Background: KLF14 has elicited attention as a master regulator of lipid metabolism.
Results: KLF14 regulates chromatin remodeling on sphingosine kinase 1 gene leading to its activation and sphingosine-1-phosphate production.
Conclusion: KLF14 acts as a transcriptional activator for the generation of lipid signaling molecules.
Significance: This new knowledge extends the functions assigned to KLF14 and contributes to understanding its role in human diseases.
Keywords: Chromatin Histone Modification, Epigenetics, Fibroblast Growth Factor (FGF), Kruppel-like Factor (KLF), Sphingosine-1-phosphate (S1P)
Abstract
Sphingosine kinase 1 (SK1) is an FGF-inducible gene responsible for generation of sphingosine-1-phosphate, a critical lipid signaling molecule implicated in diverse endothelial cell functions. In this study, we identified SK1 as a target of the canonical FGF2/FGF receptor 1 activation pathway in endothelial cells and sought to identify novel transcriptional pathways that mediate lipid signaling. Studies using the 1.9-kb SK1 promoter and deletion mutants revealed that basal and FGF2-stimulated promoter activity occurred through two GC-rich regions located within 633 bp of the transcription start site. Screening for GC-rich binding transcription factors that could activate this site demonstrated that KLF14, a gene implicated in obesity and the metabolic syndrome, binds to this region. Congruently, overexpression of KLF14 increased basal and FGF2-stimulated SK1 promoter activity by 3-fold, and this effect was abrogated after mutation of the GC-rich sites. In addition, KLF14 siRNA transfection decreased SK1 mRNA and protein levels by 3-fold. Congruently, SK1 mRNA and protein levels were decreased in livers from KLF14 knock-out mice. Combined, luciferase, gel shift, and chromatin immunoprecipitation assays showed that KLF14 couples to p300 to increase the levels of histone marks associated with transcriptional activation (H4K8ac and H3K14ac), while decreasing repressive marks (H3K9me3 and H3K27me3). Collectively, the results demonstrate a novel mechanism whereby SK1 lipid signaling is regulated by epigenetic modifications conferred by KLF14 and p300. Thus, this is the first description of the activity and mechanisms underlying the function of KLF14 as an activator protein and novel regulator of lipid signaling.
Introduction
Sphingosine kinase 1 (SK1),3 a ubiquitously expressed enzyme expressed in different mammalian tissues including endothelia, is responsible for the generation of sphingosine-1-phosphate (S1P), a lipid molecule that regulates cell growth, survival, differentiation, and motility (1–5). SK1 also regulates production of ceramide and sphingosine, additional lipid signaling molecules that critically mediate endothelial cell (EC) biology (1, 6). Although SK1 transcriptional activation may occur downstream from canonical receptor tyrosine kinases, the transcriptional and epigenetic mechanisms of SK1 regulation remain incompletely defined. We now provide evidence that KLF14 works downstream of the canonical fibroblast growth factor receptor (FGFR) tyrosine kinase pathway to transcriptionally activate SK1 expression with the consequent up-regulation in S1P lipid signaling.
The evolutionarily conserved family of Kruppel-like factor (KLF) transcription factors is composed of 17 members that regulate the expression of genes required for the proper execution of important biological processes, including proliferation, apoptosis, differentiation, and development (7). In addition, alterations in either the structure or function of several members of this family are associated with human diseases (8–10). Although some KLF genes are widely expressed, others show tissue-specific patterns of expression. Many organs and cells express a defined repertoire of KLF proteins, some of which engage in functional cooperation or antagonism dependent upon the context (7, 11, 12). This context is created by several cytokines and growth factors that induce the expression of distinct KLF proteins, as well as post-translational modification, degradation, DNA binding, and coupling to chromatin (13–15). Structurally, KLF proteins are characterized by the presence of their conserved DNA-binding domain, composed of three Cys2/His2 zinc fingers similar to Sp1 at the C terminus. The C-terminal zinc fingers recognize and bind to the GT/GC-rich cis-regulatory sites found in gene promoters and enhancers. The members of the KLF family also have a nuclear localization signal sequence and transcriptional regulatory domain localized in their N-terminal portion. This N terminus is highly variable and confers functional specificity to KLF interactions with distinct nuclear proteins leading to gene activation, gene repression, or both. Thus, KLF transcription factors form a network of proteins that is significantly important for mediating disease phenotypes in many organisms ranging from flies to humans (7).
We have previously reported the cloning and characterization of several KLF proteins, namely KLF10, KLF11, KLF13, KLF14, and KLF16, which together with KLF9 form a subfamily of proteins that regulate metabolic functions in humans. Notably, the Drosophila melanogaster ancestor for this protein, Cabut, also plays a conserved role in metabolic pathways. Moreover, select functions of some of these proteins (such as KLF11 and KLF14) can cause common human metabolic diseases, including neonatal and juvenile diabetes, as well as the obesity-insulin resistance-metabolic syndrome, which leads to adult type 2 diabetes (10, 16, 17). Thus, because of their structural conservation and function, these six proteins have become to be known as the Cabut family of metabolic regulators (9, 18). Consequently, we and others have recently focused on how these proteins regulate the expression of genes and gene networks involved in lipid metabolism and signaling. Thus, the current work aimed on defining a novel function for KLF14, involving transcriptional activation of SK1 induced by FGF2 to expand our understanding of its role in lipid biology.
Our results demonstrate, for the first time, that KLF14 is not only a regulator of lipid metabolism but also mediates lipid signaling. Furthermore, we show that KLF14 achieves this function by coupling to the p300 histone acetyl transferase to catalyze the deposition of acetylated marks on promoter-associated histones. Because previous characterization of KLF14 has only been related to its coupling to the Sin3a-HDAC complex to repress gene expression (13), the recognition of its p300-dependent activating function extends the mechanistic understanding of this protein as a transcription factor. Similarly, the identification of its role in SK1-mediated signaling helps to better delineate how this protein contributes to lipid homeostasis. Thus, the new knowledge derived from this study is relevant to the field of transcription and chromatin-mediated regulation of lipid signaling and should be taken into consideration for better understanding of diseases that are caused by dysregulation of KLF14-mediated pathways.
EXPERIMENTAL PROCEDURES
Cell Culture
Human hepatic sinusoidal endothelial cells (HHSECs), human umbilical vein endothelial cells (HUVECs), and primary murine liver endothelial cells were grown with endothelial culture media with 10% serum and 1% endothelial growth supplement. Human HEK 293 cells were grown in Dulbecco's modified Eagle's medium with 10% serum. Primary liver endothelial cells were obtained from C57Bl/6J and Klf14−/− mice as previously described (19). Klf14−/− mice were generated at the Mayo Transgenic and Gene Targeted Mouse Shared Resource facility using ES cells engineered by University of CA Davis Knock-out project to carry a 1035-bp deletion encompassing the complete, intronless, coding region of this gene. Recombined ES clones were used for the generation of chimaeras and then crossed to C57BL/6J mice. Heterozygous founders were used to generate Klf14−/− animals. The genotype of the animals used to derive the Klf14−/− endothelial liver cells was confirmed by PCR, using 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 60 s with the following primers for Klf14 or the Neomycin cassette: Klf14 forward, 5′-TCAACTAGCTGCTTCGAGCC-3′; Klf14 reverse, 5′-ACGACCTCGGTACTCGATCA-3′; NEO forward, 5′-TCATTCTCAGTATTGTTTTGCC-3′; and NEO reverse, 5′-GAGTTCATCCCTTCTCAAAGG-3′. The presence of a 323-bp product was diagnostic for the presence of the Neomycin cassette and homozygous deletion of Klf14, whereas a 540-bp product indicated an intact Klf14 when run on 1.5% agarose gel (supplemental Fig. S1). All different cells lines used in this study were maintained at 37 °C and 5% CO2.
RNA Isolation, cDNA Synthesis, and Quantitative Real Time PCR
Total RNA was extracted from cells and mouse tissue according to the manufacturer's instructions using an RNeasy kit (Qiagen), and 5 μg was used for cDNA synthesis with oligo(dT) primer using SuperScriptTM III first strand synthesis system for reverse transcription PCR (RT-PCR) (Invitrogen) per the manufacturer's protocol. Real time PCR was performed in a total 25-μl volume reaction using Sybr Green Master Mix and the 7500 real time PCR system (both from Applied Biosystems), according to the manufacturer's instructions. RT-PCR analysis was performed with the following primer sets: human SK1 forward, 5′-AATTTCAAATATTGAACAGCTCGGAA-3′; human SK1 reverse, 5′-TTTATAATGTTTGACATGGTCTCCTTT-3′; mouse SK1 forward, 5′-AATTTCAAATATTGAACAGCTCGGAA-3′; and mouse SK1 reverse, 5′-TTTATAATGTTTGACATGGTCTCCTTT-3′. Amplification of human GAPDH and mouse β-actin was performed in the same reaction for respective samples as internal controls. Each experiment was done in triplicate.
