Abstract
Background
Heterozygous human mutations of NKX2-5 are highly penetrant and associated with varied congenital heart defects. The heterozygous knockout of murine Nkx2-5, in contrast, manifests less profound cardiac malformations, with low disease penetrance. We sought to study this apparent discrepancy between human and mouse genetics. Since missense mutations in the NKX2-5 homeodomain (DNA binding domain) are the most frequently reported type of human mutation, we replicated this genetic defect in a murine knock-in model.
Methods and Results
We generated a murine model in a 129/Sv genetic background by knocking-in an Nkx2-5 homeodomain missense mutation previously identified in humans. The mutation was located at homeodomain position 52Arg→Gly (R52G). All the heterozygous neonatal Nkx2-5+/R52G mice demonstrated a prominent trabecular layer in the ventricular wall, so called noncompaction, along with diverse cardiac anomalies, including atrioventricular septal defects, Ebstein’s malformation of the tricuspid valve, and perimembranous and/or muscular ventricular septal defects. In addition, P10 Nkx2-5+/R52G mice demonstrated atrial septal anomalies, with significant increase in the size of the inter-atrial communication and fossa ovalis, and decrease in the length of the flap valve compared to control Nkx2-5+/+ or Nkx2-5+/− mice.
Conclusion
The results of our study demonstrate that heterozygous missense mutation in the murine Nkx2-5 homeodomain (R52G) are highly penetrant, and result in pleiotropic cardiac effects. Thus, in contrast to heterozygous Nkx2-5 knockout mice, the effects of the heterozygous knock-in mimic findings in humans with heterozygous missense mutation in NKX2-5 homeodomain.
Keywords: genetics, heart defects, congenital, noncompaction, knock-in
Introduction
Cardiac defects are the most prevalent congenital malformations in the fetal and neonatal periods, affecting approximately 1% of live births1-3. Advances in clinical care, such as surgical interventions, have enabled most patients to reach adulthood. Prolonged survival, however, has been achieved at a cost, as many patients suffer late complications, of which heart failure and arrhythmias are the most prominent4. Thus, alternative treatment towards correcting abnormal developing hearts, such as prenatal cellular and genetic therapy, will offer tremendous potential in the future5,6. Despite remarkable progress in understanding cardiac development, the mechanisms underlying the cardiac anomalies are largely unknown, even when related to mutation of a single gene7.
To date, nearly 40 mutations in the transcription factor NKX2-5 have been found related to familial congenital heart disease (OMIM, NCBI). Almost half of these mutations have been found in the homeodomain (DNA binding domain)8-13. Homeodomain missense mutations have a high disease penetrance, with pleiotropic cardiac effects compared to missense mutations outside the homeodomain8-13. In addition to DNA binding, the homeodomain plays a critical role in protein-protein interactions and nuclear targeting. Our previous studies showed markedly reduced DNA binding was found in homeodomain missense NKX2-5 mutations, with preserved ability in nuclear targeting, and some interaction with NKX2-5 and selected transcriptional partners14,15.
There has been considerable interest in understanding the differences between phenotypes associated with human mutations as opposed to the knockout-strategies employed in mice16. Studies in murine models of Nkx2-5 mutations have typically shown a lower incidence of cardiac anomalies than those observed in human patients17-19. There has been speculation that the differences in phenotype severity and frequency are due to species effects, in other words mouse as opposed to man. Based on predicted effects of human mutation on protein function, and some overlap of the phenotype of heterozygous Nkx2-5 knockout mice17-19, human NKX2-5 mutations have been considered due to loss-of-function mutations.
Alternatively, the effect of human mutation may differ from the effect of the haploinsufficiency resulting from heterozygous knockout. For example, mutant proteins that preserve protein-protein interactions, but fail to bind to DNA, may have broader functional consequences, including dominant negative effects. It might be anticipated that, for such diverse effects, such as haploinsufficiency compared with dominant-negative, there would be associated variation in clinical phenotypes15. In fact, earlier studies in Xenopus20, and in transgenic mice21 suggested potential dominant negative effects of Nkx2-5 homeodomain missense mutations with decreased DNA binding.
In order to address mechanistically the relationships between genotype and phenotype, focusing on missense mutation in the homeodomain, we generated a novel heterozygous knock-in mouse model by introducing the disease-causing point missense mutation in the homeodomain (52Arg→Gly). Our data demonstrate that heterozygous knock-in mice, Nkx2-5+/R52G, show diverse and highly penetrant cardiac anomalies, which are comparable to, or even more profound, than those found in human patients with heterozygous missense mutation in NKX2-5 homeodomain.
Materials and Methods
Building models and calculation of intra-protein and protein-DNA interaction energies for Nkx2-5 homeodomain mutants
We used models of protein-DNA interactions as described previously22. Briefly, an energy minimized wild-type model was mutated to reflect the point mutations, and internal and protein-DNA binding energy was calculated.
Generation of Nkx2-5+/R52G and Nkx2-5+/− mice
The targeting vector for generation of Nkx2-5+/R52G knock-in mice was constructed as follows: a genomic fragment for Nkx2-5 from a 129/Sv mouse genomic library was cloned, and a targeting vector was constructed by introduction of a CGT to GGT point mutation in exon 2, insertion of thymidine kinase (TK) gene for negative selection and floxed-neomycin-resistant gene (NeoR) for positive selection in the intergenic regions. Nkx2-5+/− mice were generated from germline excision of floxed-exon 2 including DNA binding homeodomain23,24 by crossing Cre-deleter mice (ACTB-Cre). Cre-deleter transgenes were eliminated during backcrossing to 129/Sv mice, purchased from Charles River (129/SvPasCrl) over 8 generations. Mouse genetic background was analyzed by MAX-BAX system at Charles River using 384 SNP panel. Additional experimental procedures are described in the supplement. All animal experiments were performed with approval from the University of Florida Institutional Animal Care and Use Committee.
Real-time RT-PCR, ANF-lacZ reporter mice, X-gal staining and chromatin immunoprecipitation (ChIP) assay
Real-time reverse transcription-PCR (RT-PCR) performed using inventoried TaqMan gene expression assays (Applied Biosystems), generation and characterization of the transgenic lacZ reporter mice, X-gal staining and ChIP assay followed by Taqman real-time PCR were performed as described previously25.
Western blotting and immunostaining
Western blot analyses and immunostaining were performed with the following antibodies: Nkx2-5 pAb26, GAPDH (Research Diagnostics Inc.) and troponin T (T6277, Sigma).
Histological analysis and cardiac 3-dimensional (3D) reconstruction
Serial paraffin-embedded tissue sectioning of 5 μm thickness and 3D-reconstruction were performed as described previously27. Two observers examined the digitalized images of the sections and performed quantitative histological measurements of trabecular vs. compact area size using the same analytical methods using Image J as reported previously27.
Statistical analysis
Data presented are expressed as mean values plus or minus the standard error of the mean. Results were analyzed by SPSS (version 22) using crosstabs with Fisher’s exact test, non-parametric test, ANOVA with Fisher’s post-hoc test or independent T-test. Levene’s test was utilized for equality of variance, and P values were calculated depending on the assurance of equality. Inter-observer variability assessed by interclass correlation coefficient. P values less than 0.05 were considered significant.
