Abstract
In mammals, insulin signaling regulates glucose homeostasis and plays an essential role in metabolism, organ growth, development, fertility, and lifespan. Defects in this signaling pathway contribute to various metabolic diseases such as type 2 diabetes, polycystic ovarian disease, hypertension, hyperlipidemia, and atherosclerosis. However, reducing the insulin signaling pathway has been found to increase longevity and delay the aging-associated diseases in various animals, ranging from nematodes to mice. These seemly paradoxical findings raise an interesting question as to how modulation of the insulin signaling pathway could be an effective approach to improve metabolism and aging. In this review, we summarize current understanding on tissue-specific functions of insulin signaling in the regulation of metabolism and lifespan. We also discuss potential benefits and limitations in modulating tissue-specific insulin signaling pathway to improve metabolism and healthspan.
Keywords: insulin signaling, tissue-specific, metabolism, aging
I. Introduction
Insulin is synthesized and secreted from pancreatic β cells in response to postprandial nutrient influx [1]. By suppressing glucose production in the liver and stimulating glucose uptake in muscle and fat, insulin reduces blood glucose levels to maintain glucose homeostasis in humans and animals. Insulin also regulates many important anabolic processes such as facilitating protein and glycogen synthesis in muscle and liver, promoting lipid synthesis and storage in liver and fat, as well as inhibiting fatty acid oxidation, glycogenolysis, and gluconeogenesis in insulin responsive tissues.
Great progress has been made during the past 3 decades on the mechanisms regulating the insulin signaling cascade in various species. Pharmacological or genetic manipulations of key components in the insulin signaling pathway have shown that defects in insulin signaling result in insulin resistance and contribute to metabolic dysfunctions and cardiovascular diseases [2]. However, recent studies have found that reducing or disrupting insulin signaling improves health-span and longevity in diverse model organisms such as yeast, worms, flies, and mammals [3-5]. These paradoxical findings raise an interesting question as to how the beneficial effects of insulin on energy homeostasis and longevity are achieved. In this review, we summarize recent progress on the mechanisms regulating the insulin signaling pathway. We also discuss tissue-specific functions of the insulin signaling pathway in the regulation of metabolism and/or longevity.
II. Insulin signaling and regulation
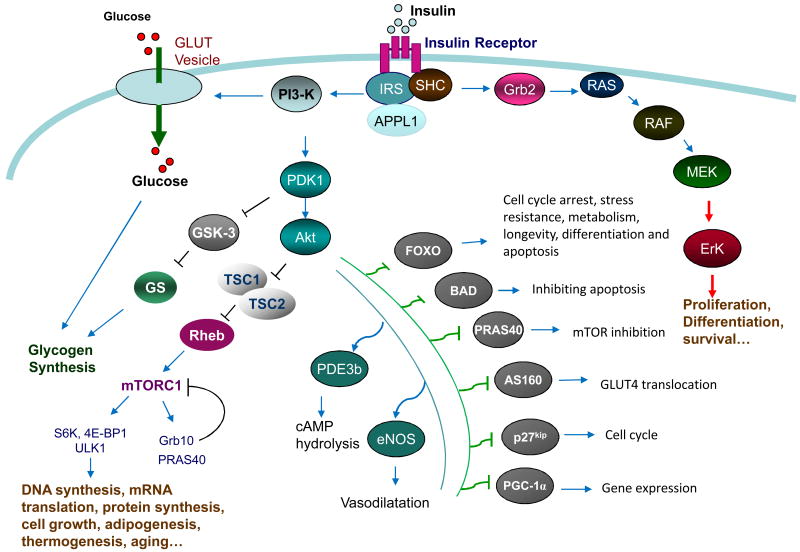
Insulin exerts its function by binding to its receptor on the cell membrane. The insulin receptor (IR) is a heterotetrameric transmembrane protein consisting of two extracellular α-subunits and two transmembrane β-subunits. The binding of insulin to the IR leads to the activation of IR tyrosine kinase and subsequent tyrosine phosphorylation of the β-subunits, the latter functions as docking sites for tyrosine phosphorylated adaptor proteins such as insulin receptor substrates 1 and 2 (IRS1/2). While the binding of IRS1/2 to IR is essential for regulating insulin signaling and function, how IRS1/2 is recruited to IR remains unclear. Very recently, Ryu and colleagues found that the interaction between IRS1/2 with IR is mediated by APPL1, an adaptor protein containing multiple function domains including the Bin1/amphiphysin/rvs167 (BAR) domain, pleckstrin homology (PH) domain, phosphotyrosine binding (PTB) domain, and coil coiled motif [6]. Under basal conditions, APPL1 forms a complex with IRS1/2 and Akt in the cytosol. In response to insulin and adiponectin stimulation, APPL1 is phosphorylated at Ser401 and this phosphorylation promotes APPL1 binding directly to the IR, allowing APPL1 to piggyback IRS proteins onto the IR. Insulin stimulation induces the dissociation of IRS proteins and Akt from the APPL1-IR complex, facilitating the binding of IRS1/2 to the activated IR near the plasma membrane [6]. The recruitment of IRS1/2 to IR leads to the activation of two major downstream signaling pathways, the phosphatidylinositol-3-kinase (PI3K) pathway and the mitogen-activated protein kinase (MAPK) pathway (Fig. 1). Activation of the PI3K signaling pathway promotes 3-phosphoinositide-dependent protein kinase 1 (PDK1)-dependent phosphorylation and activation of protein kinase B (PKB or Akt), which in turn phosphorylates a number of important downstream effectors, including glycogen synthase kinase (GSK)-3, Forkhead box protein O (FOXO), and mechanistic target of rapamycin (mTOR). Activation of the MAPK cascade, which consists of Raf, MEK, and ERK1/2, plays a critical role for insulin to regulate mitogenic events such as cell proliferation, differentiation, and survival (Fig. 1 and [7]). Insulin signaling is negatively regulated by a number of mechanisms, such as the binding of the adaptor protein Grb10 to the kinase domain of IR [8, 9], serine phosphorylation of IRS1 [10, 11], and dephosphorylation of IR and IRS1/2 by protein tyrosine phosphatase-1B (PTP-1B) [12].
Figure 1. Insulin signaling and function.
The binding of insulin to its receptor on cell membrane results in insulin receptor (IR) tyrosine kinase activation and IR tyrosine phosphorylation. APPL1 functions as a piggyback protein that promotes IRS binding to the tyrosine phosphorylated IR, leading to IRS tyrosine phosphorylation and subsequent activation of the PI3K/PDK1/Akt signaling pathway. Activation of Akt promotes the phosphorylation and inhibition of TSC1/2, a negative regulator of the mTORC1 signaling pathway. Akt also phosphorylates many other cellular proteins and plays key roles in various cellular events. Tyrosine phosphorylation of IR promotes the association of the adaptor proteins Shc and Grb2 to the IR, leading to the activation of the RAS-RAF-MEK-ERK1/2 cascade, which is essential for cell growth, differentiation and protein synthesis.
III. Tissue-specific function of insulin signaling in metabolism
1. Insulin suppresses glucose production in the liver
Insulin activates the insulin receptor in the liver, which phosphorylates IRS1 and IRS2, leading to activation of PI3K and ultimately Akt2. Activation of Akt2 promotes glycogen synthesis and inhibits gluconeogenesis and glucose production. One of the most important functions of insulin in liver is to suppress hepatic glucose production (HGP) when serum glucose levels are high, such as after a meal. Insulin suppresses HGP by inhibiting gluconeogenic enzymes and activating glycolytic and fatty acid synthetic enzymes, resulting in the switch from fatty acid oxidation to fatty acid synthesis [13]. Insulin action in the liver leads to the reduction in glycogenolysis and gluconeogenesis, increased glycogen synthesis, and enhanced lipid and glycogen storage. The regulation of hepatic glucose metabolism by insulin is mainly through a direct effect, but some evidence suggests that insulin could also indirectly regulates HGP by inhibiting glucagon secretion from pancreatic α cells [13-15], by suppressing free fatty acid production from adipose tissues via inhibition of lipolysis [16-18], and by activating hepatic IL-6-STAT3 signaling via brain insulin action through the brain-liver axis [19]. Suppression of lipolysis by insulin in adipose tissue decreases serum non-esterified fatty acids (NEFA) and glycerol levels, which is responsible for the acute inhibition of hepatic glucose production[18]. Brain insulin signaling has been shown to reduce IL-6 expression and STAT3 phosphorylation in the liver, leading to a decrease in the expression of the gluconeogenic protein Glucose 6-phosphatase (G6Pase) [20].
