Background: Outer membrane protein assembly is an incompletely understood process in Gram-negative bacteria.
Results: A Neisseria meningitidis mutant lacking the lipoprotein GNA2091 is affected in growth and accumulates unassembled outer membrane proteins.
Conclusion: We identified a novel component involved in outer membrane biogenesis.
Significance: Our findings contribute to the understanding of a fundamental process occurring in Gram-negative bacteria.
Keywords: Bacteria, Membrane Biogenesis, Membrane Protein, Microbiology, Vaccine, BAM Complex, Neisseria meningitidis, YraP
Abstract
GNA2091 of Neisseria meningitidis is a lipoprotein of unknown function that is included in the novel 4CMenB vaccine. Here, we investigated the biological function and the subcellular localization of the protein. We demonstrate that GNA2091 functions in the assembly of outer membrane proteins (OMPs) because its absence resulted in the accumulation of misassembled OMPs. Cell fractionation and protease accessibility experiments showed that the protein is localized at the periplasmic side of the outer membrane. Pulldown experiments revealed that it is not stably associated with the β-barrel assembly machinery, the previously identified complex for OMP assembly. Thus, GNA2091 constitutes a novel outer membrane-based lipoprotein required for OMP assembly. Furthermore, its location at the inner side of the outer membrane indicates that protective immunity elicited by this antigen cannot be due to bactericidal or opsonic activity of antibodies.
Introduction
The cell envelope of Gram-negative bacteria is composed of two membranes separated by the peptidoglycan-containing periplasm. The outer membrane (OM)2 constitutes a highly asymmetric bilayer containing phospholipids and lipopolysaccharides (LPSs) in the inner and outer leaflets, respectively. Furthermore, in contrast to the α-helical proteins found in most biological membranes, the integral outer membrane proteins (OMPs) are composed of amphipathic antiparallel β-strands forming a β-barrel structure. The biogenesis of the OM has been a major focus of research lately. It was boosted by the discoveries of the Lpt machinery for the transport of LPS to the OM (1–4) and of the assembly machinery for OMPs, designated the β-barrel assembly machinery (BAM) (5, 6).
The central component of the Bam, known as Omp85/NmBamA in Neisseria meningitidis or EcBamA in Escherichia coli, is conserved in all Gram-negative bacteria and even in eukaryotic cell organelles of endosymbiont origin, mitochondria, and chloroplasts (7, 8). However, the accessory proteins also present in the complex are less well conserved (9). In E. coli, the Bam complex contains four accessory lipoproteins designated BamB, BamC, BamD, and BamE (6, 10). In N. meningitidis, a homolog of BamB is missing, but the complex contains an additional component, RmpM, that is not a lipoprotein (11). With its N-terminal domain, RmpM binds and stabilizes OMP complexes while it anchors the OM via its C-terminal domain to the peptidoglycan layer (12). In Caulobacter crescentus, a BamC homolog is lacking, but two other lipoproteins, i.e. Pal, which, like RmpM of N. meningitidis, has a peptidoglycan-binding motif, and BamF, appear to be part of the complex (9, 13). The mitochondrial β-barrel OMP assembly machinery does not contain homologs of any of the bacterial lipoproteins, but it does contain two unrelated accessory proteins (7). Besides BamA, only BamD, which is also known as ComL in N. meningitidis, is essential in the bacterial systems. The other accessory components are dispensable for growth, and their deletion only results in mild OMP assembly and OM permeability defects (6, 10, 11).
In this study, we investigated the possible involvement of the meningococcal lipoprotein GNA2091 in OM biogenesis. GNA2091 is included in a novel four-component meningococcal serogroup B vaccine, 4CMenB, developed by Novartis (14). This vaccine contains three major immunogenic proteins plus an OM vesicle preparation. GNA2091 is included in this vaccine as a fusion with one of the major immunogenic antigens because it was demonstrated to elicit protective immunity against meningococcal disease in a mouse model (15). The biological function of GNA2091 is not known. Interestingly, GNA2091 is a homolog of a lipoprotein called YraP in E. coli. The yraP gene was reported to be a member of the σE regulon, which is induced when the bacteria sense envelope stress. Furthermore, its absence was shown to result in an SDS-sensitive phenotype, which is indicative of a defect in OM integrity (16). These data suggest that YraP might have a role in envelope homeostasis, but its function has never been elucidated. In this study, we demonstrate that its homolog GNA2091 has a role in OMP biogenesis.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The N. meningitidis strains used in this study are listed in Table 1. They were grown at 37 °C in candle jars on GC agar plates (Oxoid) supplemented with Vitox (Oxoid) and when necessary antibiotics (10 μg/ml chloramphenicol or 80 μg/ml kanamycin). Liquid cultures were grown in tryptic soy broth (TSB) (BD Biosciences). To achieve depletion of proteins encoded by genes cloned behind an isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible promoter, cells grown overnight on plates containing 1 μm IPTG were resuspended in TSB without IPTG to an absorbance at 550 nm (A550) of 0.1 and grown for 6 h. To induce expression of IPTG-regulated genes, 0.5 mm IPTG was added to the medium. E. coli strain DH5α, which was used for routine cloning experiments, was grown on LB agar plates at 37 °C or in liquid LB medium supplemented when appropriate with antibiotics (25 μg/ml chloramphenicol, 100 μg/ml ampicillin, or 50 μg/ml kanamycin).
TABLE 1.
N. meningitidis strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source/Ref. |
|---|---|---|
| Strains | ||
| HB-1 | Unencapsulated derivative of serogroup B strain H44/76 | 1 |
| HB-1ΔcomL(pComL) | HB-1 containing pEN11-ComL with the chromosomal comL copy replaced by a kanR cassette | 11 |
| HB-1ΔGNA2091 | HB-1 with GNA2091 replaced by a kanR cassette | This work |
| HB-1ΔrmpM | HB-1 with rmpM replaced by a kanR cassette | 11 |
| HB-1ΔGNA2091(pGNA2091) | HB-1ΔGNA2091 carrying pEN11-GNA2091 | This work |
| HB-1ΔGNA2091(pGNA2091-His) | HB-1ΔGNA2091 carrying pEN11-GNA2091-His | This work |
| HB-1ΔbamC | HB-1 with bamC replaced by a catR cassette | 18 |
| HB-1ΔsurA | HB-1 with surA replaced by a catR cassette | 18 |
| HB-1skp::cat | HB-1 with a catR cassette inserted in skp | 18 |
| HB-1ΔbamCΔGNA2091 | HB-1ΔbamC with GNA2091 replaced by a kanR cassette | This work |
| HB-1ΔsurAΔGNA2091 | HB-1ΔsurA with GNA2091 replaced by a kanR cassette | This work |
| HB-1skp::catΔGNA2091 | HB-1skp::cat with GNA2091 replaced by a kanR cassette | This work |
| Plasmids | ||
| pCRII-TOPO | TA cloning vector for PCR products | Invitrogen |
| pCRII-ΔGNA2091 | GNA2091 deletion plasmid | This work |
| pMB25 | pCRII-TOPO with imp inactivation construct containing kanR cassette | 1 |
| pEN11-Imp | Neisseria replicative plasmid containing H44/76-derived imp under lac promoter control | 1 |
| pEN11-GNA2091 | pEN11-Imp with imp replaced by HB-1 derived GNA2091 | This work |
| pEN11-GNA2091-His | pEN11-Imp derivative encoding C-terminally His-tagged GNA2091 | This work |
| pRIT15566 | E. coli expression plasmid encoding GNA2091 without signal sequence and with C-terminal His6 tag | GSKa |
a GlaxoSmithKline Biologicals S.A., Rixensart, Belgium.