Results
Characteristics of NKX2-5 homeodomain missense mutations
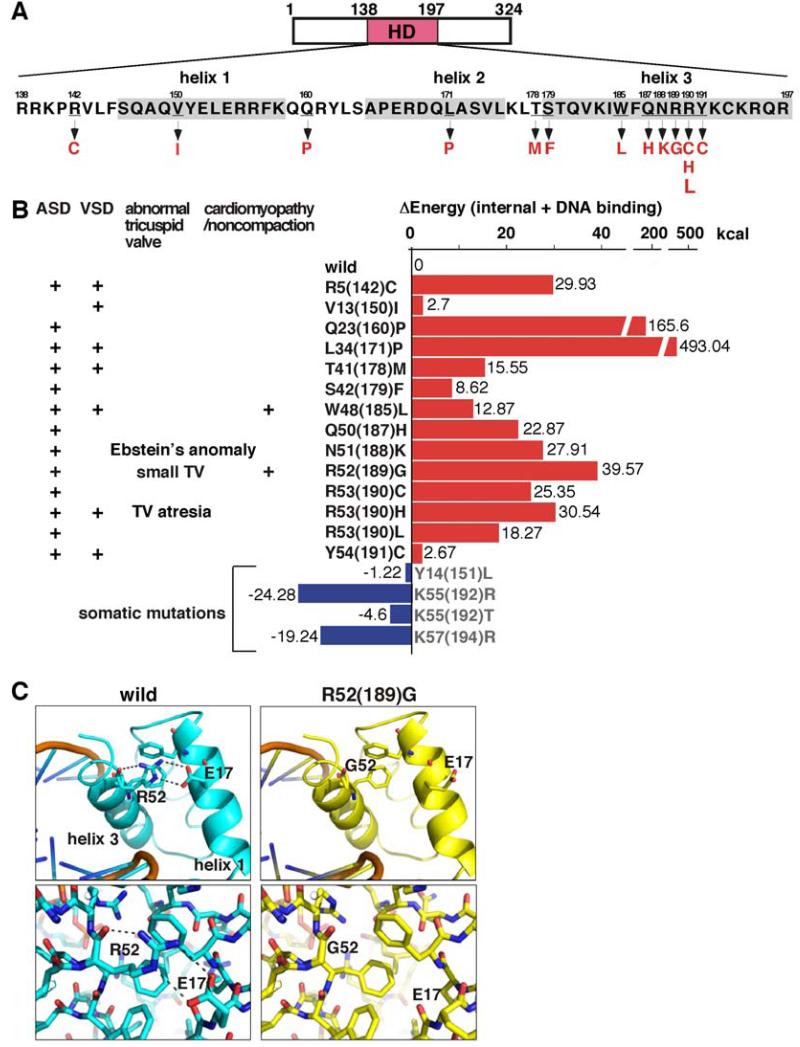
The human NKX2-5 protein is composed of 324 amino acids: the DNA binding region or homeodomain comprises 60 amino acids (Figure 1A). To our knowledge, analysis of previously reported human NKX2-5 mutations associated with familial congenital heart defects has identified a total of 37 mutations9-13; nearly half are in the homeodomain, which is the greatest in frequency when compared to the other domains. Most homeodomain mutations, 14 to be precise, are missense mutations (Figure 1A), with the remaining three (18%) being nonsense mutations8-13. Taken overall, 40% of human NKX2-5 mutations identified to date are homeodomain missense mutations. We list the cardiac anomalies reported in the pedigrees having NKX2-5 homeodomain missense mutations in the left panel of Figure 1B.
Figure 1. Position of homeodomain missense mutations and computational modeling of mutants and DNA binding.
(A) Human NKX2-5 protein structures including 60 amino acids of homeodomain (residues 138 to 197). Mutation positions are underlined with codon changes listed below. (B) Summary of genotype and phenotype (left) and calculated energy of internal protein folding and protein-DNA interaction differed from wild-type proteins (right). (C) Computational modeling of wild-type vs. R52(189)G mutant. Of note, V13(150)I mutation was found in the setting of familial transposition (concordant atrioventricular and discordant ventriculo-arterial connections) with an additional mutation in ZIC3 gene11.
Previously, we found that the best correlation between cardiac phenotype and biochemical characteristics among 8 homedomain missense mutations was reduced DNA binding, as analyzed in electrophoretic mobility shift assays15. Using the recently solved crystal structure of homeodomain and the double-stranded DNA22, we modeled mutant proteins, along with the predicted stability of proteins and protein-DNA binding. Compared to wild-type protein, higher energies are required for protein-DNA binding in all the mutants (Figure 1B, right panel), suggesting that these mutations have reduced DNA binding. In contrast, four somatic mutations found in the area of ventricular septal defect (VSD)28 demonstrate decreased energy, suggesting that these mutations potentially increase protein stability and protein-DNA binding (Figure 1B, somatic mutations).
In this study, we analyzed a mouse model having an 52Arg→Gly (R52G) in the homeodomain. Arg52 in the third helix interacts with Glu17 of the first helix, and stabilizes the structure of the homeodomain (Figure 1C). This interaction will be lost in R52G mutations, resulting in destabilization of the homeodomain structure and reduction of DNA binding. We previously showed that the mutant protein is present in the nuclei and protein-protein interaction with NKX2-5 itself (homodimerization), GATA4 and TBX5 being preserved by around 50%15.
Generation of mice with knock-in of a homeodomain missense mutation (Arg52(188)Gly, Nkx2-5+/R52G) associated with human congenital heart defects
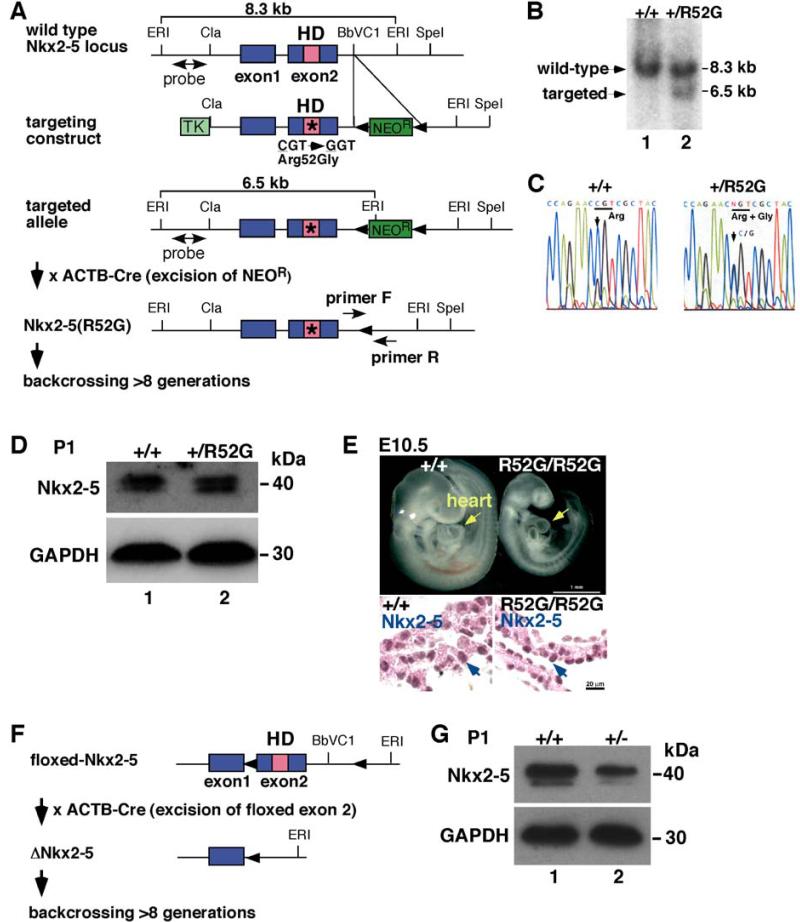
In order to develop a model to mechanistically study the high-disease penetrance of diverse cardiac anomalies associated with human homeodomain missense mutations, we generated a knock-in mouse model having R52G mutation in the homeodomain. R52G positions at 188 amino acid in mouse, as opposed to 189 amino acid in man (Figures 2A-C). To minimize non-specific genetic effects, we first eliminated the drug-selection marker, floxed-NeoR cassette that was inserted in the intergenic region of Nkx2-5, by crossing with Cre-deleter mice. Second, mice were backcrossed over 8 generations to 129/Sv mice purchased from Charles River (129/SvPasCrl). Two genomic DNAs randomly chosen from knock-in mice revealed 99.61 and 99.74% identity to 129/Sv using mouse 384 SNP panel. Thus, they have a homogenous genomic sequence. During the backcrossing, Cre-deleter transgenes were eliminated.
Figure 2. Generation of a knock-in mouse having the human NKX2-5 disease-causing missense mutation in the homeodomain at the position of 52Arg→Gly (Arg52Gly, R52G).
(A) Structure of wild-type Nkx2-5 locus, and the targeting construct. Southern blotting hybridized with the 5′ probe showed 8.3 kb EcoRI-digested genomic DNA in wild-type alleles and 6.5 kb fragments in the targeted alleles. PCR primers used for genotyping are shown. (B) Identification of wild-type (lane 1) and mutant alleles (lane 2) in ES cells using Southern blotting. (C) Sequence analyses of genomic DNA demonstrate CGT in wild-type vs. (C/G)GT in +/R52G. (D) Western blotting of total Nkx2-5 proteins and GAPDH in P1 hearts dissected from Nkx2-5+/+ (lane 1) vs. Nkx2-5+/R52G (lane 2) mice. (E) Growth retardation in E10.5 homozygous Nkx2-5R52G/R52G mouse in comparison to wild-type littermate. Nuclear localized wild-type or mutant Nkx2-5 proteins were visualized by immunohistochemistry (brown). (F) Generation of germline Nkx2-5 knockout mice with elimination of floxed-exon 2 by Cre-deleter mice. (G) Western blotting of Nkx2-5 proteins and GAPDH in P1 hearts dissected from Nkx2-5+/+ (lane 1) vs. Nkx2-5+/− (lane 2) mice.
