Abstract
The immune system has evolved mechanisms to protect the host from the deleterious effects of inflammation. The generation of immune suppressive cells like myeloid derived suppressor cells (MDSCs) that can counteract T cell responses represents one such strategy. There is an accumulation of immature myeloid cells or MDSCs in bone marrow (BM) and lymphoid organs under pathological conditions such as cancer. MDSCs represent a population of heterogeneous myeloid cells comprising of macrophages, granulocytes and dendritic cells that are at early stages of development. Although, the precise signaling pathways and molecular mechanisms that lead to MDSC generation and expansion in cancer remains to be elucidated. It is widely believed that perturbation of signaling pathways involved during normal hematopoietic and myeloid development under pathological conditions such as tumorogenesis contributes to the development of suppressive myeloid cells. In this review we discuss the role played by key signaling pathways such as PI3K, Ras, Jak/Stat and TGFb during myeloid development and how their deregulation under pathological conditions can lead to the generation of suppressive myeloid cells or MDSCs. Targeting these pathways should help in elucidating mechanisms that lead to the expansion of MDSCs in cancer and point to methods for eliminating these cells from the tumor microenvironment.
Keywords: Myeloid derived suppressor cells (MDSC), PI3K, TGFβ, Ras, CSF-1, Jak/Stat, tumor microenvironment
1. Introduction
The link between inflammation and cancer was suggested by Virchow a century ago. The presence of inflammatory cells in tumor stroma has been shown to facilitate tumor development, both in experimental models and in the pathogenesis of human cancers. The tumor stroma is composed of an extracellular matrix and that contains a milieu of different cell types including fibroblasts, endothelial cells and immune cells. During tumor progression, the tumor stroma may be altered to promote the growth of cancer cells and facilitate their dissemination [1]. Cytokines and chemokines secreted by cancer cells aid in the recruitment of immune cells into the tumor microenvironment (TME), setting up paracrine and autocrine signaling networks which promote tumor development [2]. The inflammatory cells residing within the TME promote tumor growth through two opposing mechanisms. The first type possesses pro-inflammatory functions and promotes tumor growth by releasing cytokine and chemokines into the extracellular matrix. The second type suppresses natural activation of immune cells, which are responsible for the destruction of tumor cells thus resulting in tumor progression.
MDSCs represent a cell type that can suppress the function of other immune cells by creating a suppressive environment in the tumor stroma. The interaction of MDSCs with T cells has been studied extensively. MDSCs can suppress T cell function utilizing a variety of mechanisms. This involves the generation of reactive oxygen species (ROS), nitric oxide (NO), arginase and cytokines that can lead to inhibition of signaling pathways in T cells [3].
The role of molecular and signaling pathways that are involved in the generation of MDSCs from myeloid precursors in disease conditions and cancer is not completely understood. Therefore, it is important to investigate the role of signaling pathways that are involved in MDSC generation, expansion and function. Once the molecular and signaling pathways involved in regulation of MDSC physiology have been identified, then therapies aimed at targeting these signaling pathways can help in eliminating MDSCs from the TME, thereby improving the therapeutic index of chemotherapeutic drugs and immune based therapies.
2. Myeloid cells
2.1. Macrophages and Granulocytes
Hematopoietic stem cells (HSC) residing in the bone marrow give rise to two distinct progenitors, the common lymphoid progenitor (CLP) and the common myeloid progenitor (CMP). These progenitors lose their self-renewal ability and acquire lineage makers due to the expression of distinct transcriptional activities [4, 5]. Myelopoiesis accounts for nearly two-thirds of the cells produced by the bone marrow [6]. Myeloid precursors can differentiate into macrophages and granulocytes depending on the presence of growth factors. The chemokine receptor CXCR4 and the growth factor G-CSF plays a key role in the generation of granulocytes by the bone marrow [7]. Granulocytes circulate in blood as dormant cells until they encounter a microorganism which they destroy via phagocytosis and generation of ROS [8]. Circulating monocytes emigrate from the bone marrow into the peripheral blood and give rise to resident tissue macrophages in a variety of tissues. Pro-inflammatory cues from tissue and tumors help in the recruitment of these cells which leads to tissue remodeling.
Based on their functional properties, macrophages have been broadly classified into two distinct subtypes, M1 and M2. Classically activated M1 macrophages possess cyotoxic activity and can destroy tumor cells and pathogens by generating nitric oxide [9–11]. In contrast, alternatively activated M2 macrophages possess high arginase activity and are phagocytotic in nature. M2 macrophages frequently are found in the TME and promote tumor growth and metastasis by secreting a variety of cytokine and chemokines into the extracellular matrix (ECM) [12, 13]. Although, M1 and M2 populations are derived from a common progenitor, there is immense interest in elucidating the factors and genes that are responsible for the differentiation of one cell type to another. M1 and M2 macrophages possess distinct gene expression profiles. The M1 macrophages express iNOS, IL-6 and TNF, whereas the M2 gene signature is characterized by the expression of Arg1, Ym-1, Fizz1 and CCl22 [9, 14–16]. A variety of agents can drive differentiation of macrophages towards M1 or M2 type. LPS and IFNγ can polarize macrophages to differentiate into the M1 type, while exposure to cytokines like IL-4 leads to the development of M2 phenotype [17].
2.2 Myeloid derived suppressor cells
MDSCs represent a heterogeneous population of myeloid cells composed of macrophages, granulocytes and dendritic cells that are at early stages of development. Under physiological conditions they represent up to 20–30% of BM cells and ~4% of nucleated cells in the spleen [13]. However, under pathological conditions such as infections, stress, and cancer there is an accumulation of MDSCs in lymphoid organs.
In mice, they are identified by expression of the myeloid markers Gr-1 and CD11b. Murine MDSCs are divided into two categories, granulocytic MDSC (G-MDSC) and monocytic MDSC (M-MDSC), based on their morphology and expression of Ly6G and Ly6C markers. G-MDSCs morphologically resemble granulocytes and are defined as CD11b+Ly6G+Ly6Clow, whereas M-MDSCs are similar to monocytes and are defined by the expression of CD11b+Ly6G−Ly6Chigh. Moreover, G-MDSCs produce ROS to suppress immune cells. In contrast, M-MDSCs generate NO to inactivate immune response [13].
Similarly, human MDSCs consist of the monocytic and granulocytic populations. A number of markers have been used to analyze and characterize MDSC populations in patients with cancer (Table-1). The presence of MDSCs was first reported in the blood of patients with head and neck cancer and they were identified as Lin−CD33+CD15+CD34+ cells [18];[19]. Human MDSCs are now broadly classified as CD33+HLADRlow/neg. Monocytic MDSCs express CD14 and granulocytic MDSCs express CD15 [20, 21].
Table-1.
Subsets of MDSCs in human cancers
| Melanoma | ||
| Lin− HLA-DR− CD33+ | [19] | |
| CD15+ IL4Rα+ | [21] | |
| CD14+ IL4Rα+ | ||
| [135] | ||
| CD14+ HLA-DR−/low | [136] | |
| [137] | ||
| CD14+ HLA-DR−/low Stathigh CD80+ | [136] | |
| CD83+ DC-Sign+ | ||
| CD14+ HLA-DR−/low | [135] | |
| CD11b+ HLA-DR− Lin− | [138] | |
| CD14+ HLA-DR− Lin− | ||
| CD33+ HLA-DR− Lin− | ||
| CD15+ HLA-DR− Lin− | ||
| Breast Cancer | ||
| Lin− HLA-DR− | [19] | |
| Lin−/low HLA-DR− CD33+ CD11b+ | [137] | |
| CD15+ granulocytes | [139] | |
| CD15+ FSClow SSChigh | ||
|
Central Nervous System cancers |
||
| Neuroblastoma | CD11b+ CD33+ HLA-DR− | [140] |
| Glioblastoma | CD14+ CD33+ HLA-DR−/low | [141] |
| CD15+ CD33+ HLA-DR− | ||
|
Head and Neck Squamous Cell |
Lin− HLA-DR− | [19] |
| CD11b+ CD14− CD33+ | [142] | |
| CD14+ | [125] | |
| SSChigh CD66b+ | [143] | |
| Lung | ||
| NSCLC | CD11b+ CD14− CD15+ CD33+ | [144] |
| Lung Cancer | Lin− HLA-DR− | [19] |
| CD11b+ CD33+ | [145] | |
| SSChigh CD66b+ | [143] | |
|
Gastrointestinal Cancers |
||
| Hepatocellular carcinoma | CD14+ HLA-DR−/low | [146] |
| Gastrointestinal | CD15+ CD14+ CD11b+ CD33+ | [87] |
| Cancer | HLA-DR− | |
| Colon | CD15+ IL4Rα+ | [21] |
| CD14+ IL4Rα+ | ||
| CD15+ granulocytes | [139] | |
| CD15+ FSClow SSChigh | ||
| CD14+ HLA-DR−/low S100A9+ | [147] | |
| Pancreas | CD15+ granulocytes | [139] |
| CD45+ Lin− CD33+ CD11b+ CD15+ | [148] | |
| Renal Cancers | ||
| CD14+ IL-4Rα+ | [149] | |
| CD15+ IL-4Rα+ | ||
| CD14+ HLA-DR−/low | ||
| CD11b+ CD14− CD15+ | ||
| CD15+ FSClow SSChigh | ||
| Lin− HLA-DR− CD33+ | ||
| Lin− HLA-DR− CD33+ | [150] | |
| [151] | ||
| CD33+ HLA-DR− | [152] | |
| CD15+ CD14− | ||
| [153] | ||
| CD11b+ CD14− CD15+ | [154] | |
| [155] | ||
| CD14+ HLA-DR−/low | [156] | |
| CD14+ HLA-DR−/low | [155] | |
|
Genitourinary Malignancies |
||
| Prostate | CD14+ HLA-DR−/low | [20] |
| Bladder & Uretheral | SSChigh CD66b+ | [143] |
| Ovarian | CD11b+ CD14+ CD15+ CD33+ | [123] |
| HLA-DRlow CD33+ CD80− CD83− | ||
Abbreviations: SSC: side scatter FSC: forward scatter Lin: lineage DC-Sign: Ctype lectin highly expressed on dendritic cell
MDSCs can be generated from bone marrow derived cells (BMDC) and peripheral blood monocyte cells (PBMC) using a combination of different growth factors and cytokines (GM-CSF, IL-6 etc) in vitro [22]. Since, there are small numbers of MDSC in normal subjects, in vitro generated MDSCs could be a valuable tool to compare functional properties of MDSCs from normal and cancer patients. Moreover, it would help to elucidate the molecular and signaling pathways involved in the generation and expansion of MDSCs in cancer.
3. The role of cytokines in MDSC development
Cytokines like M-CSF (CSF-1), G-CSF and GM-CSF that play a pivotal role during various stages of development of myeloid cells in both humans and mice can also affect MDSC biology.
3.1. CSF-1
Macrophage colony stimulating factor (M-CSF) or CSF-1, functions by binding to its cognate CSF-1R receptor [23]. This binding leads to dimerization and autophosphorylation of receptor tyrosine residues and results in the activation of Ras/Raf/Mapk/Erk pathway. This promotes the growth and differentiation of macrophages both in vitro and in vivo [24]. Mice lacking the CSF-1 ligand exhibit several abnormalities that result in growth retardation, low fertility, defects in osteoclast differentiation and reduced numbers of tissue macrophages [25]. Most of these defects can be restored by administration of CSF-1 or expression of the transgene in mice [26];[27].
High levels of CSF-1 can interfere with proper myeloid development, resulting in the generation of MDSCs. Further evidence of this relationship comes from studies that show high expression of CSF-1 mRNA under pathological conditions correlating with expansion of MDSCs [28]. Recruitment of macrophages to the sites of inflammation contributes to increased levels of CSF-1 that perturbs the normal homeostasis resulting in an accumulation of MDSCs [29].