Defects in insulin signaling and action in the liver lead to increased HGP, impaired glucose disposal, and reduced postprandial VLDL-TG secretion. Since insulin clearance in vivo occurs primarily in the liver, insulin resistance in the liver also results in reduced insulin degradation. Consistent with this, liver-specific insulin receptor knockout mice (LIRKO) are hyperglycemic and hyperinsulinemic, and display reduced liver size compared to control wild-type mice [21]. Liver-specific IRS1 knockout (LIRS1-KO) mice show insulin resistance after refeeding, but fail to exhibit insulin resistance during fasting. Conversely, liver-specific IRS2 knockout (LIRS2-KO) mice display insulin resistance during fasting but not after refeeding, displaying dynamic functional relay [22]. LIRS1-KO mice show significant impairment in hepatic nutrient homeostasis and fatty acid oxidation due to dysregulation of FOXA1, while IRS2 apparently controls gluconeogenesis by inhibiting FOXO1 and CREB-binding protein (CBP). Nutrient-sensitive transcripts are significantly downsregulated in LIRS1-KO mice but are normal in LIRS2-KO mice, indicating that IRS1, but not IRS2, plays a major role in regulating hepatic nutrient homeostasis [23]. However, liver-specific knockout of both IRS1 and IRS2 leads to severe glucose intolerance and impaired lipid metabolism [24].
2. Insulin promotes glucose and fatty acid uptake in skeletal muscle
Skeletal muscle accounts for 60-70% of whole body insulin-stimulated glucose uptake and thus plays an essential role in the regulation of whole body energy homeostasis [27]. Insulin regulates muscle metabolism by promoting glucose uptake, glycogen synthesis, and lipid utilization and storage. Insulin stimulates glucose uptake in skeletal muscle by promoting the membrane translocation of GLUT4, the major glucose transporter in skeletal muscle [28]. Insulin has also been shown to stimulate glucose and free fatty acid (FFA) uptake in skeletal muscle by increasing glucose flux and activating key enzymes involved in muscle glucose or fat oxidation [29]. The effects of insulin, however, are reduced in skeletal muscle of type 2 diabetic subjects [30].
Muscle is one of the major sites of insulin resistance in type 2 diabetic patients [31] and in fact, muscle insulin resistance has been considered to be one of the earliest signs in the pathogenesis of metabolic syndrome [32]. Impaired tyrosine phosphorylation of IRS1 and activation of PI3K have been found in skeletal muscle of type 2 diabetic patients [33]. Defects in glucose transport in skeletal muscle also correlate with impaired whole body glucose uptake in type 2 diabetic patients [34]. Muscle-specific knockout of IR led to increased fat mass, elevated serum triglyceride levels, and muscle insulin resistance in mice, but had no significant effects on global glucose tolerance [30]. A possible explanation for these findings is that glucose is shunted from insulin-resistant muscle to the relatively more insulin sensitive adipose tissue, where it is converted into triglyceride for storage, thus compensating for reduced muscle insulin sensitivity at the whole body level in the mice [35]. Impaired glucose uptake is also found in mice lacking both IRS1 and IRS2 in skeletal and cardiac muscle, although without hyperglycemia or hyperinsulinemia [36].
Insulin does not stimulate lipid storage in muscle under both normal and insulin resistant conditions. In contrast, palmitate, by elevating intracellular synthesis of ceramide and activation of protein kinase PKCζ [37], suppresses insulin-induced plasma membrane recruitment and phosphorylation of Akt, which is associated with a loss in insulin-stimulated glucose transport [37]. The cellular mechanisms that lead to the initial accumulation of intracellular lipid intermediates have not been completely elucidated yet, but may result from lower rates of fatty acid (FA) oxidation, higher rates of FA uptake, or both, in insulin-resistant skeletal muscle [38].
Recent studies have identified skeletal muscles as a secretory organ that secretes several hundred peptides (myokines) in response to environmental and/or metabolic changes [39]. Among them, the brain-derived neurotrophic factor (BDNF) [40] and irisin [41] have been shown to regulate glucose homeostasis and obesity by cross-talking with other organs (e.g. adipose tissue). Irisin improves energy homeostasis by promoting UCP1 expression and the browning process of white adipose tissue (WAT) [42]. Inhibition of myostatin, a member of the transforming growth factor-β (TGF-β) superfamily predominantly expressed in skeletal muscle, has been found to prevent diabetes and hyperphagia in a mouse model of lipodystrophy [43]. However, whether biosynthesis and secretion of the myokines are regulated by insulin remains to be determined.
3. Insulin inhibits lipolysis and stimulates lipid biosynthesis in adipose tissues
Adipose tissue is an important energy storage and endocrine organ that plays important roles in the maintenance of energy homeostasis, including insulation and protection of tissues and organs from heat and cold, providing protective padding, thermogenesis, adipogenesis, and the production of a variety of hormones/cytokines collectively known as adipokines [44]. The expansion of adipose tissue is known to exert a buffering effect that prevents excess lipids from being ectopically deposited in other metabolically important organs such as liver, muscle and pancreas, which are major causes of insulin resistance [45]. However, under excess nutrition, diet-induced obesity remains to be the leading cause of insulin resistance, unless the lipid storage capacity of adipose tissues is enhanced by other means, such as PPARγ stimulation [46].
Insulin stimulates the uptake of glucose into adipocytes where it converts into lipids as a more efficient form of energy storage (adipogenesis) [47]. Although adipose tissue accounts for a relatively small proportion (<10%) of the peripheral glucose utilization in response to insulin [48], it is not a passive repository for excess energy over the research in the past decades [49, 50]. Adipose tissue is the primary site for triacylglycerol storage [51], while enhancement of glucose uptake increases triacylglycerol and fatty acid synthesis in adipose tissues and might lead to obesity [52]. The major bucket of triacylglycerol in the body is in adipose tissues, which can be mobilized in the form of long-chain fatty acids to other tissues via the bloodstream [53]. Insulin regulates lipid, glucose and protein metabolism primarily in adipose tissue [49, 54]. First of all, insulin effectively decreases the rate of lipolysis in adipose tissues, leading to reduced plasma fatty acid level [55]. Insulin also has the ability to increase triglycerides uptake from the blood into adipose tissue [55]. Secondly, the major effect of insulin on adipose tissue is to increase glycolysis rate and to promote glucose transport across the cell membrane [55]. Insulin stimulates the translocation of the glucose transporter GLUT4 from intracellular pools to the surface of cell membrane to increase glucose uptake in adipose and muscle [56]. Finally, insulin increases the rate of protein synthesis in adipose tissue. The protein synthesis stimulated by insulin causes some proteins to be phosphorylated and dissociate from eIF-4E, the translation initiation factor regulator, thereby relieving the translational inhibition [57].
Insulin promotes the biosynthesis of lipids and inhibits its degradation. The action of insulin in the regulation of lipid metabolism is mediated by two main transcriptional factors: SREBP1c, which determines the transcription of many adipocyte specific genes [58, 59], and FOXO1 [60]. Down-regulation of SREBP-1 blocks the expression of gluconeogenic and lipogenic genes [61]. On the other hand, overexpression of SREBP-1 increases gluconeogenic and lipogenic gene transcription [62]. Insulin-stimulated glucose uptake in adipocytes activates ChREBP, which up-regulates de novo lipogenesis [63]. Recently studies show that the mTORC1 signaling pathway mediates insulin-stimulated processing of SREBP-1c through its substrate protein kinase S6K [64, 65]. Insulin also profoundly inhibits lipolysis in adipocytes via inhibition of phosphodiesterase 3b (PDE3b) via Akt-mediated phosphorylation, leading to reduced intracellular cAMP levels and thus PKA activity [66]. However a noncanonical pathway has been described by which insulin regulates lipid metabolism via an Akt-independent and PKA-mediated phosphorylation of a lipid droplet-associated substrate, perilipin [67]. Secondly, insulin inhibits lipolysis through phosphorylation of adipose-specific phospholipase A2, which via arachidonic acid production increases prostaglandin E2 levels and in a paracrine/autocrine manner reduces cAMP levels through inhibition of adenylate cyclase [68, 69]. Thirdly, insulin represses lipolysis by transcriptionally silencing lipase genes via repression of the transcription factors FOXO1 and IRF4 [70, 71]. FOXO1 is phosphorylated by Akt and the phosphorylation prevents FOXO1 from entering into the nucleus, leading to increased PPARγ activity [72]. Finally, insulin upregulates the transcription of lipid droplet protein FSP27 and dampens thus the downstream target of Akt, lipolysis [73] [74].
