Background: CD81-associated tetraspanins influence the invasive and metastatic processes of various cancer cell types.
Results: CD81-Akt signaling induces Sp1-mediated MT1-MMP expression, which enhances melanoma cell motility and invasiveness.
Conclusion: CD81 functions as an inducer of MT1-MMP that plays a role in melanoma invasive processes.
Significance: This study provides the first evidence that CD81 contributes to cancer malignant progression and elucidates the underlying mechanism.
Keywords: Cell Motility, Cell Signaling, Invasion, Melanoma, Sp1, Tetraspanins
Abstract
Despite the importance of multiple tetraspanin proteins in cancer invasion and metastasis, little is known about the role and significance of tetraspanin CD81 in these processes. In the present study, we examined CD81 effects on melanoma cell invasiveness and metastasis. Transfection of CD81 into melanoma cells lacking endogenous CD81 expression significantly enhanced the migrating, invasive, and metastatic abilities of melanoma cells. Interestingly, membrane type 1 matrix metalloproteinase (MT1-MMP) expression was found in CD81-expressing melanoma cells but not in CD81-deficient cells. siRNA knockdown of CD81 in melanoma cells with endogenous CD81 demonstrated decreased MT1-MMP levels and cell motility. Notably, CD81-induced cell migration was abrogated by antibody blocking and siRNA knockdown of MT1-MMP, indicating that MT1-MMP is responsible for CD81-stimulated melanoma cell migration. Promoter analysis revealed an essential role of the Sp1 transcription factor in CD81-induced MT1-MMP transcription. We also demonstrate that the Sp1-activating Akt pathway is involved in adhesion-dependent CD81 signaling to induce MT1-MMP expression and cell motility. Importantly, human skin cancer tissue specimens displayed a positive correlation of CD81 with MT1-MMP expression levels and a close association of CD81 with malignant melanomas. Taken together, these results strongly suggest that CD81 stimulates melanoma cell motility by inducing MT1-MMP expression through the Akt-dependent Sp1 activation signaling pathway, leading to increased melanoma invasion and metastasis.
Introduction
The tetraspanin family (also called tetraspan family or transmembrane 4 superfamily) is a large group of ubiquitously expressed membrane proteins that contain four transmembrane domains, short N- and C-terminal cytoplasmic domains, two extracellular loops, and a small intracellular loop (1, 2). Tetraspanin proteins are implicated not only in a broad range of physiological processes but also in pathological situations such as cancer invasion and metastasis (2, 3). One distinctive feature of tetraspanins is their ability to associate with other membrane proteins such as integrins, growth factor receptors, proteases, and other tetraspanin proteins and to form multimolecular complexes in the membrane known as tetraspanin webs or tetraspanin-enriched microdomains (TEMs)3 (1, 4). Tetraspanins are also shown to associate with signaling molecules such as protein kinase C (PKC), phosphoinositide 4-kinase (PI4K), and small GTPases including Ras, Rac1, and CDc42 at the cytoplasmic face (5–7). Upon organization into TEMs, the activity and function of tetraspanin-associated molecules become altered (8). Through this mechanism, tetraspanins influence a broad range of cellular activities including adhesion, proliferation, activation, differentiation, migration, and signaling (4, 8).
CD81, a member of the tetraspanin family, was originally identified as the target of an anti-proliferative antibody (TAPA-1) that inhibited B cell proliferation (9). CD81 functions in normal physiology have been largely documented for the immune system. On B cells, CD81 induces surface expression and maturation of CD19, a member of the immunoglobulin superfamily, and forms a complex with CD19 together with CD21 (10). This complex contributes to B cell activation by bringing together signaling molecules required for B cell receptor signaling (1). CD81 also plays a role in T cell receptor signaling and T cell development in the thymus (1). CD81 knock-out mice exhibit not only impaired immune responses but also abnormal brain development and reduced sperm-egg fusion during oocyte fertilization (11, 12), implicating the importance of CD81 in diverse physiological processes. Because CD81 was identified as a cellular receptor for the hepatitis C virus (13), the majority of pathological investigations of CD81 have focused on hepatitis C virus infection of liver cells. Meanwhile, CD81 forms TEMs with other tetraspanin proteins including CD9, CD82, CD151, and TSPAN8 together with α3β1 and α4β1 integrins (8, 14, 15). Several tetraspanins associated with CD81 in TEMs such as CD9, CD82, CD151, and TSPAN8 have been extensively investigated for their involvement and regulatory roles in the invasive and metastatic processes of various cancers. As a result, prometastatic roles for CD151 and TSPAN8 and metastasis-suppressing functions of CD82 and CD9 have been well demonstrated in a number of studies using mouse tumor models and cancer tissue specimens from patients (2, 3, 16). A previous report has described an inverse correlation of CD81 levels with distant metastasis in patients with hepatocellular carcinoma (17), but little is currently known about the involvement and significance of CD81 in these malignant processes. Because TEM constituents may affect the activities and functions of one another, identification of CD81 as a metastasis regulator will lead to greater understanding of tetraspanin complexes in cancer malignant progression.
Multiple tetraspanins in TEMs are well characterized in melanoma for their involvement in cancer invasion and metastasis. Therefore, in this study, we selected melanoma as a cancer model to determine the role and significance of CD81 in these malignant processes. Our present results indicate that CD81 functions to stimulate cell motility and hence elevates the invasive and metastatic potential of melanoma cells. In an effort to reveal the mechanism(s) by which CD81 stimulates melanoma cell motility, we found that adhesion-dependent CD81 signaling induces expression of membrane type 1 matrix metalloproteinase (MT1-MMP), which renders melanoma cells more motile. Immunohistochemical analysis of human skin cancer tissue specimens revealed a positive correlation between CD81 and MT1-MMP expression levels. Furthermore, both CD81 and MT1-MMP expressions were highly associated with malignant melanomas as compared with non-melanoma skin cancers. Together, the present data suggest that CD81 is linked to the development of malignant melanoma and in particular to melanoma invasion and metastasis through its stimulating effect on cell motility.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Mouse monoclonal antibodies to CD81 (5A6) and MMP-9 (2C3) and rabbit polyclonal antibodies to MMP-2 (H-76) and Sp1 (H-225and H-225X) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-paxillin monoclonal antibody was obtained from BD Biosciences. Polyclonal antibodies against MT1-MMP (MMP-14) were purchased from Millipore (Billerica, MA) and Abcam (EP1264Y; Cambridge, UK). Rabbit anti-phospho-AktSer-473 and anti-Akt antibodies were obtained from Cell Signaling Technology (Beverly, MA). A rabbit antibody specific to phospho-Sp1Thr-453 was obtained from Abcam. Texas Red-labeled phalloidin and FITC-conjugated goat anti-mouse secondary antibodies were from Invitrogen. All other reagents were from Sigma-Aldrich unless indicated otherwise.
Cell Culture and Generation of Stable CD81 Transfectant Cells
Human melanoma cell lines C8161, MelJuSo, SK-Mel-2, and Malme-3M were cultured in DMEM/F-12 medium supplemented with 10% fetal bovine serum (FBS) in 5% CO2 at 37 °C. To generate stable CD81 transfectant clones of MelJuSo cells, full-length CD81 cDNA subcloned into pcDNA3 vector (Invitrogen) was transfected into MelJuSo cells using Lipofectamine/PLUS reagent (Invitrogen) according to the manufacturer's instructions. pcDNA3 vector only was transfected as a control (mock transfectant). Neomycin-resistant clones were isolated by growing the cells in DMEM/F-12 medium containing 10% FBS and 0.5 mg/ml G418 (Invitrogen). CD81 expression levels in stable transfectant clones were examined by RT-PCR, immunoblotting, and flow cytometric analyses.
Quantitative Real Time RT-PCR
Total cellular RNA was extracted from the cultured cells using TRIzol reagent (Invitrogen) and reverse transcribed into cDNA using Moloney murine leukemia virus reverse transcriptase. Quantitative real time PCRs were performed with AccuPower 2× GreenStar qPCR Master Mix (Bioneer, Daejeon, Korea) using the Rotor-Gene® Q real time PCR cycler (Qiagen, Valencia, CA). Primers used for real time PCR were as follows: 5′-CTGACTGCTTTGACCACCTCA-3′ (sense) and 5′-AATGCCGATGAGGTACAGCTT-3′ (antisense) for CD81, 5′-ATGACATCTTCCTGGTGGCTG-3′ (sense) and 5′-ACCCCCATAAAGTTGCTGGAT-3′ (antisense) for MT1-MMP (MMP-14), 5′-CACTTCCCCTTCATCTTCGAG-3′ (sense) and 5′-GTGTAGAGTCTCTCGCTGGGG-3′ (antisense) for MMP-9, 5′-TTCCAGGGCACATCCTATGAC-3′ (sense) and 5′-CACCTTCTGAGTTCCCACCAA-3′ (antisense) for MMP-2, 5′-AGTCAGTCAGGGGGAACAGGT-3′ (sense) and 5′-ATTGCTGCCATTGGTACTGCT-3′ (antisense) for Sp1, and 5′-AAGGTCGGAGTCAACGGATTT-3′ (sense) and 5′-CCATGGGTGGAATCATATTGG-3′ (antisense) for GAPDH. The thermal PCR conditions were 15-s denaturation at 95 °C and 1-min annealing extension at 60 °C for 40 cycles. Fluorescence was detected at the end of each 60 °C phase. The transcript levels of genes were normalized to the transcript level of GAPDH.