Studies using animal models have shown that CSF-1 has therapeutic potential in the treatment of inflammatory diseases and cancer. Studies by Hidaka et al showed that administration of CSF-1 to patients with ovarian cancer resulted in the improvement of NK and T cell functions [30]. Similarly, infusion of recombinant CSF-1 in patients with melanoma led to an increase in the number of circulating monocytes, suggesting that blocking CSF-1 action could be therapeutically useful [31].
GW2580 is an antibody that is highly selective to CSF-1R [32]. Irvin et al showed that GW2580 was able to suppress the expression of inflammatory cytokines in macrophages [28]. There is now evidence to show that MDSCs isolated from tumor bearing mice also express the CSF-1R receptor in addition to Gr-1 [33]. In a recent study, treatment of mice with GW2580 could inhibit the infiltration of monocytic MDSCs in lung and prostate tumors. Furthermore, combining it with an anti-VEGR2 antibody resulted in a significant reduction of tumor growth and angiogenesis. CSF-1R signaling has been shown to play an important role in MDSC migration therefore, targeting this receptor together with other anti-angiogenic drugs, could be an effective strategy at combating tumor growth [34].
3.2. GM-CSF
Granulocyte macrophage colony stimulating factor (GM-CSF) was discovered as a growth factor capable of generating macrophages and granulocytes from bone marrow precursors in vitro. GM-CSF functions by binding to the high affinity GSF2R receptor, a hetrodimer composed of a unique α and a common β chain. Stimulation of the GM-CSF receptor leads to activation of a number of signals including Jak/Stat, MAPK and PI3K pathways [35, 36].
GM-CSF is required for maturation of alveolar macrophages and invariant natural killer T cells (iNKT) [37]. In addition, GM-CSF is also involved in the generation of dendritic cells that accumulate at sites of injury [38]. Studies using neutralizing antibodies against GM-CSF show that endogenous GM-CSF plays a central role in the recruitment of monocytes and granulocytes from the bone marrow to sites of inflammation [39]. Similarly, studies in murine atherosclerotic models have shown that GM-CSF is involved in the proliferation and migration of splenic monocyte precursors that contribute to the expansion of myeloid cells at sites of inflammation [40].
Under steady state conditions, the circulating levels of GM-CSF are low. But depending on the levels, GM-CSF can possess inhibitory or stimulatory effects on immune system [41]. At low concentrations, GM-CSF augments the immune response by enhancing antigen presentation by dendritic cells, whereas at high concentrations, GM-CSF leads to immune suppression due to the generation of MDSCs [42, 43].
Like CSF-1, GM-CSF is linked to tumor progression and metastasis. The presence of GM-CSF in tumor stroma is associated with development of MDSCs. There is a direct correlation between levels of GM-CSF in patients with head and neck cancers and numbers of circulating MDSCs [18]. Studies in Kras mouse models of pancreatic cancer have revealed that GM-CSF secreted by pancreatic ductal epithelial cells facilitates the recruitment of MDSCs leading to evasion of antitumor immunity by CD8+ T cells [44].
3.3. G-CSF
Granulocyte colony stimulating factor (G-CSF) is closely related in structure to GM-CSF that stimulates myeloid development. G-CSF plays a critical role during neutrophil development and in emergency granulopoiesis. Mice lacking G-CSF or G-CSF receptor (G-CSFR or CSF3R), are severely neutropenic and are prone to bacterial infection [45]. The G-CSFR is mainly expressed by cells of myeloid lineages. However, some non-hematopoietic cells, such as neuronal cells and cardiomyocytes located at the maternal fetal interface, also express this receptor [46];[47]. There are up to six different isoforms of G-CSFR which are generated due to differential splicing. Overexpression of some of these isoforms has been reported in certain myeloid leukemias [48];[49].
G-CSF is routinely administered to patients with neutropenia to boost the number of neutrophils. Also, it is used to mobilize HSC from BM into the periphery for transplantation purposes. In contrast, exogenous G–CSF can inhibit innate immune responses through the recruitment of MDSCs. Waight et al demonstrated a direct correlation between the level of G-CSF and the number of G-MDSC in tumor bearing mice. They further showed that abrogating G-CSF production using RNAi resulted in a reduced accumulation of G-MDSC that lead to an attenuation of tumor growth [50]. A recent study has shown that administration of G-CSF to mobilize stem cells is accompanied by an expansion of G-MDSC [51].
The intracellular domain of G-CSFR contains two domains referred to as box-1 and box-2. This region is critical for the binding of Jak kinases to the receptor [52]. The ligation of G-CSFR leads to the activation of numerous signaling pathways including the Jak/Stat pathway [53]. In myeloid cells, Stat expression leads to the activation of transcription factors like Myc and C/EBPβ that can promote MDSC development [54]. These studies suggest that high levels of G-CSF can hamper the innate response by promoting the expansion of MDSCs.
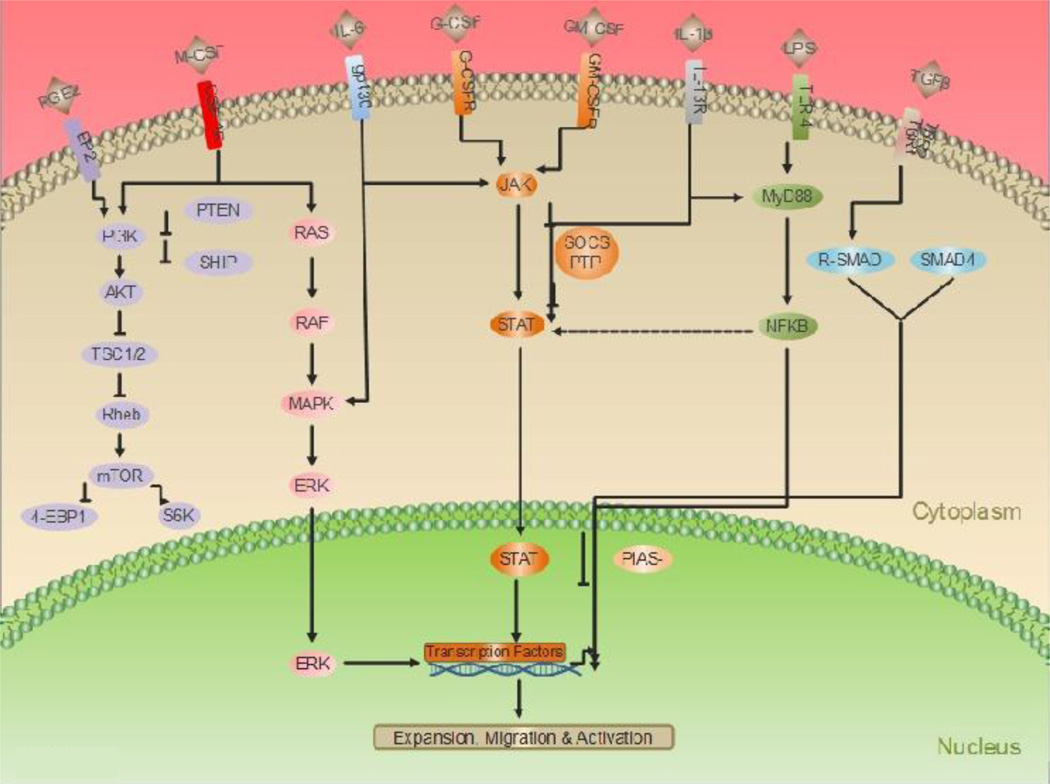
The three cytokines M-CSF, GM-CSF and G-CSF are intimately involved with the development of cells of the myeloid lineage. Phosphorylation of tyrosine residues on these receptors leads to the activation of several downstream signaling pathways. Principle among them are the Ras/Raf/MAPK, PI3K/Akt and Jak/Stat signaling pathways. These pathways are involved in the regulation of a variety of physiological processes in MDSCs (Figure 1).
Figure 1. Signaling pathways involved in regulation of MDSC function.
Cytokines produced by hematopoietic and cancer cells bind to their respective cognate receptors leading to phosphorylaton of key tyrosine residues leading to the activation of Ras/Mitogen activated protein kinase (Ras/MAPK), Phosphatidylinositol-3-kinase (PI3K), Janus kinase/Signal transducer activator of transcription (Jak/Stat) and Transforming growth factor β (TGFβ) pathways. This subsequently results in activation of transcription factors (TF). The TF’s then bind to their putative sites on gene promoters leading to activation of genes involved in proliferation, survival and migration of MDSCs. Transcription factor (TF), Stat binding sites (SBS), Smad binding elements (SBE). Macrophage colony stimulating factor (M-CSF), Granulocyte colony stimulating factor (G-CSF), Granulocyte macrophage colony stimulating factor (GM-CSF), Colony stimulating factor 1 receptor (CSF-1R), Granulocyte macrophage colony stimulating factor receptor (GM-CSFR), Granulocyte colony stimulating factor receptor (G-CSFR), Serine/threonine specific kinase (Akt), Suppressor of cytokine signaling (SOCS), Protein inhibitor of activated Stat (PIAS), Protein tyrosine phosphatase (PTP), Extracellular signal regulated kinase (ERK), Receptor regulated Smad (R-SMAD), Mammalian target of rapamycin (mTOR), Phosphatase with tensin homology (PTEN), Tuberous sclerosis complex (TSC), Ras homolog enriched in brain (Rheb), 4E binding protein-1 (4EBP1), Interleukin-6 (IL-6), Prostaglandin E2 (PGE2), Lipopolysaccharide (LPS), Interleukin-1 beta (IL-1b), Glycoprotein 130 (Gp130).
4. Signaling pathways in MDSC function
4.1. Ras signaling
The Ras family consists of low molecular weight proteins with intrinsic GTPase activity. In humans, the Ras gene encodes four distinct 21-kDa proteins: HRas, NRas, Kras4A and Kras4B [55]. The activation state of Ras is dependent on its association with GTP or GDP; when bound with GTP, they are active and can interact with other downstream targets. In contrast, when bound to GDP, they are inactive and fail to bind to other proteins. Under physiological conditions, these interactions are regulated by guanine nucleotide exchange factor (GEF) and GTPase activating proteins (GAP). GEF and GAP promote Ras activation by promoting the exchange of GDP for GTP and hydrolysis of GTP, respectively [56]. Mutations in Ras genes accounts for ~20% of all mutations found in human cancers [57]. These mutations in cancer cells promote cell proliferation, survival and migration thus enabling tumors to acquire a malignant phenotype.
Ras signaling also plays an important role during myeloid development. Transgenic mice overexpressing mutant Ras protein show alterations in macrophage proliferation and differentiation in response to growth factor stimulation. Similarly, hematopoietic cell lines can undergo differentiation into myeloid cells following overexpression of activated Ras [58]. Enhanced Ras signaling promotes granulopoiesis by increasing the binding of C/EBPα to the G-CSF receptor via phosphorylation of C/EBPα at serine 248 [59].
Ras can also promote tumor growth by altering the tumor microenvironment through cell autonomous and non-cell autonomous mechanisms. Oncogenic Ras promotes tumor angiogenesis through the activation of numerous signaling pathways that eventually lead to upregulation of pro-angiogenic factors like VEGF [60]. Evasion from the host antitumor response is another mechanism through which cancer cells can grow. Oncogenic Ras promotes tumor evasion by decreasing the expression of MHC molecules on the surface of cancer cells, which leads to reduced immunogenicity of Ras transformed cells [61];[62]. Studies using Ras driven tumor models have shown that these tumors can evade immune surveillance from the host. In a mouse model of pancreatic cancer, overexpression of constitutively active Kras resulted in the induction of cytokines like MIP-2 and MCP-1. This promoted the recruitment of macrophages and MDSCs into the tumor stroma resulting in a suppression of the anti-tumor response [63]. Similarly, in a Kras driven model of lung cancer, the immune response was significantly attenuated due to the presence of immunosuppressive cells or MDSCs in the tumor stroma [64].