Insulin resistance in adipose tissue, which is a characteristic of the obese state [75], has been recognized as one of the leading causes of type 2 diabetes [76, 77]. Key steps in the insulin signaling pathway, such as tyrosine phosphorylation of IRS1 and activation of PI3K, are greatly reduced in adipose tissue than in muscle in type 2 diabetic patients [78]. However, fat-specific knockout of IR, which led to a 50% decrease in fat pad mass and a 30% decrease in whole body triglyceride content, improves metabolism in mice [79]. There is some evidence showing that the insulin signaling pathway plays a more important role in the development and maintenance of normal triglyceride storage than in the maintenance of euglycemia in mice [52]. Knockout of IR in brown adipose tissue (BAT), which is important for thermal adaptation as well as determining peripheral insulin sensitivity [80], led to age-dependent brown fat atrophy, accompanied with deteriorated β cell function, decreased β cell mass, and hyperglycemia [81]. These findings suggest a functional link between β cell mass/function and BAT. Fat-specific knockout of GLUT4 in mice led to a 53% decrease in insulin-stimulated whole body glucose uptake and a reduction in glycolysis and glycogen synthesis [82]. Interestingly, fat-specific knockout of GLUT4 in mice also led to a 40% reduction in insulin-stimulated glucose transport into skeletal muscle and impairment in insulin-stimulated suppression of HGP, indicating that down-regulation of GLUT4 in adipose tissues can cause whole-body insulin resistance, a hallmark of type 2 diabetes. However, it remains to be determined whether insulin resistance in adipose tissues leads to dysfunction in other targeted organs and whether the effect is mediated via an endocrine or a paracrine mechanism.
4. Insulin positively regulates insulin secretion and β cell function
Pancreatic β cells, which produce and secrete insulin in response to the blood glucose concentration in order to keep glycaemia in a narrow physiological range, are of vital importance in maintaining glucose homeostasis. Glucose, the principal nutrient secretagogue of insulin, is transported into pancreatic β cells via glucose transporter 2 (GLUT2). In β cells, glucose is metabolized via glycolysis and Krebs cycle, leading to an increase in cellular ATP/ADP ratio and subsequent closure of the ATP-sensitive potassium (KATP) channels [83]. The closure of KATP channels results in β cell membrane depolarization that induces opening voltage-dependent Ca2+ channels and subsequent influx of Ca2+ into β cells. The increase of the intracellular free Ca2+ concentration constitutes the indispensable triggering signal to induce exocytosis of insulin-containing secretory granules [83]. Historically, insulin was thought to have a negative or no effect on β cell proliferation, insulin biosynthesis, and secretion [84]. However, many more recent studies have clearly demonstrated that insulin plays a positive role in transcription, translation, ion flux, β cell survival, proliferation and insulin secretion [85].
Targeted overexpression or specific ablation of key components of insulin signaling in mouse pancreatic β cells significantly affects β cell function and survival [86]. Overexpression of IR greatly promoted insulin-stimulated insulin gene expression in mouse β cells [87]. Disruption of insulin signaling in β cells by β cell-specific knocking out IRS2 [88] or the IR [89] reduced pancreatic insulin content, accompanied by the development of a phenotype similar to that of type 2 diabetes mellitus. Mice with a global knockout of IRS1 [90] or IRS2 [91], β cell-specific knockout of IR [92], or pancreas-specific deletion of IRS2 [93] all showed decreased β cell mass and a marked impairment in glucose-stimulated insulin secretion. On the other hand, constitutive activation of Akt1/PKBα increased β cell proliferation (hyperplasia) and increases β cell size (hypertrophia) [94].
While it is now well-established that insulin signaling plays critical positive role in β cell function, the autocrine effect of insulin on β cell function remains unclear [95]. A major argument is that since pancreatic β cells are exposed to such a high level of secreted insulin, the respective insulin signal transduction pathway must be desensitized [96]. Another question is whether the central nervous system (CNS) has a predominant influence on β cell function [97, 98]. While most of the intriguing data on the roles of insulin in β cell function come from knockout mice generated by using the pancreatic and duodenal homeobox 1 (PDX1) gene promoter- or the rat insulin gene promoter (RIP)-driven Cre recombination system [92, 99, 100], recently studies have revealed the caveats of these genetic knockout approaches due to non-specific knockout of the target genes in other tissues in addition to β cell or pancreas [101, 102]. Indeed, the RIP and PDX1 gene promoters have been shown to drive Cre-expression in several regions of the brain, including the hypothalamus [102]. Moreover, the RIP-Cre mice themselves display mild glucose intolerance [103]. Therefore, further studies may still be needed to elucidate the exact mechanisms underlying the phenotypes of the knockout mice generated by using the aforementioned promoter-driven Cre mice. Recently, the mouse insulin promoter (MIP)-driven Cre recombination system has been shown to disrupt gene expression exclusively in β cells [101], providing a critical tool to determine the functional roles of a protein in β cells in vivo.
5. Central insulin signaling promotes neuronal survival and regulates whole-body energy homeostasis
Brain insulin signaling has been shown to be essential for the regulation of various neuronal activities such as learning, memory, food intake, reproduction, and peripheral metabolism [104, 105]. Insulin in the brain, especially in the hippocampus, cortex, hypothalamus, olfactory bulb, and pituitary, can reach a level 10- to 100-fold greater than that in the plasma [106, 107]. The origin of central insulin remains to be a topic of debating, but some evidence suggests that insulin could cross the blood–brain barrier (BBB) by an active and saturable process [105]. Consistent with this finding, circulating insulin levels are positively correlated with the concentration of this peptide hormone in cerebrospinal fluid (CSF) [107].
Brain insulin signaling also regulates whole body energy balance by cross-talking with peripheral tissues, involving complex interactions of various hormones, neuropeptides, and other signaling molecules involved in the regulation of food intake and energy expenditure [1]. Direct administration of insulin into the brain reduces food intake and body weight gain in C. elegans, Drosophila, and baboons [108-110]. Deletion of IR in neurons of mice increases food intake and body weight in conjunction with increased body fat, plasma insulin levels, and hypertriglyceridemia [111]. Deletion of IRS2 specifically in the brain increases lifespan in the presence of peripheral insulin resistance [112]. Surprisingly, increasing neuronal-specific IRS2 expression in mice also increased fat mass, insulin resistance, and glucose intolerance during aging due to decreased locomotor activity in the presence of unaltered exploratory behavior and motor function [113]. CNS inflammation contributes to peripheral tissue insulin resistance, particularly in the liver, via a brain-liver neuronal signal [114]. These observations demonstrate that insulin signaling in the brain can influence glucose homeostasis in response to afferent input from peripheral tissues. However, the precise mechanisms underlying the cross-talk between central insulin signaling and peripheral signaling pathways remain to be further elucidated.
IV. Insulin signaling in aging and aging-associated diseases
1. Insulin signaling is conserved among species
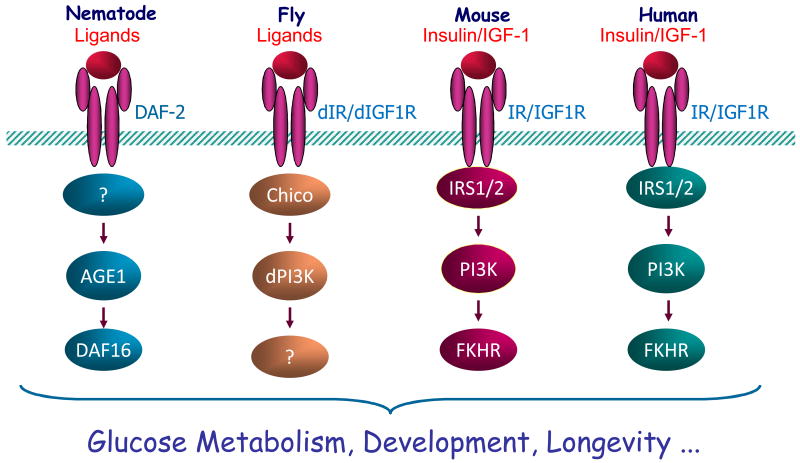
The insulin/insulin growth factor (IGF) signaling (IIS) cascade is an evolutionarily conserved pathway among diverse species, ranging from yeast to humans ([115] and Fig. 2). While defects in the insulin signaling pathway lead to insulin resistance and diabetes in rodents and humans, disruption of this signaling pathway has been shown to significantly extend lifespan in C. elegans [116], flies [117], mice [118], and humans [119, 120]. These long-lived IIS mutants share some important phenotypic characteristics including reduced insulin signaling, enhanced insulin sensitivity, and reduced serum IGF-1 levels, together with reduced oxidative damage of macromolecules and increased stress resistance. The daf-2/IR/IGFR mutant C. elegans live twice as long as wild-type, and the longevity phenotype does not need go through the dauer state [116]. PI3K-null adult C. elegans are more resistant to oxidative and electrophilic stresses and live remarkably longer under both normal and toxic environments compared to wild-type controls [121]. These effects have been shown to depend on the integrity of the dauer daf-16/FOXO, which has similarity to a family of mammalian forkhead transcriptor factors[109]. The lower level of free radicals in daf-2/IR/IGFR mutants has been shown to be essential for life span extension [122]. Indeed, the gene ctl-1, which encodes a cytosolic catalase, is required for the extension of adult life span by daf-2/IR/IGFR [123]. The expression of mitochondrial Superoxide dismutase 2 (SOD2) is required for the longevity extension caused by mutations decreasing the activity of the Ras/Cyr1/PKA and Sch9 pathways in yeast [124]. Similarly, flies homozygous for chico/IRS null mutant have increased levels of SOD, reduced body size, greatly reduced fecundity, and increased longevity [125]. These findings highlight the central position of oxidative stress in the aging-regulatory machinery and superoxide toxicity plays an important role in aging and death. C. elegans proteotoxicity models indicate that the IIS pathway directly links aging to the onset of toxic protein aggregation, and the protective effects are dependent on daf-16/FOXO, as the effects could be abolished by RNAi-mediated depletion of daf-16 [126]. Another strong link between insulin/IGF-1 signaling and life span in animal models comes from dietary restriction. Primates maintained on dietary restriction feeding regimens exhibit increased insulin sensitivity and enhanced glucose tolerance [127]. Calorically restricted rats have lower levels of IGF-1 that contributes to the protective effect against age-related pathology and resistance to p-cristine-induced bladder cancer [128]. Taken together, these results suggest that the IIS pathway is critical for regulating various aging-related disorders and longevity.