Transfection of Small Interfering RNA (siRNA)
siRNAs for CD81, Akt, MT1-MMP, and Sp1 were designed and synthesized using the software and SilencerTM siRNA construction kit from Ambion (Austin, TX) according to the manufacturer's instructions. Oligonucleotide sequences for each siRNA were as follows: 5′-GAACUUUCCUGUUACCUUUdTdT-3′ (sense) and 5′-AAAGGUAACAGGAAAGUUCdTdT-3′ (antisense) for CD81 siRNA, 5′-AAGAGACGAUGGACUUCCGAUdTdT-3′ (sense) and 5′-AUGGGAAGUCCAUCGUCUCUUdTdT-3′ (antisense) for Akt siRNA 1, 5′-GACUGACACCAGGUAUUUUdTdT-3′ (sense) and 5′-AAAAUACCUGGUGUCAGUCdTdT-3′ (antisense) for Akt siRNA 2, 5′-AAUCUGCAUCAGCUUUGCCUGdTdT-3′ (sense) and 5′-AACAGGCAAAGCUGAUGCAGAdTdT-3′ (antisense) for MT1-MMP siRNA 1, 5′-AACAGGCAAAGCUGAUGCAGAdTdT-3′ (sense) and 5′-AAUCUGCAUCAGCUUUGCCUGdTdT-3′ (antisense) for MT1-MMP siRNA 2, 5′-AAGGAGUUGUUGCUGUUCUCAdTdT-3′ (sense) and 5′-AAUGAAAACAGCAACAACUCCdTdT-3′ (antisense) for Sp1 siRNA 1, and 5′-CUAUGAACUACAGGUGUUUdTdT-3′ (sense) and 5′-AAACACCUGUAGUUCAUAGdTdT-3′ (antisense) for Sp1 siRNA 2. The siRNA control was 5′-UUCUCCGAACGUGUCACGUdTdT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAAdTdT-3′ (antisense), which bear no homology with relevant human genes (18). Transfection of siRNA was carried out using Lipofectamine/PLUS reagent as described previously (19).
Invasion Assays
In vitro invasion assay into Matrigel was performed as described previously (19). In vivo cancer cell invasion assay were conducted using 11-day-old chick embryos wherein 105 cells labeled with a fluorescent probe for long term tracing of living cells, CellTrackerTM Orange 5-(and-6)-(((4-chloromethyl)benzoyl)amino)tetramethylrhodamine (Invitrogen), were suspended in 100 μl of serum-free DMEM and seeded atop the chick chorioallantoic membrane (CAM) as described previously (20). After incubating for 3 days in a humidified stationary incubator at 38 °C, the embryos were snap frozen in liquid nitrogen and cross-sectioned with a microtome. Following staining with DAPI, CAM cryosections with 20-μm thickness were viewed under a fluorescence microscope (Olympus). Portions beneath the CAM surface were also subjected to Alu PCR analysis to detect human cells.
Spontaneous Pulmonary Metastasis Assay Using a Mouse Xenograft Model
Stable MelJuSo mock and CD81 transfectant cells (1 × 106) were injected subcutaneously into the dorsal flank region of BALB/c nu/nu mice (8 weeks old). Tumor length and width were measured every 4 days using a caliper. Seven weeks after cell inoculation, mice were sacrificed and photographed. Next, tumors were dissected out and weighed. Lungs were also harvested and stained with Bouin's solution to assess metastatic tumor lesions.
Cell Motility Assay
Chemostatic cell migration was analyzed using an OrisTM cell migration assay kit (Platypus Technologies, Madison, WI) following the manufacturer's instructions. Briefly, cells (5 × 104) suspended in culture medium were seeded onto each well (coated with or without fibronectin) of the Oris plate and incubated overnight in 5% CO2 at 37 °C. After removal of the stoppers from the Oris plate, each well was washed with PBS to remove any unattached cells and then incubated with complete culture medium for the indicated time period.
Fibrin Zymography
Fibrin zymography was performed to determine the activity of the plasminogen activators as described previously (21). The samples were subjected to SDS-PAGE using a 10% gel containing fibrinogen (2 mg/ml), plasminogen (25 μg/ml), and thrombin (1 unit/ml). The gel was washed twice with 2.5% Triton X-100 for 30 min each time at room temperature to remove SDS and then incubated with 0.1 m glycine buffer (pH 7.5) at 37 °C overnight. Following staining with 0.1% Coomassie Blue R-250 for 1 h, the gel was destained in a solution of 10% acetic acid and 50% methanol.
Human Skin Cancer/Melanoma Tissue Microarray and Immunohistochemistry
A commercially available human skin cancer/melanoma tissue microarray (AccuMaxTM arrays) was obtained from Petagen Inc. (Seoul, Korea). The tissue microarray contained 41 basal cell carcinoma, 33 squamous cell carcinoma, and 10 malignant melanoma cases of skin cancer patients along with two non-neoplastic skin tissue specimens. Immunohistochemistry for CD81 and MT1-MMP in the tissue microarrays was carried out as described previously (22). Briefly, two microarray slides containing consecutive sections of human skin tumors were deparaffinized and autoclaved for 15 min in citrate buffer (pH 6.0) and then incubated for 30 min in 0.33% hydrogen peroxide diluted in methanol to quench endogenous peroxide activity. After blocking with bovine serum albumin, the slides were incubated with anti-CD81 or anti-MT1-MMP monoclonal antibody for 3 h at room temperature. After washing with PBS, the sections were incubated with peroxidase-labeled anti-mouse IgG (Pierce) and with 3,3′-diaminobenzidine to develop the signal. Finally, counterstaining was carried out with hematoxylin.
Other Analyses/Assays
Immunoprecipitation and immunoblotting analyses, RT-PCR analysis, flow cytometry, immunocytochemistry, gelatin zymography, promoter/luciferase reporter assay, electrophoretic mobility shift assay (EMSA), chromatin immunoprecipitation (ChIP) assay, and 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium cell proliferation assay were performed as described previously (7, 19, 23).
Statistical Analysis
Different numbers between two groups were analyzed by Student's t test. Immunohistochemistry analysis was performed using Pearson's χ2 test. A value of p < 0.03 was considered statistically significant. Correlation between immunohistochemical scores of CD81 and MT1-MMP was determined using the Spearman rank correlation coefficient test.
RESULTS
CD81 Effects on Melanoma Cell Metastasis
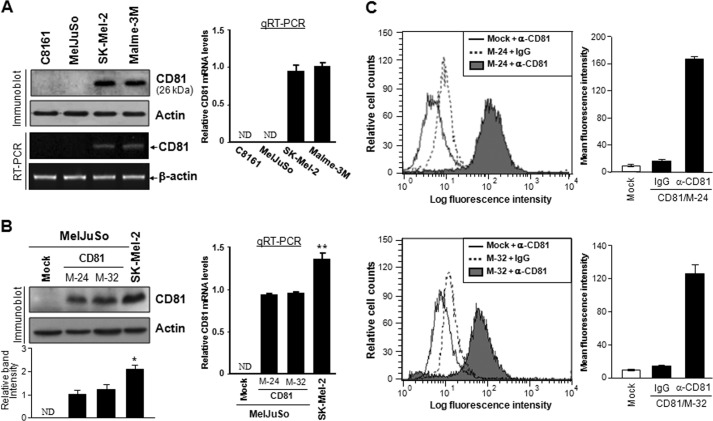
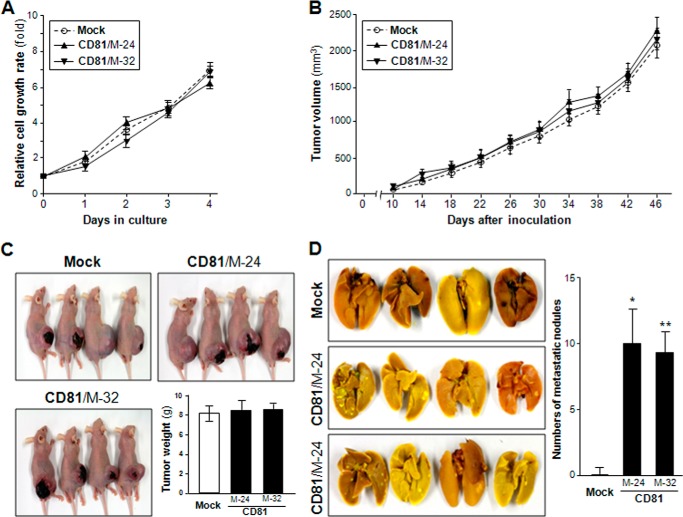
We first examined CD81 expression in four human melanoma cell lines with metastatic potential, C8161, MelJuSo, SK-Mel-2, and Malme-3M. Among these parental cell lines, SK-Mel-2 and Malme-3 M expressed CD81, whereas C8161 and MelJuSo did not (Fig. 1A). Using MelJuSo cells lacking endogenous CD81, we generated stable CD81 transfectant clones with ectopically expressed CD81 at the cell surface (Fig. 1, B and C). CD81 protein and mRNA levels in MelJuSo CD81 transfectant clones were not higher than endogenous CD81 levels in SK-Mel-2 cells, implicating that these CD81 transfectant clones express exogenous CD81 within a physiological range. We next examined the effect of CD81 on tumor growth and metastasis of MelJuSo cells using a mouse xenograft model in which mice were orthotopically injected in the dorsal flank region with melanoma cells. Minimal difference was observed in cell proliferation and tumor growth rate between mock and CD81 transfectant cells (Fig. 2, A–C). However, lungs of mice inoculated with CD82 transfectant cells exhibited a few metastatic nodules, whereas metastatic lesions could hardly be found in the lungs of mice inoculated with mock transfectant cells (Fig. 2D). These findings indicate CD81-stimulated pulmonary metastasis of MelJuSo cells. Thus, CD81 appeared to promote metastasis of melanoma cells without affecting tumor growth.
