferase gene in Saccharomyces cerevisiae. Protein Expr. Purif. 27, 357–364 [DOI] [PubMed] [Google Scholar]
- 24. Magni G., Amici A., Emanuelli M., Orsomando G., Raffaelli N., Ruggieri S. (2004) Structure and function of nicotinamide mononucleotide adenylyltransferase. Curr. Med. Chem. 11, 873–885 [DOI] [PubMed] [Google Scholar]
- 25. Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Cohen H., Lin S. S., Manchester J. K., Gordon J. I., Sinclair D. A. (2002) Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 277, 18881–18890 [DOI] [PubMed] [Google Scholar]
- 26. Emanuelli M., Carnevali F., Lorenzi M., Raffaelli N., Amici A., Ruggieri S., Magni G. (1999) Identification and characterization of YLR328W, the Saccharomyces cerevisiae structural gene encoding NMN adenylyltransferase: expression and characterization of the recombinant enzyme. FEBS Lett. 455, 13–17 [DOI] [PubMed] [Google Scholar]
- 27. Natalini P., Ruggieri S., Raffaelli N., Magni G. (1986) Nicotinamide mononucleotide adenylyltransferase: molecular and enzymatic properties of the homogeneous enzyme from baker's yeast. Biochemistry 25, 3725–3729 [DOI] [PubMed] [Google Scholar]
- 28. Emanuelli M., Carnevali F., Saccucci F., Pierella F., Amici A., Raffaelli N., Magni G. (2001) Molecular cloning, chromosomal localization, tissue mRNA levels, bacterial expression, and enzymatic properties of human NMN adenylyltransferase. J. Biol. Chem. 276, 406–412 [DOI] [PubMed] [Google Scholar]
- 29. Raffaelli N., Lorenzi T., Mariani P. L., Emanuelli M., Amici A., Ruggieri S., Magni G. (1999) The Escherichia coli NadR regulator is endowed with nicotinamide mononucleotide adenylyltransferase activity. J. Bacteriol. 181, 5509–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kurnasov O. V., Polanuyer B. M., Ananta S., Sloutsky R., Tam A., Gerdes S. Y., Osterman A. L. (2002) Ribosylnicotinamide kinase domain of NadR protein: identification and implications in NAD biosynthesis. J. Bacteriol. 184, 6906–6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singh S. K., Kurnasov O. V., Chen B., Robinson H., Grishin N. V., Osterman A. L., Zhang H. (2002) Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenyltransferase and ribosylnicotinimide kinase activities. J. Biol. Chem. 277, 33291–33299 [DOI] [PubMed] [Google Scholar]
- 32. Grose J. H., Bergthorsson U., Roth J. R. (2005) Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica. J. Bacteriol. 187, 2774–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaneko Y., Tamai Y., Toh-e A., Oshima Y. (1985) Transcriptional and post-transcriptional control of PHO8 expression by PHO regulatory genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 5, 248–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu Y. F., Létisse F., Absalan F., Lu W., Kuznetsova E., Brown G., Caudy A. A., Yakunin A. F., Broach J. R., Rabinowitz J. D. (2013) Nucleotide degradation and ribose salvage in yeast. Mol. Syst. Biol. 9, 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lenburg M. E., O'Shea E. K. (1996) Signaling phosphate starvation. Trends Biochem. Sci. 21, 383–387 [PubMed] [Google Scholar]
- 36. AbdelRaheim S. R., Cartwright J. L., Gasmi L., McLennan A. G. (2001) The NADH diphosphatase encoded by the Saccharomyces cerevisiae NPY1 nudix hydrolase gene is located in peroxisomes. Arch. Biochem. Biophys 388, 18–24 [DOI] [PubMed] [Google Scholar]
- 37. Zhai R. G., Zhang F., Hiesinger P. R., Cao Y., Haueter C. M., Bellen H. J. (2008) NAD synthase NMNAT acts as a chaperone to protect against neurodegeneration. Nature 452, 887–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ocampo A., Liu J., Barrientos A. (2013) NAD+ salvage pathway proteins suppress proteotoxicity in yeast models of neurodegeneration by promoting the clearance of misfolded/oligomerized proteins. Hum. Mol. Genet. 22, 1699–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sporty J. L., Kabir M. M., Turteltaub K. W., Ognibene T., Lin S. J., Bench G. (2008) Single sample extraction protocol for the quantification of NAD and NADH redox states in Saccharomyces cerevisiae. J. Sep. Sci. 31, 3202–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sies H. (1982) Metabolic Compartmentation, pp. 205–231, Academic Press, Orlando, FL [Google Scholar]
- 41. Young J. D., Yao S. Y., Sun L., Cass C. E., Baldwin S. A. (2008) Human equilibrative nucleoside transporter (ENT) family of nucleoside and nucleobase transporter proteins. Xenobiotica 38, 995–1021 [DOI] [PubMed] [Google Scholar]
- 42. Evans C., Bogan K. L., Song P., Burant C. F., Kennedy R. T., Brenner C. (2010) NAD+ metabolite levels as a function of vitamins and calorie restriction: evidence for different mechanisms of longevity. BMC Chem. Biol. 10, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clapper D. L., Walseth T. F., Dargie P. J., Lee H. C. (1987) Pyridine nucleotide metabolites stimulate calcium release from sea urchin egg microsomes desensitized to inositol trisphosphate. J. Biol. Chem. 262, 9561–9568 [PubMed] [Google Scholar]
- 44. Guse A. H., Lee H. C. (2008) NAADP: a universal Ca2+ trigger. Sci. Signal. 1, re10. [DOI] [PubMed] [Google Scholar]
- 45. Cheng W., Roth J. (1995) Isolation of NAD cycle mutants defective in nicotinamide mononucleotide deamidase in Salmonella typhimurium. J. Bacteriol. 177, 6711–6717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gallo C. M., Smith D. L., Jr., Smith J. S. (2004) Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol. Cell. Biol. 24, 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bitterman K. J., Anderson R. M., Cohen H. Y., Latorre-Esteves M., Sinclair D. A. (2002) Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277, 45099–45107 [DOI] [PubMed] [Google Scholar]
- 48. Revollo J. R., Grimm A. A., Imai S. (2004) The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 279, 50754–50763 [DOI] [PubMed] [Google Scholar]
- 49. Schmeisser K., Mansfeld J., Kuhlow D., Weimer S., Priebe S., Heiland I., Birringer M., Groth M., Segref A., Kanfi Y., Price N. L., Schmeisser S., Schuster S., Pfeiffer A. F., Guthke R., Platzer M., Hoppe T., Cohen H. Y., Zarse K., Sinclair D. A., Ristow M. (2013) Role of sirtuins in lifespan regulation is linked to methylation of nicotinamide. Nat. Chem. Biol. 9, 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ohashi K., Kawai S., Murata K. (2013) Secretion of quinolinic acid, an intermediate in the kynurenine pathway, for utilization in NAD+ biosynthesis in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 12, 648–653 [DOI] [PMC free article] [PubMed] [Google Scholar]