Plasmid and Mutant Constructions
Plasmids are listed in Table 1. Primers (listed in Table 2) were designed based on the genome sequence of serogroup B strain MC58 (17). A deletion construct of GNA2091 (locus tag NMB2091 in MC58 and NMBH4476_2037 in strain H44/76) was obtained by amplifying DNA fragments upstream and downstream of the gene by PCR using genomic DNA of strain HB-1, a non-encapsulated derivative of strain H44/76, as template and primer pairs NMB2090-for/NMB2091-rev-Acc and NMB2091-for-Acc/NMB2092-rev. The fragments were cloned into pCRII-TOPO and joined together in one plasmid using the AccI sites that were introduced via the primers and the XbaI and/or SpeI sites in the vector. A kanamycin resistance (kanR) cassette including the neisserial DNA uptake sequence was obtained from pMB25 and inserted into the plasmid after AccI restriction. The inactivation construct was verified to contain the kanR cassette in the same transcriptional direction as GNA2091. N. meningitidis strains were transformed as described (1) with a PCR fragment obtained from the gene replacement construct using primers M13Rev and M13For. The transformants were checked by PCR for the presence of the mutant allele using primer pair NMB2090-for and NMB2092-rev and for the absence of the wild-type allele using primer pair GNA2091-for1 and NMB2092-rev. Double skp::catΔGNA2091, ΔsurAΔGNA2091 and ΔbamCΔGNA2091 mutants were obtained by introducing the ΔGNA2091::kan allele into previously constructed ΔsurA::cat, ΔbamC::cat, and insertional skp::cat mutants (11, 18). For complementation purposes, GNA2091 was amplified by PCR using genomic DNA of HB-1 as template and primers GNA2091-for-NdeI and GNA2091-rev-AatII. The resulting PCR product was cloned into pCRII-TOPO and subcloned into the neisserial replicative plasmid pEN11-Imp via NdeI/AatII restriction and ligation. For construction of a C-terminally His-tagged version of GNA2091, we used plasmid pRIT15566, which comprises an expression construct of GNA2091 with a C-terminal His tag but without a signal sequence. This plasmid was used as template in a PCR with primers GNA2091-for2 and GNA2091-rev-His. The resulting PCR product was cloned into pCRII-TOPO, yielding pCRII-GNA2091-His. An AccI-AatII fragment from this plasmid was ligated into similarly restricted pEN11-GNA2091, yielding pEN11-GNA2091-His, allowing for expression of full-length GNA2091 including its signal sequence and with a C-terminal His6 tag.
TABLE 2.
Primers used in this study
| Primer name | Sequence (5′–3′) | Restriction sitea |
|---|---|---|
| GNA2091-for2 | GAGATATACATATGGTCAGC | NdeI |
| GNA2091-rev-His | ATGACGTCAGCAGCCGGATCTCAGTG | AatII |
| GNA2091-for1 | AAGGCTACACGCCCCAAATC | |
| NMB2091-rev-Acc | ATGTCGACGGTGTGCGGTTTGG | AccI |
| NMB2090-for | TACAGGCTGCCGAGCTGATGC | |
| NMB2092-rev | GGGTGTCCGGTTTGGTATGG | |
| NMB2091-for-Acc | ATGTCGACAGAACAGGCGCAGATTACC | AccI |
| GNA2091-for-NdeI | ATCATATGAAACCCAAACCGC | NdeI |
| GNA2091-rev-AatII | ATGACGTCAGCGTTGGACGTAGTTTTGG | AatII |
| M13-for | GTAAAACGACGGCCAGT | |
| M13-rev | AACAGCTATGACCATG |
a Restriction sites are underlined in the sequences.
Cell Fractionations
To collect proteins from extracellular media, bacterial cultures were centrifuged for 5 min at 10,000 × g in an Eppendorf centrifuge. Proteins were precipitated from 1 ml of supernatant with 7% TCA for 1 h at 4 °C. The pellet obtained after centrifugation for 30 min at 16,000 × g was washed twice with ice-cold acetone and boiled in SDS-PAGE sample buffer. Cell envelopes were prepared as described (11). Inner and outer membranes were separated by sucrose gradient centrifugation as described (19).
Protease Accessibility Experiments
One milliliter of HB-1 cells grown in TSB to an A550 of 2.0 was centrifuged for 3 min at 8000 rpm in an Eppendorf centrifuge and resuspended in 0.5 ml of 10 mm Tris-HCl, 5 mm MgCl2, pH 7.6. Proteinase K was added at 0, 0.2, or 2 μg/ml for 20 min at room temperature. After addition of 2 mm phenylmethylsulfonyl fluoride, cells were collected by centrifugation and subjected to denaturing SDS-PAGE.
Pulldown Experiments
Strains HB-1ΔGNA2091(pGNA2091)and HB-1ΔGNA2091(pGNA2091-His) were grown in 100 ml of TSB plus IPTG to an A550 of 2.5. Cells were harvested, and cell envelopes were isolated after freeze-thawing and ultrasonic disruption of the cells. These were extracted with 1.25% Elugent (Calbiochem) in 20 mm Tris-HCl, 150 mm NaCl, pH 7.5 (TBS) for 2 h at room temperature and centrifuged for 1 h at 100,000 × g. The supernatant was incubated with Ni2+-nitrilotriacetic acid-agarose beads (Qiagen) in TBS containing 0.1% Elugent and 20 mm imidazole (wash buffer) for 16 h at 4 °C. The beads were washed five times with wash buffer. Bound material was eluted with wash buffer containing 300 mm imidazole. Samples were analyzed by denaturing SDS-PAGE and immunoblotting.