Plasmid and Adenovirus Construction
The technique used to clone KLF2, KLF3, KLF4, KLF5, KLF6, KLF7, KLF10, KLF12, KLF14, KLF15, and KLF16 was previously described (13). The KLF14 mutant defective in Sin3-histone deacetylase (HDAC) coupling has been previously characterized (20, 21) The human p300 and p300 dominant-negative (p300DN) construct was from Upstate Biotechnology, Inc. (Lake Placid, NY). For FGFR1 adenovirus constructs, FGFR1 cDNA was first subcloned into the TA vector as an intermediate step. NotI and SalI restriction enzyme digestion of FGFR1-TA was carried out and ligated with an AdEASY-FLAG shuttle. The AdEASY adenovirus generation was performed as described previously (22).
SK1 Activity Assay
HUVEC cells infected with FGFR1 adenovirus were stimulated for 6 h with 25 ng/ml of FGF2 or 0.1% BSA (control). Cells were lysed in an SK1 buffer, containing 20 mm Tris-HCl, pH 7.5, 1 mm EDTA, 0.4 mm PMSF, 15 mm NaF, 1 mm Na3VO4, 1 mm β-mercaptoethanol, 40 mm β-glycerol phosphate, 50% glycerol, 0.5 mm deoxypyridoxine, and 0.1% Triton X-100. Briefly, 60 μg of protein lysate was used for the enzymatic assay. The reaction (30 min at 37 °C) was started with the addition of 5 mm sphingosine and [γ-32P]ATP (5 μCi, 1 mm) in a final volume of 90 μl. The reaction was terminated with 10 μl of 1 n HCl and with 400 μl of chloroform/methanol/HCl (100:200:1, v/v/v). After vigorous vortexing, 120 μl of chloroform and 120 μl of 2 m KCl were added. After incubation (10 min at room temperature), the suspension was centrifuged at 1000 × g for 10 min at room temperature. The organic phase was removed and dried in the hood overnight. The next day, the samples were resuspended in 40 μl of chloroform/methanol/HCl (100:200:1, v/v/v) and applied to a silica gel 60 TLC plate, which was developed in butanol/ethanol/acetic acid/water (80:20:10:20, v/v/v/v). The spot corresponding to S1P was visualized by autoradiography and quantified by densitometer.
Sphingosine-1-Phosphate ELISA
Sphingosine-1-Phosphate ELISA (Echelon Biosciences Inc.) was performed using the same FGF2 treatment condition used for SK1 activity assay. S1P levels were also measured from isolated liver endothelial cells from wild-type (C57BL/6J) and Klf14−/− mice. The ELISA was performed according to the manufacturer's instructions.
Transfection and siRNA
siRNA targeting human KLF4, KLF14, and a scrambled control were purchased from Qiagen. HUVEC, an endothelial cell line amenable to siRNA transfection, were seeded in 60-mm dishes and transfected with scrambled control and KLF14 siRNA using Oligofectamine (Invitrogen) per manufacturer's protocol. Transfected cells were incubated in medium containing 10% FBS for 72 h prior to experiments. Gene knockdown by siRNA was confirmed by PCR using specific primers. Adenoviral and plasmid transfection of cells was performed as described previously (22).
Construction of Luciferase Reporter Vector
A 1.9-kb region upstream from the first exon of SK1 gene was amplified from human genomic DNA using PCR. The primers used for the amplification were designed with HindIII and NcoI sites (forward, 5′-AAGCTTTCGTTCCTGTTTCTCGGAGT-3′; and reverse, 5′-CCATGGTGCTGGGCACGAAGTTCTG-3′). The amplified product was transferred into HindIII and NcoI site of the pGL3 basic vector (Promega). This construct was named as full promoter or −1977-bp SK1 construct promoter (+1 denotes the transcription initiation point of exon 1). The deletion mutants were designed using restriction enzymes. The full promoter was digested with HindIII + PstI, HindIII + PflMI, and HindIII + BmgBI. The fragments were filled in using Klenow fragment and Pfu DNA polymerase and then ligated individually with T4 DNA ligation given origin to −1117-, −633-, and −390-bp SK1 promoter construct, respectively. Computer analyses of SK1 promoter indicated a SP1/KLF binding site located between −516/−528 bp of promoter. To introduce mutation to this site, a primer set (forward, 5′-GGGAGCGCGATTATTACCCAGGCCG-3′; and reverse, 5′-CGGCCTGGGTAATAATCGCGCTCCC-3′) was designed, and three PCRs were performed using the −1977 SK1-pGL3, −1117 SK1-pGL3, and −633 SK1-pGL3 as a template. All mutant constructs were confirmed by sequencing.
Transcriptional Reporter Assays
Because of the low transfection efficiency of HUVEC cells, HEK 293 cells were used for the transcriptional studies. The cells (1 × 106) were transfected using Effectene transfection reagent (Qiagen) with 1 μg of reporter plasmid containing various lengths of the 5′ promoter region of the human SK1 gene, 0.5 μg of FGFR1 plasmid, and, depending on the experiment, 3 μg of other protein constructs. After 48 h of transfection, cells were stimulated for 6 h with FGF2 (25 ng/ml) and lysed, and then relative luciferase activity was measured using the luciferase assay system (Promega) and a Turner 20/20 luminometer. Each experiment was repeated at least three times. Total protein concentrations were measured and used for normalization in all experiments.
EMSAs
Gel shift assays were performed as described previously (23). Briefly, a double-stranded DNA probe containing a SP1/KLF protein DNA-binding domain (5′-CCGGGAGCGCGGGGCGGAGCCAGGCCGGCG-3′) and a double-stranded DNA probe containing a mutation in the same KLF protein DNA-binding domain (5′-CCGGGAGCGCAAAACAAAGCCAGGCCGGCG-3′) were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase according to the manufacturer's protocol (Promega). A GST fusion protein carrying the zinc finger region of KLF14 was purified as described previously (13). 8 μg of purified GST or GST fusion recombinant KLF14 were incubated in a buffer containing 20 mm HEPES (pH 7.5), 50 mm KCl, 5 mm MgCl2, 10 μm ZnCl2, 6% glycerol, 200 μg/ml bovine serum albumin, and 50 μg/ml poly(dI-dC)·poly(dI-dC) for 7 min at room temperature. End-labeled probes, excess of cold probes, or antibodies were added as indicated to each reaction and incubated at room temperature for an additional 20 min. Anti-GST antibody was added as indicated, and then samples were loaded onto a 4% nondenaturing polyacrylamide gel, run for 3 h at 200 V, vacuum-dried, and exposed to autoradiography film.
ChIP Assay
ChIP assays were performed with two different cells lines in different experiments. HUVEC cells infected with FGFR1 adenovirus were stimulated with FGF2 (25 ng/ml) for 4 h and then fixed with 1% formaldehyde. HEK 293 cells were transfected with KLF14 plasmid, and after 48 h the cells were fixed with 1% formaldehyde. ChIP assays were performed using EZ-Magna-ChIP kit (Millipore). The resulting nuclear extract was sonicated on wet ice and then immunoprecipitated with appropriate antibodies against His tag (for recombinant KLF14), p300 (Millipore), H3K4me3, H3K9me3, H4K8ac, H3K14ac, or H3K27me3 (Abcam). The following primer set targeting SK1 promoter was used for quantitative PCR: forward, 5′-TCTAGCCAGACGCCTAGGACGA-3′; and reverse, 5′-TCGCTCCCTCCGGCCTCAAA-3′. As sample reference, we utilized 1% of input for all ChIP performed, and to normalize the samples, we employed the following standard equation: 1% input = 2 ^ [(mean Ct input − 6.64) − mean Ct ChIP] * 100. The results are shown as fold change (sample/control).