Heterozygous neonatal Nkx2-5+/R52G mice were born expressing a level of Nkx2-5 proteins (wild-type and mutant) equivalent to that in Nkx2-5+/+ mice, as analyzed by Western blotting (Figure 2D). On the other hand, homozygous Nkx2-5 R52G/R52G mice were never born. A representative E10.5 Nkx2-5R52G/R52G mouse embryo demonstrates growth retardation compared to control Nkx2-5+/+ mice, in which Nkx2-5R52G proteins were expressed in the nuclei (Figure 2E).
To compare missense mutation in the homeodomain with null mutation, we generated a second mouse model, Nkx2-5+/−, to serve as a control. The floxed-exon 2 of Nkx2-5 including the DNA binding homeodomain23,24 was deleted by crossing to Cre-deleter mice (Figure 2F), which was eliminated by subsequent backcrossing to 129/Sv mice as described above. Heterozygous neonatal Nkx2-5+/− mice showed a reduction of approximately one-half of Nkx2-5 proteins in the heart lysates compared to control Nkx2-5+/+ mice (Figure 2G).
Cardiac anomalies in newborn heterozygous Nkx2-5+/R52G mutant mice
Periodically, newborn heterozygous Nkx2-5+/R52G mice were found cyanotic or dead (Figure 3A). The ratio between P1 Nkx2-5+/+ (48%) vs. Nkx2-5+/R52G (52%) mice (n=65) and P10 Nkx2-5+/+ (58%) vs. Nkx2-5+/R52G (42%) mice (n=61) suggested that approximately one-quarter of the Nkx2-5+/R52G mice die perinatally (P = 0.002). All live littermates in the same experimental group were analyzed regardless of cyanosis (n=17): examination revealed evidence of ventricular noncompaction and varied cardiac malformations.
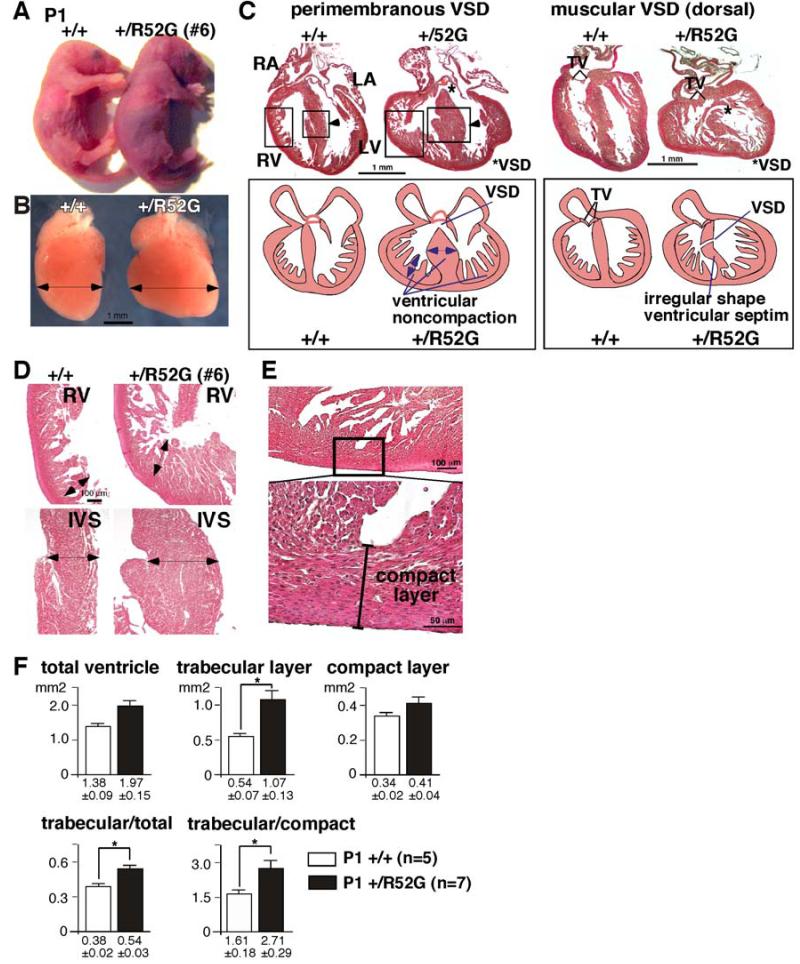
Figure 3. Representative cyanotic cardiac anomaly and noncompaction in P1 Nkx2-5+/R52G mouse in comparison to control wild-type littermate.
Control Nkx2-5+/+ (left) vs. cyanotic Nkx2-5+/R52G mouse #6 (right): (A) whole bodies, (B) dissected hearts, and (C) heart sections with simplified illustrations. (D) Enlarged images of heart sections of right ventricle (RV, top) and interventricular septum (IVS, bottom) obtained from Nkx2-5+/+ (left) vs. Nkx2-5+/R52G mice (right). (E) Enlarged images of tissue sections defining the compact layer morphologically as the appearance of a compact band of uniform tissue, while the endocardial noncompacted trabecular layer consists of trabecular meshwork with deep endomyocardial spaces29. (F) Quantification of area size of total ventricle, trabecular and compact layer. Nkx2-5+/R52G hearts demonstrating profound anomalies were excluded. RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle. *P<0.05.
Ventricular noncompaction
Representative hearts isolated from wild-type and cyanotic Nkx2-5+/R52G newborn mice (mouse #6, Table 1) demonstrated that the Nkx2-5+/R52G heart was enlarged (Figures 3A-C). A prominent trabecular layer was observed in the ventricular walls, a feature usually described as ventricular noncompaction. In addition, there was failure of compaction of the muscular ventricular septum along with perimembranous and muscular VSDs (Figures 3C, D). Of note, this is an abnormal finding, as ventricular septal formation is completed, with the interventricular communication being closed by embryonic day E13.5-14 in normal mouse embryos30.
Table 1. Summary of cardiac anomalies of P1 Nkx2-5+/R52G mice (129/Sv background) (n=17).
| mouse ID |
phenotype (excluded atrial septal defect and ventricular noncompaction) |
tricuspid valve anomalies |
|---|---|---|
| 1 | - | incomplete delamination |
| 2 | VSD (perimembranous) | - |
| 3 | VSD (perimembranous) | - |
| 4 | VSD (perimembranous) | incomplete delamination |
| 5 | VSD (muscular) | incomplete delamination |
| 6 | multiple VSDs (perimembranous + muscular) | incomplete delamination |
| 7 | multiple VSDs (perimembranous + muscular) | - |
| 8 | multiple VSDs (perimembranous + muscular | - |
| 9 | multiple VSDs (perimembranous + muscular) | incomplete delamination |
| 10 | multiple VSDs (muscular) | - |
| 11 | multiple VSDs (muscular) | - |
| 12 | atrioventricular septal defect multiple VSDs (muscular) |
hypoplastic |
| 13 | atrioventricular septal defect double outlet right ventricle multiple VSDs (muscular) |
hypoplastic |
| 14 | atrioventricular septal defect double-chambered right ventricle right isomerism |
hypoplastic |
| 15 | Ebstein’s malformation | incomplete delamination,
Ebstein’s malformation |
| 16 | Ebstein’s malformation multiple VSDs (muscular) |
tethered, Ebstein’s malformation |
| 17 | tricuspid valve atresia small right ventricular cavity VSD (muscular) |
atresia |
Ventricular noncompaction is a cardiomyopathy characterized by a thickened trabecular layer when compared to the compact layer of the ventricular walls, usually with deep inter-trabecular recesses31. All 17 newborn Nkx2-5+/R52G mice analyzed by serial tissue-sectioning demonstrated such ventricular noncompaction involving both ventricles. We quantified the total area of the trabecular and compact layers of the walls and compared the relative ratio to that of randomly selected newborn hearts (+/+ n=5, +/R52G n=7)(Figure 3E, Supplemental Figure S1). This comparison excluded Nkx2-5+/R52G hearts demonstrating profound anomalies (Table 1). A significant increase in the ratio of the trabecular layer relative to the compact layer was found in Nkx2-5+/R52G mice compared to controls. There was no significant inter-observer variability in 12 sections (Pearson Correlation 0.986, P = 0.000).