SDS-PAGE and Immunoblotting
Protein samples were analyzed by seminative or denaturing SDS-PAGE and immunoblotting as described (11). Band intensity on immunoblots was quantified by calculating the mean value of all pixels in the band after inversion of the grayscale image of the blot using Adobe Photoshop CS6. These means were used to calculate ratios between mono- and multimers (for PilQ) or percent difference in signal strength between mutant and parent (for NalP and NhhA). Equal loading of samples was verified by quantification of cross-reacting, nonspecific bands and by staining of blots with 0.1% Ponceau red before incubating with antibodies. For LPS analysis, cell lysates were treated with proteinase K before SDS-PAGE analysis, and gels were stained with silver as described (1).
Antisera
Antisera against the translocator domains of IgA protease and NalP (20) were produced in our laboratory. Antibodies against NhhA, PilQ, GNA2091, NmBamA, and fHbp were gifts from GlaxoSmithKline (Rixensart, Belgium), and mAbs directed against FbpA, PorA, and PorB were generously provided by Peter van der Ley (InTraVacc, Bilthoven, The Netherlands).
RESULTS
A GNA2091 Deletion Mutant Displays Major Defects in OM Integrity and Anchoring
To investigate the function of GNA2091, we created a deletion mutant by replacing the major part of the gene by a kanamycin resistance cassette in strain HB-1. Deletion mutants were easily obtained, showing that the GNA2091 gene is not essential. The mutant, however, grew more slowly than its parent (Fig. 1A). SDS-PAGE analyses of the total cellular proteins revealed no major differences (Fig. 1B), but the culture supernatant of the ΔGNA2091 mutant contained increased amounts of many proteins (Fig. 1C). Neisserial strains are known to release OM vesicles profusely, a process that is strongly enhanced in an rmpM mutant (21) and that can be clearly detected by the appearance of the two major meningococcal OMPs, the porins PorA and PorB, in the culture medium (11) (Fig. 1C). Increased protein levels in spent medium can also be diagnostic for periplasmic leakage, a process observed in mutants deficient in LPS biosynthesis or transport (22) or in strains depleted for essential Bam complex components such as ComL, the BamD homolog in N. meningitidis (11). The extracellular protein profile of the ΔGNA2091 mutant showed bands characteristic of both a ComL-depleted strain and a ΔrmpM mutant (Fig. 1C), demonstrating that the cells are leaky and blebbing, which is indicative for major defects in OM integrity and anchoring.
FIGURE 1.
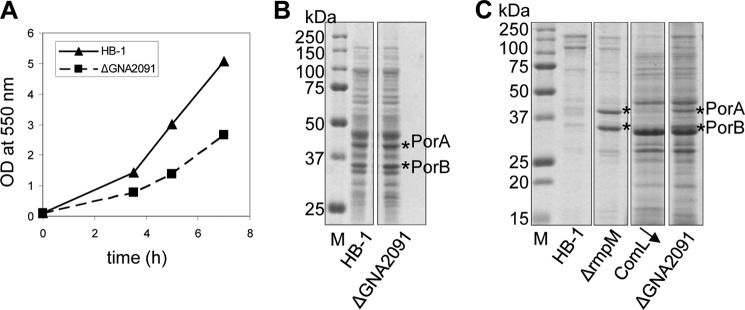
Phenotype of the ΔGNA2091 mutant. A, the growth of strain HB-1 and its ΔGNA2091 mutant derivative in TSB was monitored by A550 measurements at intervals. B, equal numbers of cells (based on A550) were collected after 6 h of growth in TSB by centrifugation and used to prepare cellular lysates for protein analysis. C, proteins were precipitated from equal volumes of supernatant with trichloroacetic acid. Samples in B and C were subjected to denaturing SDS-PAGE and Coomassie Blue staining. For comparison, the extracellular protein profiles of a ΔrmpM mutant and a ComL-depleted strain (ComL↓) are also shown. Porins are indicated by asterisks. M, molecular weight markers.
The ΔGNA2091 Strain Is Affected in Porin Assembly
The GNA2091 gene is located downstream of and possibly in the same operon as gmhA, which encodes phosphoheptose isomerase, an enzyme involved in the synthesis of an LPS precursor. Hence, we first considered the possibility that GNA2091 is involved in LPS transport. To address this possibility, we analyzed cellular LPS levels because defects in LPS transport in N. meningitidis lead to a feedback inhibition on LPS synthesis (1, 22). SDS-PAGE analysis, however, revealed similar LPS levels in parent and mutant cells (Fig. 2A).
FIGURE 2.
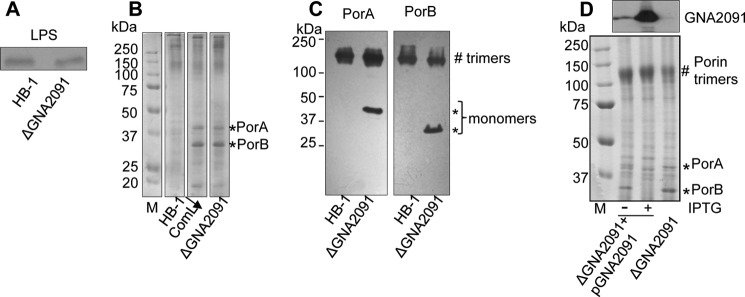
Effect of the ΔGNA2091 mutation on LPS synthesis and porin assembly. A, lysates prepared from equal numbers of cells (based on A550) were treated with proteinase K, subjected to denaturing SDS-PAGE, and stained with silver to visualize LPS. B and C, cell envelopes of the indicated strains were subjected to seminative SDS-PAGE followed by Coomassie Blue staining (B) or by immunoblotting with antibodies directed against PorA or PorB as indicated above the panels (C). For comparison, the protein profile of a ComL-depleted strain (ComL↓) is shown in B. D, the ΔGNA2091 mutant containing the GNA2091 gene on a plasmid (ΔGNA2091+pGNA2091) was grown with or without IPTG as indicated. Cell envelopes were isolated and subjected to denaturing SDS-PAGE followed by immunoblotting with anti-GNA2091 antiserum (upper panel) or to seminative SDS-PAGE followed by Coomassie Blue staining (lower panel). Monomers (*) and trimers (#) of the porins are indicated. M, molecular weight markers.