FIGURE 1.
CD81 expression levels in human melanoma cell lines and CD81 transfectant cells. A and B, human melanoma parental cell lines (A) and stable CD81 transfectant clones of MelJuSo cells (B) were examined for CD81 protein levels by immunoblotting analysis. A bar graph under the immunoblot indicates relative band intensity of CD81 normalized to the loading control (actin). CD81 gene expression was also examined by quantitative real time RT-PCR (qRT-PCR) analysis, and relative mRNA levels of CD82 normalized to the GAPDH level are shown in bar graphs next to the immunoblots. Results are the mean ± S.D. from three separate experiments. Error bars represent S.D. C, cell surface expression levels of CD81 in stable CD81 transfectant clones of MelJuSo cells were analyzed by flow cytometry using an anti-CD81 monoclonal antibody. Mean fluorescence intensity of each cell population is shown in a bar graph next to the distribution chart. Values are the mean ± S.D. from three separate experiments with triplicates per experiment (* and **, p < 0.03 versus MelJuSo CD81 transfectant clones; Student's t test). ND, not detectable.
FIGURE 2.
CD81 enhances the metastatic ability of MelJuSo melanoma cells without affecting tumor growth. A, stable MelJuSo mock and CD81 transfectant cells (clones M-24 and M-32) were measured for in vitro cell growth rate using a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium cell proliferation assay with quadruple cultures. B–D, cells (1 × 106) were inoculated subcutaneously into the dorsal flank region of athymic nude mice. B, tumor length and width were measured to assess tumor volume every 4 days after observation of tumor formation. C, 7 weeks after cell inoculation, mice were sacrificed and photographed, and tumors were dissected out and weighed. D, lungs were harvested and stained with Bouin's solution, and metastatic nodules in the lungs were counted. Values are the mean ± S.D. from two separate groups with four mice per group (* and **, p < 0.01 versus mock). Error bars represent S.D.
CD81 Effects on Melanoma Cell Invasion and Motility
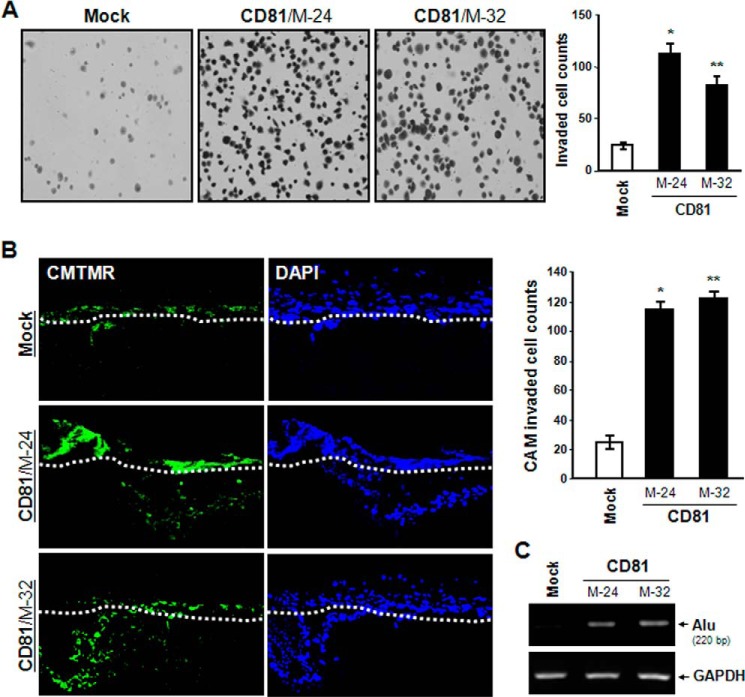
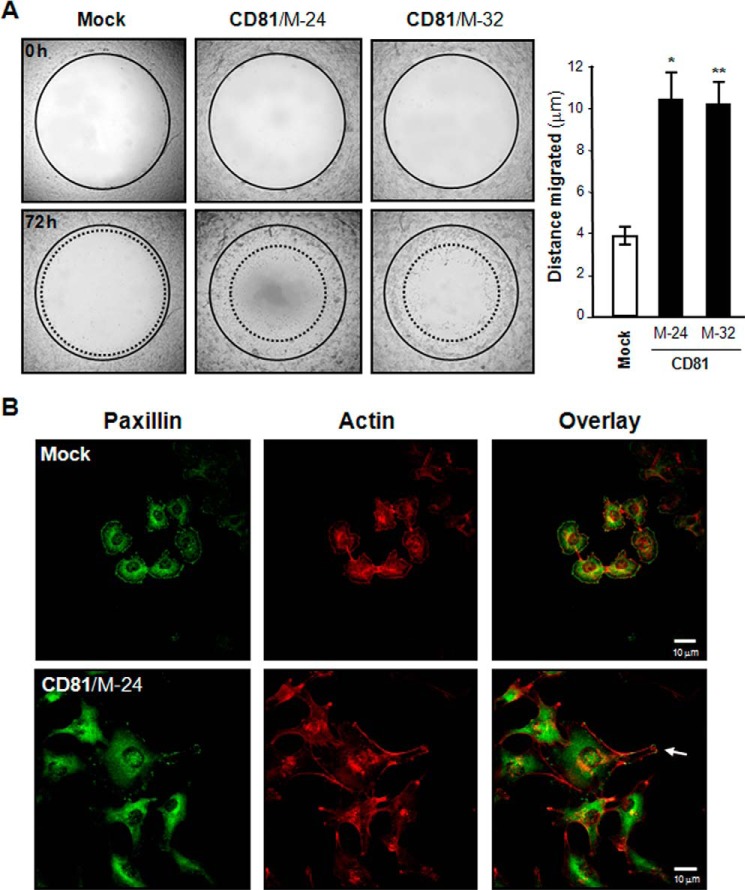
When cell invasiveness was examined in vitro using a Transwell chamber with Matrigel-coated membrane, MelJuSo CD81 transfectant cells exhibited significantly enhanced invasive ability as compared with mock transfectant cells (Fig. 3A). An in vivo invasion assay using chick embryos also revealed that the invasiveness of CD81 transfectant cells is much higher than that of mock transfectant cells (Fig. 3, B and C). Moreover, CD81-expressing cells displayed approximately a 2.5-fold higher migrating ability than CD81-deficient cells in the wound closure cell migration assay (Fig. 4A). Additionally, CD81 expression in MelJuSo cells resulted in increased cell spreading along with the development of motile subcellular structures such as membrane protrusions (Fig. 4B). CD81 expression, therefore, apparently renders melanoma cells more motile and more invasive.
FIGURE 3.
CD81 increases melanoma cell invasiveness. A, cells (2 × 104) were seeded into a Transwell chamber insert equipped with a Matrigel-coated filter. After 12 h of incubation, cells on the lower surface of the filter were stained with Gill's hematoxylin and counted. Results are the mean ± S.D. from three separate experiments with triplicates per experiment (* and **, p < 0.01 versus mock). Error bars represent S.D. B and C, cells were labeled with a fluorescent cell tracker, 5-(and-6)-(((4-chloromethyl)benzoyl)amino)tetramethylrhodamine (CMTMR) (green), and seeded atop the CAM of 11-day-old chicks. After incubating for 3 days, the embryos were frozen and cross-sectioned as described under “Experimental Procedures.” B, CAM sections stained with DAPI (blue) were photographed under a fluorescence microscope. The CAM surface is marked by a dashed line. Cells showing both green and blue fluorescence beneath the CAM surface were counted and expressed as the mean ± S.D. of three sections in each of three embryos (* and **, p < 0.01 versus mock). C, human melanoma cells that penetrated the CAM surface were detected as Alu sequence by PCR on DNA extracted from the lower CAM.