Other cardiac malformations
Next, we thoroughly analyzed for additional cardiac anomalies by serial tissue-section of 17 P1 Nkx2-5+/R52G mice. Despite a lack of genetic heterogeneity as analyzed by 384 SNP panel (>99.61% homology), these mice demonstrated diverse cardiac anomalies (Figures 4 and 5, Table 1). The most prevalent lesions were perimembranous and/or muscular VSDs found in a single or multiple positions in 80% of the mice. In representative tissue-sections of Nkx2-5+/R52G (mouse #9), the perimembranous VSD was positioned dorsally, with fibrous continuity between the developing leaflets of the aortic and tricuspid valves, while additional apical muscular defects were positioned ventrally, being enclosed within musculature of the septum, matching the human criteria for VSD classification32,33 (Figure 4A).
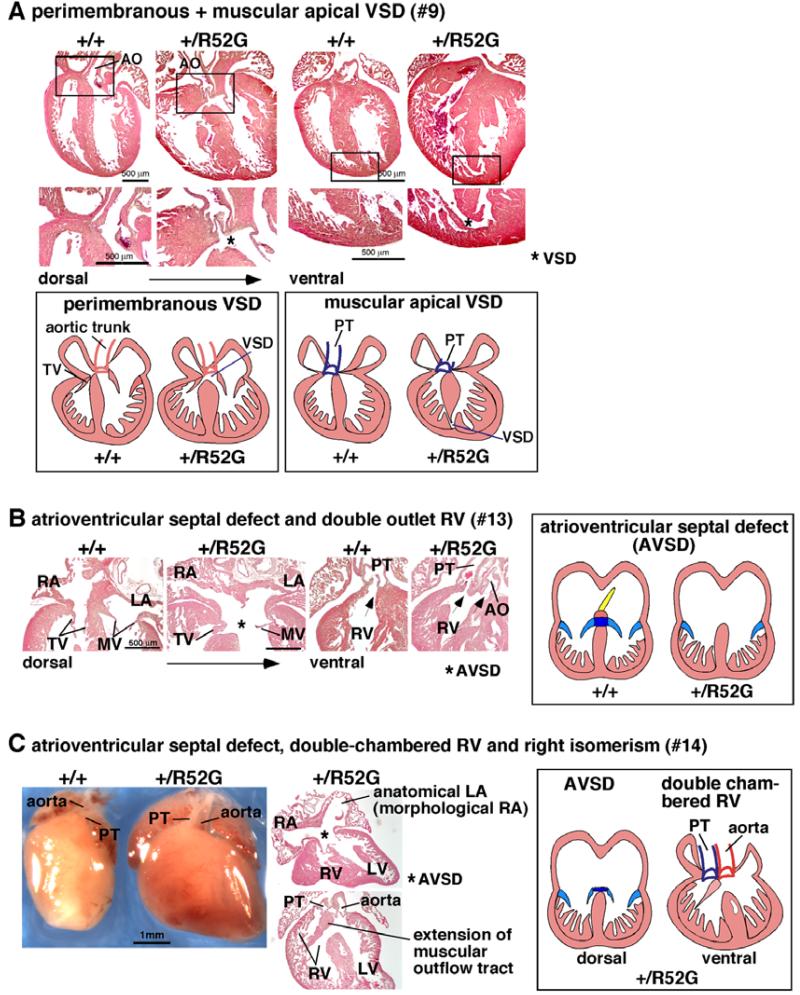
Figure 4. Representative additional cardiac anomalies found in P1 Nkx2-5+/R52G mice.
(A) Perimembranous VSD with additional muscular VSD at the apical region shown in heart sections and simplified illustrations (mouse #9). (B) Atrioventricular septal defect with double outlet right ventricle in mouse #13. (C) Atrioventricular septal defect, double-chambered right ventricle and right isomerism shown in mouse #14. AO, aorta; PT, pulmonary trunk; AVSD, atrioventricular septal defect.
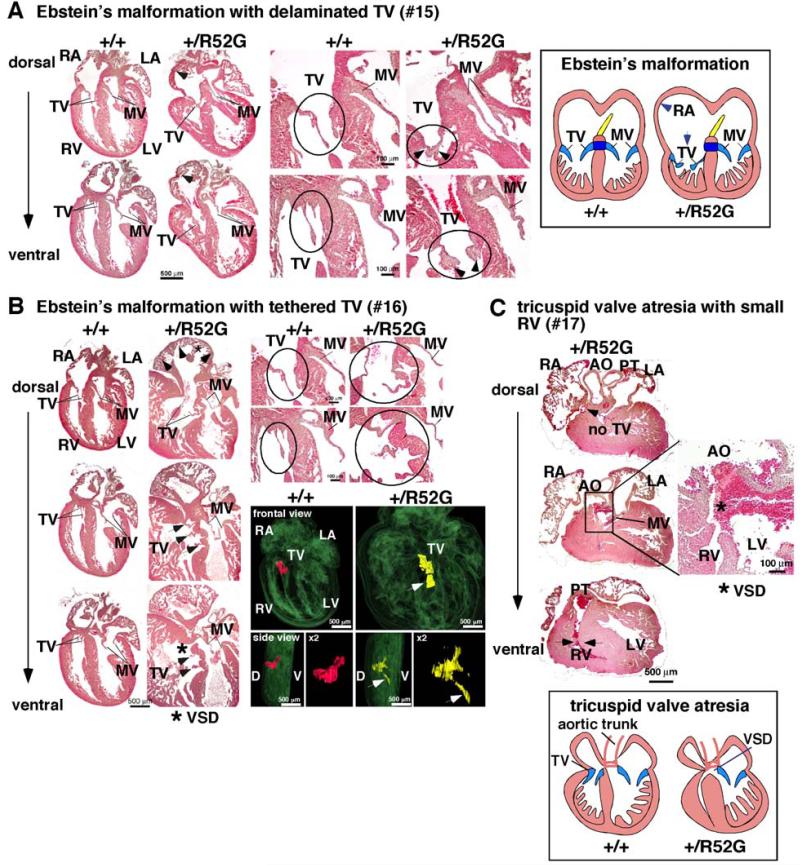
Figure 5. Representative tricuspid valve anomalies in P1 Nkx2-5+/R52G mice.
(A) Ebstein’s malformation in mouse #15. Positions of septal and mural (posterior) leaflets of Nkx2-5+/R52G mice displaced toward apex compared to control mice. Delaminated tricuspid valves are also observed. (B) Ebstein’s malformation with tethered tricuspid valves in mouse #16. 3D reconstructed heart sections including tricuspid valves highlighted in red in control or yellow in Nkx2-5+/R52G mouse. Arrows indicate displaced anterior leaflet of Nkx2-5+/R52G mice toward apex compared to control mice. Of note, septal leaflet could be displaced as well; however it was difficult to distinguish between these two leaflets. (C) Tricuspid valve atresia, muscular VSD, and small right ventricular cavity in mouse #17. See Supplemental Figure S3.
More complex cardiac anomalies were also found in Nkx2-5+/R52G mice. Three mice demonstrated atrioventricular septal defects: (Figures 4B and C). Figure 4B demonstrates an atrioventricular septal defect with common atrioventricular junction, along with double outlet right ventricle (mouse #13), and apical additional muscular VSDs (not shown). Figure 4C also demonstrates atrioventricular septal defect with common atrioventricular junction, along with double-chambered right ventricle, and isomerism of the right atrial appendages (mouse #14). Additional features of isomerism within the atrial chambers included appendages with morphology right characteristics bilaterally, and expression of atrial natriuretic factor (ANF) in the left-sided appendage (Supplemental Figure S2).
Nearly 50% of mice had an abnormality of the tricuspid valve. Profound tricuspid valve anomalies were found in three mice (Figure 5A-C). Two mice (#15, 16) demonstrated Ebstein’s malformation, in which the hinges of tricuspid valve leaflets were displaced towards the apex of the right ventricle (Figures 5A and B). In addition, tricuspid valves were inappropriately delaminated in mouse #15 (Figure 5A), and tethered in mouse #16 (Figure 5B). Figure 5C shows absence of the right atrioventricular connection, which is a feature of tricuspid valvar atresia (mouse #17): the right ventricle was incomplete and hypoplastic, lacking its inlet component (see Supplemental Figure S3). There was no direct communication with the right atrium, but there was communication with the left ventricle through a muscular VSD. Other mice exhibited a more subtle tricuspid valve abnormality characterized by abnormal delamination of the tricuspid valve leaflets (Table 1).