RESULTS
FGF2 Induces Both the Expression and Enzymatic Activity of SK1 in Endothelial Cells
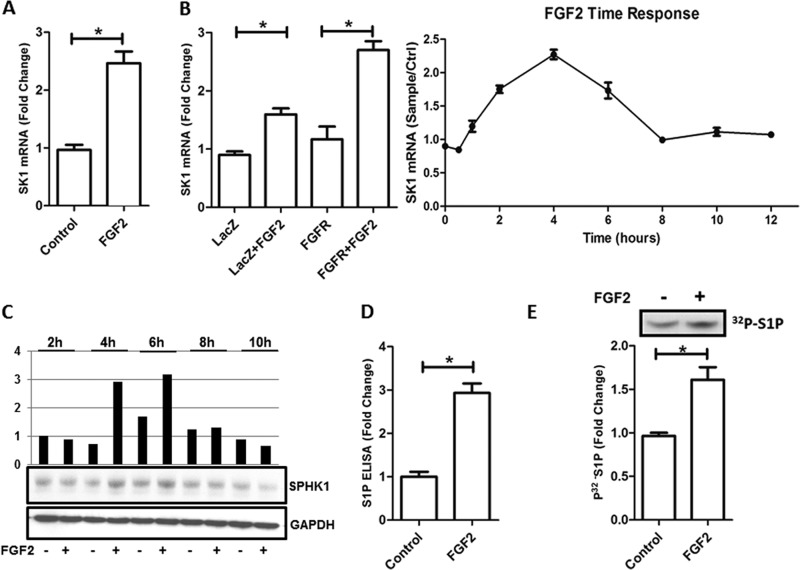
The present study was initially designed to uncover novel mechanisms underlying FGF2/FGFR1-induced activation of EC. For this purpose, we treated HHSECs, which express high levels of FGFR1, with FGF2 (25 ng/ml) and performed pathway-specific microarray studies for angiogenesis and endothelial cell enriched genes (supplemental Fig. S2). These experiments revealed that FGF2 induced a 2.4-fold (control, 0.96 ± 0.1 versus FGF2, 2.45 ± 0.2, p < 0.05) increase in expression of SK1, a signaling enzyme that catalyzes the phosphorylation of sphingosine to form S1P (24, 25). We confirmed the increase in SK1 expression in HHSECs by quantitative PCR, which showed that FGF2 up-regulated SK1 mRNA levels by 2.6 ± 0.3-fold (control, 1.1 ± 0.25 versus FGF2, 2.7 ± 0.3, p < 0.05) compared with vehicle control (Fig. 1A). Similar results were obtained in HUVEC endothelial cells (control, 0.9 ± 0.1 versus FGF2, 1.6 ± 0.2; p < 0.05). To maximize this stimulation, HUVECs were transduced with FGFR1 expressing adenovirus. FGF2 treatment increased SK1 mRNA levels (Fig. 1B, left panel) with a maximal stimulation peak of 2.6-fold at 4 h (control, 1.2 ± 0.4 versus FGF2, 2.7 ± 0.3; p < 0.05) (Fig. 1B, right panel). This result was further supported by the observation that FGF2 increases the levels of SK1 protein, which peaks at 4 and 6 h after treatment (Fig. 1C). Subsequently, we investigated whether this response at the mRNA level corresponded to elevated SK1 enzymatic activity in whole lysate obtained from FGF2-stimulated HUVECs cells. As shown in Fig. 1D, exposure to FGF2 for 6 h resulted in a 60% (control, 0.97 ± 0.3 versus FGF2, 1.6 ± 0.15; p < 0.05) increase in SK1 activity. Furthermore, we utilized ELISA assays to determine whether FGF2 stimulation also enhanced the formation of S1P. Indeed, Fig. 1E shows that treatment of HUVEC cells with this growth factor increased the level of S1P by 3-fold (control, 1.1 ± 0.15 versus FGF2, 2.9 ± 0.22; p < 0.05). These studies reveal the existence of an FGF2-FGFR1-SK1 pathway, which regulates the formation of S1P in endothelial cells. These observations led us to execute transcriptional analyses of the SK1 promoter.
FIGURE 1.
Effect of FGF2 (25 ng/ml) in SK1 mRNA level, SK1 enzyme activity, and S1P production. A, liver endothelial cells were stimulated with FGF2 for 4 h, and SK1 level was determined by quantitative PCR. B, the right panel shows the mRNA levels of SK1 in HUVEC infected with LacZ or FGFR1 adenovirus and stimulated or not with FGF2 for 4 h. In the left panel, HUVEC cells infected with FGFR1 adenovirus were used for FGF2 time response (0, 0.5, 1, 2, 4, 6, 8, 10, and 12 h). SK1 mRNA level is shown as fold change (FGF2/control). A and B, all results are expressed relative to the housekeeping (GAPDH) gene expression as arbitrary units. C, Western blotting with anti-SPHK1 antibody. SK1 protein levels were measured from HUVEC from 2, 4, 6, 8, and 10 h after FGF2 stimulation. D, S1P ELISA was used to measure the S1P production in HHSEC after 6 h of stimulation with FGF2. E, SK1 enzymatic activity of HUVEC cells stimulated with FGF2. The upper panel shows the autoradiography of 6-h FGF2 stimulation in HUVEC cell samples. A representative TLC result is shown by 32P-labeled S1P. The lower panel shows the result of densitometry of each band. The data are shown as means ± S.D. of triplicates, and are representative of three independent experiments performed. *, p < 0.05.
FGF2-mediated Induction of SK1 Gene Expression in Endothelial Cells Occurs via Distinct KLF Binding Sites
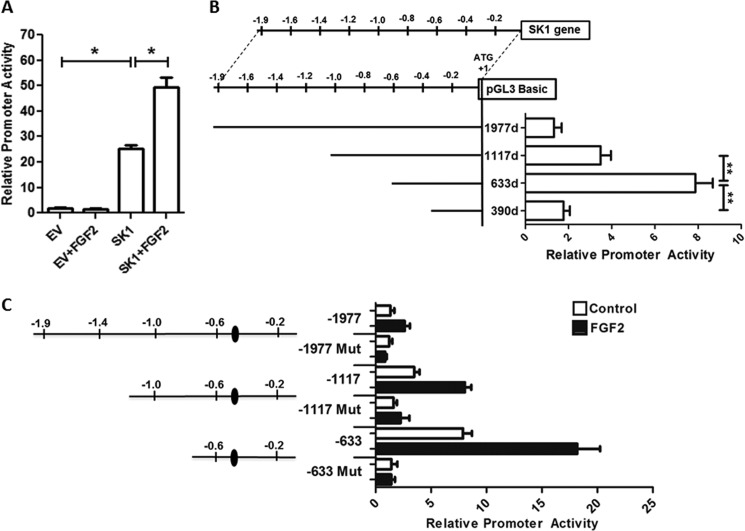
To examine regulation of the SK1 gene, we initially performed luciferase-based reporter assays in HEK 293 cells using a construct containing a 1980-bp fragment amplified from the 5′-flanking region of SK1 gene (−1977 to +3). From this experiment, we observed that the promoter activity given by this construct was ∼13-fold higher than the control empty vector pGL3-Basic (EV) (EV, 1.8 ± 0.5 versus 1980 construct, 24.9 ± 3; p < 0.05) (Fig. 2A). FGF2 stimulation does not affect the reporter activity of the control EV vector. In contrast, activation of the FGF2 receptor by 25 ng/ml of FGF2 enhanced the 1980 construct activity by 2-fold (control, 24.9 ± 3 versus FGF2, 49.7 ± 6.7; p < 0.05). Therefore, we subsequently sought to map the promoter region required for supporting transcriptional activity using deletion mutagenesis of the 1980-bp promoter to generate three contiguous reporter constructs (−1117, −633, and −390). Fig. 2B shows that the highest promoter activity was obtained with the −633 deletion construct (−1977, 1.0 ± 0.02 versus −633, 7.6 ± 0.4; p < 0.05), which was drastically decreased when the promoter length was further reduced to nucleotide −390 (−633, 7.6 ± 0.4 versus −390, 2.2 ± 0.3; p < 0.05). Thus, together, these results demonstrate that whereas the region from −1977 to −633 is not critical for achieving maximal promoter activity, removal of sequences contained between nucleotides −633 to −390 compromises the function of this promoter. The effects of FGF2 stimulation on these constructs are shown in Fig. 2C. Examination of cis-regulatory sequences located in the latter construct revealed the presence of a distinct KLF consensus site located between nucleotides −528 and −516. Thus, to characterize whether this consensus site was responsible for promoter activity, we performed site-directed mutagenesis to disrupt the putative KLF site located between nucleotides −528 through −516 upstream of the transcriptional initiation site. Similarly, we studied the effect of the same mutation in the context of the larger promoter fragments, including the −1117 construct (1117d), and the full promoter (1977d). The results of these experiments, shown in Fig. 2C, demonstrate that the disruption of this −528/−516 putative KLF site resulted in a prominent reduction of promoter activity in all constructs (−1117, 3.5 ± 0.5 versus 1117d, 1.6 ± 0.3; p < 0.05; and −633, 7.9 ± 0.8 versus 633d, 1.4 ± 0.5; p < 0.05), indicating that this region is important for the transcriptional regulation of SK1 under basal conditions. To extend our understanding of the regulation of this promoter, we also examined whether the −528/−516 putative KLF site participates in the FGF2-regulated transcriptional activation of SK1. As shown in Fig. 2C, mutation of this site significantly decreased the promoter activity induced by FGF2 (−633, 18.2 ± 2.1 versus 633d, 1.4 ± 0.33; p < 0.05). These data, therefore, reveal that disruption of the −528/−516 putative KLF site also compromises the transcriptional activation of SK1 in response to angiogenic growth factors. This information was both challenging and important because 17 different mammalian KLF proteins have the potential to bind and regulate a myriad of either GC- or GT-rich promoters. However, the defined characterization of the gene targets as well as the identity of the KLF members involved in this process remains to be fully understood. Consequently, we designed experiments that could identify which of the KLF family members are primarily responsible for this function.