Thus, overexpression of oncogenic Ras by cancer cells can help in the recruitment of MDSC into the TME, this could be an additional mechanism through which Ras can promote tumor growth.
4.2. PI3K signaling
PI3K catalyzes phosphorylation of the 3′OH on phosphotidylinositol in the plasma membrane. PI3K/Akt signaling affects cell growth, survival, migration, and cellular metabolism. Various signaling proteins including serine, threonine kinase, and G-binding protein have domains that specifically bind to phosphorylated phosphoinositols. In resting cells, these proteins are localized in cytoplasm but translocate to the plasma membrane in response to lipid phosphorylation. These activated proteins can initiate a variety of cellular processes, such as actin polymerization and assembly of signaling complexes [65].
Based on substrate specificities, PI3K has been divided into three families: class I, II and III. Class I consists of two subclasses, class IA and class IB PI3K. Class II PI3K consists of a single p110 subunit [66]. Class III PI3K consists of a single family member, VSp34. Vsp34 has been reported to be involved in autophagy and mTOR regulation [67]. Distinct members of PI3K are activated in the immune system based on cell type. Class IA PI3K can be activated by cytokines like IL-2, IL-6, IL-7, GM-CSF and interferons in dendritic and T cells. Cytokine receptors such as CSF-1R, expressed by macrophages and c-Kit receptors on mast cells, can regulate the expression of class IA PI3K [68]
Selective disruption of various PI3K isoforms has revealed that these enzymes are involved in chemotaxis and phagocytosis of leukocytes, including macrophages and neutrophils, in vivo. Leukocytes can phagocytize antibody coated cells via their FcR receptors. Class IA PI3K enzymes regulate phagocytosis by controlling the production of digestive enzymes and ROS. Class III PI3K isoforms are involved in targeting the phagosome to the membrane [69]. Yoo et al studied amoeboid like migration in neutrophils and dendritic cells using zebrafish embryos. This process involved coordinated protrusion and retraction of cells, which is observed during the migration of cells in response to chemoattractants. The cells lacking PI3Kγ isoform could not migrate and exhibited impaired polarization [70]
Generation of p110γ−/− mice further helped in elucidating the function of P13K in neutrophils. Mice lacking the PI3Kγ isoform were viable and had normal levels of macrophages and neutrophils in blood and other hematopoietic tissues. However, neutrophils isolated from the PI3Kγ deficient mice failed to activate protein kinase B and exhibited severe defects in migration and oxidative burst. Moreover, peritoneal macrophages isolated from these mice exhibited reduced migration in a mouse model of peritonitis [71]. Besides migration, PI3Kγ is also involved in the regulation of intrinsic pathways that control apoptosis in neutrophils. No PI3K activity could be detected from macrophages isolated from p110γ−/− after stimulation by a GPCR agonist. Moreover, the p110γ deficient neutrophils had high rates of apoptosis under steady state conditions and after LPS treatment. These neutrophils had low levels of Akt and other pro-apoptotic proteins like Bcl2, suggesting a role for p110γ in neutrophil survival in vivo [72];[73].
Given that PI3k is involved in regulating physiological functions in neutrophils, this would suggest that the PI3k pathway could modulate MDSC function. A study by Enioutina et al has suggested involvement of PI3K in the regulation of MDSC function in aging mice. They observed an accumulation of MDSCs in the BM and secondary lymphoid organs of aging mice. The molecular basis of this phenomenon was linked to a defect in the PI3K/AKT signaling pathway in MDSCs, which compromised the normal immune functions in aging mice and contributed to immune senescence [74].
In addition to PI3K, the SH2 domain containing-5-inositol phosphatase (SHIP) and phosphatase and tensin homolog (PTEN) are involved in the regulation of phosphoinoside metabolism in immune cells. SHIP and PTEN are negative regulators of the PI3K signaling pathway [75]. To ensure PI3K is appropriately suppressed, SHIP suppresses proliferation and survival of cells by translocating to the cell membrane following extracellular stimulation, resulting in the hydrolysis of PIP3 into PI3,4-P2 [76]. SHIP null mice have higher number of macrophages and monocytes due to the increased survival and proliferation of myeloid progenitors. Interestingly, peritoneal and alveolar macrophages from SHIP−/− mice have reduced NO production due to high arginase I activity, which competes with iNOS for the arginine substrate [77]. Furthermore, SHIP−/− mice show polarization of macrophages from M1 populations towards M2 populations. Studies from the Kyrstal group have demonstrated that macrophages from SHIP−/− mice are polarized towards an M2 phenotype due to high levels of PIP3 [78].
SHIP functions as a tumor suppressor in hematological malignancies in humans and its deficiency is associated with myleoproliferative diseases in mice. Cancer cells secrete factors that can downregulate SHIP expression, thereby contributing to the expansion of MDSCs. Moreover, there is an increase in the number of MDSCs in SHIP−/− tumor mice. Therefore, strategies aimed at increasing SHIP activity could be beneficial in treatment of cancer, as this would mitigate the immunosupressive affects of MDSCs [79].
Mammalian target of rapamycin or mTOR is another component of the PI3k/Akt pathway which is primarily involved in cell proliferation and nutrient sensing. Phosphorylation of S6k and 4EBP1 proteins by mTOR results in increased mRNA and protein synthesis, stimulating cell growth and proliferation [80]. There is evidence to suggest that the mTOR pathway regulates inflammatory responses by affecting NFkB and Stat3 activity in myeloid cells [81]. Recently, it has been shown that mTOR pathway is also involved in the differentiation of monocytes into TAM [82]. The ability of mTOR to affect myelopoiesis suggests that this pathway could be involved MDSC generation.
The role of PI3K signaling in the regulation of the physiological processes of macrophages and neutrophils is well established. Moreover, the PI3K pathway can control the expression of transcription factors and genes that are involved in proliferation and survival of MDSC’s. Based on the results from the above studies suggests, that PI3K could play a central role in regulation of in MDSC biology and strategies aimed at targeting the PI3K pathway could be an effective way of eliminating MDSCs in cancers.
4.3. JAK /STAT pathway
Inflammation plays a critical role during various stages of tumor progression. The Janus kinase/Signal transducer and activator of transcription (Jak/Stat) pathway has a central role in mediating inflammatory response. The mammalian Jak family consists of four members: Jak1, Jak2, Jak3 and tyrosine kinase2 (Tyk2). The Stat family is composed of seven family members: Stat1, Stat2, Stat3, Stat4, Stat5a, Stat5b and Stat6 [83]. Gene knockout studies have helped elucidate various components of this pathway, which was originally discovered as part of the IFN signaling system [84]. It was shown that Jak could be activated by IFNα/β and γ, and that Stat served as substrate for these kinases. Binding of cytokine to its receptor leads to receptor dimerization, and subsequent activation of Jak. Specific tyrosine residues on receptors are phosphorylated by Jak, and these receptors then serve as docking sites for the Stat family of transcriptional factors. Once phosphorylated by Jak, Stat translocates to the nucleus and activates genes by binding to putative sites on their promoters. Hamilton and colleagues showed that Jak phosphorylation and Stat activation occurs in macrophages following CSF-1 stimulation and that this effect was abrogated when a key tyrosine residue on the CSF-1R receptor was mutated [85, 86].
Studies from our group have shown that MDSCs can interfere with host immune responses in tumors by inhibiting immune cell responsiveness to IFNs. Patients with advanced cancers also exhibit reduced activation of IFN signaling pathways. In fact, the cytokine profile of GI cancer patients was associated with unique subsets of MDSC populations that impacted their cellular response to clinically relevant cytokines. We also observed an inverse correlation between the percentage CD15+ MDSCs and levels of STAT1 phosphorylation in CD4+ T cells. Furthermore, co-culture of in vitro generated MDSCs with CD4+ T cells isolated from normal donors also led to reduced IFN-responsiveness [87].
Similarly, our studies in mice have demonstrated that nitric oxide generated by MDSCs can lead to nitration of tyrosine residues, thereby impairing the ability of immune effector cells to release stimulatory signals. Using a mouse model of murine adenocarcinoma, an expansion of splenic Gr1+CD11b+ MDSCs was observed. Splenocytes isolated from these mice exhibited reduced phosphorylation of Stat1 in response to IFN-γ stimulation. Treatment of mice with gemcitabine, which leads to depletion of MDSCs, could restore IFN responsiveness. Furthermore, splenocytes isolated from iNOS−/− tumor bearing mice exhibited a significantly elevated IFN-response compared to the control mice. We demonstrated that MDSCs produce a variety of chemical compounds that can suppress immune response by interfering with interferon signaling in immune cells [88].
Although Stat molecules can be activated by a variety of cytokines, they nevertheless possess cytokine specificity. For example, Stat1 is activated by IFNα/β, and Stat6 is activated by IL-4 and IL-13. Stat proteins play a central role during tumor initiation, and later during maintenance. Stat3, together with NFkB, is activated in cancers that facilitate in transducing signals from extracellular stimuli. Stats function as transcriptional activators of genes involved in cell proliferation, survival, angiogenesis, and migration. Stat3 plays an important role in the development of pancreatic ductal carcinoma (PDAC), and ablation of Stat3 could reduce the incidence of pancreatic intraepithelial neoplasia (PIN) formation in mice [89, 90]. Studies by Lesina et al demonstrate that TAMs are major source of Stat3 activating cytokines during PDAC development [44]. Therefore, drugs that target Stat3, such as activating kinases like Jak2 or proinflammatory cytokines (IL-6), could have therapeutic potential. The role of Stat3 in the regulation of MDSC function is further supported by studies in head and neck cancer patients which show that Stat3 can control MDSC suppressive functions by regulating arginase 1 activity [91].
Activation of Stat3 in immune cells enables antitumor responses and promotes development and recruitment of TAMs and MDSCs. A number of studies have shown the role of Stat3 in expansion of MDSCs in mice. MDSCs isolated from tumor bearing mice showed high levels of activated Stat3, which led to an increase in the levels of the proangiogenic factor VEGF. Conversely, inhibition of Stat3 resulted in a reduction of tumor angiogenesis [92, 93]. In another study, inhibition of Stat3 by sutinib, a tyrosine kinase inhibitor, blocked expansion of MDSCs in tumor bearing mice [94]. Activation of Stat3 by API6 overexpression in myeloid cells leads to an expansion of MDSCs, resulting in inflammation and a higher incidence of adenocarcinomas in the lung. Similarly, overexpression of the constitutively active form of Stat3 in lung alveolar cells significantly enhances levels of MDSCs in the lungs, due to an increased level of MDSC-inducing cytokines like IL-6 and IL-1β. These studies suggest that persistent Stat3 activation promotes inflammation and tumor growth by promoting MDSC expansion [95].
Activation of Stat3 can also affect a number of downstream targets which include the proinflammatory proteins S100A8 and S100A9 [96]. Overexpression of S100A9 inhibits the differentiation of dendritic cells and macrophages, and promotes MDSC formation. In contrast, inhibition of S100A9 results in reduction of MDSCs in the spleens of tumor bearing mice. Although the precise mechanism of this phenomenon remains to be determined, it is postulated that the S100A8 and S100A9 hetrodimers assist in the formation of NADPH oxidase complex. This complex is responsible for the generation of ROS in myeloid cells, which can interfere with the differentiation of myeloid cells [97].
Stat3 also interacts with transcriptional factors like C/EBPβ that have a central role during myeloid development. Stat3 deficiency makes myeloid progenitors refractory to growth stimulation by G-CSF. Furthermore, Stat3 regulates Myc expression by increasing the duration of occupancy of C/EBPβ on the Myc promoter [54]. Recently, it has been shown that MDSCs isolated from cancer patients treated with amiloride, which inhibits exosome formation, had reduced suppressive functions [98, 99].