We next assessed OMP assembly by analyzing cell envelopes of wild type and mutant in seminative SDS-PAGE. In the wild-type strain, the porins PorA and PorB migrate as trimers when analyzed under such conditions (5, 11). In contrast, a considerable proportion of the porins in cell envelopes from the ΔGNA2091 mutant strain migrated as monomers (Fig. 2, B and C), indicating that porin assembly is compromised. A similar porin assembly defect was detected in cell envelopes from a ComL-depleted strain (11) (Fig. 2B). To confirm that the assembly defect was caused by the absence of GNA2091, a plasmid with the GNA2091 gene under an IPTG-inducible promoter was introduced into the ΔGNA2091 mutant. Porin assembly was indeed completely restored in the complemented strain upon induction of expression of GNA2091 with IPTG (Fig. 2D, lower panel). Even in the absence of IPTG, the porin assembly defect appeared somewhat reduced in the complemented strain, which is likely due to leakiness of the promoter as revealed by immunoblotting with GNA2091-specific antiserum (Fig. 2D, upper panel). Thus, GNA2091 is required for the efficient assembly of porins into their native trimeric conformation.
The Absence of GNA2091 Affects Assembly of Multiple OMPs
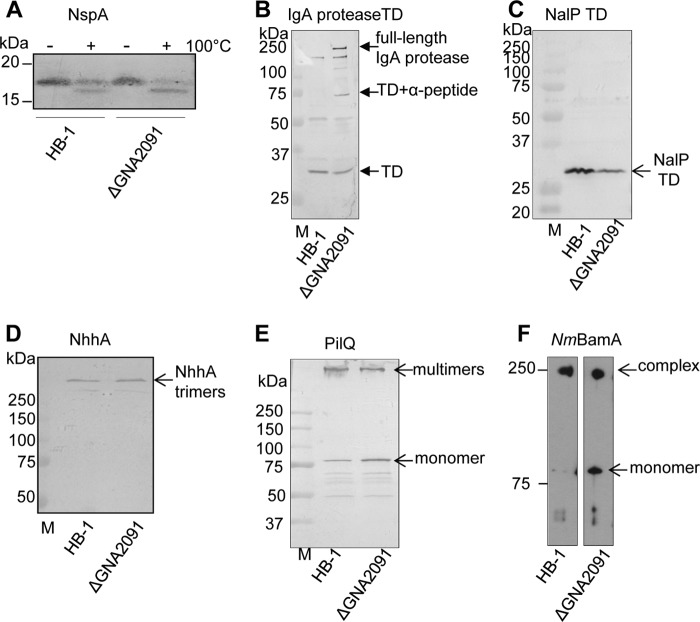
To test whether GNA2091 has a general role in OMP assembly, we analyzed the assembly of several other OMPs in the ΔGNA2091 mutant. NspA is a small eight-stranded β-barrel OMP whose correctly folded form migrates more slowly in seminative SDS-PAGE than the denatured form (18). Using this assay, we found that all NspA was folded in the ΔGNA2091 mutant (Fig. 3A). Because NspA is a very stable β-barrel, boiled samples still showed some folded NspA protein (Fig. 3A). Autotransporters consist of a C-terminal translocator domain that inserts as a β-barrel into the OM and facilitates the translocation of the associated passenger domain to the cell surface. In the case of IgA protease, the passenger is then released from the cell surface either by autocatalytic cleavage or by the activity of another autotransporter protease, NalP. When NalP is expressed, only the translocator domain of IgA protease remains behind in the OM. When NalP is not active, autocatalytic processing of IgA protease leads to the release of a smaller passenger domain (23), and a larger fragment consisting of the translocator domain and a so-called α-peptide remain detectable in the cell envelopes.3 Immunoblotting of cell envelopes from ΔGNA2091 mutant cells demonstrated the accumulation of significant amounts of unprocessed, full-length IgA protease (Fig. 3B), suggesting a defect in the biogenesis of this autotransporter. Furthermore, a band corresponding to the translocator domain associated with the α-peptide was detected (Fig. 3B), suggesting a defect also in the biogenesis of NalP. Accordingly, although we did not detect accumulation of unprocessed NalP in the cell envelopes of the ΔGNA2091 mutant, considerably lower levels of its translocator domain were found (Fig. 3C); quantification of the blots showed that this amount was reduced to 62% of the wild-type level. The misassembled full-length NalP protein is probably degraded in this case. The autotransporter NhhA forms very stable trimers that resist boiling in SDS-containing sample buffer. Assembly of NhhA apparently did not require GNA2091 as we detected similar levels (96%) of the trimeric protein in the ΔGNA2091 mutant as in the parent strain and no accumulation of the monomeric form (Fig. 3D). The secretin PilQ forms large stable complexes in the OM whose assembly was demonstrated to be affected by the absence of NmBamA or ComL (BamD) (5, 11). Also in the ΔGNA2091 mutant, we found relative accumulation of monomeric PilQ and decreased amounts of multimers (Fig. 3E; multimer:monomer ratio, 1.79 for parent and 1.06 for the mutant strain), indicating suboptimal assembly of this OMP. Under seminative SDS-PAGE conditions, a stable oligomeric subcomplex of the Bam complex can be detected that consists of NmBamA and the RmpM protein but not the accessory lipoproteins (11). The formation of this complex was also affected by the absence of GNA2091 (Fig. 3F). Thus, the absence of GNA2091 affects the assembly of many but not all OMPs.
FIGURE 3.
Role of GNA2091 in assembly of various OMPs. Cell envelopes of strain HB-1 and its ΔGNA2091 mutant derivative were subjected to seminative (A and F) or denaturing (B–E) SDS-PAGE followed by immunoblotting with antibodies against the proteins indicated above the blots. TD, translocator domain; M, molecular weight markers. A, boiled (+) and unboiled (−) samples were run in seminative SDS-PAGE conditions. C, mean pixel values for the NalP TD bands: 90.4 for HB-1 and 56.4 for the ΔGNA2091 mutant. D, mean pixel values for the NhhA trimers: 49.2 for HB-1 and 50.0 for the ΔGNA2091 mutant. E, mean pixel values for PilQ multimer and monomer bands: 65.8 and 36.8, respectively, for HB-1 and 61.02 and 57.71, respectively, for the ΔGNA2091 mutant.