FIGURE 4.
CD81 stimulates melanoma cell migration. A, cell motility was determined using Oris cell migration assay kit as described under “Experimental Procedures.” Shown are representative images taken under a microscope with 40× magnification. Results are the mean ± S.D. from three separate experiments with triplicates per experiment (* and **, p < 0.01 versus mock). Error bars represent S.D. B, cells were permeabilized and incubated with mouse anti-paxillin antibody and Texas Red-labeled phalloidin. Following incubation with Alexa Fluor 488 (green)-conjugated anti-mouse IgG, fluorescence images of cells were taken under a confocal microscope. Shown are the representative images of three separate experiments. An arrow indicates membrane protrusion linked to cell migration.
CD81-induced Expression of MT1-MMP in Melanoma Cells
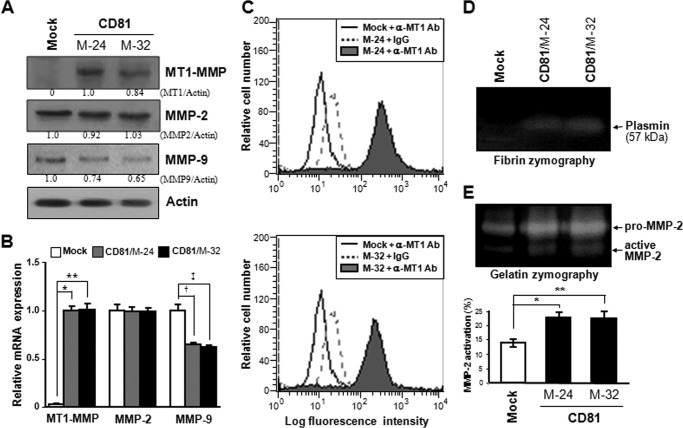
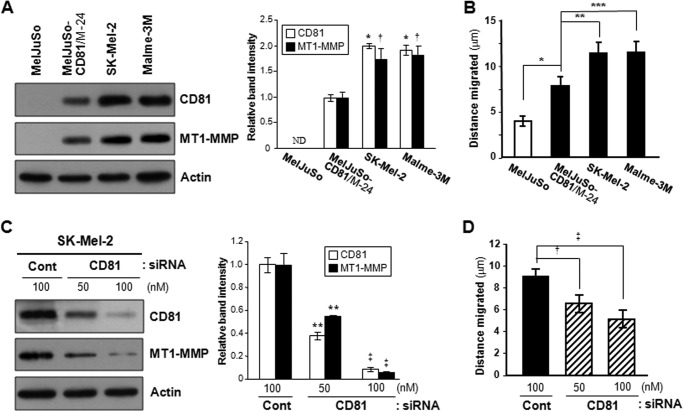
To identify the molecule(s) that mediates CD81-induced cell motility, we screened 84 key genes involved in the cellular movement using the RT2 ProfilerTM Human Cell Motility PCR Array from SABiosciences/Qiagen. The gene expression profiling with quantitative RT-PCR illustrated that MT1-MMP/MMP-14 exhibited a distinct difference in expression levels between CD81-expressing and CD81-deficient cells. As shown in Fig. 5 (A and B), MT1-MMP was expressed in MelJuSo CD81 transfectant cells but not in mock transfectant cells. FACS analysis data also indicated that MT1-MMP is present at the surface of CD81-expressing cells only (Fig. 5C). We next compared the biochemical activity of MT1-MMP between CD81-expressing and CD81-deficient cells by assessing its enzymatic activity as a plasminogen and pro-MMP-2 activator. In fibrin zymography, which shows the proteolytic conversion of plasminogen to plasmin, fibrinogen degradation by plasmin was observed in the lysate of CD81 transfectant cells but not in mock transfectant cells (Fig. 5D). In gelatin zymography, CD81 transfectant cells also exhibited higher proteolytic activity toward pro-MMP-2 than mock transfectant cells (Fig. 5E). In addition to MelJuSo CD81 transfectant cells, two human melanoma parental cell lines with endogenous CD81, SK-Mel-2 and Malme-3M, also expressed MT1-MMP (Fig. 6A). Furthermore, both SK-Mel-2 and Malme-3 M cell lines exhibited significantly greater migrating ability than MelJuSo parental cells without both CD81 and MT1-MMP (Fig. 6B). Importantly, SK-Mel-2 and Malme-3 M cells did not only contain more CD81 but also expressed more MT1-MMP than MelJuSo CD81 transfectant cells. Also, higher cell motility was observed in SK-Mel-2 and Malme-3 M cells relative to MelJuSo CD81 transfectant cells. Therefore, it seems likely that CD81 is closely associated with MT1-MMP expression and cell motility in melanoma cells. Meanwhile, siRNA-mediated knockdown of endogenous CD81 in SK-Mel-2 cells decreased both the MT1-MMP level and cell motility (Fig. 6, C and D). CD81 knockdown also attenuated the invasive ability of SK-Mel-2 cells (data not shown). Collectively, these results strongly suggest that CD81 increases MT1-MMP expression in human melanoma cells together with cell motility and invasiveness.
FIGURE 5.
CD81 induces expression of MT1-MMP in MelJuSo melanoma cells. A and B, stable CD81 transfectant clones of MelJuSo cells were examined for protein (A) and mRNA (B) levels of MT1-MMP, MMP-2, and MMP-9 using immunoblotting and quantitative real time RT-PCR analyses, respectively. Numbers under the immunoblot (A) represent relative band intensity of protein normalized to the actin level. A bar graph (B) indicates the relative mRNA levels of genes normalized to the GAPDH level. Values are the mean ± S.D. from three separate experiments with three determinations per experiment (* and **, p < 0.01; † and ‡, p < 0.03). Error bars represent S.D. C, cell surface expression level of MT1-MMP was assessed by flow cytometry using anti-MT1-MMP antibody (Ab). Shown are the representative data of three independent experiments. D and E, enzymatic activity of MT1-MMP in the CD81 transfectant cells was determined by zymographies using fibrin (D) and gelatin (E) as substrates. Proteolytic activation of MMP-2 in gelatin zymography was calculated from the relative band intensity of active MMP-2 to the sum of pro-MMP2 and active MMP-2. Data shown are the mean ± S.D. from three separate experiments with three determinations per experiment (* and **, p < 0.03).
FIGURE 6.
CD81 levels in melanoma cells are closely associated with MT1-MMP expression and cell motility. A and B, human melanoma parental cell lines MelJuSo, SK-Mel-2, and Malme-3M and a stable MelJuSo CD81 transfectant clone, M-24, were examined for CD81 and MT1-MMP protein levels (A) and cell motility (B). C and D, SK-Mel-2 cells with endogenous CD81 were transfected with CD81-specific siRNA and then examined for MT1-MMP protein levels (C) and cell motility (D). Shown are representative immunoblots of three independent experiments (* and †, p < 0.03 versus a MelJuSo CD81 transfectant clone, M-24; **, p < 0.03; ‡, p < 0.01 versus control (Cont) siRNA-transfected cells). Migration data shown are the mean ± S.D. from three separate experiments with triplicates per experiment (*, **, ***, †, and ‡, p < 0.03). Error bars represent S.D.
Role of MT1-MMP in CD81-induced Melanoma Cell Motility
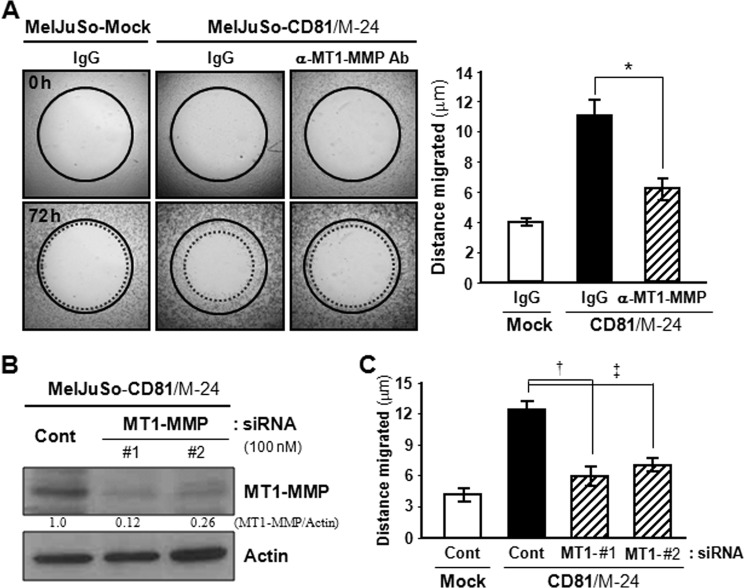
MT1-MMP has been shown to play an important role in the process of cell migration (24). To examine whether the effect of CD81 on melanoma cell motility is dependent on MT1-MMP, CD81-transfected MelJuSo cells were pretreated with an anti-MT1-MMP blocking antibody prior to cell migration. As shown in Fig. 7A, antibody blocking of MT1-MMP significantly interfered with the migration of CD81 transfectant cells. CD81-induced cell motility was also abrogated by siRNA knockdown of MT1-MMP (Fig. 7, B and C). Additionally, MT1-MMP-specific siRNAs decreased invasiveness of SK-Mel-2 cells with endogenous CD81 and MT1-MMP (data not shown). Because CD81 increases both MT1-MMP levels and cell motility in melanoma cells, it is very likely that CD81-induced cell motility is attributed to increased MT1-MMP expression by CD81.