F1 crossing 129 with C57Bl/6 mice
To minimize non-specific genetic effects, congenic mice were generated and analyzed as described above. To determine if the high disease penetrance of R52G is maintained in an outbred background, 129 congenic Nkx2-5+/R52G mice were crossed with C57Bl/6, and F1 newborn mice were analyzed (n=16)(Supplemental Figure S4, Supplemental Table 1). No complex cardiac malformations (e.g. atrioventricular septal defects) were observed. However, all but one F1 mice demonstrated ventricular noncompaction (94%, n=15), and half (n=8) of the mice demonstrated perimembranous and/or muscular VSDs found in a single or multiple positions. Subtle tricuspid valve leaflet abnormalities were seen in ~50% of mice, but neither tricuspid atresia or Ebstein’s malformation were observed. As anticipated, the mice with mixed genetic backgrounds had reduced phenotypic severity for most cardiac phenotypes. However, noncompaction continued to be highly penetrant.
Cardiac anomalies in newborn heterozygous knockout mice (Nkx2-5+/−)
We examined 18 hearts from P1 Nkx2-5+/− mutants (Supplemental Figure S5, Table S2). In agreement with previous studies17-19, cardiac anomalies were observed with lower prevalence compared to Nkx2-5+/R52G mice. Perimembranous or muscular VSDs were observed in 33%, noncompaction in 33%, and incomplete delamination of tricuspid valve (11%) or apical displacement of septal leaflet of tricuspid valve (6%). Only two mice had both a VSD and abnormal tricuspid valve. The incidence of cardiac malformations fall within the range of a wide variety of those in multiple heterozygous Nkx2-5+/− mice, in which the incidence of VSD can vary from zero to 20 to 40%17-19,34. Control Nkx2-5+/+ (n=13) did not demonstrate notable cardiac anomalies. Further, the ratio between P10 Nkx2-5+/+ (49%) vs. Nkx2-5+/− (51%) mice (n=41) was almost equal, suggesting perinatal death is not prevalent in Nkx2-5+/− mice.
Atrial septal abnormalities in P10 Nkx2-5+/R52G mutant mice
The most prevalent cardiac anomaly found in human patients with missense mutations in the NKX2-5 homeodomain is atrial septal defect15, characterized by deficient, persistent communications between the atrial chambers permitting postnatal shunting from left-to-right. The fossa ovalis, in normal development, is closed by attachment of the flap valve, in other words, the foreshortened primary septum, to its rims, and the rims formed by infolding and formation of the secondary basal septum35.
We did not assess atrial septal anomalies in P1 mice, because all the mice, including wild-type controls demonstrate interatrial communications during the transition from the embryonic circulation, when right-to-left shunting of blood through the foramen ovale remains essential36. In FVB mice, such morphological closure of the inter-atrial communication is progressive, and is completed 1 week after birth36. In 129/Sv mice, in contrast, half of the wild-type mice still demonstrated small interatrial communications, with an average size of ~40 μm (8 serial sections, 5 μm thickness) at P10 (n=10). The incidence of interatrial communications was 86% in Nkx2-5+/R52G (n=7) and 70% in Nkx2-5+/− (n=10) at P10, and both were not statistically significantly increased from the wild-type mice (P=0.30 and 0.65 respectively).
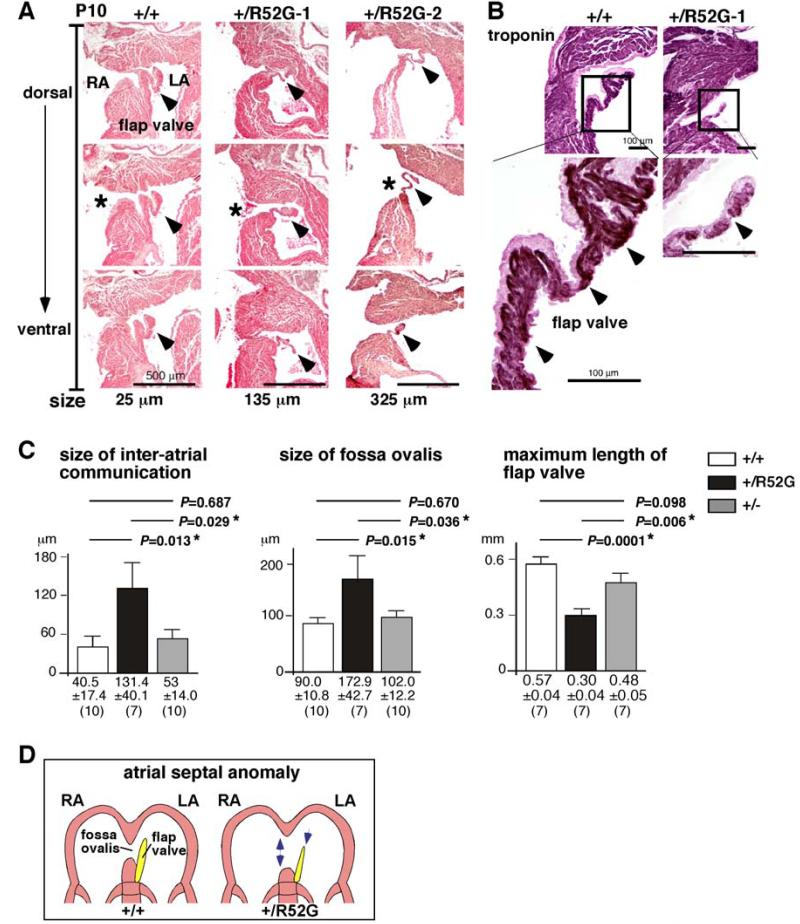
Representative serial tissue sections of P10 wild-type mice (Figure 6A, +/+) demonstrate that a small area of the fossa ovalis was not sealed by the flap valve, with the distance from the opening to the closure being 25 μm (marked with *), while in Nkx2-5+/R52G mice, the distance from the opening to the closure was increased to 135 μm in mouse #1, and 325 μm in mouse #2 (Figure 6A, +/R52G, marked with *). Immunostaining of the troponin T positive muscular structure in the flap valve showed it was poorly developed in Nkx2-5+/R52G mice compared to age-matched Nkx2-5+/+ mice (Figure 6B, arrowheads). Quantitative analyses from a total of 27 hearts showed an increase in the size of the interatrial communication and the foramen ovalis, with the maximum length of the septum primum being decreased in Nkx2-5+/R52G compared to age-matched Nkx2-5+/+ or Nkx2-5+/− mice with statistical significance (Figure 6C). A cartoon depicting the atrial septal anomaly found in Nkx2-5+/R52G mice is presented in Figure 6D.
Figure 6. Inter-atrial communication in P10 Nkx2-5+/R52G hearts compared to age-matched Nkx2-5+/+ or Nkx2-5+/− hearts.
(A) Representative histological sections from P10 mouse hearts selected for a total of 25 μm (5 sections, 5 μm thickness) from Nkx2-5+/+ mouse, and for 135 μm or 325 μm from two Nkx2-5+/R52G mice. Asterisks indicate open inter-atrial communication. Arrowheads indicate flap valve. Dorsal/posterior (top panel) to ventral/anterior sections (bottom panel) are shown. (B) Immunostaining of troponin T (brown) with counter-staining with eosin (pink). (C) Size of inter-atrial communication, fossa ovalis and maximum length of flap valve calculated from a number of tissue sections (5 μm per section). The number of mice examined is indicated. ANOVA demonstrated significant differences among the three groups (F = 4.04, 3.77, 12.03; P = 0.031, 0.038, 0.000, respectively). Of note, maximum length of flap valve from three heart sections each from Nkx2-5+/+ and Nkx2-5+/− could not be measured because flap valve and septum secundum were fused. (D) Simplified illustrations of the atrial septal structure in Nkx2-5+/+ vs. Nkx2-5+/R52G.
A summary of phenotypic comparison between Nkx2-5+/R52G vs Nkx2-5+/+ or Nkx2-5+/− is listed in Table 2.