GNA2091 Is Localized to the Periplasmic Side of the OM
GNA2091 is likely a lipoprotein because it contains a so-called lipobox at the end of the signal sequence that directs proteins to the lipoprotein-processing enzymes (Fig. 4). Fluorescence-activated cell sorting analysis of intact meningococci stained with anti-GNA2091 antibodies showed only slight surface exposure of the protein compared with established surface-exposed antigens such as fHbp (15). Furthermore, only some but not all strains were positive with anti-GNA2091 in this analysis (15), making its exact subcellular location unclear. To address this matter, we first assessed whether GNA2091 is associated with the inner or the outer membrane by subjecting cell envelopes to sucrose density gradient centrifugation. Analysis of gradient fractions (Fig. 5A) showed that GNA2091 co-fractionated with the porins in the OM fractions and not with the DsbA proteins, which in N. meningitidis are two co-migrating inner membrane lipoproteins (19). Next, we treated intact cells with limited amounts of protease to test whether GNA2091 is cell surface-exposed. As a control, we assessed the protease accessibility of PorA and the lipoprotein fHbp, which are known to be surface-exposed (18, 24), and of the periplasmic iron-binding protein FbpA. Whereas fHbp and PorA were readily accessible to proteinase K, no degradation of GNA2091 was detected (Fig. 5B). This was not due to intrinsic protease resistance because it was readily degraded when cell envelopes were treated with proteinase K (Fig. 5C). Thus, GNA2091 is not exposed at the bacterial cell surface.
FIGURE 4.
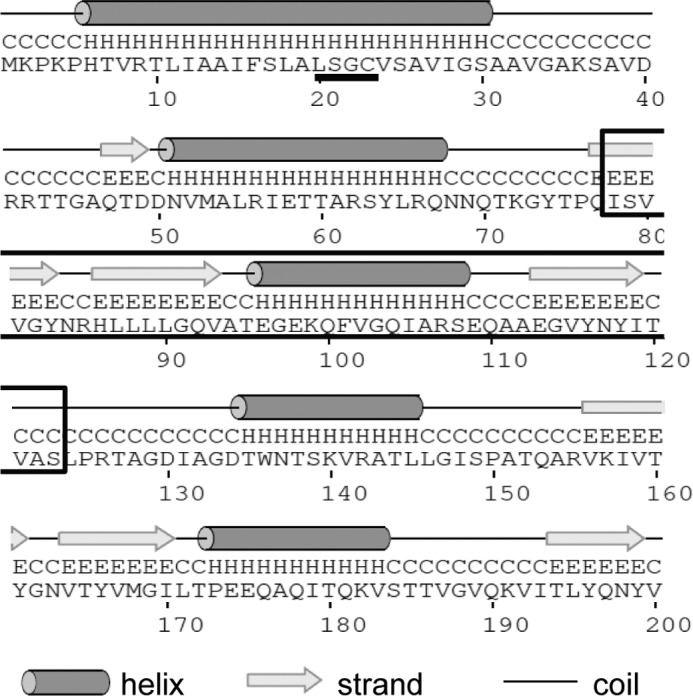
Secondary structure prediction of GNA2091. The amino acid sequence of GNA2091 was submitted to the PSIPRED server for secondary structure analysis and to the Conserved Domain Database for protein domain identification. The lipobox sequence (LSGC) is underlined, and the BON domain is boxed.
FIGURE 5.
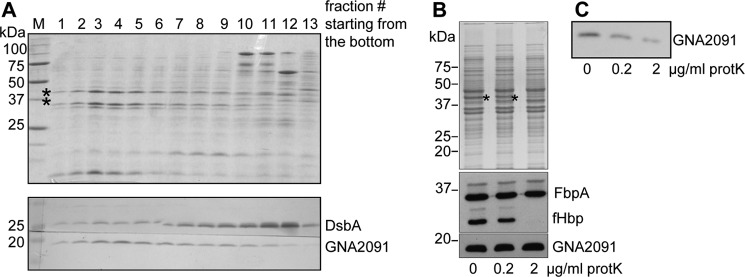
Subcellular localization of GNA2091. A, cell envelope preparations were subjected to centrifugation on sucrose density gradients. The porins (indicated by asterisks) were used as markers for the OM and are visualized on the Coomassie Blue-stained SDS-PAGE gel (upper panel). The DsbA lipoproteins were used as a marker for the inner membrane. The localization of DsbA and GNA2091 proteins in the different fractions was revealed by immunoblotting (lower panels). B, HB-1 cells were treated with the indicated concentrations of proteinase K and processed for denaturing SDS-PAGE followed by Coomassie Blue staining (upper panel) or immunoblotting (lower panels) with antibodies directed against the proteins indicated at the right side of the blots. The proteinase K-sensitive PorA is indicated with an asterisk. C, cell envelopes derived from strain HB-1 were treated with the indicated concentrations of proteinase K and processed for denaturing SDS-PAGE followed by immunoblotting with anti-GNA2091 antiserum. M, molecular weight markers; protK, proteinase K.
GNA2091 Is Not Stably Associated with the Bam Complex
To test whether GNA2091 might be a novel component of the Bam complex, we constructed a plasmid encoding a C-terminally His-tagged version of GNA2091 and verified that it complemented the GNA2091 deletion in a similar way as the untagged GNA2091 (results not shown). Cell envelopes isolated from the ΔGNA2091 mutant strain containing this plasmid or as a control a plasmid encoding an untagged version of GNA2091 were extracted with a mild detergent, and the extracted proteins were subjected to Ni2+ affinity chromatography. The His-tagged GNA2091 was pulled down using this procedure (Fig. 6, A and B), but SDS-PAGE analysis of the elution fractions did not reveal any other specifically co-purifying proteins (Fig. 6A). Immunoblotting with anti-NmBamA antibodies did not reveal any association of NmBamA with GNA2091 (Fig. 6B). Hence, GNA2091 does not constitute a stably associated component of the Bam complex, although transient interactions cannot be excluded at this stage.
FIGURE 6.

GNA2091 is not present in stable complexes. A and B, pulldown experiments were performed using cells expressing His-tagged GNA2091 (lanes 1 and 3) or untagged GNA2091 (lanes 2 and 4) as control. Lanes 1 and 2 represent the cell envelopes used as starting material; lanes 3 and 4 show the elution fractions from Ni2+-nitrilotriacetic acid-agarose purification. Samples were subjected to denaturing SDS-PAGE and stained with Coomassie Blue (A) or blotted and probed with antibodies against the proteins indicated on the right side of the blots (B). The arrow indicates His-tagged GNA2091. M, molecular weight markers.
