Abstract
The changes from normal cells to cancer cells are primarily regulated by genome instability, which foster hallmark functions of cancer through multiple mechanisms including protein mislocalization. Mislocalization of these proteins, including oncoproteins, tumor suppressors, and other cancer-related proteins, can interfere with normal cellular function and cooperatively drive tumor development and metastasis. This review describes the cancer-related effects of protein subcellular mislocalization, the related mislocalization mechanisms, and the potential application of this knowledge to cancer diagnosis, prognosis, and therapy.
Keywords: subcellular localization, cancer, oncogene, tumor suppressor
1. Introduction
All eukaryotic cells are surrounded by a plasma cell membrane and contain a membrane-bound nucleus. The compartment between the plasma and nuclear membranes, called the cytosol, contains numerous other membrane-enclosed organelles, such as endoplasmic reticulum, Golgi complex, mitochondria, and peroxisomes. Numerous proteins are localized inside cellular spaces or are embedded into membranes, where they play various functions to regulate cell survival, proliferation, differentiation, or death. Protein translation occurs primarily in the cytosol. Accurate trafficking and translocation of proteins from the cytosol to their ultimate destinations is essential for maintaining proper cellular function and activities. It is estimated that about half of proteins have to be transported to their functional destination [1].
Cancer has hallmarks including “sustaining proliferative signaling, evading growth suppressors, resisting cell death, enabling replicative immortality, inducing angiogenesis, activating invasion and metastasis, reprogramming of energy metabolism, and evading immune destruction” [2]. These changes from normal cells to tumor cells are primarily regulated by genome instability, which fosters hallmark functions via different mechanisms such as aberrant expression and function of tumor suppressors and oncogenes. Protein mislocalization is a less emphasized mechanism in cancer development; however, aberrant subcellular localization of certain proteins including tumor suppressor proteins and oncoproteins has been frequently reported in various cancers. The mislocalization of these proteins can alter their function such that their ability to suppress tumor cells is diminished or their ability to induce cancer development, metastasis, or drug resistance is increased. Thus, the mislocalization of such proteins could serve as novel diagnostic markers or therapeutic targets of cancer. This review describes the mechanisms and functional diversity of protein mislocalization associated with cancer and the potential application of this knowledge in the clinical setting.
2. Mislocalization of oncoproteins
The mutation or overexpression of oncogenes can cause a cell to be transformed to a malignant state. Many oncoproteins are mislocalized in tumor cells compared to normal cells (Table 1). The well-known tyrosine kinase epidermal growth factor receptor (EGFR), which is normally localized in the plasma membrane, can be trafficked to the nucleus, causing cancer [3]. Nuclear EGFR can bind to the promoters of cyclin D1 and B-Myb, inducing the expression of these cell cycle-promoting genes [4]. Because the EGFR nuclear translocation mechanism and function have been reviewed extensively elsewhere [5], we focus here on a less well known oncoprotein, mucin 1 (MUC1), as an example on the scope of subcellular mislocalization and function in cancers. The complex role of MUC1 at different subcellular localizations is depicted in Figure 1.
Table 1.
Protein localization in normal cells and in cancer cells
| Protein | Subcellular localization | Mislocalization mechanisms in cancer | Function of protein mislocalization | Reference | |
|---|---|---|---|---|---|
| Normal cells | Cancer cells | ||||
| Breast Cancer Gene 1 (BRCA1) | Nucleus | Cytoplasm | Mutations on exon 11 result in splicing variants lacking NLS; p53 may promote BRCA1 nuclear export by interrupting the association of BRCA1 with BARD 1 | Nuclear BRCA1 functions in DNA repair and cell-cycle checkpoints, whereas cytoplasmic BRCA1 regulates centrosome function and p53-independent apoptosis | [137–142] |
| p53 | Nucleus | Nucleus Mitochondria, Cytoplasm | Mutations; interaction with MDM2; CRM1-mediated nuclear export | Loss of tumor suppressor function; p53 mutants function as dominant-negative in the nucleus | [57, 58, 143] |
| Retinoblastoma protein (Rb) | Nucleus | Cytoplasm | CDK phosphorylation-dependent nuclear export | Loss of tumor suppressor role | [144, 145] |
| Sirtuin 1 (SIRT1) | Nucleus | Cytoplasm | Deregulation of PI3K/IGF-1R signaling | Loss of tumor suppressor function | [146, 147] |