A recent study by the Abrams lab has helped in elucidating the role of IRF8 (interferon regulatory factor 8), a transcriptional factor that acts downstream of Stat 3 in the regulation of MDSC development. They demonstrated that overexpression of IRF8 in mice lead to a reduction in the number of tumor MDSC. In addition, they observed a reduction in the expression of IRF8 with a concomitant increase in the number of MDSC in breast cancer patients, suggesting that IFR8 is a negative regulator of MDSCs [100] .
Other Stat family members are also involved in the generation and development of MDSCs. Stat1 is involved in the activation of IFNγ and IL-1β. Stat1 deficient MDSCs fail to inhibit T cell activation due to their inability to upregulate iNOS and arginase activities. Binding of IL-4 and IL-13 to the CD124 receptor results in the activation of Stat6 in MDSCs which leads to the activation of TGFβ and arginase expression in these cells [101, 102].
There is experimental evidence that suggests a type 1 immune response is involved in tumor rejection [103]. Deletion of Stat6 in mice leads to a generation of a potent type 1 immune response that leads to tumor rejection. One of the underlying mechanisms for this rejection is the presence of reactive T cells that can recognize the Stat6 peptide presented by tumor cells. However, this study could not explicitly state whether the rejection of tumors was because of the presence of reactive T cells or due to polarization towards type-1 phenotype [104]. Studies from the Rosenberg laboratory further elucidated the mechanism through which Stat6 suppresses tumor growth. They demonstrated that removal of mammary tumors from Stat6−/− mice reduced the number of MDSCs resulting in a higher T cell response resulting in an attenuation of tumor growth. Moreover, the Stat6−/− tumor mice had higher numbers of nitric oxide producing M1 macrophages that slowed tumor growth [101].
The suppressor of cytokine signaling (SOCS) is a negative regulator of the Jak/Stat pathway [105]. Socs is also a major regulator of various cytokine mediated signaling pathways in macrophages and is a negative regulator of IL-6 signaling in macrophages. Macrophages isolated from Socs3 deficient mice showed prolonged activation of Stat1 and Stat3 after IL-6 stimulation. In contrast, Stat activation was normal in Socs3−/− macrophages following IL-6 stimulation [106]. Together, these studies highlight the role of Stat3 and related family members in the regulation of immunosuppressive properties of MDSC.
4.4. TGFβ pathway
TGFβ is a pleiotropic cytokine that affects cell proliferation, survival and migration. It also promotes the differentiation of various cell types, including immune cells during development and in disease conditions. TGFβ can function both as a positive or negative regulator of transcription depending on the target genes and the cellular context. TGFβ belongs to TGFβ super family. Mammals express three TGFβ isoforms: TGFβ1, TGFβ2 and TGFβ3, which are encoded by three different genes [107]. Proteolytic cleavage and/or interaction with integrins leads to the formation of latent TGFβ which then binds to its receptor. TGFβ activation leads to the assembly of a hetrotrimeric receptor complex, consisting of one type I and two type II receptors. In this complex, the type II receptor phosphorylates type I components, which helps propagate the signals. Upon phosphorylation by the receptor, R-Smads together with other Smad co-regulators like Smad4, translocate to the nucleus to activate transcription factors.
The generation of TGFβ1−/− mice helped establish the role of this growth factor during inflammation and in autoimmune diseases. TGFβ is a potent regulator of the inflammatory response which operates by affecting the activity of cells of the innate and adaptive immune system. The TGFβ receptor is expressed by most immune cells including T cells, NK cells and myeloid cells [108]. Early studies by Keller et al showed that TGFβ can act as a bifunctional growth regulator of hematopoietic cells. It can inhibit the growth of hematopoietic progenitor cells. TGFβ functions as a chemoattractant, facilitating the recruitment of macrophages to the site of inflammation. It also promotes polarization of macrophages from M1 towards the M2 phenotype. Treatment of macrophages with TGFβ can inhibit the expression of several inflammatory cytokines, as well as production of ROS in LPS activated macrophages in vitro [109]. Furthermore, TGFβ impairs the ability of macrophages to produce IL-12 and inhibits the expression of co-stimulatory molecule CD40, which leads to the inhibition of the antigen presentation capacity of macrophages [110].
The cells of the innate immune system are the principle source of TGFβ in tumors. TGFβ can attenuate NK cytolytic activity by inhibiting the production of IFNγ through transcriptional regulation of Smad3. Downregulation of NK2GD expression was observed in cancer patients with elevated levels of TGFβ [111]. An inverse correlation between MDSC number and loss of NK cytotoxic activity was observed in the livers of tumor bearing mice. NK2GD expression and IFNγ levels were also significantly reduced after incubation of NK cells with MDSCs. Membrane bound TGFβ1 on the surface of MDSC was critical in mediating this suppression [112]. Studies by Young et al demonstrated that immature myeloid cells (iMC) derived from tumor bearing mice produce TGFβ. The TGF together with NO produced by iMC, could inhibit T cell proliferation in vitro [113]. Blocking TGFβ or depleting MDSCs prevented tumor recurrence, demonstrating that IL-13 and CD4+ CDid restricted cells are required for the generation of TGFβ by MDSCs in vivo [114]. Studies by Yang et al helped to consolidate the role of TGFβ signaling in MDSC and its role in tumor progression. Ablation of type II TGFβ receptors in mammary cancer cells increased infiltration of MDSCs, promoting tumor growth and metastasis. They further demonstrated that MDSCs were a major source of TGFβ and metalloproteinases in the tumors that enhanced the invasive properties of cancer cells [115].
In addition, TGFβ can regulate MDSC function indirectly by altering microRNA (miR) expression. Analysis of MDSCs isolated from tumor bearing mice has revealed that TGFβ1 can regulate MDSC proliferation by inducing miR494 expression. Deletion of miR494 in MDSCs resulted in an attenuation of tumor growth and metastasis [116].
Tumor exosomes are small micro-vesicles secreted by cells, including tumor cells. They appear to be involved in cell communication and the transport of bioactive molecules. Exosomes are packed with a variety of immune-suppressive molecules like TGFβ and PGE2, which can inhibit immune responses by acting on various signaling pathways. MDSC tumor promoting properties were dependent on the presence of these molecules in tumor exosomes. Therefore, blocking exosomal TGFβ and PGE2 could reduce MDSC induction, delaying tumor growth [117].
MDSCs together with Tregs are the major suppressor cells in the tumor stroma that contribute to the suppression antitumor responses in cancer. Recent studies have helped elucidate the cross talk between these two immune suppressive cells. G-MDSCs isolated from tumor bearing mice were capable of inhibiting TGFβ1 induced polarization of naïve T cells into Tregs demonstrating that G-MDSC plays an important role in generation and expansion of Treg during tumor development [118].
4.5. PGE2 and Cox2 pathways
PGE2 is one of the best-characterized and studied isoforms of eicosanoids it possesses both proinflammatory and immunosuppressive properties. This eicosanoid is synthesized by Cox2, which converts arachidonic acid into prostaglandin H2 (PGH2) and prostaglandin synthase 1 (PGES1), which isomerizes PGH2 to PGE2. PGE2 signals through the PGE2 receptor E-prostanoid (EP) 4 was found to induce arginase 1 in MDSC [119, 120]. EP receptor agonists, including PGE2, induced the generation of MDSC from bone marrow stem cells, whereas receptor antagonists blocked differentiation. Kusmartsev et al showed that tumor derived factors could induce the expression of Cox2 and PGES1 in BM-derived MDSC. Moreover, this secretion correlated with arginase overexpression and phosphylation of Stat proteins in the MDSC a phenotype typically associated with MDSC suppressive activity. Cox2 inhibition resulted in the reversal of immune suppression and partially restores the differentiation of BM cells into myeloid DCs [121]
Furthermore, in vivo administration of the Cox2 inhibitor could significantly reduced MDSC accumulation in the Fas-over-expressing murine tumors [120]. BALB/c Ptger2 (EP2) knockout mice inoculated with 4T1 mammary carcinoma cells had delayed tumor growth and reduced numbers of MDSC, suggesting that PGE2 partially mediates MDSC induction through the EP2 receptor. Similarly, treatment of 4T1 tumor-bearing mice with the Cox2 inhibitor SC58236 delayed tumor growth and reduced MDSC accumulation [122].
The Cxcl12-Cxcr4 axis has been shown to a play a central role during tumor progression. Studies from the Kalinski lab have shown that inhibition of the Cox2 and PGE2 receptors EP2/EP4 on MDSC suppressed the expression of CxCr4 and resulted in reduced migration of MDSC in response to Cxcl12 in ovarian cancer. These results demonstrate the role of this signaling pathway in the regulation of MDSC function [123].
Tumor angiogenesis has been shown to correlate with the intratumoral concentration of prostaglandin. PGE2 can affect VEGF mobilization and the incorporation of fibroblast and endothelial cells into a tumor vessel through the binding of EP-2 receptors [124, 125]. MDSCs can regulate VEGF bioavailability though MMP9 expression and promote tumor angiogenesis. Together, these studies suggest that Cox2 and PGE2 represent key signaling pathways that are involved in the regulating of both MDSC function and differentiation.
4.6. IL-1β pathway
Tumor-derived IL-1β secreted into the tumor microenvironment has been shown to induce the accumulation of MDSC that promotes tumor growth. Elkabets et al found that the enhanced suppressive potential of IL-1β-induced MDSC was due to the activity of a novel subset of MDSC lacking Ly6C expression. This subset was present at low frequency in tumor-bearing mice in the absence of IL-1β-induced inflammation. However, under inflammatory conditions Ly6Cneg MDSC were predominant population and were able to impair NK cell development and functions [126]. Similarly, IL-1R-deficient mice exhibit a delayed accumulation of MDSCs, which can be partially restored by IL-6, indicating that IL-6 is a downstream mediator of the IL-1β-induced expansion of MDSCs [127].
IL-1beta transgenic mice develop gastric dysplasia that is accompanied by a marked increase in MDSCs in the stomach. IL-1β stimulation of MDSCs led to increased NF-κB activity both in vitro and in vivo. Tu et al further showed using an IL-1β;NF-κBEGFP mouse model that overexpression of IL-1β directly activated NF-κB in immune cells including MDSCs. Thereby linking IL-1β directly to NF-κB activation in MDSCs and to the downstream targets IL-6 and TNF-α. Suggesting that IL-1β activates MDSCs through IL-1RI/NF-kappaB signaling pathway and that antagonism of IL-1 receptor signaling can inhibit the development of gastric pre-neoplasia [128].
5. Conclusions
There is immense interest in elucidating the role of signaling pathways that are involved in the regulation of MDSC function. Most signaling pathways function by activating transcription factors and genes that are involved in cell proliferation, survival, and differentiation. Therefore, it is imperative to examine the role of cell cycle regulators during MDSC development and function. Our recent studies show that besides regulating cell proliferation in vitro, cell cycle regulators also play an essential role in the development of myeloid cells in vivo [129, 130]. Further evidence in support of this hypothesis has been demonstrated by the analysis of gene expression arrays from tumor MDSC that showed a significant alteration in expression of genes involved in cell cycle control.
These findings are further corroborated by recent findings from the Gabrilvoich lab that showed epigenetic silencing of Rb promotes the generation of G-MDSC from monocytes in cancer [131, 132]. This highlights the engagement of cell cycle genes in regulation of MDSC biology and myeloid development in vivo [129, 133, 134]. Therefore, strategies aimed at targeting signaling pathways should help in eliminating MDSCs from the tumor microenvironment, increasing the therapeutic potential of drugs and render immunotherapies more effective.
6. Summary
Myeloid cells, including MDSCs and TAMs, form a significant component of the tumor stroma. They promote tumor growth and metastasis through opposing mechanisms. TAMs secrete cytokines and proteases that facilitate tumor growth and angiogenesis. In contrast, MDSCs promote tumor growth by suppressing the function of immune cells. It is widely believed that perturbation of signaling pathways that are required for myeloid development leads to the generation of MDSC.