Table 2. Summary of phenotype of Nkx2-5+/R52G in comparison to Nkx2-5+/− and/or Nkx2-5+/+ (129/Sv background).
| 2a: Cardiovascular malformations (P1). | ||||
|---|---|---|---|---|
| W/R52G (n=17) | W/− (n=18) | P(W/R52G vs. W/−) | X 2 | |
| any malformations | 17 (100%) | 8 (44%) | 0.000* | 13.2 |
| ventricular noncompaction | 17 (100%) | 6 (33%) | 0.000* | 17.2 |
| ventricular septal defects | 14 (82%) | 6 (33%) | 0.006* | 8.6 |
| atrioventricular septal defects | 3 (18%) | 0 | 0.104 | 3.5 |
| tricuspid valve anomalies | 8 (47%) | 3 (17%) | 0.006* | 8.4 |
| 2b: Quantitative measurement of atrial septum morphology (P10). | |||||
|---|---|---|---|---|---|
| W/R52G | W/− | W/W | P (W/R52G vs. W/−) | P (W/− vs. W/W) | |
| size of inter-atrial communication | 131.4 ± 40.1 | 53 ± 14.0 | 40.5 ± 17.4 | 0.029* | 0.687 |
| size of fossa ovalis | 172.9 ± 42.7 | 102.0 ± 12.2 | 90.0 ± 10.8 | 0.036* | 0.37 |
| maximum length of flap valve | 0.30 ± 0.04 | 0.48± 0.05 | 0.57 ± 0.04 | 0.006* | 0.098 |
| 2c: Survival (P10). | |||
|---|---|---|---|
| W/R52G vs. W/W | W/- vs. W/W | W/R52G vs. W/− | |
| number of litters | 26 vs. 35 (total 61) | 21 vs. 20 (total 41) | |
| survival rate of mutant mice | 43% | 51% (P=1.00) | 43% vs. 51% |
| P | 0.002* | 1 | 0.009* |
| X 2 | 10.3 | 1 | 6.4 |
See Figure 6C for detail (n=7 to 10).
P, P value; X2, Pearson’s chi-square value.
P < 0.05.
Downregulation of ANF mRNA, reporter lacZ expression and Nkx2-5 binding to the ANF promoter region in Nkx2-5+/R52G mice
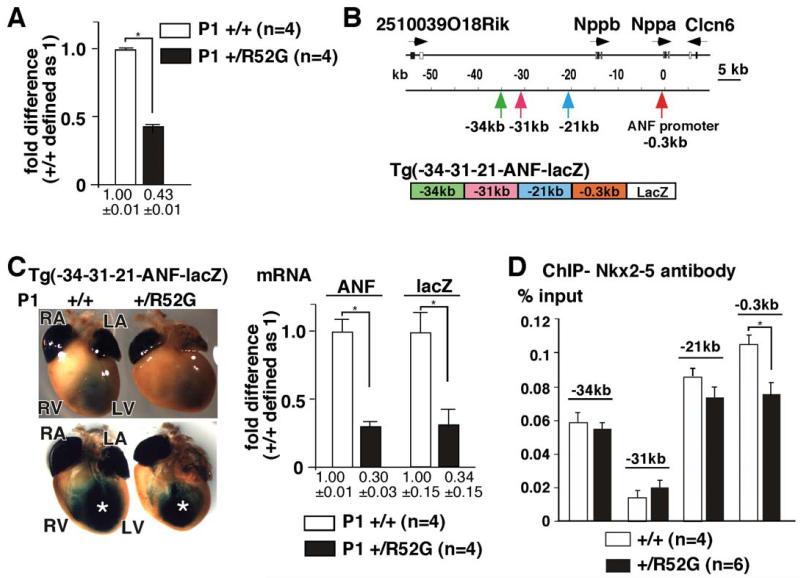
ANF gene is a direct target of Nkx2-5, and is markedly reduced in homozygous germline and conditional Nkx2-5 knock-out mice25,27,37, but not in Nkx2-5+/− mice38. In contrast, mRNA expression in P1 Nkx2-5+/R52G heart was reduced by approximately 60% compared to control Nkx2-5+/+ heart using Taqman RT PCR (Figure 7A). We previously reported Nkx2-5 binding at three 5′ regulatory elements (kb −34, −31, and −21) and at the proximal ANF promoter by chromatin immunoprecipitation (ChIP), which were sufficient for transactivating a reporter lacZ gene (-34-31-21ANF-lacZ) in the trabecular layers mirroring endogenous ANF gene expression (Figure 7B)25. In inducible homozygous Nkx2-5 knockout hearts, lacZ expression was markedly reduced at E13.5 despite the non-compaction25,27. Similarly, intensity of lacZ expression was reduced in Nkx2-5+/R52G compared to the Nkx2-5+/+ littermates with X-gal staining performed side by side. Quantitative measurements of ANF and lacZ mRNA using whole hearts confirmed approximately 70% reduction in Nkx2-5+/R52G mice relative to the control Nkx2-5+/+ mice, indicating that the genomic element(s) responsible for ANF mRNA reduction in Nkx2-5+/R52G mice are indeed present within the four genomic regions (Figure 7C, supplemental Figure S6).
Figure 7. Reduction of ANF and X-gal staining of -34-31-21-ANF-lacZ transgenic mice in Nkx2-5+/R52G hearts relative to those from Nkx2-5+/+.
(A) Difference of ANF mRNA expression normalized to β-actin between Nkx2-5+/+ and Nkx2-5+/R52G hearts. ANF expression in Nkx2-5+/+ hearts defined as 1. Number of animals analyzed is shown. (B) Organization of the genomic locus including ANF (Nppa). Positions of three regulatory elements and the proximal promoter of ANF, and schematic of transgenic lacZ reporter construct are indicated. (C) Representative images of X-gal staining from a total of 4 sets of Nkx2-5+/+ and Nkx2-5+/R52G heterozygous -34-31-21-ANF-lacZ-positive P1 littermates are shown. Decreased lacZ staining in the left ventricle is marked (*). Difference, as determined by TaqMan real-time RT-PCR, between ANF and lacZ mRNA expression normalized to β-actin, with expression in Nkx2-5+/+ defined as 1. Number of animals analyzed is shown. (D) ChIP analysis of Nkx2-5 in percentage of input DNA recovered from 2-3 independent experiments with TaqMan PCR performed in duplicate.
To examine occupancy of Nkx2-5 proteins at these elements, ChIP assay was performed using two different cell populations of neonatal cardiomyocytes isolated either from Nkx2-5+/R52G or Nkx2-5+/+ mice. Among the four elements described above, 29% reduction in immunoprecipitation was demonstrated at the proximal promoter region in Nkx2-5+/R52G relative to Nkx2-5+/+ mice (Figure 7D). Taken together, these studies demonstrate a 60-70% reduction of ANF mRNA and X-gal staining in -34-31-21ANF-lacZ reporter mice, with a 29% reduction Nkx2-5 protein occupancy at the proximal promoter region in Nkx2-5+/R52G relative to the control Nkx2-5+/R52G littermates.
Discussion
Heterozygous missense mutations in the homeodomain of human NKX2-5 are known to lead to a high penetrance of diverse cardiac anomalies when compared to mutations outside of the homeodomain15. A novel heterozygous knock-in mouse, having a disease-causing missense mutation in the homeodomain, Nkx2-5+/R52G demonstrates a high incidence of diverse forms of congenital cardiac malformations. The mutant mice displayed complete penetrance of ventricular noncompaction, and a high incidence of VSDs and atrial septal anomalies, as well as perinatal lethal complex cardiac anomalies, such as common atrioventricular junction, tricuspid atresia, or Ebstein’s malformation of tricuspid valve. Overall, introduction of a missense mutation (Arg52Gly) in the mouse genome resulted in similar, or even more profound, cardiac anomalies than those seen in humans.
Ventricular noncompaction is a cardiomyopathy characterized by persistence of the trabecular layer of the ventricular walls, in some patients producing both diastolic and systolic dysfunction31,39-41. During normal cardiac development, the ventricular trabecular layer is formed around murine embryonic day (E) 10.5, coinciding with an increase in the ventricular myocardial mass prior to development of a discrete coronary circulation42. As normal cardiac development progresses, the compact layer thickens, with trabecular myocardium becoming less obvious. Pathogenesis of ventricular noncompaction is largely unknown, but it has been speculated that it reflects an early arrest of myocardial maturation, with cessation of proliferation of the trabecular meshwork31. Noncompaction was evident in P1 Nkx2-5+/R52G mutant mice, suggesting that heterozygous Nkx2-5 homeodomain missense mutation dysregulates these processes, as was shown in homozygous Nkx2-5 knockout using ventricular myocyte-specific (myosin light chain 2v) Cre23 as well as tamoxifen-inducible Cre beginning from E10.527. Although the discrimination between the compact and trabecular layers is somewhat subjective, we followed the consistent criteria throughout the analyses.