Figure 2. Conserved insulin/IGF-1 regulation in longevity.
The insulin/IGF-1-like pathway is conserved among various species and suppressing this pathway extends lifespan in species such as worm, fly, and mouse.
2. Impaired neuronal insulin/IGF-1 signaling (IIS) is associated with aging and aging-related neuronal degenerative diseases
Human aging is associated with neurophysiological changes in the brain and variable degrees of cognitive decline. While systemic disruption of the IIS pathway has been shown to extend lifespan in diverse species, abnormality in insulin signaling in peripheral neurons has been shown to contribute to diabetic neuropathy [129]. Noteworthy, aging has been found to be associated with a decrease in brain IR number and the binding capacity of insulin, especially in hippocampus, cortex, and choroid plexus [130]. Defects in neuronal IIS pathway such as reduced Akt and GSK3 activities are implicated in the development of Alzheimer's disease (AD) [131], one of the most important aging-related neuronal disorders. AD is associated with a decrease in insulin level in the CSF and CSF/plasma insulin ratio [106] and insulin signaling impairment is more severe in individuals with both T2DM and AD [132, 133], suggesting that T2DM may be a risk factor for AD. Consistent with this, brains of T2DM and AD mice show similar pathophysiological changes [134]. In addition, reversing diabetes-induced neuronal mitochondrial dysfunction is beneficial for neuronal survival [135]. These studies clearly suggest a connection between AD and diabetes, and reinforce the idea that AD can be considered as a form of diabetes in the brain (also known as type 3 diabetes or brain-type diabetes) [133]. However, neuron-specific knocking out IRS2 has been found to extend lifespan in mice [112]. Interestingly, knockout of IRS2, but not neuronal IR or IGF-1R, prevents premature death and delays amyloid accumulation in a mouse model of AD [136]. Thus, it remains to be established whether the neuronal IIS pathway has a protective role in aging and aging-associated neuronal degeneration and various other diseases.
3. Disruption of insulin signaling in adipose tissues improves healthy aging and extends lifespan
Insulin lowers plasma fatty acid levels by inhibiting lipolysis and stimulating fatty acid and triacylglycerol synthesis and by promoting triglyceride uptake in adipose tissues [53]. Fat-specific knockout of the IR led to a 50% reduction in adipose tissue mass despite normal food intake [52, 79]. The fat-specific IR knockout mice (FIRKO) have a significantly lower insulin level and extended lifespan compared to control mice [52, 79], demonstrating an essential role of fat-specific insulin signaling in aging.
How disrupting insulin signaling in adipose tissues leads to extended lifespan remains unknown. The FIRKO mice showed increased metabolic rate, suggesting that reducing free radical damage may not be an important factor to extend lifespan. Another possibility is that the extended lifespan of the FIRKO mice is caused by reduced fat mass. Consistent with this, caloric restriction, a well-established strategy to extend lifespan in rodents; reduces fat mass, particularly visceral fat, in mice [79, 137]. However, calorie restriction also greatly extends the maximum lifespan of genetically obese ob/ob mice [138], suggesting that reduced adiposity per se may not be the major component in extending longevity of the obese mice. Lastly, the increased longevity in FIRKO mice may be the direct result of altered insulin signaling in adipose tissues, which is in agreement with the findings that reducing IIS increases lifespan in C. elegans and Drosophila [110, 116, 125, 139, 140]. However, how altering adipose insulin signaling leads extended longevity remains unclear.
4. mTORC1: The downstream target of insulin/IGF-1 signaling
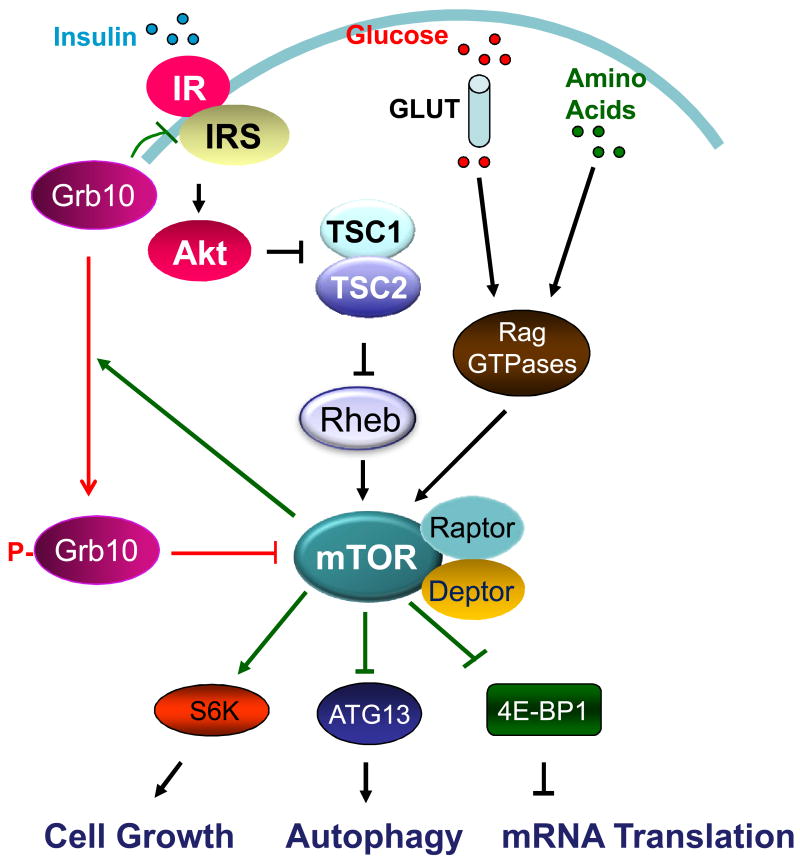
mTOR (mechanistic/mammalian target of rapamycin) is an evolutionarily conserved serine/threonine kinase that acts downstream of the IIS pathway (Fig. 3). mTOR exists in two functionally distinct complexes, mTOR complex 1 (mTORC1) and complex 2 (mTORC2). As an energy sensor, mTORC1 is activated by nutrients such as amino acids and glucose, cellular energy levels, and many growth factors/hormones such as insulin and IGF-1, and plays critical roles in the regulation of key cellular events, including cell growth, proliferation, differentiation, gene transcription, mRNA translation, autophagy, survival, and metabolism [141-148]. mTORC2 phosphorylates Akt and plays an important role in insulin signaling. Suppression of mTOR signaling extends lifespan in yeast [149, 150], worms [151], Drosophila [152], and mice [153]. mTOR signaling suppression has also been found to attenuate various age-related diseases such as obesity, neurodegenerative diseases, osteoporosis, osteoarthritis, and age-related macular degeneration [154].
Figure 3. mTORC1 signaling and regulation.
mTORC1, which regulates various cellular events such as cell growth, autophagy, and mRNA translation, is activated by insulin-stimulated and Akt-mediated phosphorylation and inhibition of TSC1/2, a negative regulator of mTOR. Inhibition of TSC1/2 leads to the activation of small G protein Rheb and thus enhanced mTORC1 signaling. mTORC1-mediated phosphorylation of Grb10 at Ser501/503 switches the binding preference of Grb10 from the insulin receptor to Raptor, leading to the dissociation of Raptor from mTOR and thus down-regulation of mTORC1 signaling.