Genetic Interactions of GNA2091
The simultaneous inactivation of two non-essential genes encoding proteins involved in the same process may lead to synthetic defects. In a search for such synthetic defects, we combined the ΔGNA2091 mutation with mutations in the genes for the non-essential Bam complex component BamC and for the periplasmic chaperones Skp and SurA. All double mutants were easily obtained, and their growth was indistinguishable from that of the ΔGNA2091 single mutant (results not shown). Furthermore, the ΔbamC and ΔsurA mutations did not exacerbate the porin assembly defect of the ΔGNA2091 single mutant (Fig. 7). However, the ΔGNA2091 and skp::cat mutations clearly showed additive effects. Whereas the ΔGNA2091 mutant showed accumulation of unassembled monomeric porins (Fig. 2, B and C), N. meningitidis strains lacking Skp produce lower levels of trimeric porins without concomitant accumulation of porin monomers (18). The skp::catΔGNA2091 double mutant demonstrated a more drastic reduction in assembled trimeric porins than the single skp::cat mutant and an amount of unassembled monomeric porins very similar to that in the ΔGNA2091 single mutant (Fig. 7). Thus, the relative amount of unassembled porins in the skp::catΔGNA2091 double mutant is much higher than in the strain lacking only GNA2091.
FIGURE 7.
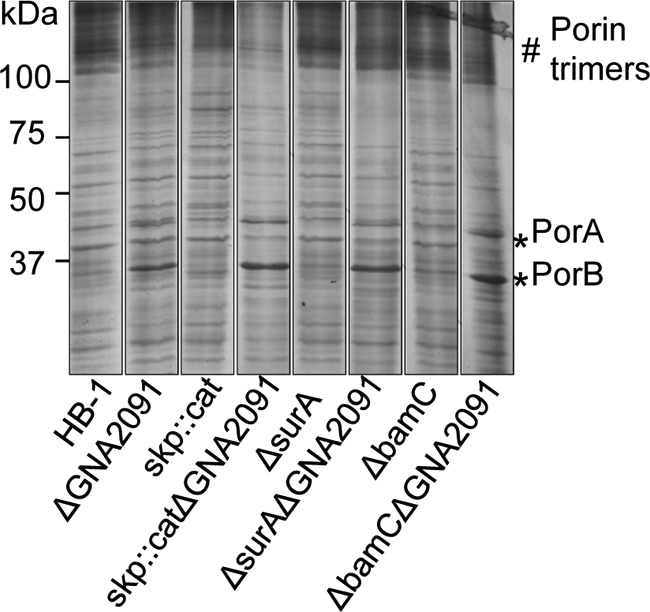
Genetic interactions of GNA2091. Cell envelopes of the indicated strains were subjected to seminative SDS-PAGE followed by Coomassie Blue staining. Monomers (*) and trimers (#) of the porins are indicated.
DISCUSSION
GNA2091 is a component of the novel 4CMenB vaccine of hitherto unknown function. We demonstrate here that this lipoprotein is localized at the periplasmic side of the OM. This conclusion is corroborated by the amino acid sequence of GNA2091, which is highly conserved among N. meningitidis strains (25). N. meningitidis is notorious for its ability to vary the amino acid sequence of surface-exposed proteins to avoid immune recognition by the host. Therefore, a low level of sequence variation in a neisserial protein is often diagnostic of an intracellular location. Furthermore, anti-GNA2091 antibodies raised in mice were not bactericidal (15). Thus, all evidence points to an intracellular location of GNA2091. GNA2091 was selected for inclusion in the 4CMenB vaccine for its ability to induce protection in a mouse intraperitoneal infection model (15). Given the intracellular location of GNA2091, this protective activity cannot be due to bactericidal or opsonic activity of anti-GNA2091 antibodies. GNA2091 possibly has a general immune stimulating activity and could function as an adjuvant in the 4CMenB vaccine.
We investigated the function of GNA2091 and demonstrate that it is required for the efficient assembly of a subset of OMPs in N. meningitidis. Accordingly, a GNA2091 mutant was reported previously to display increased sensitivity toward SDS and Tween, suggesting infringement of the OM barrier function (26). Similarly, inactivation of the GNA2091 homolog YraP of E. coli resulted in an SDS-sensitive phenotype (16). Therefore, it seems likely that also YraP in E. coli has a role in OMP assembly, although this has not yet been demonstrated.
The yraP gene in E. coli is a member of the σE regulon; its expression is induced when the cells sense envelope stress (16). Other members of this regulon include the genes for the periplasmic OMP chaperones Skp and SurA and for the periplasmic protease DegP, which degrades misfolded OMPs (27). A similar stress response system is lacking in N. meningitidis (28). σE is a member of the extracellular function family of σ factors. N. meningitidis contains only a single extracellular function σ factor, which regulates the expression of a small regulon that neither includes GNA2091 nor any other gene known to be involved in envelope biogenesis (29). Hence, it remains to be determined whether and how expression of GNA2091 is regulated. GNA2091 up-regulation was found in a N. meningitidis hfq mutant, suggesting that it can be regulated by small RNAs (30).
Remarkably, our pulldown experiments did not reveal stable association of GNA2091 with the Bam complex or with any other complex. These findings are consistent with several studies that addressed the composition of outer and inner membrane complexes in E. coli. Using various approaches, these studies identified all the known Bam complex components consistently together in one complex, but none of them reported on the presence of GNA2091 homolog YraP in any complex (31–33). In this respect, it is noteworthy that also a stable interaction between BamA and BamD was recently reported to be unnecessary for proper functioning of the Bam complex in E. coli (34).
To study possible genetic interactions of GNA2091, we constructed a series of double mutants. A ΔsurA mutation did not aggravate the OMP assembly defect of the ΔGNA2091 mutant. In contrast, studies in E. coli where SurA is considered to be the major OMP chaperone revealed synthetic lethality between yraP and surA mutations (16, 35), suggesting that these proteins may have overlapping functions. However, in N. meningitidis, a surA single mutant is not noticeably affected in OMP assembly (18). Thus, the lack of synthetic defects of the ΔsurA and ΔGNA2091 mutations confirms the different roles of SurA in N. meningitidis and in E. coli. Also, a ΔbamC mutation, which showed only mild, if any, OMP assembly defects on its own (11), did not exacerbate the defects of the ΔGNA2091 mutation. In contrast, the skp::cat and ΔGNA2091 mutations did demonstrate additive effects. Previously, we demonstrated that all porins detected in a skp mutant of N. meningitidis were correctly assembled into their native oligomeric conformation; however, their total amounts were decreased due to reduced synthesis (18). We hypothesized that Skp stimulates the release of OMPs from the Sec machinery in the inner membrane as has been reported in E. coli (36) and that the occupancy of the Sec channel in a skp mutant results in feedback inhibition on OMP synthesis (18). Obviously, such a defect is not compensated or aggravated by defects downstream in the OMP assembly pathway. Because GNA2091 is localized in the OM and its absence results in the accumulation of unassembled porins, it indeed likely acts downstream of Skp in the pathway. The amounts of unassembled porins that accumulated in the skp::catΔGNA2091 double mutant were similar to the amounts in the ΔGNA2091 single mutant. Apparently, the reduced synthesis of porins due to the skp mutation does not compensate for the impaired assembly due to the GNA2091 mutation. This is probably due to the additional role of Skp as a chaperone that prevents periplasmic aggregation of OMPs (37). Thus, in a skp single mutant, the absence of this chaperone function is compensated by the reduced synthesis of porins. However, when GNA2091 also is lacking, OMPs will misfold and aggregate in the periplasm despite the reduced synthesis.