FIGURE 7.
Antibody blocking and siRNA knockdown of MT1-MMP attenuate CD81-induced melanoma cell motility. A, CD81-transfected MelJuSo cells attached to the Oris cell migration assay plate were pretreated with an anti-MT1-MMP antibody (Ab) or normal mouse IgG for 6 h before removal of the stopper from the Oris plate to initiate cell migration. B and C, CD81 transfectant MelJuSo cells were transfected with siRNAs targeting MT1-MMP. MT1-MMP protein level (B) and motility (C) of siRNA-transfected cells were determined by immunoblotting and Oris cell migration analyses, respectively. Numbers under the immunoblot indicate relative band intensity of MT1-MMP normalized to the actin level. Data shown are the mean ± S.D. from three separate experiments with triplicates per experiment (*, †, and ‡, p < 0.03). Error bars represent S.D. Cont, control.
Functional Involvement of the Sp1 Transcription Factor in CD81-induced MT1-MMP Gene Transcription
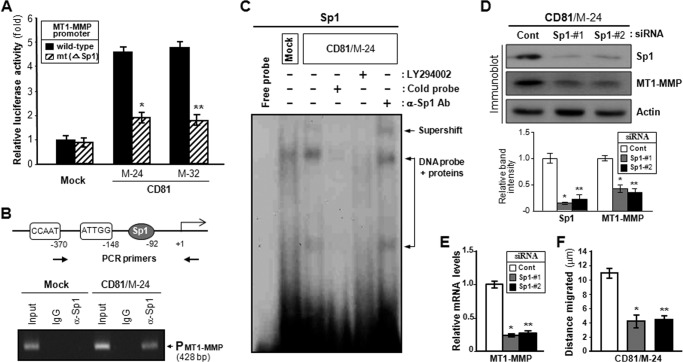
We next investigated the transcriptional regulation of MT1-MMP gene by using a mutant of its 5′-proximal promoter region (25). When a reporter vector containing a wild-type promoter of MT1-MMP gene was transiently transfected into MelJuSo cells, CD81 transfectant cells exhibited about a 5-fold higher promoter activity than mock transfectant cells (Fig. 8A). However, a promoter with a deletion at the Sp1 binding site exhibited minimal difference in transcriptional activity between CD81 and mock transfectant cells. In a ChIP assay, an anti-SP1 antibody was able to precipitate a Sp1 site-containing MT1-MMP promoter region obtained from CD81-expressing cells but not that from CD81-deficient cells, indicating CD81-dependent Sp1 binding to the MT1-MMP promoter (Fig. 8B). Furthermore, EMSA data illustrated that CD81 transfectant cells had more nuclear proteins bound to the Sp1 recognition oligonucleotide than mock transfectant cells (Fig. 8C). In EMSA, incubation with an anti-Sp1 antibody resulted in a partial supershift of the nuclear protein-DNA complex, indicating that the Sp1 protein was one of the nuclear proteins bound to the MT1-MMP promoter in CD81-expressing cells. Additionally, the nuclear protein-DNA complex was significantly decreased by pretreatment of CD81 transfectant cells with the PI3K-Akt pathway inhibitor LY294002, suggesting involvement of the PI3K-Akt pathway in Sp1-dependent MT1-MMP transcription by CD81 signaling. To further confirm the transcriptional dependence of MT1-MMP upon the Sp1 factor, we transfected CD81-expressing cells with Sp1-specific siRNA. As shown in Fig. 8 (D and E), Sp1 knockdown in CD81 transfectant cells resulted in a decrease in MT1-MMP mRNA and protein levels. Also, the migrating ability of CD81 transfectant cells was suppressed by Sp1-targeting siRNAs (Fig. 8F). Thus, CD81-induced expression of MT1-MMP appeared to be dependent on the Sp1 transcription factor.
FIGURE 8.
Sp1 transcription factor mediates CD81-dependent expression of MT1-MMP. A, wild-type promoter of the human MT1-MMP gene and the mutant promoter (mt) without Sp1 binding site were fused to a luciferase reporter gene of the pGL3 vector and transfected into MelJuSo mock and CD81 transfectant cells. The transcriptional activity of wild-type and mutant promoters was determined by a Dual-Luciferase assay. Normalized luciferase activities ±S.D. from triplicate experiments are plotted (* and **, p < 0.01). Error bars represent S.D. B, MelJuSo mock and CD81 transfectant cells were sonicated to shear DNA. Following incubation with an anti-Sp1 antibody or normal mouse IgG, the immunocomplex was subjected to 25 PCR cycles using primers specific to the Sp1 binding region of the human MT1-MMP gene promoter. C, 32P-labeled oligonucleotides containing Sp1 recognition sequences were incubated with nuclear extracts obtained from CD81 transfectant cells pretreated with or without the PI3K-Akt signaling blocker LY294002. Specific DNA binding activity of Sp1 factor was determined by an electrophoretic mobility shift assay in the absence or presence of an 8-fold excess of cold competitor. Anti-Sp1 antibody (Ab) was used for supershift analysis. D and E, following transfection of two Sp1-specific siRNAs, CD81 transfectant cells were examined for MT1-MMP protein (D) and mRNA levels (E) using immunoblotting and quantitative real time RT-PCR analyses, respectively. F, Sp1 siRNA-transfected cells were examined for cell motility. Data shown are the mean ± S.D. from three separate experiments with triplicates per experiment (* and **, p < 0.03 versus control (Cont)).
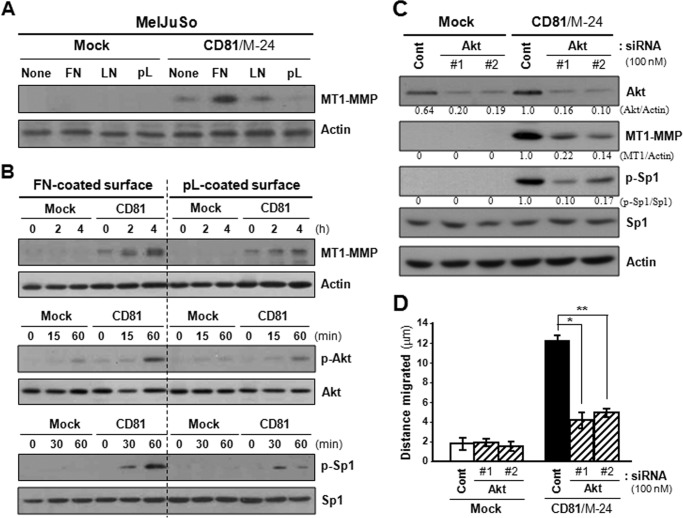
Integrin-dependent Akt Signaling Pathways Involved in CD81-induced MT1-MMP Expression
To reveal the CD81 signaling pathways leading to MT1-MMP transcriptional up-regulation, we first examined the effect of several signaling inhibitors on the MT1-MMP expression in CD81-expressing cells. Among the inhibitors examined, LY294002, an inhibitor targeting the PI3K-Akt pathway, significantly suppressed MT1-MMP expression in CD81 transfectant cells (data not shown). Moreover, the migrating ability of CD81 transfectant cells was also impaired by treatment with LY294002. These results suggest that the PI3K-Akt pathway is involved in CD81 signaling cascades to increase MT1-MMP expression and cell motility. Because CD81 was revealed to associate with α3β1 and α5β1 integrins in MelJuSo cell membranes, we next examined the effect of integrin activation on CD81 signaling for MT1-MMP expression. Fibronectin and to a lesser extent laminin increased MT1-MMP levels in CD81 transfectant cells, whereas poly-l(+)-lysine, which minimally activates integrins (7), did not (Fig. 9A). Besides MT1-MMP expression levels, phosphorylation levels of Akt and Sp1 in MelJuSo cells were increased not only by CD81 expression but also by cell adhesion to fibronectin (Fig. 9B). Additionally, CD81-expressing cells exhibited increased migrating ability on fibronectin-coated plates as compared with uncoated plates (data not shown). Thus, CD81 appeared to work together with integrins to induce the phosphorylation of Akt and Sp1 followed by expression of MT1-MMP. Meanwhile, siRNA knockdown of Akt significantly decreased MT1-MMP expression and Sp1 phosphorylation in CD81-expressing cells (Fig. 9C). The positive effect of CD81 on cell motility was also attenuated by Akt knockdown (Fig. 9D). Taken together, CD81 seems likely to collaborate with the associated α3β1/α5β1 integrins to stimulate Akt-dependent Sp1 activation signaling pathways, which lead to an increase in MT1-MMP expression and cell motility.