The activation of the mTORC1 signaling pathway by insulin or IGF-1 is mediated by Akt-mediated phosphorylation and inhibition of Rheb, a negative upstream regulator of mTORC1 (Fig. 3). mTORC1 is inhibited by rapamycin, a small molecular immunosuppressant drug used to prevent rejection in organ transplantation. Although rapamycin is generally considered as a specific inhibitor of mTORC1, extended treatment with rapamycin can also effectively inhibit mTORC2 [155]. There is some evidence that suggests that the negative effects of rapamycin on glucose tolerance and hepatic insulin are mediated by inhibition of mTORC2, whereas lifespan extension is considered to result from mTORC1 inhibition [156]. Very recently, we identified Grb10, a Src-homology 2 (SH2) and pleckstrin homology (PH)-domain containing adaptor protein that interacts with tyrosine phosphorylated IR and inhibits insulin/IGF-1 signaling [8], as a negative feedback regulator of mTORC1 [157]. mTOR-mediated phosphorylation at Ser501/503 switches the binding preference of Grb10 from the insulin receptor to raptor, leading to the dissociation of raptor from mTOR and thus down-regulation of mTORC1 signaling [157], demonstrating a feedback mechanism of regulation.
5. Will suppressing mTORC1 in adipose tissues improve healthspan?
Disrupting insulin signaling in adipose tissues improved metabolism and extended lifespan in mice [52, 79], yet the precise mechanism remains unknown. Since mTORC1 is a downstream target of insulin signaling (Fig.1), fat-specific knockout of IR may improve healthspan and longevity by suppressing mTORC1 signaling in adipose tissues. Interestingly, adipose-specific disrupting mTORC1 signaling by knocking out Raptor prevented diet-induced obesity in mice [158]. On the other hand, overactivation of mTORC1 signaling in adipose tissues by fat-specific knockout of Grb10 led to obesity as well as glucose and insulin intolerance [157], suggesting that reducing mTORC1 signaling in adipose tissues has a beneficial effect on metabolism. However, it is currently unknown if suppressing mTORC1 signaling in adipose tissues is sufficient to improve healthspan and lifespan.
V. Conclusion
The insulin-signaling cascade is an evolutionally conserved signaling pathway among diverse species that plays a critical role in the regulation of metabolism and longevity. In mammals, insulin regulates lipid and glucose metabolism and energy homeostasis by initiating its signaling events in target tissues such as liver, skeletal muscle, adipose tissues, and the brain. Defects in the insulin signaling pathway may cause insulin resistance, leading to various metabolic disorders such as type 2 diabetes. However, numerous studies have demonstrated that suppressing insulin and/or IGF-1 signaling pathway may lead to extended lifespan. An important question is whether targeting the IIS signaling pathway is a valid strategy to improve healthspan and lifespan. Interestingly, disrupting the insulin signaling pathway in adipose tissues improves metabolism and extends lifespan [52, 79], suggesting tissue-specific suppression of insulin signaling may be an effective approach to improve healthspan and longevity. The mechanisms by which altering adipose tissue insulin signaling leads to extended lifespan remain unknown, but reduction of mTORC1 signaling could play a role. Generation and characterization of adipose tissue-specific transgenic and knockout animal models in which the mTORC1 signaling pathway is specifically altered should enable us to test this hypothesis and elucidate the mechanistic links between aging and metabolic disease. Results from these studies should provide insights into the development of new therapeutic approaches aimed at alleviating aging-associated disorders and improving healthspan.
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (2014CB910500), National Nature Science Foundation of China (81130015, 81000316 and 81370017), Natural Science Foundation of Hunan Province, China (14JJ3034), and the NIH (RO1 DK100697).
Abbreviations
- AD
Alzheimer's disease
- APPL1
adaptor protein, phosphotyrosine interaction
- BAR
Bin1/amphiphysin/rvs167
- BAT
brown adipose tissue
- BBB
blood–brain barrier
- BDNF
Brain-derived neurotrophic factor
- CBP
CREB-binding protein
- ChREBP
carbohydrate response element binding protein
- CNS
central nervous system
- CREB
cAMP response element-binding
- CSF
cerebrospinal fluid
- ERK1/2
extracellular signal-regulated kinases1 and 2
- FA
fatty acid
- FFA
free fatty acid
- FIRKO
fat-specific insulin receptor gene knockout
- FOXA1
Forkhead box protein A1
- FOXO
Forkhead box protein O
- FSP27
fat specific protein 27
- HGP
hepatic glucose production
- G6Pase
Glucose 6-phosphatase
- GLUT
Glucose transporter
- Grb10
Growth factor receptor-bound protein 10
- GSK3
Glycogen synthase kinase 3
- IGF-1
Insulin-like growth factor 1
- IGFR
Insulin-like growth factorreceptor
- IL-6
Interleukin-6
- IIS
insulin/insulin-like growth factor signaling
- IR
insulin receptor
- IRF4
interferon regulatory factor 4
- IRS
insulin receptor substrates
- KATP
ATP-sensitive potassium
- LIRKO
liver-specific insulin receptor gene knockout
- LIRS1-KO
liver-specific IRS1 knockout
- LIRS2-KO
liver-specific IRS2 knockout
- MAPK
Mitogen-activated Protein Kinase
- MEK
Mitogen-activated protein/extracellular signal-regulated kinase kinase
- MIP
mouse insulin promoter
- mTOR
mammalian target of rapamycin
- mTORC1/2
mTOR complex 1/2
- NEFA
non-esterified fatty acids
- PDK1
Phosphoinositide- dependent kinase-1
- PDE3b
phosphodiesterase 3b
- PDX1
Pancreatic and duodenal homeobox 1
- PH
pleckstrin homology
- PI3K
Phosphatidylinositol-3-Kinase
- PKA
protein kinase A
- PKB
protein kinase B
- PKC
protein kinase C
- PPARγ
Peroxisome proliferator-activated receptor
- PTB
phosphotyrosine binding
- PTP-1B
protein tyrosine phosphatase-1B
- RIP
Rat insulin gene promoter
- Rheb
Ras homolog enriched in brain
- S6K
S6-kinase
- SH2
Src-homology 2
- SOD
Superoxide dismutase
- SREBPs
Sterol Regulatory Element-Binding Proteins
- STAT3
Signal transducer and activator of transcription 3
- T2DM
type 2 diabetes mellitus
- TG
triglyceride
- TGF-β
transforming growth factor-β
- UCP1
uncoupling protein 1
- VLDL
Very-low-density lipoprotein
- WAT
white adipose tissue
References
- 1.Sharma MD, Garber AJ, Farmer JA. Role of insulin signaling in maintaining energy homeostasis. Endocr Pract. 2008;14(3):373–80. doi: 10.4158/EP.14.3.373. [DOI] [PubMed] [Google Scholar]
- 2.Kadowaki T. Insights into insulin resistance and type 2 diabetes from knockout mouse models. J Clin Invest. 2000;106(4):459–65. doi: 10.1172/JCI10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katic M, Kahn CR. The role of insulin and IGF-1 signaling in longevity. Cell Mol Life Sci. 2005;62(3):320–43. doi: 10.1007/s00018-004-4297-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porte D, Jr, Baskin DG, Schwartz MW. Insulin signaling in the central nervous system: a critical role in metabolic homeostasis and disease from C. elegans to humans. Diabetes. 2005;54(5):1264–76. doi: 10.2337/diabetes.54.5.1264. [DOI] [PubMed] [Google Scholar]
- 5.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414(6865):799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 6.Ryu J, et al. APPL1 Potentiates Insulin Sensitivity by Facilitating the Binding of IRS1/2 to the Insulin Receptor. Cell Report. 2014 doi: 10.1016/j.celrep.2014.04.006. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochim Biophys Acta. 2010;1802(4):396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Lim MA, Riedel H, Liu F. Grb10: more than a simple adaptor protein. Front Biosci. 2004;9:387–403. doi: 10.2741/1226. [DOI] [PubMed] [Google Scholar]
- 9.Holt LJ, Siddle K. Grb10 and Grb14: enigmatic regulators of insulin action--and more? Biochem J. 2005;388(Pt 2):393–406. doi: 10.1042/BJ20050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copps KD, White MF. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia. 2012;55(10):2565–82. doi: 10.1007/s00125-012-2644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo S. Insulin signaling, resistance, and the metabolic syndrome: insights from mouse models into disease mechanisms. J Endocrinol. 2014;220(2):T1–T23. doi: 10.1530/JOE-13-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiganis T. PTP1B and TCPTP--nonredundant phosphatases in insulin signaling and glucose homeostasis. FEBS J. 2013;280(2):445–58. doi: 10.1111/j.1742-4658.2012.08563.x. [DOI] [PubMed] [Google Scholar]
- 13.Lewis GF, Vranic M, Giacca A. Glucagon enhances the direct suppressive effect of insulin on hepatic glucose production in humans. Am J Physiol. 1997;272(3 Pt 1):E371–8. doi: 10.1152/ajpendo.1997.272.3.E371. [DOI] [PubMed] [Google Scholar]
- 14.Liljenquist JE, et al. Evidence for an important role of glucagon in the regulation of hepatic glucose production in normal man. J Clin Invest. 1977;59(2):369–74. doi: 10.1172/JCI108649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrick GK, et al. Effect of hyperglucagonemia on hepatic glycogenolysis and gluconeogenesis after a prolonged fast. Am J Physiol. 1990;258(5 Pt 1):E841–9. doi: 10.1152/ajpendo.1990.258.5.E841. [DOI] [PubMed] [Google Scholar]