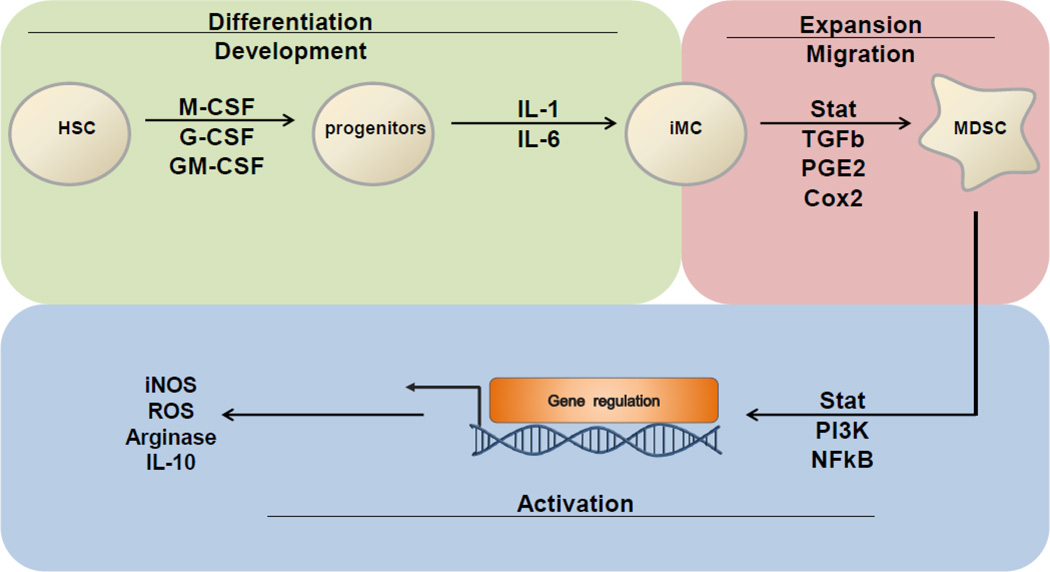
Ras/MAPK, PI3K/Akt, Jak/Stat, TGFb signaling pathways together with the cytokines M-CSF, GM-CSF and G-CSF are involved in the regulation various aspects of MDSC biology (Figure 2). Increased production of these cytokine during tumorogenesis can interfere with normal myeloid development in the bone marrow that leads to the generation and development of immature myeloid cells. Furthermore, cancer cells secrete factors like PGE2 and Cxcl12 that help in the recruitment of MDSC to the TME. Finally, the activation of downstream PI3K/Akt and Jak/Stat signaling pathways regulate the expression of genes like iNOS, IL-10 and arginase that are involved in mediating the immune suppressive function of MDSCs.
Figure 2. Role of signaling pathways involved in generation, expansion and functional regulation of MDSCs in cancer.
Cytokines like macrophage colony stimulating factors (M-CSF) and granulocyte macrophage colony stimulation factor (GM-CSF) are involved during normal myeloid development from hematopoietic stem cells (HSC). Increased production of these cytokines during tumorogenesis interferes with normal myeloid development resulting in the generation of immature myeloid cells (iMC). These iMC generally differentiate into macrophages and granulocytes. However, in presence of factors like IL-6 and IL-1β, iMC differentiate into myeloid derived suppressor cells (MDSCs). Furthermore, cancer cells secrete factors like PGE2 and Cxcl12 that help in the recruitment of MDSC to the TME. Finally, the activation of downstream PI3K/Akt and Jak/Stat signaling pathways regulate the expression of genes like iNOS, IL-10 and arginase that are involved in mediating the immune suppressive function of MDSCs.
Acknowledgement
We thank the members of Carson Lab for comments. and Ian Landi for his help with the graphic. This work is supported by NIH grant P01CA095426 to WEC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
Authors declare no conflict of interest.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17(3):320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9(3):162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 5.Kondo M, Scherer DC, Miyamoto T, King AG, Akashi K, Sugamura K, Weissman IL. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature. 2000;407(6802):383–386. doi: 10.1038/35030112. [DOI] [PubMed] [Google Scholar]
- 6.Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26(6):726–740. doi: 10.1016/j.immuni.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Semerad CL, Liu F, Gregory AD, Stumpf K, Link DC. G-CSF is an essential regulator of neutrophil trafficking from the bone marrow to the blood. Immunity. 2002;17(4):413–423. doi: 10.1016/s1074-7613(02)00424-7. [DOI] [PubMed] [Google Scholar]
- 8.Dang PM, Stensballe A, Boussetta T, Raad H, Dewas C, Kroviarski Y, Hayem G, Jensen ON, Gougerot-Pocidalo MA, El-Benna J. A specific p47phox -serine phosphorylated by convergent MAPKs mediates neutrophil NADPH oxidase priming at inflammatory sites. J Clin Invest. 2006;116(7):2033–2043. doi: 10.1172/JCI27544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177(10):7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 10.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 11.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 12.Condeelis J, Pollard J. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124(2):263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181(8):5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5(12):953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 15.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 16.Mantovani A, Allavena P, Sica A. Tumour-associated macrophages as a prototypic type II polarised phagocyte population: role in tumour progression. Eur J Cancer. 2004;40(11):1660–1667. doi: 10.1016/j.ejca.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176(1):287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pak AS, Wright MA, Matthews JP, Collins SL, Petruzzelli GJ, Young MR. Mechanisms of immune suppression in patients with head and neck cancer: presence of CD34(+) cells which suppress immune functions within cancers that secrete granulocyte-macrophage colony-stimulating factor. Clin Cancer Res. 1995;1(1):95–103. [PubMed] [Google Scholar]
- 19.Almand B, Clark JI, Nikitina E, van Beynen J, English NR, Knight SC, Carbone DP, Gabrilovich DI. Increased production of immature myeloid cells in cancer patients: a mechanism of immunosuppression in cancer. J Immunol. 2001;166(1):678–689. doi: 10.4049/jimmunol.166.1.678. [DOI] [PubMed] [Google Scholar]
- 20.Vuk-Pavlović S, Bulur PA, Lin Y, Qin R, Szumlanski CL, Zhao X, Dietz AB. Immunosuppressive CD14+HLA-DRlow/− monocytes in prostate cancer. Prostate. 2010;70(4):443–455. doi: 10.1002/pros.21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandruzzato S, Solito S, Falisi E, Francescato S, Chiarion-Sileni V, Mocellin S, Zanon A, Rossi CR, Nitti D, Bronte V, et al. IL4Ralpha+ myeloid-derived suppressor cell expansion in cancer patients. J Immunol. 2009;182(10):6562–6568. doi: 10.4049/jimmunol.0803831. [DOI] [PubMed] [Google Scholar]
- 22.Lechner MG, Liebertz DJ, Epstein AL. Characterization of cytokine-induced myeloid-derived suppressor cells from normal human peripheral blood mononuclear cells. J Immunol. 2010;185(4):2273–2284. doi: 10.4049/jimmunol.1000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherr C, Rettenmier C, Sacca R, Roussel M, Look A, Stanley E. The c-fms proto-oncogene product is related to the receptor for the mononuclear phagocyte growth factor, CSF-1. Cell. 1985;41(3):665–676. doi: 10.1016/s0092-8674(85)80047-7. [DOI] [PubMed] [Google Scholar]
- 24.Stanley E, Guilbert L, Tushinski R, Bartelmez S. CSF-1--a mononuclear phagocyte lineage-specific hemopoietic growth factor. J Cell Biochem. 1983;21(2):151–159. doi: 10.1002/jcb.240210206. [DOI] [PubMed] [Google Scholar]
- 25.Begg S, Radley J, Pollard J, Chisholm O, Stanley E, Bertoncello I. Delayed hematopoietic development in osteopetrotic (op/op) mice. J Exp Med. 1993;177(1):237–242. doi: 10.1084/jem.177.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei S, Dai XM, Stanley ER. Transgenic expression of CSF-1 in CSF-1 receptor-expressing cells leads to macrophage activation, osteoporosis, and early death. J Leukoc Biol. 2006;80(6):1445–1453. doi: 10.1189/jlb.0506304. [DOI] [PubMed] [Google Scholar]
- 27.Dai XM, Zong XH, Sylvestre V, Stanley ER. Incomplete restoration of colony-stimulating factor 1 (CSF-1) function in CSF-1-deficient Csf1op/Csf1op mice by transgenic expression of cell surface CSF-1. Blood. 2004;103(3):1114–1123. doi: 10.1182/blood-2003-08-2739. [DOI] [PubMed] [Google Scholar]
- 28.Irvine KM, Burns CJ, Wilks AF, Su S, Hume DA, Sweet MJ. A CSF-1 receptor kinase inhibitor targets effector functions and inhibits pro-inflammatory cytokine production from murine macrophage populations. FASEB J. 2006;20(11):1921–1923. doi: 10.1096/fj.06-5848fje. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Z, French DL, Ma G, Eisenstein S, Chen Y, Divino CM, Keller G, Chen SH, Pan PY. Development and function of myeloid-derived suppressor cells generated from mouse embryonic and hematopoietic stem cells. Stem Cells. 2010;28(3):620–632. doi: 10.1002/stem.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hidaka T, Akada S, Teranishi A, Morikawa H, Sato S, Yoshida Y, Yajima A, Yaegashi N, Okamura K, Saito S. Mirimostim (macrophage colony-stimulating factor; M-CSF) improves chemotherapy-induced impaired natural killer cell activity, Th1/Th2 balance, and granulocyte function. Cancer Sci. 2003;94(9):814–820. doi: 10.1111/j.1349-7006.2003.tb01524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jakubowski AA, Bajorin DF, Templeton MA, Chapman PB, Cody BV, Thaler H, Tao Y, Filippa DA, Williams L, Sherman ML, et al. Phase I study of continuous-infusion recombinant macrophage colony-stimulating factor in patients with metastatic melanoma. Clin Cancer Res. 1996;2(2):295–302. [PubMed] [Google Scholar]
- 32.Conway JG, McDonald B, Parham J, Keith B, Rusnak DW, Shaw E, Jansen M, Lin P, Payne A, Crosby RM, et al. Inhibition of colony-stimulating-factor-1 signaling in vivo with the orally bioavailable cFMS kinase inhibitor GW2580. Proc Natl Acad Sci U S A. 2005;102(44):16078–16083. doi: 10.1073/pnas.0502000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang B, Pan PY, Li Q, Sato AI, Levy DE, Bromberg J, Divino CM, Chen SH. Gr-1+CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res. 2006;66(2):1123–1131. doi: 10.1158/0008-5472.CAN-05-1299. [DOI] [PubMed] [Google Scholar]
- 34.Priceman SJ, Sung JL, Shaposhnik Z, Burton JB, Torres-Collado AX, Moughon DL, Johnson M, Lusis AJ, Cohen DA, Iruela-Arispe ML, et al. Targeting distinct tumor-infiltrating myeloid cells by inhibiting CSF-1 receptor: combating tumor evasion of antiangiogenic therapy. Blood. 2010;115(7):1461–1471. doi: 10.1182/blood-2009-08-237412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamilton JA. GM-CSF in inflammation and autoimmunity. Trends Immunol. 2002;23(8):403–408. doi: 10.1016/s1471-4906(02)02260-3. [DOI] [PubMed] [Google Scholar]
- 36.Hamilton JA, Achuthan A. Colony stimulating factors and myeloid cell biology in health and disease. Trends Immunol. 2013;34(2):81–89. doi: 10.1016/j.it.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 37.Bezbradica JS, Gordy LE, Stanic AK, Dragovic S, Hill T, Hawiger J, Unutmaz D, Van Kaer L, Joyce S. Granulocyte-macrophage colony-stimulating factor regulates effector differentiation of invariant natural killer T cells during thymic ontogeny. Immunity. 2006;25(3):487–497. doi: 10.1016/j.immuni.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 38.Vremec D, Lieschke GJ, Dunn AR, Robb L, Metcalf D, Shortman K. The influence of granulocyte/macrophage colony-stimulating factor on dendritic cell levels in mouse lymphoid organs. Eur J Immunol. 1997;27(1):40–44. doi: 10.1002/eji.1830270107. [DOI] [PubMed] [Google Scholar]
- 39.Cook AD, Turner AL, Braine EL, Pobjoy J, Lenzo JC, Hamilton JA. Regulation of systemic and local myeloid cell subpopulations by bone marrow cell-derived granulocyte-macrophage colony-stimulating factor in experimental inflammatory arthritis. Arthritis Rheum. 2011;63(8):2340–2351. doi: 10.1002/art.30354. [DOI] [PubMed] [Google Scholar]