FIGURE 2.
Promoter structure of the 5′-region of human SK1 gene and promoter activity assay. For all experiments, HEK 293 cells were transfected with 1.0 μg of SK1 constructs and 0.5 μg of FGFR1 plasmid. A, to evaluate the role of FGF2 in the transcriptional activation of human SK1 gene, HEK 293 cells were transfected with full promoter (SK1) or pGL3-Basic vector (EV) and stimulated for 12 h with FGF2 (25 ng/ml). The relative luciferase values were normalized to EV control. B, the upper panel illustrates the genome structure of the human SK1 gene. The position of the first ATG was designated as +1. Deletion mutants of various lengths of the 5′ promoter of human SK1 are shown in the left lower panel, designed as 1977d, 1117d, 633d, and 390d, respectively. The right lower panel shows the luciferase reporter assay for the corresponding constructs. C, the left panel shows constructs of SK1 promoter and a SP1/KLF binding site located between −516 and −528 bp. Relative luciferase activity is shown in the right panel. Solid columns are samples treated with FGF2, and open columns are samples without treatment. For all experiments, means ± S.D. were calculated from at least three separate experiments, each performed in triplicate. The relative luciferase activity of 1977d nonstimulated construct was regarded as 1.0. Total protein concentrations were measured and used for normalization in all experiments. *, p < 0.05.
KLF14 Potentiates FGF2 Induction of Endothelial SK1 Expression
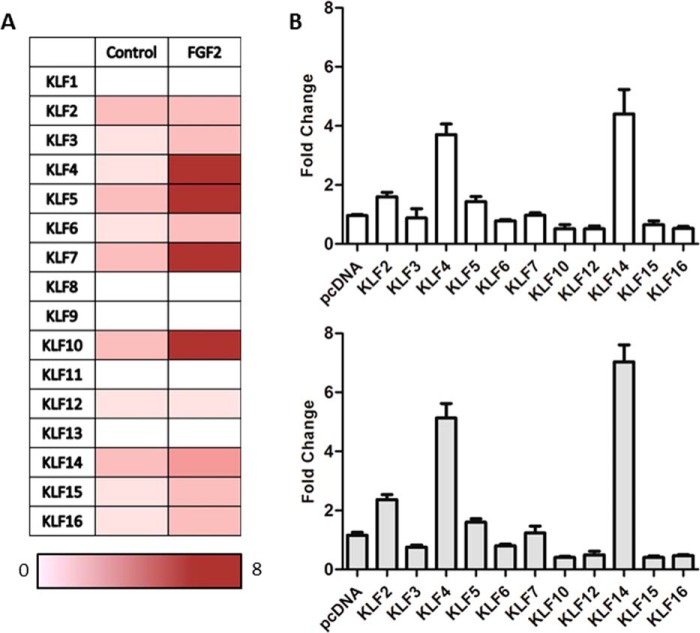
To identify KLF proteins involved in the regulation of SK1 expression, we designed a multitier screening approach that began with executing expression studies aimed at ruling out the candidacy of members of this protein family that are not expressed in the cells used for these studies. Fig. 3A demonstrates that among all KLF proteins, KLF1, KLF8, KLF9, KLF11, and KLF13 are not expressed in HHSECs. In contrast, KLF2–7, KLF10, KLF12, KLF14, KLF15, and KLF16 are significantly expressed in this cell population. Thus, through this first step in our screening approach, we were able to eliminate five KLF proteins as candidates for the regulation of the SK1 −528/−516 KLF site in this cell population. Notably, treatment of HHSEC with FGF2 for 4 h further increased the expression of KLF4, KLF5, KLF6, KLF10, KLF12, and KLF14 in HHSEC cells, thereby leading us to hypothesize that at least one of these factors is involved in modulating the basal and growth factor regulated activity of the SK1 promoter, a hypothesis that was tested in an unbiased manner by performing reporter experiments. For this purpose, these candidate KLFs, found in HHSECs, were cotransfected with the −633 SK1 promoter luciferase construct, and luciferase values were normalized to measurements from cells transfected with the empty vector control. As shown in Fig. 3B, we found that among the tested transcription factors, KLF2, KLF4, and KLF14 significantly increased the promoter activity of the −633 construct (pcDNA, 0.96 ± 0.1; KLF2, 1.6 ± 0.3; KLF4, 3.7 ± 0.6; and KLF14, 4.4 ± 0.8; p < 0.05). In addition, our experiments showed that treatment with FGF2 further increased promoter activity, with the most prominent activation achieved by KLF14 (pcDNA, 1.2 ± 0.2 versus KLF14, 7.03 ± 1.0; p < 0.05). Thus, the data suggest that KLF4 or KLF14 possibly regulates the SK1 expression.
FIGURE 3.
KLF14 is up-regulated upon FGF2 stimulation and increases SK1 promoter activity. A, in HHSEC, the mRNA fold changes of KLF family members upon FGF2 (25 ng/ml) stimulation were measured using quantitative real time PCR. Fold changes of mRNA were calculated as the ratio of KLF levels in FGF2-stimulated cells to those in cells without FGF2 treatment. KLF1, KLF8, KLF9, KLF11, and KLF13 were not amplified. The fold changes in mRNA levels are color-coded according to scale at the bottom of the chart. B, HEK 293 cells were transfected with the 633d SK1 construct, FGFR1 plasmid, along with the control pcDNA empty vector or one of the KLF members found to be expressed in HUVEC cells. The upper panel (white bars) shows the basal activation of the 633d construct with overexpression of the different KLF members, whereas the lower panel (gray bars) shows same conditions, but in the presence of FGF2. The relative luciferase activity of 633d construct transfected with control empty vector and without FGF2 stimulation was regarded as 1.0. For all experiments, means ± S.D. were calculated from at least three separate experiments, each performed in triplicate.
KLF14 Is a Transcriptional Activator of SK1 Gene Expression in Endothelial Cells
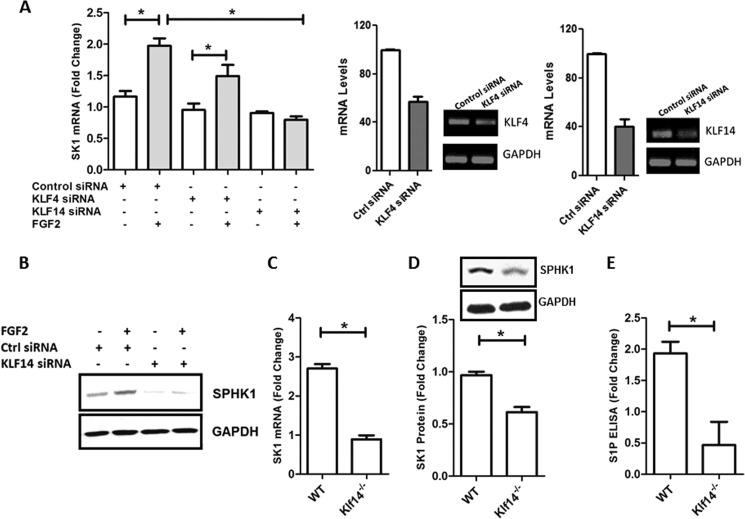
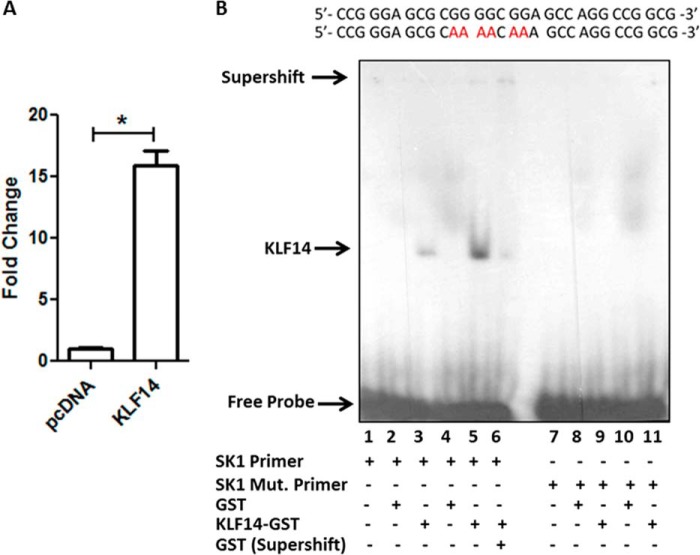
To gain insight as to whether endogenous KLF4 or KLF14 regulates SK1 promoter activity in response to FGF2 treatment, we first utilized siRNA technology (Fig. 4A, right panel). We found that transfection of HUVEC, with KLF4 siRNA did not significantly reduce SK1 mRNA after FGF2 stimulation (control siRNA, 2.0 ± 0.14 versus KLF4 siRNA, 1.49 ± 0.3; not significant). On the other hand, KLF14 siRNA decreased basal SK1 mRNA levels (control siRNA, 1.2 ± 0.03 versus KLF14 siRNA, 0.89 ± 0.05; not significant), and more importantly, the reduction in SK1 expression was significantly greater after FGF2 stimulation (control siRNA, 2.0 ± 0.14 versus KLF14 siRNA, 0.79 ± 0.05; p < 0.05) (Fig. 4A, left panel). Consistent with this result, KLF14 siRNA reduced the levels of SK1 protein after FGF2 stimulation (densitometry: control siRNA, 1.89 ± 0.03 versus KLF14 siRNA, 0.65 ± 0.04; p < 0.05) (Fig. 4B). These results led us to evaluate KLF14 inactivation in mice by homologous recombination to study the regulation of SK1 by this transcription factor in vivo in endothelial cells. For this purpose, we generated the first KLF14 knock-out (Klf14−/−) mouse (see “Experimental