FIGURE 9.
Integrin-dependent CD81 signaling activates Akt, which up-regulates Sp1-mediated MT1-MMP expression and cell motility. A, MelJuSo mock and CD81 transfectant cells were suspended in serum-free medium for 60 min and seeded onto plates precoated with fibronectin (FN), laminin (LN), or poly-l(+)-lysine (pL). Following 24-h culture, cells were examined for MT1-MMP expression using immunoblotting analysis. B, cells in suspension were seeded onto fibronectin- or poly-l(+)-lysine-coated plate for the indicated time period and then examined for protein levels of MT1-MMP, Akt, and Sp1. Phosphorylation levels of Akt and Sp1 were also assessed by immunoblotting analyses using anti-phospho-AktSer-473 and phospho-Sp1Thr-453 antibodies, respectively. Shown are representative immunoblots of three independent experiments. C and D, cells grown on fibronectin-coated plate were transfected with two siRNAs targeting Akt and then examined for MT1-MMP protein and Sp1 phosphorylation levels (C) and cell motility (D). The protein levels of Akt and MT1-MMP and phosphorylation levels of Sp1 were normalized to actin and Sp1 protein levels, respectively. Mean values of relative band intensity are shown under the immunoblot. Migration data shown are the mean ± S.D. from three separate experiments with three determinations per experiment (* and **, p < 0.01). Error bars represent S.D. Cont, control.
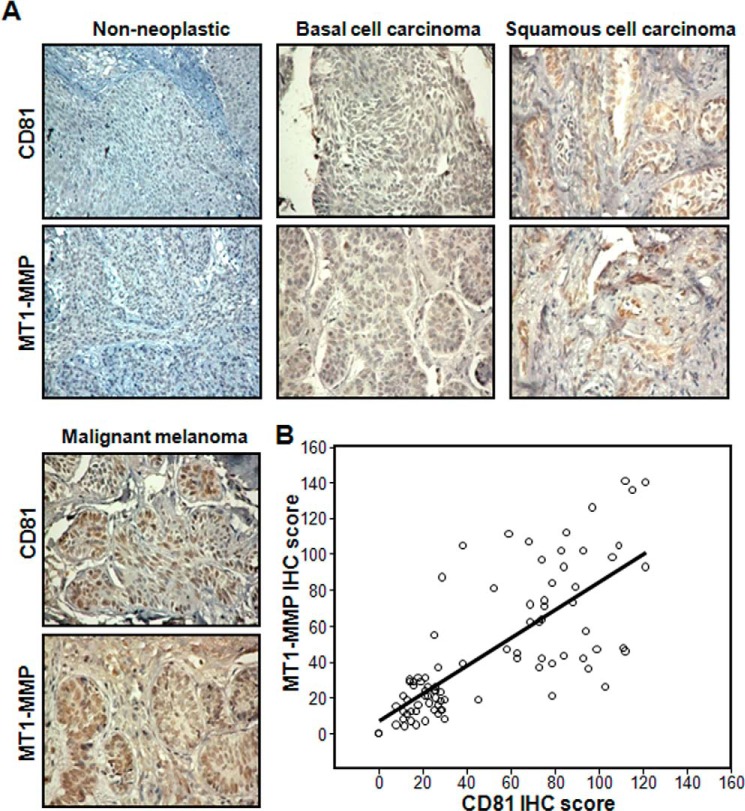
Elevated Expression of CD81 and MT1-MMP in Malignant Melanomas
We examined the expression levels of CD81 and MT1-MMP in human skin cancer tissues from patients through immunohistochemical analysis of tissue microarray sections comprising 41 basal cell carcinoma, 33 squamous cell carcinoma, 10 malignant melanoma, and two normal human skin tissue samples. Both CD81 and MT1-MMP proteins were detected in the majority of the skin cancer tissue samples but not in normal skin tissues (Fig. 10A and Table 1). Importantly, CD81 expression was positively correlated with MT1-MMP expression (ρ = 0.774, p = 0.000001; Fig. 10B), indicating a close link in expression between CD81 and MT1-MMP in human skin cancers. Moreover, malignant melanomas contained more CD81- and MT1-MMP-positive cells than non-melanoma skin cancers such as basal and squamous cell carcinomas, suggesting a close association of these two proteins with the development/progression of malignant melanoma (Table 1). Collectively, our findings suggest that CD81 is co-overexpressed with MT1-MMP during malignant melanoma development, which contributes to melanoma invasion and metastasis by enhancing cell migrating ability.
FIGURE 10.
Expression levels of CD81 and MT1-MMP in human skin cancer tissue samples. Two microarray slides containing consecutive sections of human skin cancer tissues with different histopathological status were subjected to immunohistochemical (IHC) staining using either anti-CD81 or anti-MT1-MMP antibody as described under “Experimental Procedures.” A, following counterstaining with hematoxylin, the tissue sections were scanned at low power to identify areas that were evenly stained. Shown are representative images of the tissue immunohistochemical staining. B, CD81 and MT1-MMP immunohistochemical scores represent the number of CD81- and MT1-MMP-positive cells among a total of 150 cells in a microscopic view. Expression levels of CD81 were significantly proportional to those of MT1-MMP (Spearman rank correlation coefficient test, ρ = 0.774, p = 0.000001).
TABLE 1.
Expression of CD81 and MT1-MMP in human skin cancer tissue samples
| Target protein staining intensitya | Percentage of cases of histologic type (n, total number)b |
p valuec | |||
|---|---|---|---|---|---|
| Non-neoplastic (n = 2) | Basal cell carcinoma (n = 41) | Squamous cell carcinoma (n = 33) | Malignant melanoma (n = 10) | ||
| CD81 | |||||
| 0 | 100% (2) | 0% (0) | 0% (0) | 0% (0) | <0.0001 |
| 1 | 0% (0) | 92.7% (38) | 6.1% (2) | 0% (0) | |
| 2 | 0% (0) | 7.3% (3) | 87.8% (29) | 40% (4) | |
| 3 | 0% (0) | 0% (0) | 6.1% (2) | 60% (6) | |
| MT1-MMP | |||||
| 0 | 100% (2) | 0% (0) | 0% (0) | 0% (0) | <0.0001 |
| 1 | 0% (0) | 90.2% (37) | 6.1% (2) | 0% (0) | |
| 2 | 0% (0) | 9.8% (4) | 87.8% (29) | 50% (5) | |
| 3 | 0% (0) | 0% (0) | 6.1% (2) | 50% (5) | |
a Staining intensity was scored from estimates of the numbers of positive cells as follows: 0, <1%; 1, 1–19%; 2, 20–70%; 3, >70% positive.
b Proportion of tumors with respective score out of total tissue number (n) examined is given.
c Statistical significance was determined using χ2 test.
DISCUSSION
Cancer cell migration into the disorganized regions of surrounding stroma is a key process in cancer invasion and metastasis. This process occurs through continual changes in interactions between integrins and extracellular matrix followed by dynamic changes in the actin cytoskeleton. Multiple tetraspanins associated with integrins such as CD9, CD63, CD82, and CD151 were shown to regulate cell migration primarily by influencing extracellular matrix binding and initial signaling of integrins functionally relevant to cell motility (3, 4, 26). Although tetraspanin CD81 is also associated with integrins in the cell membrane, studies addressing the functional involvement of CD81 in cell motility are still scarce and illustrate opposite effects on cell motility. In hepatic carcinoma cells, CD81 was shown to inhibit cell motility through complex formation with type II PI4K (27). The interaction of CD81 with type II PI4K impaired proper assembly of the actin cytoskeleton by generating intracellular vesicles that sequester actinin-4 from the plasma membrane. In contrast, Quast et al. (28) demonstrated an essential role of CD81 in the migration of adherent dendritic cells. This study revealed that CD81 gene knock-out and CD81-selective knockdown resulted in an inability of cells to form actin protrusions such as lamellipodia at the cell front. Moreover, a defect in Rac signaling, which is involved in integrin-dependent migration, was also observed. CD81-dependent migration of dendritic cells is suggested to involve CD81 function, which mobilizes preformed integrin clusters into the leading edge of migrating cells. A recent study using a breast cancer model also demonstrated the significance of CD81 in the stimulation of protrusive activity and motility of breast cancer cells in which CD81 present in fibroblast-secreted exosomes contributed to this process (29). In this study, we also demonstrated that in human melanoma cell lines cell motility was positively correlated with CD81 expression levels. Furthermore, ectopic expression of CD81 in a melanoma cell line lacking endogenous CD81 resulted in increased cell motility, whereas knockdown of CD81 in a cell line with endogenous CD81 displayed the opposite effect. Thus, CD81 appeared to function as a positive regulator for cell motility in melanoma cells, which is in agreement with its role in breast cancer and dendritic cells but in opposition to its role in hepatic carcinoma cells. Taken together, we speculate here that CD81 may exert differential effects on cancer cell motility depending on the developmental origin of the cells. Specifically, both melanoma and breast carcinoma arise from cells of ectodermal origins, whereas hepatic carcinoma is an endoderm derivative.