- 40.Robbins CS, Chudnovskiy A, Rauch PJ, Figueiredo JL, Iwamoto Y, Gorbatov R, Etzrodt M, Weber GF, Ueno T, van Rooijen N, et al. Extramedullary hematopoiesis generates Ly-6C(high) monocytes that infiltrate atherosclerotic lesions. Circulation. 2012;125(2):364–374. doi: 10.1161/CIRCULATIONAHA.111.061986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen G, Hercus TR, McClure BJ, Stomski FC, Dottore M, Powell J, Ramshaw H, Woodcock JM, Xu Y, Guthridge M, et al. The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell. 2008;134(3):496–507. doi: 10.1016/j.cell.2008.05.053. [DOI] [PubMed] [Google Scholar]
- 42.Dranoff G, Jaffee E, Lazenby A, Golumbek P, Levitsky H, Brose K, Jackson V, Hamada H, Pardoll D, Mulligan RC. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc Natl Acad Sci U S A. 1993;90(8):3539–3543. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serafini P, Carbley R, Noonan KA, Tan G, Bronte V, Borrello I. High-dose granulocyte-macrophage colony-stimulating factor-producing vaccines impair the immune response through the recruitment of myeloid suppressor cells. Cancer Res. 2004;64(17):6337–6343. doi: 10.1158/0008-5472.CAN-04-0757. [DOI] [PubMed] [Google Scholar]
- 44.Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Klöppel G, Yoshimura A, Reindl W, Sipos B, Akira S, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19(4):456–469. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 45.Demetri GD, Griffin JD. Granulocyte colony-stimulating factor and its receptor. Blood. 1991;78(11):2791–2808. [PubMed] [Google Scholar]
- 46.McCracken S, Layton JE, Shorter SC, Starkey PM, Barlow DH, Mardon HJ. Expression of granulocyte-colony stimulating factor and its receptor is regulated during the development of the human placenta. J Endocrinol. 1996;149(2):249–258. doi: 10.1677/joe.0.1490249. [DOI] [PubMed] [Google Scholar]
- 47.Harada M, Qin Y, Takano H, Minamino T, Zou Y, Toko H, Ohtsuka M, Matsuura K, Sano M, Nishi J, et al. G-CSF prevents cardiac remodeling after myocardial infarction by activating the Jak-Stat pathway in cardiomyocytes. Nat Med. 2005;11(3):305–311. doi: 10.1038/nm1199. [DOI] [PubMed] [Google Scholar]
- 48.Larsen A, Davis T, Curtis BM, Gimpel S, Sims JE, Cosman D, Park L, Sorensen E, March CJ, Smith CA. Expression cloning of a human granulocyte colony-stimulating factor receptor: a structural mosaic of hematopoietin receptor, immunoglobulin, and fibronectin domains. J Exp Med. 1990;172(6):1559–1570. doi: 10.1084/jem.172.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong F, Brynes RK, Tidow N, Welte K, Löwenberg B, Touw IP. Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N Engl J Med. 1995;333(8):487–493. doi: 10.1056/NEJM199508243330804. [DOI] [PubMed] [Google Scholar]
- 50.Waight JD, Hu Q, Miller A, Liu S, Abrams SI. Tumor-derived G-CSF facilitates neoplastic growth through a granulocytic myeloid-derived suppressor cell-dependent mechanism. PLoS One. 2011;6(11):e27690. doi: 10.1371/journal.pone.0027690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luyckx A, Schouppe E, Rutgeerts O, Lenaerts C, Fevery S, Devos T, Dierickx D, Waer M, Van Ginderachter JA, Billiau AD. G-CSF stem cell mobilization in human donors induces polymorphonuclear and mononuclear myeloid-derived suppressor cells. Clin Immunol. 2012;143(1):83–87. doi: 10.1016/j.clim.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 52.Barge RM, de Koning JP, Pouwels K, Dong F, Löwenberg B, Touw IP. Tryptophan 650 of human granulocyte colony-stimulating factor (G-CSF) receptor, implicated in the activation of JAK2, is also required for G-CSF-mediated activation of signaling complexes of the p21ras route. Blood. 1996;87(6):2148–2153. [PubMed] [Google Scholar]
- 53.Nicholson SE, Oates AC, Harpur AG, Ziemiecki A, Wilks AF, Layton JE. Tyrosine kinase JAK1 is associated with the granulocyte-colony-stimulating factor receptor and both become tyrosine-phosphorylated after receptor activation. Proc Natl Acad Sci U S A. 1994;91(8):2985–2988. doi: 10.1073/pnas.91.8.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang H, Nguyen-Jackson H, Panopoulos AD, Li HS, Murray PJ, Watowich SS. STAT3 controls myeloid progenitor growth during emergency granulopoiesis. Blood. 2010;116(14):2462–2471. doi: 10.1182/blood-2009-12-259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 56.Scheffzek K, Ahmadian MR, Kabsch W, Wiesmüller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277(5324):333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 57.Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ, Vogelstein B. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327(6120):293–297. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- 58.Hibi S, Löhler J, Friel J, Stocking C, Ostertag W. Induction of monocytic differentiation and tumorigenicity by v-Ha-ras in differentiation arrested hematopoietic cells. Blood. 1993;81(7):1841–1848. [PubMed] [Google Scholar]
- 59.Behre G, Singh SM, Liu H, Bortolin LT, Christopeit M, Radomska HS, Rangatia J, Hiddemann W, Friedman AD, Tenen DG. Ras signaling enhances the activity of C/EBP alpha to induce granulocytic differentiation by phosphorylation of serine 248. J Biol Chem. 2002;277(29):26293–26299. doi: 10.1074/jbc.M202301200. [DOI] [PubMed] [Google Scholar]
- 60.Rak J, Mitsuhashi Y, Bayko L, Filmus J, Shirasawa S, Sasazuki T, Kerbel RS. Mutant ras oncogenes upregulate VEGF/VPF expression: implications for induction and inhibition of tumor angiogenesis. Cancer Res. 1995;55(20):4575–4580. [PubMed] [Google Scholar]
- 61.Lohmann S, Wollscheid U, Huber C, Seliger B. Multiple levels of MHC class I down-regulation by ras oncogenes. Scand J Immunol. 1996;43(5):537–544. doi: 10.1046/j.1365-3083.1996.d01-73.x. [DOI] [PubMed] [Google Scholar]
- 62.Seliger B, Harders C, Lohmann S, Momburg F, Urlinger S, Tampé R, Huber C. Down-regulation of the MHC class I antigen-processing machinery after oncogenic transformation of murine fibroblasts. Eur J Immunol. 1998;28(1):122–133. doi: 10.1002/(SICI)1521-4141(199801)28:01<122::AID-IMMU122>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 63.Clark CE, Hingorani SR, Mick R, Combs C, Tuveson DA, Vonderheide RH. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res. 2007;67(19):9518–9527. doi: 10.1158/0008-5472.CAN-07-0175. [DOI] [PubMed] [Google Scholar]
- 64.DuPage M, Cheung AF, Mazumdar C, Winslow MM, Bronson R, Schmidt LM, Crowley D, Chen J, Jacks T. Endogenous T cell responses to antigens expressed in lung adenocarcinomas delay malignant tumor progression. Cancer Cell. 2011;19(1):72–85. doi: 10.1016/j.ccr.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7(8):606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 66.Didichenko SA, Thelen M. Phosphatidylinositol 3-kinase c2alpha contains a nuclear localization sequence and associates with nuclear speckles. J Biol Chem. 2001;276(51):48135–48142. doi: 10.1074/jbc.M104610200. [DOI] [PubMed] [Google Scholar]
- 67.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280(38):33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 68.Okkenhaug K, Bilancio A, Farjot G, Priddle H, Sancho S, Peskett E, Pearce W, Meek SE, Salpekar A, Waterfield MD, et al. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 2002;297(5583):1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 69.Suire S, Hawkins P, Stephens L. Activation of phosphoinositide 3-kinase gamma by Ras. Curr Biol. 2002;12(13):1068–1075. doi: 10.1016/s0960-9822(02)00933-8. [DOI] [PubMed] [Google Scholar]
- 70.Yoo SK, Deng Q, Cavnar PJ, Wu YI, Hahn KM, Huttenlocher A. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev Cell. 2010;18(2):226–236. doi: 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hirsch E, Katanaev VL, Garlanda C, Azzolino O, Pirola L, Silengo L, Sozzani S, Mantovani A, Altruda F, Wymann MP. Central role for G protein-coupled phosphoinositide 3-kinase gamma in inflammation. Science. 2000;287(5455):1049–1053. doi: 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- 72.Lindemans CA, Coffer PJ. Regulation of granulocyte apoptosis by phosphatidylinositol 3-kinase. Biochem Soc Trans. 2004;32(Pt3):480–484. doi: 10.1042/BST0320480. [DOI] [PubMed] [Google Scholar]
- 73.Yang KY, Arcaroli J, Kupfner J, Pitts TM, Park JS, Strasshiem D, Perng RP, Abraham E. Involvement of phosphatidylinositol 3-kinase gamma in neutrophil apoptosis. Cell Signal. 2003;15(2):225–233. doi: 10.1016/s0898-6568(02)00063-3. [DOI] [PubMed] [Google Scholar]
- 74.Enioutina EY, Bareyan D, Daynes RA. A role for immature myeloid cells in immune senescence. J Immunol. 2011;186(2):697–707. doi: 10.4049/jimmunol.1002987. [DOI] [PubMed] [Google Scholar]
- 75.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6(3):184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 76.Damen JE, Ware MD, Kalesnikoff J, Hughes MR, Krystal G. SHIP's C-terminus is essential for its hydrolysis of PIP3 and inhibition of mast cell degranulation. Blood. 2001;97(5):1343–1351. doi: 10.1182/blood.v97.5.1343. [DOI] [PubMed] [Google Scholar]
- 77.Helgason CD, Damen JE, Rosten P, Grewal R, Sorensen P, Chappel SM, Borowski A, Jirik F, Krystal G, Humphries RK. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 1998;12(11):1610–1620. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rauh MJ, Ho V, Pereira C, Sham A, Sly LM, Lam V, Huxham L, Minchinton AI, Mui A, Krystal G. SHIP represses the generation of alternatively activated macrophages. Immunity. 2005;23(4):361–374. doi: 10.1016/j.immuni.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 79.Ghansah T. A novel strategy for modulation of MDSC to enhance cancer immunotherapy. Oncoimmunology. 2012;1(6):984–985. doi: 10.4161/onci.20201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ballou LM, Lin RZ. Rapamycin and mTOR kinase inhibitors. J Chem Biol. 2008;1(1–4):27–36. doi: 10.1007/s12154-008-0003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weichhart T, Costantino G, Poglitsch M, Rosner M, Zeyda M, Stuhlmeier KM, Kolbe T, Stulnig TM, Hörl WH, Hengstschläger M, et al. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity. 2008;29(4):565–577. doi: 10.1016/j.immuni.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 82.Chen W, Ma T, Shen XN, Xia XF, Xu GD, Bai XL, Liang TB. Macrophage-induced tumor angiogenesis is regulated by the TSC2-mTOR pathway. Cancer Res. 2012;72(6):1363–1372. doi: 10.1158/0008-5472.CAN-11-2684. [DOI] [PubMed] [Google Scholar]