Procedures” and supplemental Fig. S1). Subsequently, we isolated primary liver ECs from both Klf14−/− and WT littermate mice and measured the expression of SK1 by real time PCR. As shown in Fig. 4C, liver ECs from Klf14−/− mice displayed reduced levels of SK1 mRNA when compared with wild-type animals. Similarly, the levels of SK1 protein were significantly reduced in Klf14−/− liver EC lysates levels (Fig. 4D). The reduction in both SK1 mRNA and protein levels correlated with a reduction in S1P levels, as measured by a specific ELISA assay (WT, 1.9 ± 0.3 versus Klf14−/−, 0.5 ± 0.4; p < 0.05) (Fig. 4E). Notably, whereas KLF14 has been recently described as a master regulator of fat metabolism (10), its role in the synthesis of signaling lipid molecules, such as S1P remains unknown. Thus, because of the potential novelty and biological importance of characterizing the role of KLF14 in this process, we subsequently tested the hypothesis that this transcription factor functions as a regulator of the putative −528/−516 KLF binding site in the SK1 promoter. Hence, we evaluated the ability of KLF14 to bind to the SK1 promoter by ChIP. Fig. 5A shows that epitope-tagged KLF14 binds to the endogenous promoter region of SK1 in 239HEK cells. We complemented these studies with EMSAs using probes containing the −528/−516 KLF site of SK1 promoter or oligonucleotides carrying a mutation in this same region. As shown in Fig. 5B, KLF14 forms a complex with the WT but not the mutant −528/−516 probe, in a manner that supershifted with specific antibodies. Thus, together, the data generated from using cells with genetic inactivation of KLF14 by either siRNA or mouse knock-out, along with EMSA and promoter assays, demonstrate that this transcription factor is involved in the regulation of SK1 expression.
FIGURE 4.
KLF14 is crucial for FGF2 simulation of SK1. A, KLF14 siRNA blocks the FGF2 stimulation of SK1 mRNA level. Left panel, real time PCR analyses of SK1 mRNA levels. HUVEC cells infected with FGFR1 adenovirus were transfected with 100 μm of scramble control siRNA, KLF4 siRNA, or KLF14 siRNA. After 48 h, where indicated, the cells were stimulated with FGF2 (25 ng/ml). Right panel, the knockdown efficiency of KLF4 and KLF14 was analyzed by quantitative PCR and conventional PCR. B, HUVEC cells infected with FGFR1 adenovirus were transfected with 100 μm of scramble control siRNA, or KLF14 siRNA were used to measure the SK1 protein level. C, SK1 mRNA levels were measured from primary liver endothelial cells isolated from WT and KLF14 knock-out (Klf14−/−) mice. D, SK1 protein levels were measured from primary liver endothelial cells isolated from WT and KLF14 knock-out (Klf14−/−) mice. E, S1P ELISA was used to measure S1P levels from primary liver endothelial cells isolated from WT and KLF14 knock-out (Klf14−/−) mice. All experiments were performed in triplicate. Ctrl, control. *, p < 0.05.
FIGURE 5.
KLF14 binds to SK1 promoter. A, epitope-tagged KLF14 or tagged empty vector were transfected in HEK 293 cells. ChIP assay using FLAG antibody demonstrates that KLF14 binds to the SK1 promoter, whereas empty vector-transfected cells serve as control. B, EMSA was performed using KLF14 recombinant protein (lanes 3, 5, 6, 9, and 11) or control GST protein (lanes 2, 4, 8, and 10) with radiolabeled double-stranded probe with intact −516/−528 KLF binding site (lanes 1–6) or a probe with a mutation in the same KLF binding site (lanes 7–11). Specific complexes between KLF14 and probe and the free probe are indicated by arrows on the left. A GST antibody shifted recombinant KLF14-SK1 probe complex (lane 6), indicating specificity. *, p < 0.05.
KLF14 Activates SK1 via a Coactivation Complex with the p300 Histone Acetyl Transferase
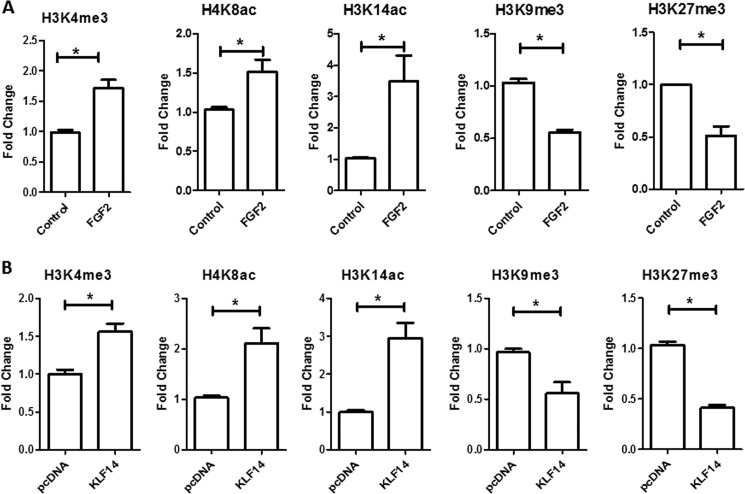
To extend our mechanistic understanding of how FGF2 regulates the transcription of SK1 through KLF14, we monitored the type of histone marks on the chromatin associated with the promoter of this gene after FGF2 treatment in HUVEC cells by ChIP assay followed by quantitative PCR. Fig. 6A shows that this treatment increased marks associated with significant transcriptional activation on the SK1 promoter, including trimethylation of histone 3 at lysine 4 (H3K4me3) (control, 1.02 ± 0.05 versus FGF2, 1.72 ± 0.15; p < 0.05), acetylation of histone 4 at lysine 8 (H4K8ac) (control, 1.03 ± 0.05 versus FGF2, 1.52 ± 0.15; p < 0.05) and acetylation of histone 3 at lysine 14 (H3K14ac) (control, 1.13 ± 0.07 versus FGF2, 3.48 ± 0.83; p < 0.05). In contrast, no significant changes were found in the levels of acetylation of histone 3 at lysine 9 (H3K9ac) (control, 0.96 ± 0.05 versus FGF2, 1.08 ± 0.1), acetylation of histone 3 at lysine 18 (H3K18ac) (control, 1.05 ± 0.03 versus FGF2, 1.17 ± 0.14), acetylation of histone 3 at lysine 12 (H4K12ac) (control, 0.98 ± 0.03 versus FGF2, 1.55 ± 0.20), and methylation of histone 3 at lysine 4 (H3K4me1) (control, 0.96 ± 0.05 versus FGF2, 1.50 ± 0.27) marks. However, both trimethylation of histone 3 at lysine 9 (H3K9me3) (control, 1.03 ± 0.03 versus FGF2, 0.55 ± 0.07; p < 0.05) and trimethylation of histone 3 at lysine 27 (H3K27me3) (control, 1.2 ± 0.12 versus FGF2, 0.51 ± 0.10; p < 0.05), which are marks of transcriptional repression, were significantly decreased in this region after FGF2 treatment. Notably, similar changes were observed in HEK 293 cells transfected with KLF14, but not with the empty vector control (Fig. 6B). These changes included a significant increase in H3K4me3 (pcDNA, 0.99 ± 0.05 versus KLF14, 1.59 ± 0.15; p < 0.05), H4K8ac (pcDNA, 1.0 ± 0.02 versus KLF14, 2.30 ± 0.1; p < 0.05), and H3K14ac (pcDNA, 1.03 ± 0.04 versus KLF14, 2.95 ± 0.41; p < 0.05) with a concomitant decrease in the levels of H3K9me3 (pcDNA, 1.01 ± 0.02 versus KLF14, 0.56 ± 0.11; p < 0.05) and H3K27me3 (pcDNA, 1.06 ± 0.03 versus KLF14, 0.41 ± 0.03; p < 0.05).
FIGURE 6.