In the present study, we demonstrated that CD81 effects on melanoma cell motility are largely dependent on MT1-MMP induced by CD81 signaling. A recent report also showed that MT1-MMP and MMP-2 induced by hemokinin-1 contribute to glioma cell migration (30). MT1-MMP degrades extracellular matrix to make a path for cells to migrate not only by utilizing its own proteolytic activity but also by converting pro-MMP-2 to active MMP-2, one of the highly active matrix-degrading proteases (24). Moreover, MT1-MMP sheds cell surface proteins to give migratory signals and activates signaling pathways enhancing cell migration (24). MT1-MMP is known to form complexes with tetraspanins including CD9, CD63, CD81, CD82, and CD151 in various cell types (31–34). Here we also found MT1-MMP co-immunoprecipitated with CD81 in human melanoma cells (data not shown). Previous reports showed that interactions of MT1-MMP with tetraspanins regulate its cell surface expression by modulating its intracellular trafficking, although those interactions exert conflicting effects on MT1-MMP functions (31, 32, 34, 35). In particular, Lafleur et al. (35) showed that CD81 in collaboration with CD9 and TSPAN12 protects the newly synthesized MT1-MMP from lysosomal degradation in breast cancer cells and thereby increases levels of MT1-MMP at the cell surface. They observed a minimal effect of CD81 on the initial biosynthesis of MT1-MMP in human breast cancer cells. However, our present results clearly indicate that CD81 is able to induce MT1-MMP expression at the transcriptional level in human melanoma cells. CD81 cDNA transfection into melanoma cells devoid of both CD81 and MT1-MMP increased expression levels of MT1-MMP as well as CD81. siRNA-mediated CD81 knockdown in CD81-expressing cells diminished MT1-MMP levels. Importantly, a positive correlation between CD81 and MT1-MMP levels was observed in human skin cancer tissue specimens. Additionally, among human melanoma parental cell lines, cell lines with CD81 contained MT1-MMP, whereas those without CD81 did not. Thus, CD81 appeared to up-regulate MT1-MMP expression in human melanoma cells. Because each of the tetraspanin family members is differentially expressed depending on the cell types, it is possible that tetraspanin proteins associated and/or collaborated with CD81 in melanoma cells are different from those in breast cancer cells. This could be the reason for the different effects of CD81 on MT1-MMP gene transcription between melanoma and breast cancer cells. Collectively, increased expression and cell surface trafficking of MT1-MMP both contribute to MT1-MMP-dependent pericellular matrix proteolysis to allow migration of cancer cells during cancer invasion.
Here we demonstrated functional involvement of the Sp1 transcription factor in CD81-induced MT1-MMP gene transcription. A previous study also showed Sp1-dependent MT1-MMP expression in human prostate cancer cells where the signaling pathways to modulate MT1-MMP expression affected protein levels of Sp1 (36). In the present study, CD81 was found to induce phosphorylation of Sp1 at the threonine 453 residue in melanoma cells without altering its protein level. Because threonine 453 phosphorylation of Sp1 was shown to increase its DNA binding and/or transcriptional activities toward target genes including VEGF (37–39), increased threonine phosphorylation of Sp1 by CD81 signaling is likely a mechanism of CD81-induced MT1-MMP transcription in melanoma cells. Meanwhile, the ERK, Akt, and JNK pathways are implicated in increasing Sp1 phosphorylation in various cell systems (38, 40). Interestingly, Carloni et al. (41) found that CD81 increases synthesis of phosphoinositides by activating the associated PI4K, leading to phosphoinositide-facilitated membrane translocation of Shc in hepatic tumor cells. Although they paid attention to Shc-mediated ERK activation only, it is possible that phosphoinositides increased by CD81 lead to activation of the PI3K-Akt pathway. Here we provide evidence for the involvement of the PI3K-Akt pathway in CD81 signaling events. In particular, adhesion-dependent phosphorylation of Akt and Sp1 was observed in CD81-expressing melanoma cells but not in CD81-deficient cells. Notably, the stimulating effect of CD81 on phosphorylation of Akt and Sp1 became more prominent when integrins were activated by cell adhesion to fibronectin. A previous report also showed Akt activation by integrin interaction with fibronectin (42). Because CD81 associates with fibronectin-binding α3β1/α5β1 integrins in the membrane of melanoma cells, it is likely that CD81 and α3β1/α5β1 integrins interact to function cooperatively as a complex with signal-transducing activity, which leads to Akt activation followed by Sp1 phosphorylation to up-regulate MT1-MMP transcription.
The functional effects of CD81 on cancer cell invasiveness and metastasis have been poorly investigated. Previously, Mazzocca et al. (27) reported that CD81-enriched vesicles inhibited intrahepatic spreading of liver cancer cells. However, they did not show secondary tumors formed by metastasized hepatocarcinoma cells in organs other than the liver. A recent report also showed that CD81 in exosomes secreted from fibroblasts plays a role in stimulating breast cancer cells to acquire invasive and metastatic behaviors (29). However, CD81 in fibroblasts, but not in breast cancer cells, is functionally involved in this intercellular communication pathway. Meanwhile, our present in vitro and in vivo data clearly indicate that CD81 present in melanoma cells plays a role in regulating melanoma cell motility and invasiveness together with distant metastasis. Furthermore, human skin cancer tissue specimens displayed a significantly increased CD81 expression in malignant melanomas as compared with non-melanocytic skin cancers. MT1-MMP also exhibited a strong expression in malignant melanoma tissues. The ONCOMINE cancer microarray database also revealed that both CD81 and MT1-MMP levels were significantly increased in cutaneous melanomas as compared with benign melanocytic skin nevi (data not shown). These findings support a close association of CD81 with the malignant progression of human melanocytic tumors. In conclusion, tetraspanin CD81 is highly likely to promote melanoma invasion and metastasis by increasing cell motility via induction of MT1-MMP expression.
Acknowledgment
We thank Elaine Por for corrections of the manuscript.
This work was supported by the Basic Science Research Program Grant 2011-0024380 through the National Research Foundation of Korea funded by the Ministry of Education and by Grant HI13C1819 of the Korean Health Technology Research and Development Project, Ministry of Health and Welfare, Republic of Korea.
- TEM
- tetraspanin-enriched microdomain
- CAM
- chick chorioallantoic membrane
- MMP
- matrix metalloproteinase
- MT1-MMP
- membrane type 1 matrix metalloproteinase
- PI4K
- phosphoinositide 4-kinase.