Malformations of the tricuspid valve are found in both patients with NKX2-5 mutations6,8, and Nkx2-5+/R52G mice. Abnormal formation of the tricuspid valve, or abnormal remodeling, seen in Nkx2-5+/R52G knock-in mice, raises interesting questions as to how a mutation in a myocardial-specific gene results in valvar maldevelopment. During the early stages of heart development, myocardial signals induce a subset of endocardial cells in the atrioventricular canal, and later in the outflow tract regions, to undergo an epithelial-to-mesenchymal transformation, resulting in formation of endocardial cushions. These cushions are the primordia of the leaflets of the valves of the four-chambered heart. Proper formation of the tricuspid valve also requires expansion of the right-side of the atrioventricular canal, so that the cavity of the right atrium is in direct communication with that of the right ventricle. Formation of the tricuspid valvar leaflets then involves myocardial, epicardial, and endocardial cushion tissues43-46. It has been suggested that the myocardium forms part of the primordium of the developing tricuspid valvar leaflets43-45. In support of this notion, we found there are prominent myocardial layers at mid embryonic stage (E12.5 and E15.5) confined mostly to the mural leaflet of the tricuspid valve. This could partly explain the high-incidence of the tricuspid valve anomalies related to Nkx2-5 mutations.
In humans, ventricular noncompaction is often accompanied by diverse forms of congenital cardiac malformations, including Ebstein’s malformation of the tricuspid valve40,47. This suggests a potential pathogenetic link of two seemingly unrelated cardiac phenotypes40. In addition, the ventricular septum in Nkx2-5+/R52G mice is wide, irregular in shape, or shows multiple muscular defects. Considering that the ventricular septum is itself formed by compaction of the trabecular layer42, this offers an obvious correlation between the noncompaction and the multiple muscular defects.
A signature phenotype of humans harboring a heterozygous homeodomain missense NKX2-5 mutation is the development of progressive atrioventricular block during post-natal life. We anticipate that Nkx2-5+/R52G mice will manifest this phenotype. Studies to investigate this possibility are currently underway.
In in vitro studies, the best correlation with clinical phenotypes and homeodomain missense mutation resulted from the markedly reduced DNA binding15. We report computational modeling that confirms this biochemical property in all 14 NKX2-5 homeodomain missense mutations found to date. Expression of ANF mRNA and -34-31-21ANF-lacZ reporter gene was reduced by 60-70% in Nkx2-5+/R52G mice, while Nkx2-5 protein occupancy was demonstrated only at the proximal promoter region and was reduced by 29%. Thus, reduction of DNA binding in Nkx2-5+/R52G relative to Nkx2-5+/+ cannot fully explain the 60-70% reduction of ANF mRNA. In consideration of preserved protein-protein interactions of R52G mutant with Nkx2-5 co-factors selected from among various factors15,48-52, this mutant has broader functional consequences including dominant negative effects. Further molecular mechanisms underlying high disease penetrance demonstrated in Nkx2-5+/R52G mice remain to be analyzed.
In summary, we report an initial characterization of a novel mouse model having a heterozygous Nkx2-5 missense mutation in the homeodomain that demonstrates a high penetrance of diverse cardiac anomalies, similar to or more profound than those observed in human patients. Just as importantly this mouse model is likely to provide insights into the molecular mechanisms underlying these cardiac anomalies. Such insights would allow potentially for the future development of specific therapeutic strategies for patients suffering from congenital cardiac anomalies.
Supplementary Material
Acknowledgments
We are grateful to T. Sanders, K.R. Chien, S.P. Oh, Y. Sugi and K. Fortin for valuable suggestions, critical reagents and technical supports.
Source of Funding:
This work was supported by the NIH Recovery Act Grant (HL081577-05S1 to HK) and American Heart Association Grant 13BGIA13960001 (to H.-J. N.).
Footnotes
Disclosures:
None.
References
- 1.Hoffman JI, Kaplan S, Liberthson RR. Prevalence of congenital heart disease. Am Heart J. 2004;147:425–439. doi: 10.1016/j.ahj.2003.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Capozzi G, Caputo S, Pizzuti R, Martina L, Santoro M, Santoro G, et al. Congenital heart disease in live-born children: Incidence, distribution, and yearly changes in the campania region. J Cardiovasc Med (Hagerstown) 2008;9:368–374. doi: 10.2459/JCM.0b013e3282eee866. [DOI] [PubMed] [Google Scholar]
- 3.Dolk H, Loane M, Garne E. Congenital heart defects in europe: Prevalence and perinatal mortality, 2000 to 2005. Circulation. 2011;123:841–849. doi: 10.1161/CIRCULATIONAHA.110.958405. [DOI] [PubMed] [Google Scholar]
- 4.van der Bom T, Zomer AC, Zwinderman AH, Meijboom FJ, Bouma BJ, Mulder BJ. The changing epidemiology of congenital heart disease. Nat Rev Cardiol. 2011;8:50–60. doi: 10.1038/nrcardio.2010.166. [DOI] [PubMed] [Google Scholar]
- 5.Roybal JL, Santore MT, Flake AW. Stem cell and genetic therapies for the fetus. Semin Fetal Neonatal Med. 2010;15:46–51. doi: 10.1016/j.siny.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Benson DW. Genetic origins of pediatric heart disease. Pediatr Cardiol. 2010;31:422–429. doi: 10.1007/s00246-009-9607-y. [DOI] [PubMed] [Google Scholar]
- 7.Andelfinger G. Genetic factors in congenital heart malformation. Clin Genet. 2008;73:516–527. doi: 10.1111/j.1399-0004.2008.01009.x. [DOI] [PubMed] [Google Scholar]
- 8.Benson DW, Silberbach GM, Kavanaugh-McHugh A, Cottrill C, Zhang Y, Riggs S, et al. Mutations in the cardiac transcription factor nkx2.5 affect diverse cardiac developmental pathways. J Clin Invest. 1999;104:1567–1573. doi: 10.1172/JCI8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reamon-Buettner SM, Borlak J. Nkx2-5: An update on this hypermutable homeodomain protein and its role in human congenital heart disease (chd) Hum Mutat. 2010;31:1185–1194. doi: 10.1002/humu.21345. [DOI] [PubMed] [Google Scholar]
- 10.Stallmeyer B, Fenge H, Nowak-Gottl U, Schulze-Bahr E. Mutational spectrum in the cardiac transcription factor gene nkx2.5 (csx) associated with congenital heart disease. Clin Genet. 2010;78:533–540. doi: 10.1111/j.1399-0004.2010.01422.x. [DOI] [PubMed] [Google Scholar]
- 11.De Luca A, Sarkozy A, Consoli F, Ferese R, Guida V, Dentici ML, et al. Familial transposition of the great arteries caused by multiple mutations in laterality genes. Heart. 2010;96:673–677. doi: 10.1136/hrt.2009.181685. [DOI] [PubMed] [Google Scholar]
- 12.Liu XY, Wang J, Yang YQ, Zhang YY, Chen XZ, Zhang W, et al. Novel nkx2-5 mutations in patients with familial atrial septal defects. Pediatr Cardiol. 2011;32:193–201. doi: 10.1007/s00246-010-9859-6. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Liu XY, Yang YQ. Novel nkx2-5 mutations responsible for congenital heart disease. Genet Mol Res. 2011;10:2905–2915. doi: 10.4238/2011.November.29.1. [DOI] [PubMed] [Google Scholar]
- 14.Kasahara H, Lee B, Schott JJ, Benson DW, Seidman JG, Seidman CE, et al. Loss of function and inhibitory effects of human csx/nkx2.5 homeoprotein mutations associated with congenital heart disease. J Clin Invest. 2000;106:299–308. doi: 10.1172/JCI9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasahara H, Benson DW. Biochemical analyses of eight nkx2.5 homeodomain missense mutations causing atrioventricular block and cardiac anomalies. Cardiovasc Res. 2004;64:40–51. doi: 10.1016/j.cardiores.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Hacking DF. ‘Knock, and it shall be opened’: Knocking out and knocking in to reveal mechanisms of disease and novel therapies. Early Hum Dev. 2008;84:821–827. doi: 10.1016/j.earlhumdev.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Biben C, Weber R, Kesteven S, Stanley E, McDonald L, Elliott DA, et al. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene nkx2-5. Circ Res. 2000;87:888–895. doi: 10.1161/01.res.87.10.888. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka M, Berul CI, Ishii M, Jay PY, Wakimoto H, Douglas P, et al. A mouse model of congenital heart disease: Cardiac arrhythmias and atrial septal defect caused by haploinsufficiency of the cardiac transcription factor csx/nkx2.5. Cold Spring Harb Symp Quant Biol. 2002;67:317–325. doi: 10.1101/sqb.2002.67.317. [DOI] [PubMed] [Google Scholar]