ChIP assay of H3K4me3, H4K8ac, H3K14ac, H3K9me3, and H3K27me3 on the human SK1 promoter in HUVEC and HEK 293 cells. A, stimulation of HUVEC with FGF2 leads to an increase in H3K4me3, H4K8ac, and H3K14ac and a decrease in H3K9me3 and H3K27me3 on the SK1 promoter relative to untreated cells. B, KLF14 His tag or His tag vector were transfected in HEK 293 cells. ChIP assay was performed and showed an increase in H3K4me3, H4K8ac, and H3K14ac and a decrease in H3K9me3 and H3K27me3 histone marks on the SK1 promoter in cells transfected with KLF14. All experiments were performed in triplicate, and the samples were normalized to the respective inputs. *, p < 0.05.
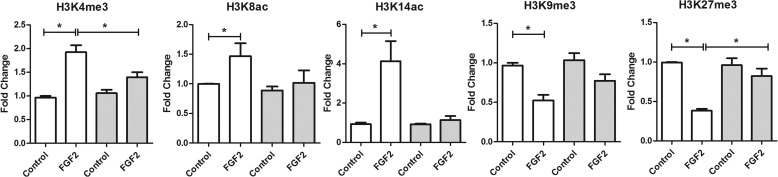
To verify the effects of FGF2 on KLF14 deficient cells, HUVEC cells were transfected with either KLF14 or control siRNA and subsequently stimulated with FGF2 (Fig. 7). The results of these experiments showed that KLF14 siRNA decreased the effects of FGF2 on gene activation histone marks, including H3K4me3 (control siRNA: control, 0.96 ± 0.33 versus FGF2, 1.9 ± 0.25; KLF14 siRNA: control, 1.1 ± 0.12 versus FGF2, 1.4 ± 0.18; p < 0.05), H3K8ac (control siRNA: control, 0.97 ± 0.04 versus FGF2, 1.56 ± 0.27; KLF14 siRNA: control, 0.89 ± 0.11 versus FGF2, 1.02 ± 0.36; p < 0.05), and H3K14ac (control siRNA: control, 0.96 ± 0.06 versus FGF2, 4.77 ± 2.42; KLF14 siRNA: control, 1.05 ± 0.3 versus FGF2, 1.71 ± 1.34; p < 0.05). No significant changes were observed in repressive histone marks after FGF2 stimulation of cells transfected with KLF14 siRNA, as shown by the levels of H3K9me3 (control siRNA: control, 0.97 ± 0.06 versus FGF2, 0.52 ± 0.13; KLF14 siRNA: control, 1.03 ± 0.15 versus FGF2, 0.77 ± 0.14; p < 0.05) and H3K27me3 (control siRNA: control, 0.99 ± 0.12 versus FGF2, 0.38 ± 0.04; KLF14 siRNA: control, 0.96 ± 0.15 versus FGF2, 0.82 ± 0.16; p < 0.05).
FIGURE 7.
Effect of FGF2 in KLF14 knockdown cells. HUVEC cells infected with FGFR1 adenovirus were transfected with 100 μm of scramble control siRNA or KLF14 siRNA. Where indicated, the cells were stimulated with FGF2 (25 ng/ml). ChIP assays for H3K4me3, H4K8ac, H3K14ac, H3K9me3, and H3K27me3 marks were performed. Stimulation of HUVEC with FGF2 in control siRNA cells leads to an increase in H3K4me3, H4K8ac, and H3K14ac and a decrease in H3K9me3 and H3K27me3 marks. These changes were not observed in KLF14 siRNA transfected cells. All experiments were performed in triplicate, and the samples were normalized to the respective inputs. *, p < 0.05.
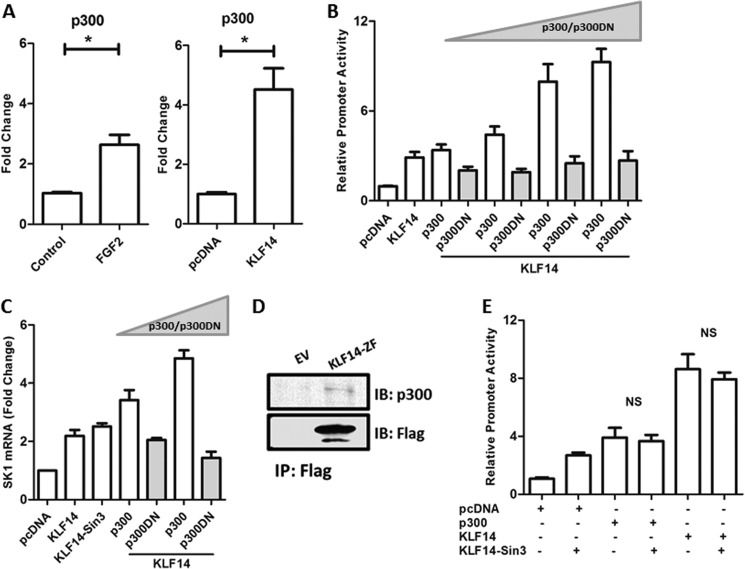
These results were mechanistically informative, because they suggested that the chromatin modifying activity of p300, which has been previously shown to acetylate both H4K8ac and H3K14ac (26, 27), might be involved in the KLF14-mediated transcriptional regulation of SK1. In support of this idea, ChIP assays demonstrated that p300 is recruited to the SK1 promoter region in KLF14-transfected cells (Fig. 8A). Similar results were obtained in cells treated with FGF2 (Fig. 8A). Thus, to further evaluate the role of p300 in this phenomenon, we performed luciferase assays in cells transfected with a constant amount of KLF14 along with increasing concentrations of this histone acetyl transferase. The results of these experiments, shown in Fig. 8B, demonstrate that p300 increased the effect of transcriptional activation of SK1 by KLF14 in a dose-dependent manner. Consistent with these results, we found that a dominant-negative form of p300 (p300DN) decreased the KLF14-mediated activation of the SK1 promoter (Fig. 8B). Congruent with this result, the overexpression of KLF14 with p300 led to an increase of SK1 mRNA level that is abrogated with the overexpression of p300DN (Fig. 8C). Lastly, using coimmunoprecipitation, we found that KLF14 binds the p300 coactivator complex (Fig. 8D). Though unlikely, as a control, we evaluated whether, in the context of our experiments, the effects observed with KLF14 could also be influenced by the previously described interaction of this transcription factor with the transcriptional corepressor Sin3-HDAC complex. For this purpose, we utilized a well characterized KLF14 construct carrying a mutation on the Sin3-HDAC binding site. As expected, we observed no changes in SK1 activation upon overexpression of this KLF14 mutant versus the wild-type control construct. Also, no changes were observed when we overexpressed p300 with this construct (Fig. 8E). Thus, we conclude that the contribution of the Sin3-HDAC complex is not relevant to the results observed with KLF14 in this study. Therefore, our overall results indicate that FGF2/FGFR1-mediated regulation of SK1 expression in endothelial cells involves a transcriptional activation pathway utilizing a novel KLF14-p300 complex (Fig. 9). Interestingly, despite the recently underscored biological importance of KLF14 in lipid metabolism, these results constitute the first description of a KLF14-mediated pathway involved in gene activation.
FIGURE 8.
Effects of the histone acetyltransferase, p300, in activation of the SK1 promoter. A, ChIP assay showing the recruitment of p300 to promoter region of SK1. The left panel shows an increase of p300 binding at the SK1 promoter of HUVEC cells after stimulation with FGF2. The right panel shows that in HEK 293 cells transfected with KLF14, p300 is recruited to the promoter region of SK1. B, luciferase assay performed with 633d SK1 construct. HEK 293 cells were transfected with the 633d promoter construct, KLF14 plasmid, and different concentrations of p300 plasmid or p300 dominant-negative (p300 DN) plasmid, as indicated. The presence of exogenous p300 increases the activation of SK1 promoter in a dose-dependent manner, whereas the transfection of p300 DN abrogates this activation. The relative luciferase activity of the 633d construct transfected with pcDNA empty vector was regarded as 1.0. Total protein concentrations were measured and used for normalization in the experiment. C, the overexpression of KLF14 wild type construct and KLF14 construct carrying mutation on the Sin3-HDAC binding site (KLF14-Sin3) increases the SK1 mRNA levels. The concomitant expression of different doses of p300 further increased these levels, whereas the overexpression of p300 DN abrogates this effect. D, HEK 293 cells were cotransfected with His tag-KLF14 or His tag-EV and p300. Immunoprecipitation was performed using anti-His tag-agarose beads, and Western blot using anti-p300 showed that KLF14 binds p300. E, no changes were observed in the SK1 promoter activity when KLF14 construct carrying mutation on the Sin3-HDAC binding site (KLF14-Sin3) was compared with KLF14 wild-type construct. All experiments were performed in triplicate. IB, immunoblot; IP, immunoprecipitation; NS, not significant. *, p < 0.05.