- 16.Mittelman SD, Bergman RN. Inhibition of lipolysis causes suppression of endogenous glucose production independent of changes in insulin. Am J Physiol Endocrinol Metab. 2000;279(3):E630–7. doi: 10.1152/ajpendo.2000.279.3.E630. [DOI] [PubMed] [Google Scholar]
- 17.Rebrin K, et al. Causal linkage between insulin suppression of lipolysis and suppression of liver glucose output in dogs. J Clin Invest. 1996;98(3):741–9. doi: 10.1172/JCI118846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sindelar DK, et al. The role of fatty acids in mediating the effects of peripheral insulin on hepatic glucose production in the conscious dog. Diabetes. 1997;46(2):187–96. doi: 10.2337/diab.46.2.187. [DOI] [PubMed] [Google Scholar]
- 19.Inoue H, et al. Role of hepatic STAT3 in brain-insulin action on hepatic glucose production. Cell Metab. 2006;3(4):267–75. doi: 10.1016/j.cmet.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Konner AC, et al. Insulin action in AgRP-expressing neurons is required for suppression of hepatic glucose production. Cell Metab. 2007;5(6):438–49. doi: 10.1016/j.cmet.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Michael MD, et al. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol Cell. 2000;6(1):87–97. [PubMed] [Google Scholar]
- 22.Kubota N, et al. Dynamic functional relay between insulin receptor substrate 1 and 2 in hepatic insulin signaling during fasting and feeding. Cell Metab. 2008;8(1):49–64. doi: 10.1016/j.cmet.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Guo S, et al. The Irs1 branch of the insulin signaling cascade plays a dominant role in hepatic nutrient homeostasis. Mol Cell Biol. 2009;29(18):5070–83. doi: 10.1128/MCB.00138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Z, et al. Foxo1 integrates insulin signaling with mitochondrial function in the liver. Nat Med. 2009;15(11):1307–1311. doi: 10.1038/nm.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leavens KF, et al. Akt2 is required for hepatic lipid accumulation in models of insulin resistance. Cell Metab. 2009;10(5):405–18. doi: 10.1016/j.cmet.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13(4):376–88. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeFronzo RA, et al. The effect of insulin on the disposal of intravenous glucose. Results from indirect calorimetry and hepatic and femoral venous catheterization. Diabetes. 1981;30(12):1000–7. doi: 10.2337/diab.30.12.1000. [DOI] [PubMed] [Google Scholar]
- 28.Kristiansen S, Hargreaves M, Richter EA. Exercise-induced increase in glucose transport, GLUT-4, and VAMP-2 in plasma membrane from human muscle. Am J Physiol. 1996;270(1 Pt 1):E197–201. doi: 10.1152/ajpendo.1996.270.1.E197. [DOI] [PubMed] [Google Scholar]
- 29.Blaak EE. Metabolic fluxes in skeletal muscle in relation to obesity and insulin resistance. Best Pract Res Clin Endocrinol Metab. 2005;19(3):391–403. doi: 10.1016/j.beem.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Bruning JC, et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell. 1998;2(5):559–69. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 31.DeFronzo RA, et al. Effects of insulin on peripheral and splanchnic glucose metabolism in noninsulin-dependent (type II) diabetes mellitus. J Clin Invest. 1985;76(1):149–55. doi: 10.1172/JCI111938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabol R, et al. Reversal of muscle insulin resistance with exercise reduces postprandial hepatic de novo lipogenesis in insulin resistant individuals. Proc Natl Acad Sci U S A. 2011;108(33):13705–9. doi: 10.1073/pnas.1110105108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krook A, et al. Characterization of signal transduction and glucose transport in skeletal muscle from type 2 diabetic patients. Diabetes. 2000;49(2):284–92. doi: 10.2337/diabetes.49.2.284. [DOI] [PubMed] [Google Scholar]
- 34.Zierath JR, et al. Insulin action on glucose transport and plasma membrane GLUT4 content in skeletal muscle from patients with NIDDM. Diabetologia. 1996;39(10):1180–9. doi: 10.1007/BF02658504. [DOI] [PubMed] [Google Scholar]
- 35.Kim JK, et al. Redistribution of substrates to adipose tissue promotes obesity in mice with selective insulin resistance in muscle. J Clin Invest. 2000;105(12):1791–7. doi: 10.1172/JCI8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long YC, et al. Insulin receptor substrates Irs1 and Irs2 coordinate skeletal muscle growth and metabolism via the Akt and AMPK pathways. Mol Cell Biol. 2011;31(3):430–41. doi: 10.1128/MCB.00983-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powell DJ, et al. Intracellular ceramide synthesis and protein kinase Czeta activation play an essential role in palmitate-induced insulin resistance in rat L6 skeletal muscle cells. Biochem J. 2004;382(Pt 2):619–29. doi: 10.1042/BJ20040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turcotte LP, Fisher JS. Skeletal muscle insulin resistance: roles of fatty acid metabolism and exercise. Phys Ther. 2008;88(11):1279–96. doi: 10.2522/ptj.20080018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedersen BK, Febbraio MA. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat Rev Endocrinol. 2012;8(8):457–65. doi: 10.1038/nrendo.2012.49. [DOI] [PubMed] [Google Scholar]
- 40.Pedersen BK. The diseasome of physical inactivity--and the role of myokines in muscle--fat cross talk. J Physiol. 2009;587(Pt 23):5559–68. doi: 10.1113/jphysiol.2009.179515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hee Park K, et al. Circulating irisin in relation to insulin resistance and the metabolic syndrome. J Clin Endocrinol Metab. 2013;98(12):4899–907. doi: 10.1210/jc.2013-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bostrom P, et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481(7382):463–8. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo T, et al. Myostatin inhibition prevents diabetes and hyperphagia in a mouse model of lipodystrophy. Diabetes. 2012;61(10):2414–23. doi: 10.2337/db11-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156(1-2):20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yki-Jarvinen H. Fat in the liver and insulin resistance. Ann Med. 2005;37(5):347–56. doi: 10.1080/07853890510037383. [DOI] [PubMed] [Google Scholar]
- 46.Chao L, et al. Adipose tissue is required for the antidiabetic, but not for the hypolipidemic, effect of thiazolidinediones. J Clin Invest. 2000;106(10):1221–8. doi: 10.1172/JCI11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Penkov DN, et al. Insulin resistance and adipogenesis: role of transcription and secreted factors. Biochemistry (Mosc) 2013;78(1):8–18. doi: 10.1134/S0006297913010021. [DOI] [PubMed] [Google Scholar]
- 48.DeFronzo RA, Bonadonna RC, Ferrannini E. Pathogenesis of NIDDM. A balanced overview. Diabetes Care. 1992;15(3):318–68. doi: 10.2337/diacare.15.3.318. [DOI] [PubMed] [Google Scholar]
- 49.Frayn KN. Adipose tissue as a buffer for daily lipid flux. Diabetologia. 2002;45(9):1201–10. doi: 10.1007/s00125-002-0873-y. [DOI] [PubMed] [Google Scholar]
- 50.Coppack SW, Patel JN, Lawrence VJ. Nutritional regulation of lipid metabolism in human adipose tissue. Exp Clin Endocrinol Diabetes. 2001;109(Suppl 2):S202–14. doi: 10.1055/s-2001-18582. [DOI] [PubMed] [Google Scholar]
- 51.Czech MP, et al. Insulin signalling mechanisms for triacylglycerol storage. Diabetologia. 2013;56(5):949–64. doi: 10.1007/s00125-013-2869-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bluher M, et al. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev Cell. 2002;3(1):25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- 53.Dimitriadis G, et al. Insulin effects in muscle and adipose tissue. Diabetes Res Clin Pract. 2011;93(Suppl 1):S52–9. doi: 10.1016/S0168-8227(11)70014-6. [DOI] [PubMed] [Google Scholar]
- 54.Dimitriadis G, et al. Glucose and lipid fluxes in the adipose tissue after meal ingestion in hyperthyroidism. J Clin Endocrinol Metab. 2006;91(3):1112–8. doi: 10.1210/jc.2005-0960. [DOI] [PubMed] [Google Scholar]
- 55.Newsholme EA, Dimitriadis G. Integration of biochemical and physiologic effects of insulin on glucose metabolism. Exp Clin Endocrinol Diabetes. 2001;109(Suppl 2):S122–34. doi: 10.1055/s-2001-18575. [DOI] [PubMed] [Google Scholar]
- 56.Shepherd PR, Kahn BB. Glucose transporters and insulin action--implications for insulin resistance and diabetes mellitus. N Engl J Med. 1999;341(4):248–257. doi: 10.1056/NEJM199907223410406. [DOI] [PubMed] [Google Scholar]