- 19.Winston JB, Erlich JM, Green CA, Aluko A, Kaiser KA, Takematsu M, et al. Heterogeneity of genetic modifiers ensures normal cardiac development. Circulation. 2010;121:1313–1321. doi: 10.1161/CIRCULATIONAHA.109.887687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grow MW, Krieg PA. Tinman function is essential for vertebrate heart development: Elimination of cardiac differentiation by dominant inhibitory mutants of the tinman-related genes, xnkx2-3 and xnkx2-5. Dev Biol. 1998;204:187–196. doi: 10.1006/dbio.1998.9080. [DOI] [PubMed] [Google Scholar]
- 21.Kasahara H, Wakimoto H, Liu M, Maguire CT, Converso KL, Shioi T, et al. Progressive atrioventricular conduction defects and heart failure in mice expressing a mutant csx/nkx2.5 homeoprotein. J Clin Invest. 2001;108:189–201. doi: 10.1172/JCI12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pradhan L, Genis C, Scone P, Weinberg EO, Kasahara H, Nam HJ. Crystal structure of the human nkx2.5 homeodomain in complex with DNA target. Biochemistry. 2012;51:6312–6319. doi: 10.1021/bi300849c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S, et al. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117:373–386. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- 24.Briggs LE, Takeda M, Cuadra AE, Wakimoto H, Marks MH, Walker AJ, et al. Perinatal loss of nkx2-5 results in rapid conduction and contraction defects. Circ Res. 2008;103:580–590. doi: 10.1161/CIRCRESAHA.108.171835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warren SA, Terada R, Briggs LE, Cole-Jeffrey CT, Chien WM, Seki T, et al. Differential role of nkx2-5 in activation of the atrial natriuretic factor gene in the developing versus failing heart. Mol Cell Biol. 2011;31:4633–4645. doi: 10.1128/MCB.05940-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasahara H, Bartunkova S, Schinke M, Tanaka M, Izumo S. Cardiac and extracardiac expression of csx/nkx2.5 homeodomain protein. Circ Res. 1998;82:936–946. doi: 10.1161/01.res.82.9.936. [DOI] [PubMed] [Google Scholar]
- 27.Terada R, Warren S, Lu JT, Chien KR, Wessels A, Kasahara H. Ablation of nkx2-5 at mid-embryonic stage results in premature lethality and cardiac malformation. Cardiovasc Res. 2011;91:289–299. doi: 10.1093/cvr/cvr037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reamon-Buettner SM, Borlak J. Somatic nkx2-5 mutations as a novel mechanism of disease in complex congenital heart disease. J Med Genet. 2004;41:684–690. doi: 10.1136/jmg.2003.017483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenni R, Oechslin E, Schneider J, Attenhofer Jost C, Kaufmann PA. Echocardiographic and pathoanatomical characteristics of isolated left ventricular non-compaction: A step towards classification as a distinct cardiomyopathy. Heart. 2001;86:666–671. doi: 10.1136/heart.86.6.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savolainen SM, Foley JF, Elmore SA. Histology atlas of the developing mouse heart with emphasis on e11.5 to e18.5. Toxicol Pathol. 2009;37:395–414. doi: 10.1177/0192623309335060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paterick TE, Umland MM, Jan MF, Ammar KA, Kramer C, Khandheria BK, et al. Left ventricular noncompaction: A 25-year odyssey. J Am Soc Echocardiogr. 2012;25:363–375. doi: 10.1016/j.echo.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 32.Soto B, Becker AE, Moulaert AJ, Lie JT, Anderson RH. Classification of ventricular septal defects. Br Heart J. 1980;43:332–343. doi: 10.1136/hrt.43.3.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minette MS, Sahn DJ. Ventricular septal defects. Circulation. 2006;114:2190–2197. doi: 10.1161/CIRCULATIONAHA.106.618124. [DOI] [PubMed] [Google Scholar]
- 34.Jay PY, Harris BS, Maguire CT, Buerger A, Wakimoto H, Tanaka M, et al. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest. 2004;113:1130–1137. doi: 10.1172/JCI19846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson RH, Brown NA, Webb S. Development and structure of the atrial septum. Heart. 2002;88:104–110. doi: 10.1136/heart.88.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cole-Jeffrey CT, Terada R, Neth MR, Wessels A, Kasahara H. Progressive anatomical closure of foramen ovale in normal neonatal mouse hearts. Anat Rec (Hoboken) 2012;295:764–768. doi: 10.1002/ar.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene csx/nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 38.Jay PY, Rozhitskaya O, Tarnavski O, Sherwood MC, Dorfman AL, Lu Y, et al. Haploinsufficiency of the cardiac transcription factor nkx2-5 variably affects the expression of putative target genes. FASEB J. 2005;19:1495–1497. doi: 10.1096/fj.04-3064fje. [DOI] [PubMed] [Google Scholar]
- 39.Zambrano E, Marshalko SJ, Jaffe CC, Hui P. Isolated noncompaction of the ventricular myocardium: Clinical and molecular aspects of a rare cardiomyopathy. Lab Invest. 2002;82:117–122. doi: 10.1038/labinvest.3780404. [DOI] [PubMed] [Google Scholar]
- 40.Sinkovec M, Kozelj M, Podnar T. Familial biventricular myocardial noncompaction associated with ebstein’s malformation. Int J Cardiol. 2005;102:297–302. doi: 10.1016/j.ijcard.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 41.Freedom RM, Yoo SJ, Perrin D, Taylor G, Petersen S, Anderson RH. The morphological spectrum of ventricular noncompaction. Cardiol Young. 2005;15:345–364. doi: 10.1017/S1047951105000752. [DOI] [PubMed] [Google Scholar]
- 42.Sedmera D, Pexieder T, Vuillemin M, Thompson RP, Anderson RH. Developmental patterning of the myocardium. Anat Rec. 2000;258:319–337. doi: 10.1002/(SICI)1097-0185(20000401)258:4<319::AID-AR1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 43.Lamers WH, Viragh S, Wessels A, Moorman AF, Anderson RH. Formation of the tricuspid valve in the human heart. Circulation. 1995;91:111–121. doi: 10.1161/01.cir.91.1.111. [DOI] [PubMed] [Google Scholar]
- 44.de Lange FJ, Moorman AF, Anderson RH, Manner J, Soufan AT, de Gier-de Vries C, et al. Lineage and morphogenetic analysis of the cardiac valves. Circ Res. 2004;95:645–654. doi: 10.1161/01.RES.0000141429.13560.cb. [DOI] [PubMed] [Google Scholar]
- 45.de Vlaming A, Sauls K, Hajdu Z, Visconti RP, Mehesz AN, Levine RA, et al. Atrioventricular valve development: New perspectives on an old theme. Differentiation. 2012;84:103–116. doi: 10.1016/j.diff.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu B, Baldwin HS, Zhou B. Nfatc1 directs the endocardial progenitor cells to make heart valve primordium. Trends Cardiovasc Med. 2013 doi: 10.1016/j.tcm.2013.04.003. doi: 10.1016/j.tcm.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Attenhofer Jost CH, Connolly HM, O’Leary PW, Warnes CA, Tajik AJ, Seward JB. Left heart lesions in patients with ebstein anomaly. Mayo Clin Proc. 2005;80:361–368. doi: 10.4065/80.3.361. [DOI] [PubMed] [Google Scholar]
- 48.Habets PE, Moorman AF, Clout DE, van Roon MA, Lingbeek M, van Lohuizen M, et al. Cooperative action of tbx2 and nkx2.5 inhibits anf expression in the atrioventricular canal: Implications for cardiac chamber formation. Genes Dev. 2002;16:1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thattaliyath BD, Firulli BA, Firulli AB. The basic-helix-loop-helix transcription factor hand2 directly regulates transcription of the atrial naturetic peptide gene. J Mol Cell Cardiol. 2002;34:1335–1344. doi: 10.1006/jmcc.2002.2085. [DOI] [PubMed] [Google Scholar]
- 50.Stennard FA, Costa MW, Elliott DA, Rankin S, Haast SJ, Lai D, et al. Cardiac t-box factor tbx20 directly interacts with nkx2-5, gata4, and gata5 in regulation of gene expression in the developing heart. Dev Biol. 2003;262:206–224. doi: 10.1016/s0012-1606(03)00385-3. [DOI] [PubMed] [Google Scholar]
- 51.Akazawa H, Kudoh S, Mochizuki N, Takekoshi N, Takano H, Nagai T, et al. A novel lim protein cal promotes cardiac differentiation by association with csx/nkx2-5. J Cell Biol. 2004;164:395–405. doi: 10.1083/jcb.200309159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuasa S, Onizuka T, Shimoji K, Ohno Y, Kageyama T, Yoon SH, et al. Zac1 is an essential transcription factor for cardiac morphogenesis. Circ Res. 2010;106:1083–1091. doi: 10.1161/CIRCRESAHA.109.214130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.