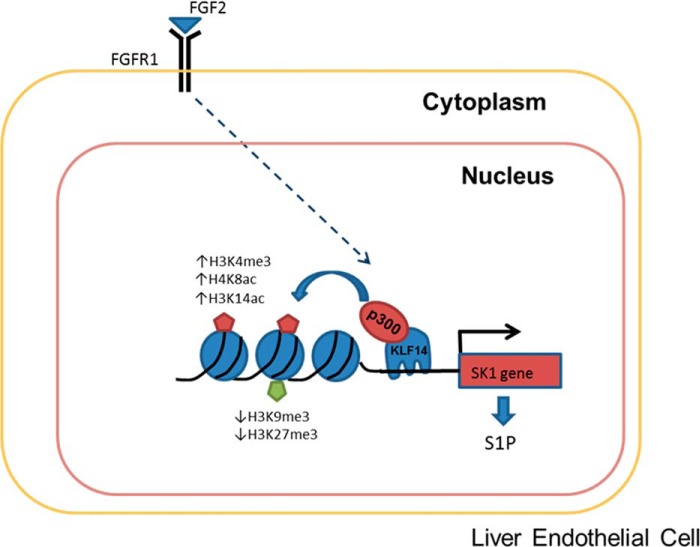
FIGURE 9.
Model of KLF14 transcriptional activation of SK1 gene. Activation of SK1 gene and S1P production result from FGF2 binding to FGFR1 receptor leading to up-regulation of SK1 promoter by KLF14. In our model, activation of FGFR1 leads to the binding of KLF14 to SK1 promoter through its GC-rich site. It then recruits p300 and promotes chromatin remodeling from a transcriptionally “inactive” to “active” state by an increase in H4K8ac, H3K14ac, and H3K4me3 marks and a decrease in H3K9me3 and H3K27me3 marks.
DISCUSSION
KLF14 has recently elicited significant attention since extensive genetic studies in humans identified a central role of this protein in the development of metabolic diseases, in particular those that regulate lipid metabolism. In fact, because of its contribution to metabolic diseases, KLF14 has been recently referred to as a “conductor of the metabolic syndrome orchestra” (28). However, despite its importance, little is known about the mechanisms by which KLF14 regulates lipid biology at the molecular and cellular levels. The current study significantly advances this field of research by providing novel insights into the role of KLF14 in the transcriptional regulation of lipid signaling, as well as outlining a novel FGF2-FGFR1-KLF14-p300-SK1-S1P pathway. These findings are of relevance to the areas of KLF proteins, growth factor signaling, and lipid-mediated signaling in normal biology and have the potential to contribute to a better understanding of diseases that associate with alterations in KLF14.
The FGF family of ligands consists of 23 growth factors that control diverse cellular processes including proliferation, differentiation, and migration. FGFs induce their biological responses by binding to and activating a family of cell surface receptors with intrinsic tyrosine kinase activity called FGF receptors (FGFRs). The binding of FGF2 to its cognate receptor, FGFR1, leads to endothelial cell activation (29–31). Our present studies were initiated by a desire to explore FGF targets in EC and to interrogate how FGF induces these targets. Because of its biological and pathophysiological importance, one target that became the focus of this study was the FGF-induced production of S1P, a lipid signaling molecule involved in EC maintenance homeostasis as well as proliferation, differentiation, and migration (25, 32). Thus, the present studies link FGF activation with EC lipid signaling through SK1 and S1P production. The additional link with KLF14 provides evidence that this metabolic master regulator acts not only in traditional metabolic epithelia, but in endothelial cells as well.
It is noteworthy that this report represents the first functional characterization of KLF14 as a novel FGF-inducible protein that mediates the up-regulation of SK1 leading to the production of S1P. In particular, because KLF14 was previously known only as a repressor protein, acting via Sin3a-HDAC (13), the data presented here unravel a novel function for this protein, namely its ability to activate a target promoter via coupling with the histone methyl transferase, p300. Previous promoter studies had revealed that cis-regulatory regions within the proximal rat and human SK1 promoter are critical for its activation (5, 33, 34). Nakade et al. (33) have shown that PMA stimulation of a leukemia cell line leads to an increase in SK1 activity. They suggest that this activation is due to the binding of Sp1 protein and unknown proteins to CG-rich regions of human SK1 promoter. In searching for candidate proteins that can regulate GC-rich sites, we performed an unbiased screening for the FGF inducibility of each of 16 KLF transcription factors. We found evidence that KLF14 is up-regulated upon FGF2-mediated FGFR1 activation to induce SK1 gene expression, its corresponding protein level, the associated enzymatic activity, and the subsequent production of its downstream signal, S1P. When combined with deletion and site-directed mutagenesis, promoter reporter studies allowed us to identify a critical role for a −323 GC-rich, consensus KLF site in the activation of SK1. ChIP assays demonstrated that the KLF14-mediated induction of SK1 expression correlates with a reduction in repressive histone methylation marks and a concomitant increase activating histone acetylation marks. Additional work using overexpression of p300 or its dominant-negative form established that this protein could mediate these effects of KLF14. Combined, these results support the existence of a novel pathway whereby a KLF14/p300 complex can activate the SK1 gene in response to FGF stimulation.
KLF14 is suggested to be an ancient retransposed copy of KLF16 because of its homology to this KLF member (12). Together, with KLF9, KLF10, KLF11, and KLF13, these proteins form a subfamily of transcription factors involved in the regulation of metabolism and metabolic diseases. Mechanistically, extensive work from our group and others has determined that these proteins couple to HDAC-containing chromatin remodeling complexes through preserved Sin3 interacting domains at their N termini (13). Interestingly, in the current study, we demonstrate, for the first time, that KLF14 binds to the histone acetyl transferase, p300, which is important for the activation of SK1 gene. In this regard, KLF14 appears to work by coupling to both silencing and activating chromatin-remodeling complexes in specific contexts. These mechanistic insights extend our understanding of how this important member of the KLF family works to regulate gene expression and metabolism. Furthermore, this is the first study to demonstrate that KLF14 participates in FGF-mediated signaling, which is important because previous reports had only associated KLF14-regulated pathways to TGF-β (13). In fact, a recent study showed that TGF-β1, acting in conjunction with progesterone, increased the proliferation of TM3 Leydig cells, implying that KLF14 may bind to and activate the endoglin promoter (35). Therefore, the mechanistic information derived from the current study, which shows how KLF14 works as a transcriptional activator, may help to clarify its role in other growth factor-mediated cascades as well. In summary, the work reported here provides new insight on the biochemical and cell biological role of KLF14, which, as an important disease-causing gene in humans, thus bears both mechanistic and biomedical relevance.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DK59015, AA021171, and P30 DL 084567 (to V. H. S.); DK52913 (to R. U.); and DK100575-01 (to R. C. H.). This work was also supported by funds from the Mayo Center for Regenerative Medicine (to R. C. H.).

This article contains supplemental Figs. S1 and S2.
- SK1
- sphingosine kinase 1
- EC
- endothelial cell
- S1P
- sphingosine-1-phosphate
- KLF
- Kruppel-like factor
- FGFR
- FGF receptor
- HHSEC
- human sinusoidal endothelial cell
- HUVEC
- human umbilical vein endothelial cell
- HDAC
- histone deacetylase.