- 57.Pause A, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371(6500):762–7. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 58.Duvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39(2):171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JB, et al. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Invest. 1998;101(1):1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu M, et al. Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nat Med. 2012;18(3):388–95. doi: 10.1038/nm.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Foretz M, et al. ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose. Mol Cell Biol. 1999;19(5):3760–8. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shimomura I, et al. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci U S A. 1999;96(24):13656–61. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herman MA, et al. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484(7394):333–8. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bae EJ, et al. Liver-specific p70 S6 kinase depletion protects against hepatic steatosis and systemic insulin resistance. J Biol Chem. 2012;287(22):18769–80. doi: 10.1074/jbc.M112.365544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li S, et al. Role of S6K1 in regulation of SREBP1c expression in the liver. Biochem Biophys Res Commun. 2011;412(2):197–202. doi: 10.1016/j.bbrc.2011.07.038. [DOI] [PubMed] [Google Scholar]
- 66.Ahmad F, et al. Differential regulation of adipocyte PDE3B in distinct membrane compartments by insulin and the beta3-adrenergic receptor agonist CL316243: effects of caveolin-1 knockdown on formation/maintenance of macromolecular signalling complexes. Biochem J. 2009;424(3):399–410. doi: 10.1042/BJ20090842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi SM, et al. Insulin regulates adipocyte lipolysis via an Akt-independent signaling pathway. Mol Cell Biol. 2010;30(21):5009–20. doi: 10.1128/MCB.00797-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duncan RE, et al. Identification and functional characterization of adipose-specific phospholipase A2 (AdPLA) J Biol Chem. 2008;283(37):25428–36. doi: 10.1074/jbc.M804146200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jaworski K, et al. AdPLA ablation increases lipolysis and prevents obesity induced by high-fat feeding or leptin deficiency. Nat Med. 2009;15(2):159–68. doi: 10.1038/nm.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chakrabarti P, Kandror KV. FoxO1 controls insulin-dependent adipose triglyceride lipase (ATGL) expression and lipolysis in adipocytes. J Biol Chem. 2009;284(20):13296–300. doi: 10.1074/jbc.C800241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eguchi J, et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 2011;13(3):249–59. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Armoni M, et al. FOXO1 represses peroxisome proliferator-activated receptor-gamma1 and -gamma2 gene promoters in primary adipocytes. A novel paradigm to increase insulin sensitivity. J Biol Chem. 2006;281(29):19881–91. doi: 10.1074/jbc.M600320200. [DOI] [PubMed] [Google Scholar]
- 73.Puri V, et al. Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. J Biol Chem. 2007;282(47):34213–8. doi: 10.1074/jbc.M707404200. [DOI] [PubMed] [Google Scholar]
- 74.Kim JY, et al. Assessment of fat-specific protein 27 in the adipocyte lineage suggests a dual role for FSP27 in adipocyte metabolism and cell death. Am J Physiol Endocrinol Metab. 2008;294(4):E654–67. doi: 10.1152/ajpendo.00104.2007. [DOI] [PubMed] [Google Scholar]
- 75.Kopelman PG. Obesity as a medical problem. Nature. 2000;404(6778):635–43. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- 76.Nishimura S, et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. 2009;15(8):914–20. doi: 10.1038/nm.1964. [DOI] [PubMed] [Google Scholar]
- 77.Minamino T, et al. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat Med. 2009;15(9):1082–7. doi: 10.1038/nm.2014. [DOI] [PubMed] [Google Scholar]
- 78.Smith U. Impaired (‘diabetic’) insulin signaling and action occur in fat cells long before glucose intolerance--is insulin resistance initiated in the adipose tissue? Int J Obes Relat Metab Disord. 2002;26(7):897–904. doi: 10.1038/sj.ijo.0802028. [DOI] [PubMed] [Google Scholar]
- 79.Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299(5606):572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 80.Lowell BB, et al. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366(6457):740–2. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- 81.Guerra C, et al. Brown adipose tissue-specific insulin receptor knockout shows diabetic phenotype without insulin resistance. J Clin Invest. 2001;108(8):1205–13. doi: 10.1172/JCI13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abel ED, et al. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409(6821):729–733. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- 83.Wang Q, Jin T. The role of insulin signaling in the development of beta-cell dysfunction and diabetes. Islets. 2009;1(2):95–101. doi: 10.4161/isl.1.2.9263. [DOI] [PubMed] [Google Scholar]
- 84.Best CH, Haist RE. The effect of insulin administration on the insulin content of the pancreas. J Physiol. 1941;100(2):142–6. doi: 10.1113/jphysiol.1941.sp003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leibiger IB, Leibiger B, Berggren PO. Insulin feedback action on pancreatic beta-cell function. FEBS Lett. 2002;532(1-2):1–6. doi: 10.1016/s0014-5793(02)03627-x. [DOI] [PubMed] [Google Scholar]
- 86.Leibiger IB, Leibiger B, Berggren PO. Insulin signaling in the pancreatic beta-cell. Annu Rev Nutr. 2008;28:233–51. doi: 10.1146/annurev.nutr.28.061807.155530. [DOI] [PubMed] [Google Scholar]
- 87.Xu GG, Rothenberg PL. Insulin receptor signaling in the beta-cell influences insulin gene expression and insulin content: evidence for autocrine beta-cell regulation. Diabetes. 1998;47(8):1243–1252. doi: 10.2337/diab.47.8.1243. [DOI] [PubMed] [Google Scholar]
- 88.Kubota N, et al. Insulin receptor substrate 2 plays a crucial role in beta cells and the hypothalamus. J Clin Invest. 2004;114(7):917–927. doi: 10.1172/JCI21484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kulkarni RN, et al. Tissue-specific knockout of the insulin receptor in pancreatic beta cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell. 1999;96(3):329–339. doi: 10.1016/s0092-8674(00)80546-2. [DOI] [PubMed] [Google Scholar]
- 90.Kulkarni RN, et al. Altered function of insulin receptor substrate-1-deficient mouse islets and cultured beta-cell lines. J Clin Invest. 1999;104(12):R69–75. doi: 10.1172/JCI8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Withers DJ, et al. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391(6670):900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 92.Otani K, et al. Reduced beta-cell mass and altered glucose sensing impair insulin-secretory function in betaIRKO mice. Am J Physiol Endocrinol Metab. 2004;286(1):E41–9. doi: 10.1152/ajpendo.00533.2001. [DOI] [PubMed] [Google Scholar]
- 93.Cantley J, et al. Pancreatic deletion of insulin receptor substrate 2 reduces beta and alpha cell mass and impairs glucose homeostasis in mice. Diabetologia. 2007;50(6):1248–56. doi: 10.1007/s00125-007-0637-9. [DOI] [PubMed] [Google Scholar]
- 94.Tuttle RL, et al. Regulation of pancreatic beta-cell growth and survival by the serine/threonine protein kinase Akt1/PKBalpha. Nat Med. 2001;7(10):1133–1137. doi: 10.1038/nm1001-1133. [DOI] [PubMed] [Google Scholar]
- 95.Rhodes CJ, et al. Direct autocrine action of insulin on beta-cells: does it make physiological sense? Diabetes. 2013;62(7):2157–63. doi: 10.2337/db13-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zick Y. Ser/Thr phosphorylation of IRS proteins: a molecular basis for insulin resistance. Sci STKE. 2005;2005(268):pe4. doi: 10.1126/stke.2682005pe4. [DOI] [PubMed] [Google Scholar]
- 97.Miller RE. Pancreatic neuroendocrinology: peripheral neural mechanisms in the regulation of the Islets of Langerhans. Endocr Rev. 1981;2(4):471–94. doi: 10.1210/edrv-2-4-471. [DOI] [PubMed] [Google Scholar]
- 98.Porte D., Jr Sympathetic regulation of insulin secretion. Its relation to diabetes mellitus. Arch Intern Med. 1969;123(3):252–60. [PubMed] [Google Scholar]
- 99.Kulkarni RN, et al. beta-cell-specific deletion of the Igf1 receptor leads to hyperinsulinemia and glucose intolerance but does not alter beta-cell mass. Nat Genet. 2002;31(1):111–115. doi: 10.1038/ng872. [DOI] [PubMed] [Google Scholar]
- 100.Ueki K, et al. Total insulin and IGF-I resistance in pancreatic beta cells causes overt diabetes. Nat Genet. 2006;38(5):583–588. doi: 10.1038/ng1787. [DOI] [PubMed] [Google Scholar]
- 101.Wicksteed B, et al. Conditional gene targeting in mouse pancreatic ss-Cells: analysis of ectopic Cre transgene expression in the brain. Diabetes. 2010;59(12):3090–8. doi: 10.2337/db10-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Song J, et al. Brain expression of Cre recombinase driven by pancreas-specific promoters. Genesis. 2010;48(11):628–34. doi: 10.1002/dvg.20672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee JY, et al. RIP-Cre revisited, evidence for impairments of pancreatic beta-cell function. J Biol Chem. 2006;281(5):2649–2653. doi: 10.1074/jbc.M512373200. [DOI] [PubMed] [Google Scholar]