As for the known Bam complex lipoprotein components, it remains to be determined how GNA2091 facilitates OMP assembly. Interestingly, submission of the GNA2091 amino acid sequence to the Conserved Domain Database at NCBI revealed that the protein contains one so-called bacterial OsmY and nodulation (BON) domain (Fig. 4), which was postulated to be a membrane-interacting, phospholipid-binding domain (38). The E. coli homolog YraP even contains two BON domains. The putative function of these BON domains was inferred from their presence in a family of osmotic shock protection proteins, some secretins, and a group of potential hemolysins (38). However, experimental evidence for such a role is lacking. The only study addressing the role of a BON domain experimentally that we know of solved the structure of the central region of the OmpATb protein of Mycobacterium tuberculosis that comprises two consecutive BON domains (39). This polypeptide was expressed in E. coli where it fractionated as a soluble protein and not with any membrane fraction. Furthermore, this protein domain did not show any binding activity for a number of lipids and detergents (39). Thus, the function of the BON domain remains elusive. Further mechanistic studies on GNA2091 or YraP will likely aid in understanding the role of this widespread BON domain family.
Acknowledgments
We are grateful to Virginie Roussel-Jazédé for provision of antisera. We thank GlaxoSmithKline for plasmids and antisera.
V. Roussel-Jazédé, J. Arenas, J. D. Langereis, J. Tommassen, and P. van Ulsen, submitted manuscript.
- OM
- outer membrane
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- OMP
- outer membrane protein
- cat
- chloramphenicol acetyltransferase
- TSB
- tryptic soy broth
- BAM
- β-barrel assembly machinery
- Nm
- N. meningitidis
- Ec
- E. coli
- BON
- bacterial OsmY and nodulation.
REFERENCES
- 1. Bos M. P., Tefsen B., Geurtsen J., Tommassen J. (2004) Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl. Acad. Sci. U.S.A. 101, 9417–9422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sperandeo P., Cescutti R., Villa R., Di Benedetto C., Candia D., Dehò G., Polissi A. (2007) Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J. Bacteriol. 189, 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu T., McCandlish A. C., Gronenberg L. S., Chng S. S., Silhavy T. J., Kahne D. (2006) Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 103, 11754–11759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ruiz N., Gronenberg L. S., Kahne D., Silhavy T. J. (2008) Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 105, 5537–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Voulhoux R., Bos M. P., Geurtsen J., Mols M., Tommassen J. (2003) Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265 [DOI] [PubMed] [Google Scholar]
- 6. Wu T., Malinverni J., Ruiz N., Kim S., Silhavy T. J., Kahne D. (2005) Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245 [DOI] [PubMed] [Google Scholar]
- 7. Walther D. M., Rapaport D., Tommassen J. (2009) Biogenesis of β-barrel membrane proteins in bacteria and eukaryotes: evolutionary conservation and divergence. Cell. Mol. Life Sci. 66, 2789–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tommassen J. (2010) Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology 156, 2587–2596 [DOI] [PubMed] [Google Scholar]
- 9. Anwari K., Webb C. T., Poggio S., Perry A. J., Belousoff M., Celik N., Ramm G., Lovering A., Sockett R. E., Smit J., Jacobs-Wagner C., Lithgow T. (2012) The evolution of new lipoprotein subunits of the bacterial outer membrane BAM complex. Mol. Microbiol. 84, 832–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sklar J. G., Wu T., Gronenberg L. S., Malinverni J. C., Kahne D., Silhavy T. J. (2007) Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 104, 6400–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Volokhina E. B., Beckers F., Tommassen J., Bos M. P. (2009) The β-barrel assembly complex of Neisseria meningitidis. J. Bacteriol. 191, 7074–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grizot S., Buchanan S. K. (2004) Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol. Microbiol. 51, 1027–1037 [DOI] [PubMed] [Google Scholar]
- 13. Anwari K., Poggio S., Perry A., Gatsos X., Ramarathinam S. H., Williamson N.A., Noinaj N., Buchanan S., Gabriel K., Purcell A. W., Jacobs-Wagner C., Lithgow T. (2010) A modular BAM complex in the outer membrane of the α-proteobacterium Caulobacter crescentus. PLoS One 5, e8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Serruto D., Bottomley M. J., Ram S., Giuliani M. M., Rappuoli R. (2012) The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: immunological, functional and structural characterization of the antigens. Vaccine 30, Suppl. 2, B87–B97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giuliani M. M., Adu-Bobie J., Comanducci M., Aricò B., Savino S., Santini L., Brunelli B., Bambini S., Biolchi A., Capecchi B., Cartocci E., Ciucchi L., Di Marcello F., Ferlicca F., Galli B., Luzzi E., Masignani V., Serruto D., Veggi D., Contorni M., Morandi M., Bartalesi A., Cinotti V., Mannucci D., Titta F., Ovidi E., Welsch J. A., Granoff D., Rappuoli R., Pizza M. (2006) A universal vaccine for serogroup B meningococcus. Proc. Natl. Acad. Sci. U.S.A. 103, 10834–10839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Onufryk C., Crouch M. L., Fang F. C., Gross C. A. (2005) Characterization of six lipoproteins in the σE regulon. J. Bacteriol. 187, 4552–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tettelin H., Saunders N. J., Heidelberg J., Jeffries A. C., Nelson K. E., Eisen J. A., Ketchum K. A., Hood D. W., Peden J. F., Dodson R. J., Nelson W. C., Gwinn M. L., DeBoy R., Peterson J. D., Hickey E. K., Haft D. H., Salzberg S. L., White O., Fleischmann R. D., Dougherty B. A., Mason T., Ciecko A., Parksey D. S., Blair E., Cittone H., Clark E. B., Cotton M. D., Utterback T. R., Khouri H., Qin H., Vamathevan J., Gill J., Scarlato V., Masignani V., Pizza M., Grandi G., Sun L., Smith H. O., Fraser C. M., Moxon E. R., Rappuoli R., Venter J. C. (2000) Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287, 1809–1815 [DOI] [PubMed] [Google Scholar]