REFERENCES
- 1. Maceyka M., Harikumar K. B., Milstien S., Spiegel S. (2012) Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol. 22, 50–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bao M., Chen Z., Xu Y., Zhao Y., Zha R., Huang S., Liu L., Chen T., Li J., Tu H., He X. (2012) Sphingosine kinase 1 promotes tumour cell migration and invasion via the S1P/EDG1 axis in hepatocellular carcinoma. Liver Int. 32, 331–338 [DOI] [PubMed] [Google Scholar]
- 3. Schwalm S., Pfeilschifter J., Huwiler A. (2013) Sphingosine-1-phosphate: a Janus-faced mediator of fibrotic diseases. Biochim. Biophys. Acta 1831, 239–250 [DOI] [PubMed] [Google Scholar]
- 4. Venkataraman K., Thangada S., Michaud J., Oo M. L., Ai Y., Lee Y. M., Wu M., Parikh N. S., Khan F., Proia R. L., Hla T. (2006) Extracellular export of sphingosine kinase-1a contributes to the vascular S1P gradient. Biochem. J. 397, 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sobue S., Hagiwara K., Banno Y., Tamiya-Koizumi K., Suzuki M., Takagi A., Kojima T., Asano H., Nozawa Y., Murate T. (2005) Transcription factor specificity protein 1 (Sp1) is the main regulator of nerve growth factor-induced sphingosine kinase 1 gene expression of the rat pheochromocytoma cell line, PC12. J. Neurochem. 95, 940–949 [DOI] [PubMed] [Google Scholar]
- 6. Rigogliuso S., Donati C., Cassarà D., Taverna S., Salamone M., Bruni P., Vittorelli M. L. (2010) An active form of sphingosine kinase-1 is released in the extracellular medium as component of membrane vesicles shed by two human tumor cell lines. J. Oncol. 2010, 509329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lomberk G., Urrutia R. (2005) The family feud: turning off Sp1 by Sp1-like KLF proteins. Biochem. J. 392, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McConnell B. B., Yang V. W. (2010) Mammalian Kruppel-like factors in health and diseases. Physiol. Rev. 90, 1337–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lomberk G., Grzenda A., Mathison A., Escande C., Zhang J. S., Calvo E., Miller L. J., Iovanna J., Chini E. N., Fernandez-Zapico M. E., Urrutia R. (2013) Kruppel-like factor 11 regulates the expression of metabolic genes via an evolutionarily conserved protein interaction domain functionally disrupted in maturity onset diabetes of the young. J. Biol. Chem. 288, 17745–17758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Small K. S., Hedman A. K., Grundberg E., Nica A. C., Thorleifsson G., Kong A., Thorsteindottir U., Shin S. Y., Richards H. B., GIANT Consortium, MAGIC Investigators, DIAGRAM Consortium,Soranzo N., Ahmadi K. R., Lindgren C. M., Stefansson K., Dermitzakis E. T., Deloukas P., Spector T. D., McCarthy M. I. (2011) Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat. Genet. 43, 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bureau C., Hanoun N., Torrisani J., Vinel J. P., Buscail L., Cordelier P. (2009) Expression and function of Kruppel-like factors (KLF) in carcinogenesis. Curr. Genomics 10, 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parker-Katiraee L., Carson A. R., Yamada T., Arnaud P., Feil R., Abu-Amero S. N., Moore G. E., Kaneda M., Perry G. H., Stone A. C., Lee C., Meguro-Horike M., Sasaki H., Kobayashi K., Nakabayashi K., Scherer S. W. (2007) Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution. PLoS Genet. 3, e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Truty M. J., Lomberk G., Fernandez-Zapico M. E., Urrutia R. (2009) Silencing of the transforming growth factor-beta (TGFβ) receptor II by Kruppel-like factor 14 underscores the importance of a negative feedback mechanism in TGFβ signaling. J. Biol. Chem. 284, 6291–6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chew Y. C., Adhikary G., Xu W., Wilson G. M., Eckert R. L. (2013) Protein kinase Cδ increases Kruppel-like factor 4 protein, which drives involucrin gene transcription in differentiating keratinocytes. J. Biol. Chem. 288, 17759–17768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deaton R. A., Gan Q., Owens G. K. (2009) Sp1-dependent activation of KLF4 is required for PDGF-BB-induced phenotypic modulation of smooth muscle. Am. J. Physiol. Heart Circ. Physiol. 296, H1027–H1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Neve B., Fernandez-Zapico M. E., Ashkenazi-Katalan V., Dina C., Hamid Y. H., Joly E., Vaillant E., Benmezroua Y., Durand E., Bakaher N., Delannoy V., Vaxillaire M., Cook T., Dallinga-Thie G. M., Jansen H., Charles M. A., Clément K., Galan P., Hercberg S., Helbecque N., Charpentier G., Prentki M., Hansen T., Pedersen O., Urrutia R., Melloul D., Froguel P. (2005) Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic beta cell function. Proc. Natl. Acad. Sci. U.S.A. 102, 4807–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bonnefond A., Lomberk G., Buttar N., Busiah K., Vaillant E., Lobbens S., Yengo L., Dechaume A., Mignot B., Simon A., Scharfmann R., Neve B., Tanyolaç S., Hodoglugil U., Pattou F., Cavé H., Iovanna J., Stein R., Polak M., Vaxillaire M., Froguel P., Urrutia R. (2011) Disruption of a novel Kruppel-like transcription factor p300-regulated pathway for insulin biosynthesis revealed by studies of the c.−331 INS mutation found in neonatal diabetes mellitus. J. Biol. Chem. 286, 28414–28424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Belacortu Y., Weiss R., Kadener S., Paricio N. (2012) Transcriptional activity and nuclear localization of Cabut, the Drosophila ortholog of vertebrate TGF-β-inducible early-response gene (TIEG) proteins. PloS one 7, e32004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huebert R. C., Jagavelu K., Hendrickson H. I., Vasdev M. M., Arab J. P., Splinter P. L., Trussoni C. E., Larusso N. F., Shah V. H. (2011) Aquaporin-1 promotes angiogenesis, fibrosis, and portal hypertension through mechanisms dependent on osmotically sensitive microRNAs. Am. J. Pathol. 179, 1851–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daftary G. S., Lomberk G. A., Buttar N. S., Allen T. W., Grzenda A., Zhang J., Zheng Y., Mathison A. J., Gada R. P., Calvo E., Iovanna J. L., Billadeau D. D., Prendergast F. G., Urrutia R. (2012) Detailed structural-functional analysis of the Kruppel-like factor 16 (KLF16) transcription factor reveals novel mechanisms for silencing Sp/KLF sites involved in metabolism and endocrinology. J. Biol. Chem. 287, 7010–7025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang J. S., Moncrieffe M. C., Kaczynski J., Ellenrieder V., Prendergast F. G., Urrutia R. (2001) A conserved α-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol. Cell. Biol. 21, 5041–5049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yaqoob U., Cao S., Shergill U., Jagavelu K., Geng Z., Yin M., de Assuncao T. M., Cao Y., Szabolcs A., Thorgeirsson S., Schwartz M., Yang J. D., Ehman R., Roberts L., Mukhopadhyay D., Shah V. H. (2012) Neuropilin-1 stimulates tumor growth by increasing fibronectin fibril assembly in the tumor microenvironment. Cancer Res. 72, 4047–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lomberk G., Mathison A. J., Grzenda A., Seo S., DeMars C. J., Rizvi S., Bonilla-Velez J., Calvo E., Fernandez-Zapico M. E., Iovanna J., Buttar N. S., Urrutia R. (2012) Sequence-specific recruitment of heterochromatin protein 1 via interaction with Kruppel-like factor 11, a human transcription factor involved in tumor suppression and metabolic diseases. J. Biol. Chem. 287, 13026–13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ancellin N., Colmont C., Su J., Li Q., Mittereder N., Chae S. S., Stefansson S., Liau G., Hla T. (2002) Extracellular export of sphingosine kinase-1 enzyme. Sphingosine 1-phosphate generation and the induction of angiogenic vascular maturation. J. Biol. Chem. 277, 6667–6675 [DOI] [PubMed] [Google Scholar]
- 25. Panetti T. S., Nowlen J., Mosher D. F. (2000) Sphingosine-1-phosphate and lysophosphatidic acid stimulate endothelial cell migration. Arterioscler. Thromb. Vasc. Biol. 20, 1013–1019 [DOI] [PubMed] [Google Scholar]
- 26. Henry R. A., Kuo Y. M., Andrews A. J. (2013) Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4. Biochemistry 52, 5746–5759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karmodiya K., Krebs A. R., Oulad-Abdelghani M., Kimura H., Tora L. (2012) H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Civelek M., Lusis A. J. (2011) Conducting the metabolic syndrome orchestra. Nat. Genet. 43, 506–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Turner N., Grose R. (2010) Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer 10, 116–129 [DOI] [PubMed] [Google Scholar]
- 30. Beenken A., Mohammadi M. (2009) The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 8, 235–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Presta M., Dell'Era P., Mitola S., Moroni E., Ronca R., Rusnati M. (2005) Fibroblast growth factor/fibroblast growth factor receptor system in angiogenesis. Cytokine Growth Factor Rev. 16, 159–178 [DOI] [PubMed] [Google Scholar]
- 32. Garcia J. G., Liu F., Verin A. D., Birukova A., Dechert M. A., Gerthoffer W. T., Bamberg J. R., English D. (2001) Sphingosine 1-phosphate promotes endothelial cell barrier integrity by Edg-dependent cytoskeletal rearrangement. J. Clin. Invest. 108, 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakade Y., Banno Y., T-Koizumi K., Hagiwara K., Sobue S., Koda M., Suzuki M., Kojima T., Takagi A., Asano H., Nozawa Y., Murate T. (2003) Regulation of sphingosine kinase 1 gene expression by protein kinase C in a human leukemia cell line, MEG-O1. Biochim. Biophys. Acta 1635, 104–116 [DOI] [PubMed] [Google Scholar]
- 34. Huang K., Huang J., Chen C., Hao J., Wang S., Huang J., Liu P., Huang H. (2014) AP-1 regulates sphingosine kinase 1 expression in a positive feedback manner in glomerular mesangial cells exposed to high glucose. Cell Signal. 26, 629–638 [DOI] [PubMed] [Google Scholar]
- 35. Gonzalez C. R., Vallcaneras S. S., Calandra R. S., Gonzalez Calvar S. I. (2013) Involvement of KLF14 and egr-1 in the TGF-β1 action on Leydig cell proliferation. Cytokine 61, 670–675 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.