REFERENCES
- 1. Levy S., Shoham T. (2005) The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 5, 136–148 [DOI] [PubMed] [Google Scholar]
- 2. Hemler M. E. (2008) Targeting of tetraspanin proteins—potential benefits and strategies. Nat. Rev. Drug Discov. 7, 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Richardson M. M., Jennings L. K., Zhang X. A. (2011) Tetraspanins and tumor progression. Clin. Exp. Metastasis 28, 261–270 [DOI] [PubMed] [Google Scholar]
- 4. Hemler M. E. (2005) Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801–811 [DOI] [PubMed] [Google Scholar]
- 5. Zhang X. A., Bontrager A. L., Hemler M. E. (2001) Transmembrane-4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific β1 integrins. J. Biol. Chem. 276, 25005–25013 [DOI] [PubMed] [Google Scholar]
- 6. Yauch R. L., Berditchevski F., Harler M. B., Reichner J., Hemler M. E. (1998) Highly stoichiometric, stable, and specific association of integrin α3β1 with CD151 provides a major link to phosphatidylinositol 4-kinase, and may regulate cell migration. Mol. Biol. Cell 9, 2751–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hong I. K., Jeoung D. I., Ha K. S., Kim Y. M., Lee H. (2012) Tetraspanin CD151 stimulates adhesion-dependent activation of Ras, Rac, and Cdc42 by facilitating molecular association between β1 integrins and small GTPases. J. Biol. Chem. 287, 32027–32039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yáñez-Mó M., Barreiro O., Gordon-Alonso M., Sala-Valdés M., Sánchez-Madrid F. (2009) Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 19, 434–446 [DOI] [PubMed] [Google Scholar]
- 9. Oren R., Takahashi S., Doss C., Levy R., Levy S. (1990) TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Mol. Cell. Biol. 10, 4007–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shoham T., Rajapaksa R., Kuo C. C., Haimovich J., Levy S. (2006) Building of the tetraspanin web: distinct structural domains of CD81 function in different cellular compartments. Mol. Cell. Biol. 26, 1373–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Geisert E. E., Jr., Williams R. W., Geisert G. R., Fan L., Asbury A. M., Maecker H. T., Deng J., Levy S. (2002) Increased brain size and glial cell number in CD81-null mice. J. Comp. Neurol. 453, 22–32 [DOI] [PubMed] [Google Scholar]
- 12. Rubinstein E., Ziyyat A., Prenant M., Wrobel E., Wolf J. P., Levy S., Le Naour F., Boucheix C. (2006) Reduced fertility of female mice lacking CD81. Dev. Biol. 290, 351–358 [DOI] [PubMed] [Google Scholar]
- 13. Pileri P., Uematsu Y., Campagnoli S., Galli G., Falugi F., Petracca R., Weiner A. J., Houghton M., Rosa D., Grandi G., Abrignani S. (1998) Binding of hepatitis C virus to CD81. Science 282, 938–941 [DOI] [PubMed] [Google Scholar]
- 14. Charrin S., Le Naour F., Oualid M., Billard M., Faure G., Hanash S. M., Boucheix C., Rubinstein E. (2001) The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J. Biol. Chem. 276, 14329–14337 [DOI] [PubMed] [Google Scholar]
- 15. Serru V., Le Naour F., Billard M., Azorsa D. O., Lanza F., Boucheix C., Rubinstein E. (1999) Selective tetraspan-integrin complexes (CD81/α4β1, CD151/α3β1, CD151/α6β1) under conditions disrupting tetraspan interactions. Biochem. J. 340, 103–111 [PMC free article] [PubMed] [Google Scholar]
- 16. Hemler M. E. (2014) Tetraspanin proteins promote multiple cancer stages. Nat. Rev. Cancer 14, 49–60 [DOI] [PubMed] [Google Scholar]
- 17. Inoue G., Horiike N., Onji M. (2001) The CD81 expression in liver in hepatocellular carcinoma. Int. J. Mol. Med. 7, 67–71 [DOI] [PubMed] [Google Scholar]
- 18. Duxbury M. S., Ito H., Zinner M. J., Ashley S. W., Whang E. E. (2004) CEACAM6 gene silencing impairs anoikis resistance and in vivo metastatic ability of pancreatic adenocarcinoma cells. Oncogene 23, 465–473 [DOI] [PubMed] [Google Scholar]
- 19. Hong I. K., Jin Y. J., Byun H. J., Jeoung D. I., Kim Y. M., Lee H. (2006) Homophilic interactions of tetraspanin CD151 up-regulate motility and matrix metalloproteinase-9 expression of human melanoma cells through adhesion-dependent c-Jun activation signaling pathways. J. Biol. Chem. 281, 24279–24292 [DOI] [PubMed] [Google Scholar]
- 20. Kim J., Yu W., Kovalski K., Ossowski L. (1998) Requirement for specific proteases in cancer cell intravasation as revealed by a novel semiquantitative PCR-based assay. Cell 94, 353–362 [DOI] [PubMed] [Google Scholar]
- 21. Choi N. S., Kim S. H. (2000) Two fibrin zymography methods for analysis of plasminogen activators on gels. Anal. Biochem. 281, 236–238 [DOI] [PubMed] [Google Scholar]
- 22. Taniuchi K., Nakagawa H., Hosokawa M., Nakamura T., Eguchi H., Ohigashi H., Ishikawa O., Katagiri T., Nakamura Y. (2005) Overexpressed P-cadherin/CDH3 promotes motility of pancreatic cancer cells by interacting with p120ctn and activating rho-family GTPases. Cancer Res. 65, 3092–3099 [DOI] [PubMed] [Google Scholar]
- 23. Por E., Byun H. J., Lee E. J., Lim J. H., Jung S. Y., Park I., Kim Y. M., Jeoung D. I., Lee H. (2010) The cancer/testis antigen CAGE with oncogenic potential stimulates cell proliferation by up-regulating cyclins D1 and E in an AP-1- and E2F-dependent manner. J. Biol. Chem. 285, 14475–14485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Itoh Y. (2006) MT1-MMP: a key regulator of cell migration in tissue. IUBMB Life 58, 589–596 [DOI] [PubMed] [Google Scholar]
- 25. Lohi J., Lehti K., Valtanen H., Parks W. C., Keski-Oja J. (2000) Structural analysis and promoter characterization of the human membrane-type matrix metalloproteinase-1 (MT1-MMP) gene. Gene 242, 75–86 [DOI] [PubMed] [Google Scholar]
- 26. Lazo P. A. (2007) Functional implications of tetraspanin proteins in cancer biology. Cancer Sci. 98, 1666–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mazzocca A., Liotta F., Carloni V. (2008) Tetraspanin CD81-regulated cell motility plays a critical role in intrahepatic metastasis of hepatocellular carcinoma. Gastroenterology 135, 244–256.e1 [DOI] [PubMed] [Google Scholar]
- 28. Quast T., Eppler F., Semmling V., Schild C., Homsi Y., Levy S., Lang T., Kurts C., Kolanus W. (2011) CD81 is essential for the formation of membrane protrusions and regulates Rac1-activation in adhesion-dependent immune cell migration. Blood 118, 1818–1827 [DOI] [PubMed] [Google Scholar]
- 29. Luga V., Zhang L., Viloria-Petit A. M., Ogunjimi A. A., Inanlou M. R., Chiu E., Buchanan M., Hosein A. N., Basik M., Wrana J. L. (2012) Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 151, 1542–1556 [DOI] [PubMed] [Google Scholar]
- 30. Mou L., Kang Y., Zhou Y., Zeng Q., Song H., Wang R. (2013) Neurokinin-1 receptor directly mediates glioma cell migration by up-regulation of matrix metalloproteinase-2 (MMP-2) and membrane type 1-matrix metalloproteinase (MT1-MMP). J. Biol. Chem. 288, 306–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takino T., Miyamori H., Kawaguchi N., Uekita T., Seiki M., Sato H. (2003) Tetraspanin CD63 promotes targeting and lysosomal proteolysis of membrane-type 1 matrix metalloproteinase. Biochem. Biophys. Res. Commun. 304, 160–166 [DOI] [PubMed] [Google Scholar]
- 32. Yañez-Mó M., Barreiro O., Gonzalo P., Batista A., Megías D., Genís L., Sachs N., Sala-Valdés M., Alonso M. A., Montoya M. C., Sonnenberg A., Arroyo A. G., Sánchez-Madrid F. (2008) MT1-MMP collagenolytic activity is regulated through association with tetraspanin CD151 in primary endothelial cells. Blood 112, 3217–3226 [DOI] [PubMed] [Google Scholar]
- 33. Kolesnikova T. V., Kazarov A. R., Lemieux M. E., Lafleur M. A., Kesari S., Kung A. L., Hemler M. E. (2009) Glioblastoma inhibition by cell surface immunoglobulin protein EWI-2, in vitro and in vivo. Neoplasia 11, 77–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schröder H. M., Hoffmann S. C., Hecker M., Korff T., Ludwig T. (2013) The tetraspanin network modulates MT1-MMP cell surface trafficking. Int. J. Biochem. Cell Biol. 45, 1133–1144 [DOI] [PubMed] [Google Scholar]
- 35. Lafleur M. A., Xu D., Hemler M. E. (2009) Tetraspanin proteins regulate membrane type-1 matrix metalloproteinase-dependent pericellular proteolysis. Mol. Biol. Cell 20, 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sroka I. C., Nagle R. B., Bowden G. T. (2007) Membrane-type 1 matrix metalloproteinase is regulated by sp1 through the differential activation of AKT, JNK, and ERK pathways in human prostate tumor cells. Neoplasia 9, 406–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Milanini-Mongiat J., Pouysségur J., Pagès G. (2002) Identification of two Sp1 phosphorylation sites for p42/p44 mitogen-activated protein kinases: their implication in vascular endothelial growth factor gene transcription. J. Biol. Chem. 277, 20631–20639 [DOI] [PubMed] [Google Scholar]
- 38. Reisinger K., Kaufmann R., Gille J. (2003) Increased Sp1 phosphorylation as a mechanism of hepatocyte growth factor (HGF/SF)-induced vascular endothelial growth factor (VEGF/VPF) transcription. J. Cell Sci. 116, 225–238 [DOI] [PubMed] [Google Scholar]
- 39. Chu S., Ferro T. J. (2005) Sp1: regulation of gene expression by phosphorylation. Gene 348, 1–11 [DOI] [PubMed] [Google Scholar]
- 40. Benasciutti E., Pagès G., Kenzior O., Folk W., Blasi F., Crippa M. P. (2004) MAPK and JNK transduction pathways can phosphorylate Sp1 to activate the uPA minimal promoter element and endogenous gene transcription. Blood 104, 256–262 [DOI] [PubMed] [Google Scholar]
- 41. Carloni V., Mazzocca A., Ravichandran K. S. (2004) Tetraspanin CD81 is linked to ERK/MAPKinase signaling by Shc in liver tumor cells. Oncogene 23, 1566–1574 [DOI] [PubMed] [Google Scholar]
- 42. Fornaro M., Plescia J., Chheang S., Tallini G., Zhu Y. M., King M., Altieri D. C., Languino L. R. (2003) Fibronectin protects prostate cancer cells from tumor necrosis factor-α-induced apoptosis via the AKT/survivin pathway. J. Biol. Chem. 278, 50402–50411 [DOI] [PubMed] [Google Scholar]