- 104.Gerozissis K. Brain insulin: regulation, mechanisms of action and functions. Cell Mol Neurobiol. 2003;23(1):1–25. doi: 10.1023/A:1022598900246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Plum L, Schubert M, Bruning JC. The role of insulin receptor signaling in the brain. Trends Endocrinol Metab. 2005;16(2):59–65. doi: 10.1016/j.tem.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 106.Duarte AI, Moreira PI, Oliveira CR. Insulin in central nervous system: more than just a peripheral hormone. J Aging Res. 2012;2012:384017. doi: 10.1155/2012/384017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van der Heide LP, Ramakers GM, Smidt MP. Insulin signaling in the central nervous system: learning to survive. Prog Neurobiol. 2006;79(4):205–21. doi: 10.1016/j.pneurobio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 108.Woods SC, et al. Chronic intracerebroventricular infusion of insulin reduces food intake and body weight of baboons. Nature. 1979;282(5738):503–5. doi: 10.1038/282503a0. [DOI] [PubMed] [Google Scholar]
- 109.Kim YI, et al. Nucleolar GTPase NOG-1 Regulates Development, Fat Storage, and Longevity through Insulin/IGF Signaling in C. elegans. Mol Cells. 2014;37(1):51–8. doi: 10.14348/molcells.2014.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tatar M, et al. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292(5514):107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 111.Bruning JC, et al. Role of brain insulin receptor in control of body weight and reproduction. Science. 2000;289(5487):2122–5. doi: 10.1126/science.289.5487.2122. [DOI] [PubMed] [Google Scholar]
- 112.Taguchi A, Wartschow LM, White MF. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science. 2007;317(5836):369–72. doi: 10.1126/science.1142179. [DOI] [PubMed] [Google Scholar]
- 113.Zemva J, et al. Neuronal overexpression of insulin receptor substrate 2 leads to increased fat mass, insulin resistance, and glucose intolerance during aging. Age (Dordr) 2013;35(5):1881–97. doi: 10.1007/s11357-012-9491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Milanski M, et al. Inhibition of hypothalamic inflammation reverses diet-induced insulin resistance in the liver. Diabetes. 2012;61(6):1455–62. doi: 10.2337/db11-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Barbieri M, et al. Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am J Physiol Endocrinol Metab. 2003;285(5):E1064–71. doi: 10.1152/ajpendo.00296.2003. [DOI] [PubMed] [Google Scholar]
- 116.Kenyon C, et al. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366(6454):461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 117.Hwangbo DS, et al. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429(6991):562–6. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 118.Holzenberger M, et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421(6919):182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 119.Roth GS, et al. Biomarkers of caloric restriction may predict longevity in humans. Science. 2002;297(5582):811. doi: 10.1126/science.1071851. [DOI] [PubMed] [Google Scholar]
- 120.Suh Y, et al. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci U S A. 2008;105(9):3438–42. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ayyadevara S, et al. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell. 2008;7(1):13–22. doi: 10.1111/j.1474-9726.2007.00348.x. [DOI] [PubMed] [Google Scholar]
- 122.Murakami S, Johnson TE. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics. 1996;143(3):1207–1218. doi: 10.1093/genetics/143.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Taub J, et al. A cytosolic catalase is needed to extend adult lifespan in C. elegans daf-C and clk-1 mutants. Nature. 1999;399(6732):162–6. doi: 10.1038/20208. [DOI] [PubMed] [Google Scholar]
- 124.Fabrizio P, et al. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics. 2003;163(1):35–46. doi: 10.1093/genetics/163.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Clancy DJ, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292(5514):104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 126.Cohen E, et al. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313(5793):1604–10. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 127.Wanagat J, Allison DB, Weindruch R. Caloric intake and aging: mechanisms in rodents and a study in nonhuman primates. Toxicol Sci. 1999;52(2 Suppl):35–40. doi: 10.1093/toxsci/52.2.35. [DOI] [PubMed] [Google Scholar]
- 128.Dunn SE, et al. Dietary restriction reduces insulin-like growth factor I levels, which modulates apoptosis, cell proliferation, and tumor progression in p53-deficient mice. Cancer Res. 1997;57(21):4667–4672. [PubMed] [Google Scholar]
- 129.Brussee V, Cunningham FA, Zochodne DW. Direct insulin signaling of neurons reverses diabetic neuropathy. Diabetes. 2004;53(7):1824–30. doi: 10.2337/diabetes.53.7.1824. [DOI] [PubMed] [Google Scholar]
- 130.Zhao WQ, et al. Insulin and the insulin receptor in experimental models of learning and memory. Eur J Pharmacol. 2004;490(1-3):71–81. doi: 10.1016/j.ejphar.2004.02.045. [DOI] [PubMed] [Google Scholar]
- 131.Schubert M, et al. Role for neuronal insulin resistance in neurodegenerative diseases. Proc Natl Acad Sci U S A. 2004;101(9):3100–3105. doi: 10.1073/pnas.0308724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu Y, et al. Deficient brain insulin signalling pathway in Alzheimer's disease and diabetes. J Pathol. 2011;225(1):54–62. doi: 10.1002/path.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bomfim TR, et al. An anti-diabetes agent protects the mouse brain from defective insulin signaling caused by Alzheimer's disease- associated Abeta oligomers. J Clin Invest. 2012;122(4):1339–53. doi: 10.1172/JCI57256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Carvalho C, et al. Metabolic alterations induced by sucrose intake and Alzheimer's disease promote similar brain mitochondrial abnormalities. Diabetes. 2012;61(5):1234–42. doi: 10.2337/db11-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huang TJ, et al. Insulin prevents depolarization of the mitochondrial inner membrane in sensory neurons of type 1 diabetic rats in the presence of sustained hyperglycemia. Diabetes. 2003;52(8):2129–36. doi: 10.2337/diabetes.52.8.2129. [DOI] [PubMed] [Google Scholar]
- 136.Freude S, et al. Neuronal IGF-1 resistance reduces Abeta accumulation and protects against premature death in a model of Alzheimer's disease. FASEB J. 2009;23(10):3315–24. doi: 10.1096/fj.09-132043. [DOI] [PubMed] [Google Scholar]
- 137.Heilbronn LK, Ravussin E. Calorie restriction and aging: review of the literature and implications for studies in humans. Am J Clin Nutr. 2003;78(3):361–9. doi: 10.1093/ajcn/78.3.361. [DOI] [PubMed] [Google Scholar]
- 138.Harrison DE, Archer JR, Astle CM. Effects of food restriction on aging: separation of food intake and adiposity. Proc Natl Acad Sci U S A. 1984;81(6):1835–8. doi: 10.1073/pnas.81.6.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wolkow CA, et al. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290(5489):147–50. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- 140.Kimura KD, et al. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277(5328):942–6. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 141.Blagosklonny MV, Hall MN. Growth and aging: a common molecular mechanism. Aging (Albany NY) 2009;1(4):357–62. doi: 10.18632/aging.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang X, Proud CG. The mTOR pathway in the control of protein synthesis. Physiology (Bethesda) 2006;21:362–9. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 143.Mizushima N, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–63. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 145.Kaeberlein M, Kennedy BK. Hot topics in aging research: protein translation and TOR signaling, 2010. Aging Cell. 2011;10(2):185–90. doi: 10.1111/j.1474-9726.2010.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kaeberlein M, Kennedy BK. Protein translation, 2007. Aging Cell. 2007;6(6):731–4. doi: 10.1111/j.1474-9726.2007.00341.x. [DOI] [PubMed] [Google Scholar]
- 147.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441(1):1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 149.Bonawitz ND, et al. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5(4):265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kaeberlein M, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310(5751):1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 151.Vellai T, et al. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426(6967):620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 152.Kapahi P, et al. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14(10):885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Blagosklonny MV. Prospective treatment of age-related diseases by slowing down aging. Am J Pathol. 2012;181(4):1142–6. doi: 10.1016/j.ajpath.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 155.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22(2):159–68. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 156.Kaeberlein M. mTOR Inhibition: From Aging to Autism and Beyond. Scientifica (Cairo) 2013;2013:849186. doi: 10.1155/2013/849186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu M, et al. Grb10 Promotes Lipolysis and Thermogenesis by Phosphorylationdependent Feedback Inhibition of mTORC1 Principal. Cell Metab. 2014 doi: 10.1016/j.cmet.2014.03.018. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Polak P, et al. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 2008;8(5):399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]