- 83.Stark GR, Darnell JE. The JAK-STAT pathway at twenty. Immunity. 2012;36(4):503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Darnell JE, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264(5164):1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 85.Novak U, Harpur AG, Paradiso L, Kanagasundaram V, Jaworowski A, Wilks AF, Hamilton JA. Colony-stimulating factor 1-induced STAT1 and STAT3 activation is accompanied by phosphorylation of Tyk2 in macrophages and Tyk2 and JAK1 in fibroblasts. Blood. 1995;86(8):2948–2956. [PubMed] [Google Scholar]
- 86.Novak U, Ward AC, Hertzog PJ, Hamilton JA, Paradiso L. Aberrant activation of JAK/STAT pathway components in response to G-CSF, interferon-alpha/beta and interferon-gamma in NFS-60 cells. Growth Factors. 1996;13(3–4):251–260. doi: 10.3109/08977199609003226. [DOI] [PubMed] [Google Scholar]
- 87.Mundy-Bosse BL, Young GS, Bauer T, Binkley E, Bloomston M, Bill MA, Bekaii-Saab T, Carson WE, Lesinski GB. Distinct myeloid suppressor cell subsets correlate with plasma IL-6 and IL-10 and reduced interferon-alpha signaling in CD4+ T cells from patients with GI malignancy. Cancer Immunol Immunother. 2011;60(9):1269–1279. doi: 10.1007/s00262-011-1029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mundy-Bosse BL, Lesinski GB, Jaime-Ramirez AC, Benninger K, Khan M, Kuppusamy P, Guenterberg K, Kondadasula SV, Chaudhury AR, La Perle KM, et al. Myeloid-derived suppressor cell inhibition of the IFN response in tumor-bearing mice. Cancer Res. 2011;71(15):5101–5110. doi: 10.1158/0008-5472.CAN-10-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li N, Grivennikov SI, Karin M. The unholy trinity: inflammation, cytokines, and STAT3 shape the cancer microenvironment. Cancer Cell. 2011;19(4):429–431. doi: 10.1016/j.ccr.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fukuda A, Wang SC, Morris JP, Folias AE, Liou A, Kim GE, Akira S, Boucher KM, Firpo MA, Mulvihill SJ, et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19(4):441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vasquez-Dunddel D, Pan F, Zeng Q, Gorbounov M, Albesiano E, Fu J, Blosser RL, Tam AJ, Bruno T, Zhang H, et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest. 2013;123(4):1580–1589. doi: 10.1172/JCI60083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kujawski M, Kortylewski M, Lee H, Herrmann A, Kay H, Yu H. Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice. J Clin Invest. 2008;118(10):3367–3377. doi: 10.1172/JCI35213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mace TA, Bloomston M, Lesinski GB. Pancreatic cancer-associated stellate cells: A viable target for reducing immunosuppression in the tumor microenvironment. Oncoimmunology. 2013;2(7):e24891. doi: 10.4161/onci.24891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xin H, Zhang C, Herrmann A, Du Y, Figlin R, Yu H. Sunitinib inhibition of Stat3 induces renal cell carcinoma tumor cell apoptosis and reduces immunosuppressive cells. Cancer Res. 2009;69(6):2506–2513. doi: 10.1158/0008-5472.CAN-08-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu L, Du H, Li Y, Qu P, Yan C. Signal transducer and activator of transcription 3 (Stat3C) promotes myeloid-derived suppressor cell expansion and immune suppression during lung tumorigenesis. Am J Pathol. 2011;179(4):2131–2141. doi: 10.1016/j.ajpath.2011.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sinha P, Okoro C, Foell D, Freeze HH, Ostrand-Rosenberg S, Srikrishna G. Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J Immunol. 2008;181(7):4666–4675. doi: 10.4049/jimmunol.181.7.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng P, Corzo CA, Luetteke N, Yu B, Nagaraj S, Bui MM, Ortiz M, Nacken W, Sorg C, Vogl T, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. 2008;205(10):2235–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chalmin F, Mignot G, Ghiringhelli F. Myeloid-derived suppressor cells: a key player in cancer. Med Sci (Paris) 2010;26(6–7):576–579. doi: 10.1051/medsci/2010266-7576. [DOI] [PubMed] [Google Scholar]
- 99.Chalmin F, Ladoire S, Mignot G, Vincent J, Bruchard M, Remy-Martin JP, Boireau W, Rouleau A, Simon B, Lanneau D, et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest. 2010;120(2):457–471. doi: 10.1172/JCI40483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Waight JD, Netherby C, Hensen ML, Miller A, Hu Q, Liu S, Bogner PN, Farren MR, Lee KP, Liu K, et al. Myeloid-derived suppressor cell development is regulated by a STAT/IRF-8 axis. J Clin Invest. 2013;123(10):4464–4478. doi: 10.1172/JCI68189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sinha P, Clements VK, Ostrand-Rosenberg S. Reduction of myeloid-derived suppressor cells and induction of M1 macrophages facilitate the rejection of established metastatic disease. J Immunol. 2005;174(2):636–645. doi: 10.4049/jimmunol.174.2.636. [DOI] [PubMed] [Google Scholar]
- 102.Bronte V, Serafini P, De Santo C, Marigo I, Tosello V, Mazzoni A, Segal DM, Staib C, Lowel M, Sutter G, et al. IL-4-induced arginase 1 suppresses alloreactive T cells in tumor-bearing mice. J Immunol. 2003;170(1):270–278. doi: 10.4049/jimmunol.170.1.270. [DOI] [PubMed] [Google Scholar]
- 103.Kacha AK, Fallarino F, Markiewicz MA, Gajewski TF. Cutting edge: spontaneous rejection of poorly immunogenic P1.HTR tumors by Stat6-deficient mice. J Immunol. 2000;165(11):6024–6028. doi: 10.4049/jimmunol.165.11.6024. [DOI] [PubMed] [Google Scholar]
- 104.Jensen SM, Meijer SL, Kurt RA, Urba WJ, Hu HM, Fox BA. Regression of a mammary adenocarcinoma in STAT6−/− mice is dependent on the presence of STAT6-reactive T cells. J Immunol. 2003;170(4):2014–2021. doi: 10.4049/jimmunol.170.4.2014. [DOI] [PubMed] [Google Scholar]
- 105.Alexander WS. Suppressors of cytokine signalling (SOCS) in the immune system. Nat Rev Immunol. 2002;2(6):410–416. doi: 10.1038/nri818. [DOI] [PubMed] [Google Scholar]
- 106.Croker BA, Krebs DL, Zhang JG, Wormald S, Willson TA, Stanley EG, Robb L, Greenhalgh CJ, Förster I, Clausen BE, et al. SOCS3 negatively regulates IL-6 signaling in vivo. Nat Immunol. 2003;4(6):540–545. doi: 10.1038/ni931. [DOI] [PubMed] [Google Scholar]
- 107.Wahl SM, Wen J, Moutsopoulos N. TGF-beta: a mobile purveyor of immune privilege. Immunol Rev. 2006;213:213–227. doi: 10.1111/j.1600-065X.2006.00437.x. [DOI] [PubMed] [Google Scholar]
- 108.Ghiringhelli F, Ménard C, Terme M, Flament C, Taieb J, Chaput N, Puig PE, Novault S, Escudier B, Vivier E, et al. CD4+CD25+ regulatory T cells inhibit natural killer cell functions in a transforming growth factor-beta-dependent manner. J Exp Med. 2005;202(8):1075–1085. doi: 10.1084/jem.20051511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tsunawaki S, Sporn M, Ding A, Nathan C. Deactivation of macrophages by transforming growth factor-beta. Nature. 1988;334(6179):260–262. doi: 10.1038/334260a0. [DOI] [PubMed] [Google Scholar]
- 110.Kosiewicz MM, Alard P, Streilein JW. Alterations in cytokine production following intraocular injection of soluble protein antigen: impairment in IFN-gamma and induction of TGF-beta and IL-4 production. J Immunol. 1998;161(10):5382–5390. [PubMed] [Google Scholar]
- 111.Lee JC, Lee KM, Kim DW, Heo DS. Elevated TGF-beta1 secretion and down-modulation of NKG2D underlies impaired NK cytotoxicity in cancer patients. J Immunol. 2004;172(12):7335–7340. doi: 10.4049/jimmunol.172.12.7335. [DOI] [PubMed] [Google Scholar]
- 112.Li H, Han Y, Guo Q, Zhang M, Cao X. Cancer-expanded myeloid-derived suppressor cells induce anergy of NK cells through membrane-bound TGF-beta 1. J Immunol. 2009;182(1):240–249. doi: 10.4049/jimmunol.182.1.240. [DOI] [PubMed] [Google Scholar]
- 113.Young MR, Wright MA, Matthews JP, Malik I, Prechel M. Suppression of T cell proliferation by tumor-induced granulocyte-macrophage progenitor cells producing transforming growth factor-beta and nitric oxide. J Immunol. 1996;156(5):1916–1922. [PubMed] [Google Scholar]
- 114.Terabe M, Matsui S, Park JM, Mamura M, Noben-Trauth N, Donaldson DD, Chen W, Wahl SM, Ledbetter S, Pratt B, et al. Transforming growth factor-beta production and myeloid cells are an effector mechanism through which CD1d-restricted T cells block cytotoxic T lymphocyte-mediated tumor immunosurveillance: abrogation prevents tumor recurrence. J Exp Med. 2003;198(11):1741–1752. doi: 10.1084/jem.20022227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, Carbone DP, Matrisian LM, Richmond A, Lin PC, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13(1):23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu Y, Lai L, Chen Q, Song Y, Xu S, Ma F, Wang X, Wang J, Yu H, Cao X, et al. MicroRNA-494 is required for the accumulation and functions of tumor-expanded myeloid-derived suppressor cells via targeting of PTEN. J Immunol. 2012;188(11):5500–5510. doi: 10.4049/jimmunol.1103505. [DOI] [PubMed] [Google Scholar]
- 117.Xiang X, Poliakov A, Liu C, Liu Y, Deng ZB, Wang J, Cheng Z, Shah SV, Wang GJ, Zhang L, et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int J Cancer. 2009;124(11):2621–2633. doi: 10.1002/ijc.24249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Centuori SM, Trad M, LaCasse CJ, Alizadeh D, Larmonier CB, Hanke NT, Kartchner J, Janikashvili N, Bonnotte B, Larmonier N, et al. Myeloid-derived suppressor cells from tumor-bearing mice impair TGF-β-induced differentiation of CD4+CD25+FoxP3+ Tregs from CD4+CD25−FoxP3− T cells. J Leukoc Biol. 2012;92(5):987–997. doi: 10.1189/jlb.0911465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rodriguez PC, Hernandez CP, Quiceno D, Dubinett SM, Zabaleta J, Ochoa JB, Gilbert J, Ochoa AC. Arginase I in myeloid suppressor cells is induced by COX-2 in lung carcinoma. J Exp Med. 2005;202(7):931–939. doi: 10.1084/jem.20050715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Y, Liu Q, Zhang M, Yu Y, Liu X, Cao X. Fas signal promotes lung cancer growth by recruiting myeloid-derived suppressor cells via cancer cell-derived PGE2. J Immunol. 2009;182(6):3801–3808. doi: 10.4049/jimmunol.0801548. [DOI] [PubMed] [Google Scholar]
- 121.Eruslanov E, Daurkin I, Ortiz J, Vieweg J, Kusmartsev S. Pivotal Advance: Tumor-mediated induction of myeloid-derived suppressor cells and M2-polarized macrophages by altering intracellular PGE2 catabolism in myeloid cells. J Leukoc Biol. 2010;88(5):839–848. doi: 10.1189/jlb.1209821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sinha P, Clements VK, Fulton AM, Ostrand-Rosenberg S. Prostaglandin E2 promotes tumor progression by inducing myeloid-derived suppressor cells. Cancer Res. 2007;67(9):4507–4513. doi: 10.1158/0008-5472.CAN-06-4174. [DOI] [PubMed] [Google Scholar]