- 18. Volokhina E. B., Grijpstra J., Stork M., Schilders I., Tommassen J., Bos M. P. (2011) Role of the periplasmic chaperones Skp, SurA, and DegQ in outer membrane protein biogenesis in Neisseria meningitidis. J. Bacteriol. 193, 1612–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tinsley C. R., Voulhoux R., Beretti J. L., Tommassen J., Nassif X. (2004) Three homologues, including two membrane-bound proteins, of the disulfide oxidoreductase DsbA in Neisseria meningitidis: effects on bacterial growth and biogenesis of functional type IV pili. J. Biol. Chem. 279, 27078–27087 [DOI] [PubMed] [Google Scholar]
- 20. Oomen C. J., van Ulsen P., van Gelder P., Feijen M., Tommassen J., Gros P. (2004) Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23, 1257–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Steeghs L., Berns M., ten Hove J., de Jong A., Roholl P., van Alphen L., Tommassen J., van der Ley P. (2002) Expression of foreign LpxA acyltransferases in Neisseria meningitidis results in modified lipid A with reduced toxicity and retained adjuvant activity. Cell. Microbiol. 4, 599–611 [DOI] [PubMed] [Google Scholar]
- 22. Bos M. P., Tommassen J. (2011) The LptD chaperone LptE is not directly involved in lipopolysaccharide transport in Neisseria meningitidis. J. Biol. Chem. 286, 28688–28696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Ulsen P., van Alphen L., ten Hove J., Fransen F., van der Ley P., Tommassen J. (2003) A neisserial autotransporter NalP modulating the processing of other autotransporters. Mol. Microbiol. 50, 1017–1030 [DOI] [PubMed] [Google Scholar]
- 24. Madico G., Welsch J. A., Lewis L. A., McNaughton A., Perlman D. H., Costello C. E., Ngampasutadol J., Vogel U., Granoff D. M., Ram S. (2006) The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J. Immunol. 177, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jacobsson S., Hedberg S. T., Mölling P., Unemo M., Comanducci M., Rappuoli R., Olcén P. (2009) Prevalence and sequence variations of the genes encoding the five antigens included in the novel 5CVMB vaccine covering group B meningococcal disease. Vaccine 27, 1579–1584 [DOI] [PubMed] [Google Scholar]
- 26. Seib K. L., Oriente F., Adu-Bobie J., Montanari P., Ferlicca F., Giuliani M. M., Rappuoli R., Pizza M., Delany I. (2010) Influence of serogroup B meningococcal vaccine antigens on growth and survival of the meningococcus in vitro and in ex vivo and in vivo models of infection. Vaccine 28, 2416–2427 [DOI] [PubMed] [Google Scholar]
- 27. Meltzer M., Hasenbein S., Mamant N., Merdanovic M., Poepsel S., Hauske P., Kaiser M., Huber R., Krojer T., Clausen T., Ehrmann M. (2009) Structure, function and regulation of the conserved serine proteases DegP and DegS of Escherichia coli. Res. Microbiol. 160, 660–666 [DOI] [PubMed] [Google Scholar]
- 28. Bos M. P., Robert V., Tommassen J. (2007) Biogenesis of the Gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61, 191–214 [DOI] [PubMed] [Google Scholar]
- 29. Huis in 't Veld R. A., Willemsen A. M., van Kampen A. H., Bradley E. J., Baas F., Pannekoek Y., van der Ende A. (2011) Deep sequencing whole transcriptome exploration of the σE regulon in Neisseria meningitidis. PLoS One 6, e29002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pannekoek Y., Huis in 't Veld R., Hopman C. T., Langerak A. A., Speijer D., van der Ende A. (2009) Molecular characterization and identification of proteins regulated by Hfq in Neisseria meningitidis. FEMS Microbiol. Lett. 294, 216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maddalo G., Stenberg-Bruzell F., Götzke H., Toddo S., Björkholm P., Eriksson H., Chovanec P., Genevaux P., Lehtiö J., Ilag L. L., Daley D. O. (2011) Systematic analysis of native membrane protein complexes in Escherichia coli. J. Proteome Res. 10, 1848–1859 [DOI] [PubMed] [Google Scholar]
- 32. Pan J. Y., Li H., Ma Y., Chen P., Zhao P., Wang S. Y., Peng X. X. (2010) Complexome of Escherichia coli envelope proteins under normal physiological conditions. J. Proteome Res. 9, 3730–3740 [DOI] [PubMed] [Google Scholar]
- 33. Stenberg F., Chovanec P., Maslen S. L., Robinson C. V., Ilag L. L., von Heijne G., Daley D. O. (2005) Protein complexes of the Escherichia coli cell envelope. J. Biol. Chem. 280, 34409–34419 [DOI] [PubMed] [Google Scholar]
- 34. Ricci D. P., Hagan C. L., Kahne D., Silhavy T. J. (2012) Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc. Natl. Acad. Sci. U.S.A. 109, 3487–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Typas A., Nichols R. J., Siegele D. A., Shales M., Collins S. R., Lim B., Braberg H., Yamamoto N., Takeuchi R., Wanner B. L., Mori H., Weissman J. S., Krogan N. J., Gross C. A. (2008) High-throughput, quantitative analyses of genetic interactions in E. coli. Nat. Methods 5, 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schäfer U., Beck K., Müller M. (1999) Skp, a molecular chaperone of Gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 274, 24567–24574 [DOI] [PubMed] [Google Scholar]
- 37. Walton T. A., Sandoval C. M., Fowler C. A., Pardi A., Sousa M. C. (2009) The cavity-chaperone Skp protects its substrate from aggregation but allows independent folding of substrate domains. Proc. Natl. Acad. Sci. U.S.A. 106, 1772–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yeats C., Bateman A. (2003) The BON domain: a putative membrane-binding domain. Trends Biochem. Sci. 28, 352–355 [DOI] [PubMed] [Google Scholar]
- 39. Teriete P., Yao Y., Kolodzik A., Yu J., Song H., Niederweis M., Marassi F. M. (2010) Mycobacterium tuberculosis Rv0899 adopts a mixed α/β-structure and does not form a transmembrane β-barrel. Biochemistry 49, 2768–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]