- 123.Obermajer N, Muthuswamy R, Odunsi K, Edwards RP, Kalinski P. PGE(2)-induced CXCL12 production and CXCR4 expression controls the accumulation of human MDSCs in ovarian cancer environment. Cancer Res. 2011;71(24):7463–7470. doi: 10.1158/0008-5472.CAN-11-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Legler DF, Bruckner M, Uetz-von Allmen E, Krause P. Prostaglandin E2 at new glance: novel insights in functional diversity offer therapeutic chances. Int J Biochem Cell Biol. 2010;42(2):198–201. doi: 10.1016/j.biocel.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 125.Serafini P, Meckel K, Kelso M, Noonan K, Califano J, Koch W, Dolcetti L, Bronte V, Borrello I. Phosphodiesterase-5 inhibition augments endogenous antitumor immunity by reducing myeloid-derived suppressor cell function. J Exp Med. 2006;203(12):2691–2702. doi: 10.1084/jem.20061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Elkabets M, Ribeiro VS, Dinarello CA, Ostrand-Rosenberg S, Di Santo JP, Apte RN, Vosshenrich CA. IL-1β regulates a novel myeloid-derived suppressor cell subset that impairs NK cell development and function. Eur J Immunol. 2010;40(12):3347–3357. doi: 10.1002/eji.201041037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bunt SK, Yang L, Sinha P, Clements VK, Leips J, Ostrand-Rosenberg S. Reduced inflammation in the tumor microenvironment delays the accumulation of myeloid-derived suppressor cells and limits tumor progression. Cancer Res. 2007;67(20):10019–10026. doi: 10.1158/0008-5472.CAN-07-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tu S, Bhagat G, Cui G, Takaishi S, Kurt-Jones EA, Rickman B, Betz KS, Penz-Oesterreicher M, Bjorkdahl O, Fox JG, et al. Overexpression of interleukin-1beta induces gastric inflammation and cancer and mobilizes myeloid-derived suppressor cells in mice. Cancer Cell. 2008;14(5):408–419. doi: 10.1016/j.ccr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Trikha P, Sharma N, Opavsky R, Reyes A, Pena C, Ostrowski MC, Roussel MF, Leone G. E2f1-3 Are Critical for Myeloid Development. Journal of Biological Chemistry. 2011;286(6):4783–4795. doi: 10.1074/jbc.M110.182733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chong J-L, Wenzel PL, Saenz-Robles MT, Nair V, Ferrey A, Hagan JP, Gomez YM, Sharma N, Chen H-Z, Ouseph M, et al. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 2009;462(7275):930–934. doi: 10.1038/nature08677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Youn JI, Kumar V, Collazo M, Nefedova Y, Condamine T, Cheng P, Villagra A, Antonia S, McCaffrey JC, Fishman M, et al. Epigenetic silencing of retinoblastoma gene regulates pathologic differentiation of myeloid cells in cancer. Nat Immunol. 2013;14(3):211–220. doi: 10.1038/ni.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Youn JI, Gabrilovich DI. New roles of Rb1 in expansion of MDSCs in cancer. Cell Cycle. 2013;12(9):1329–1330. doi: 10.4161/cc.24577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Walkley C, Shea J, Sims N, Purton L, Orkin S. Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell. 2007;129(6):1081–1095. doi: 10.1016/j.cell.2007.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Viatour P, Somervaille T, Venkatasubrahmanyam S, Kogan S, McLaughlin M, Weissman I, Butte A, Passegué E, Sage J. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell. 2008;3(4):416–428. doi: 10.1016/j.stem.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Filipazzi P, Valenti R, Huber V, Pilla L, Canese P, Iero M, Castelli C, Mariani L, Parmiani G, Rivoltini L. Identification of a new subset of myeloid suppressor cells in peripheral blood of melanoma patients with modulation by a granulocyte-macrophage colony-stimulation factor-based antitumor vaccine. J Clin Oncol. 2007;25(18):2546–2553. doi: 10.1200/JCO.2006.08.5829. [DOI] [PubMed] [Google Scholar]
- 136.Poschke I, Mougiakakos D, Hansson J, Masucci GV, Kiessling R. Immature immunosuppressive CD14+HLA−DR−/low cells in melanoma patients are Stat3hi and overexpress CD80, CD83, and DC-sign. Cancer Res. 2010;70(11):4335–4345. doi: 10.1158/0008-5472.CAN-09-3767. [DOI] [PubMed] [Google Scholar]
- 137.Diaz-Montero CM, Salem ML, Nishimura MI, Garrett-Mayer E, Cole DJ, Montero AJ. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol Immunother. 2009;58(1):49–59. doi: 10.1007/s00262-008-0523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kotsakis A, Harasymczuk M, Schilling B, Georgoulias V, Argiris A, Whiteside TL. Myeloid-derived suppressor cell measurements in fresh and cryopreserved blood samples. J Immunol Methods. 2012;381(1–2):14–22. doi: 10.1016/j.jim.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schmielau J, Finn OJ. Activated granulocytes and granulocyte-derived hydrogen peroxide are the underlying mechanism of suppression of t-cell function in advanced cancer patients. Cancer Res. 2001;61(12):4756–4760. [PubMed] [Google Scholar]
- 140.Gowda M, Godder K, Kmieciak M, Worschech A, Ascierto ML, Wang E, Marincola FM, Manjili MH. Distinct signatures of the immune responses in low risk versus high risk neuroblastoma. J Transl Med. 2011;9:170. doi: 10.1186/1479-5876-9-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Raychaudhuri B, Rayman P, Ireland J, Ko J, Rini B, Borden EC, Garcia J, Vogelbaum MA, Finke J. Myeloid-derived suppressor cell accumulation and function in patients with newly diagnosed glioblastoma. Neuro Oncol. 2011;13(6):591–599. doi: 10.1093/neuonc/nor042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Corzo CA, Cotter MJ, Cheng P, Cheng F, Kusmartsev S, Sotomayor E, Padhya T, McCaffrey TV, McCaffrey JC, Gabrilovich DI. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J Immunol. 2009;182(9):5693–5701. doi: 10.4049/jimmunol.0900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Brandau S, Trellakis S, Bruderek K, Schmaltz D, Steller G, Elian M, Suttmann H, Schenck M, Welling J, Zabel P, et al. Myeloid-derived suppressor cells in the peripheral blood of cancer patients contain a subset of immature neutrophils with impaired migratory properties. J Leukoc Biol. 2011;89(2):311–317. doi: 10.1189/jlb.0310162. [DOI] [PubMed] [Google Scholar]
- 144.Liu CY, Wang YM, Wang CL, Feng PH, Ko HW, Liu YH, Wu YC, Chu Y, Chung FT, Kuo CH, et al. Population alterations of L-arginase- and inducible nitric oxide synthase-expressed CD11b+/CD14−/CD15+/CD33+ myeloid-derived suppressor cells and CD8+ T lymphocytes in patients with advanced-stage non-small cell lung cancer. J Cancer Res Clin Oncol. 2010;136(1):35–45. doi: 10.1007/s00432-009-0634-0. [DOI] [PubMed] [Google Scholar]
- 145.Srivastava MK, Bosch JJ, Thompson JA, Ksander BR, Edelman MJ, Ostrand-Rosenberg S. Lung cancer patients' CD4(+) T cells are activated in vitro by MHC II cell-based vaccines despite the presence of myeloid-derived suppressor cells. Cancer Immunol Immunother. 2008;57(10):1493–1504. doi: 10.1007/s00262-008-0490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hoechst B, Ormandy LA, Ballmaier M, Lehner F, Krüger C, Manns MP, Greten TF, Korangy F. A new population of myeloid-derived suppressor cells in hepatocellular carcinoma patients induces CD4(+)CD25(+)Foxp3(+) T cells. Gastroenterology. 2008;135(1):234–243. doi: 10.1053/j.gastro.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 147.Zhao F, Hoechst B, Duffy A, Gamrekelashvili J, Fioravanti S, Manns MP, Greten TF, Korangy F. S100A9 a new marker for monocytic human myeloid-derived suppressor cells. Immunology. 2012;136(2):176–183. doi: 10.1111/j.1365-2567.2012.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sanford DE, Porembka MR, Panni RZ, Mitchem JB, Belt BA, Plambeck-Suess SM, Lin G, Denardo DG, Fields RC, Hawkins WG, et al. A Study of Zoledronic Acid as Neo-Adjuvant, Perioperative Therapy in Patients with Resectable Pancreatic Ductal Adenocarcinoma. J Cancer Ther. 2013;4(3):797–803. doi: 10.4236/jct.2013.43096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Walter S, Weinschenk T, Stenzl A, Zdrojowy R, Pluzanska A, Szczylik C, Staehler M, Brugger W, Dietrich PY, Mendrzyk R, et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med. 2012 doi: 10.1038/nm.2883. [DOI] [PubMed] [Google Scholar]
- 150.Mirza N, Fishman M, Fricke I, Dunn M, Neuger AM, Frost TJ, Lush RM, Antonia S, Gabrilovich DI. All-trans-retinoic acid improves differentiation of myeloid cells and immune response in cancer patients. Cancer Res. 2006;66(18):9299–9307. doi: 10.1158/0008-5472.CAN-06-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kusmartsev S, Su Z, Heiser A, Dannull J, Eruslanov E, Kübler H, Yancey D, Dahm P, Vieweg J. Reversal of myeloid cell-mediated immunosuppression in patients with metastatic renal cell carcinoma. Clin Cancer Res. 2008;14(24):8270–8278. doi: 10.1158/1078-0432.CCR-08-0165. [DOI] [PubMed] [Google Scholar]
- 152.Ko JS, Zea AH, Rini BI, Ireland JL, Elson P, Cohen P, Golshayan A, Rayman PA, Wood L, Garcia J, et al. Sunitinib mediates reversal of myeloid-derived suppressor cell accumulation in renal cell carcinoma patients. Clin Cancer Res. 2009;15(6):2148–2157. doi: 10.1158/1078-0432.CCR-08-1332. [DOI] [PubMed] [Google Scholar]
- 153.Zea AH, Rodriguez PC, Atkins MB, Hernandez C, Signoretti S, Zabaleta J, McDermott D, Quiceno D, Youmans A, O'Neill A, et al. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res. 2005;65(8):3044–3048. doi: 10.1158/0008-5472.CAN-04-4505. [DOI] [PubMed] [Google Scholar]
- 154.Rodriguez PC, Ernstoff MS, Hernandez C, Atkins M, Zabaleta J, Sierra R, Ochoa AC. Arginase I-producing myeloid-derived suppressor cells in renal cell carcinoma are a subpopulation of activated granulocytes. Cancer Res. 2009;69(4):1553–1560. doi: 10.1158/0008-5472.CAN-08-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Finke JH, Rini B, Ireland J, Rayman P, Richmond A, Golshayan A, Wood L, Elson P, Garcia J, Dreicer R, et al. Sunitinib reverses type-1 immune suppression and decreases T-regulatory cells in renal cell carcinoma patients. Clin Cancer Res. 2008;14(20):6674–6682. doi: 10.1158/1078-0432.CCR-07-5212. [DOI] [PubMed] [Google Scholar]
- 156.van Cruijsen H, van der Veldt AA, Vroling L, Oosterhoff D, Broxterman HJ, Scheper RJ, Giaccone G, Haanen JB, van den Eertwegh AJ, Boven E, et al. Sunitinib-induced myeloid lineage redistribution in renal cell cancer patients: CD1c+ dendritic cell frequency predicts progression-free survival. Clin Cancer Res. 2008;14(18):5884–5892. doi: 10.1158/1078-0432.CCR-08-0656. [DOI] [PubMed] [Google Scholar]