| Tumor-specific cyclin E isoforms | Nucleus | Cytosol | Deletion of the NLS portion of the aminoterminus | Oncogenic role | [148] |
| V-Erb-B2 Erythroblastic Leukemia Viral Oncogene (ErbB3) | Plasma membrane, Cytoplasm | Nucleus | Bone microenvironment and androgen signaling; alternative transcription initiation producing a functional NLS | Oncogenic role | [149] |
| Mucin-13 | Transmembrane | Cytoplasm, Nucleus | Unclear | Oncogenic role | [150] |
| Inhibitor of Growth Family, Member 1 (ING1/p33) | Nucleus | Cytoplasm | Mutations | Loss of tumor suppressor function | [151] |
| SET-CAN fusion protein | Nucleus | Cytoplasm | Unclear | May be involved in oncogenesis of acute undifferentiated leukemia | [152] |
| Mucin 1 C-terminal (MUC1-C) | Apical membrane | Cytoplasm, Nucleus, Mitochondria | Interaction with β-catenin and EGFR | Oncogenic signal transduction; induce oncogene expression | [32, 95, 153] |
| KL-6 mucin | Cytoplasm, Cell surface | Nucleus | Unclear | Oncogenic role | [154] |
| Epidermal growth factor receptor | Membrane | Cytoplasm, Nucleus | MUC1-dependent, c-Src signaling | Oncogenic signal transduction; induce oncogene expression | [19] |
| Androgen receptor | Cytosol | Nucleus | Nuclear import is facilitated by the f-actin cross-linking protein filamin | Oncogenic role | [155] |
| Nuclear receptor corepressor (N-CoR) | Nucleus | Cytoplasm | Aberrant IKK activation, IKK-induced phosphorylation and binding with 14-3-3 | Loss of tumor suppressor function | [156] |
| Nucleophosmin (NPM1) | Nucleus | Cytosol | Mutations create new NES and impair NLS | Interfere with tumor suppressor ARF | [67, 68, 157] |
| β-Catenin | Membrane | Nucleus, Cytoplasm | Mutations in exon 3 (loss of GSK3β-targeted phosphorylation) | Oncogenic role | [158–161] |
| Adenomatous polyposis coli (APC) | Plasma membrane, Nucleus | Cytoplasm | Mutations; CRM1-dependent nuclear export | Loss of tumor suppressor function; important for colon cancer development | [162, 163] |
| p27 | Nucleus | Cytoplasm | Oncogenic activation of PI3K- and MEK-dependent kinases | Loss of tumor suppressor function | [164, 165] |
| p44/WDR77 | Nucleus | Cytoplasm | Loss of NLS function | Promote proliferation of prostate epithelial cancer cells | [166] |
| E-cadherin | Membrane | Nucleus, Cytoplasm | C-terminal fragment of β-cadherin is translocated into the nucleus by p120 | Loss of tumor suppressor role; induce oncogene expression | [167] [168] |
| ERM Binding Protein 50 (EBP50) | Cytoplasm | Nucleus | Interaction with β-catenin | Oncogenic role; promote colorectal carcinogenesis | [169] |
| Epithelial Cell Adhesion Molecule (EPCAM) | Cell surface | Cytoplasm, Nucleus | Unclear, concomitant with nuclear import of β catenin | A marker for aggressive thyroid cancer and poor prognosis | [170] |
| B7-H1 | Cell surface | Nucleus | Induced by doxorubicin treatment | A possible anti-apoptotic function | [61, 171] |
| B7-H3 | Cell surface | Nucleus | Unclear | Predict poor outcome in colon cancer | [63] |
| B7-H4 | Cell surface | Nucleus | Unclear | promote tumor progression and cell proliferation | [62] |
| Cyclin D1 | Cytosol | Nucleus | KRAS and PIK3CA mutations lead to increased AKT activity and inactivation of GSK3β, loss-of-function mutation of F-box protein 4, and missense mutations in cyclin D1 gene | Oncogenic role | [172–174] |
| Vascular Endothelial Growth Factor Receptor (VEGFR) | Transmembrane | Cytosol, Nucleus | unclear | Angiogenesis; induce VEGFR expression | [175] |
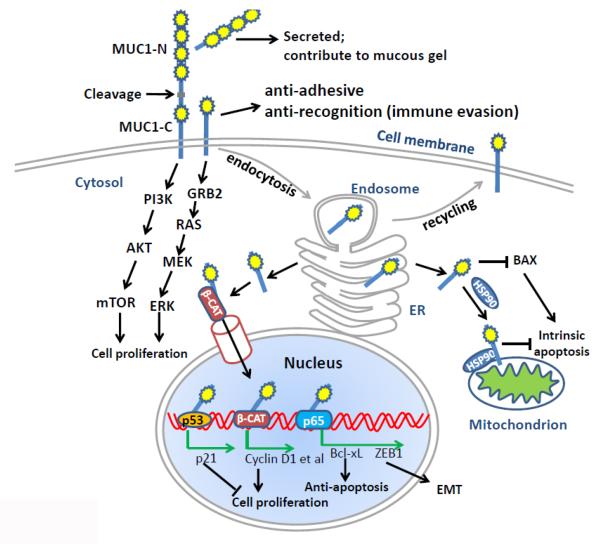
Figure 1. The complex role of MUC1 at different subcellular localizations in cancer.
MUC1 is cleaved at the SEA domain to generate two subunits, MUC1-N and MUC1-C. MUC1-N is secreted to extracellular compartments and contributes to the mucous gel. MUC1-C is a transmembrane subunit. The extracellular portion of MUC1-C is thought to play anti-adhesive and anti-recognition functions, which may be involved in the immune evasion. The intracellular portion of MUC1-C can interact with multiple signaling molecules such as PI3K p85 subunit and GRB2, activating AKT/mTOR and RAS/ERK pathways, respectively. The activation of these signaling pathways is important for cell survival and proliferation. MUC1-C constitutively undergoes endocytosis for receptor recycling. MUC1-C in the ER can be released to cytosol by the Sec61 translocon. The cytoplasmic MUC-C1 can be further imported into nucleus, where it associates with multiple transcriptional factors or nuclear receptors, such as p53, β-catenin and NF-κB p65, inducing the expression of targeted genes that are important to tumor cell proliferation or survival in the adverse microenvironment. Hsp90 can assist the transport of cytoplasmic MUC1-C onto mitochondria, where it suppresses intrinsic apoptosis. The cytoplasmic MUC1-C can also bind to proapoptotic protein BAX, antagonizing its function.
Mucins are large proteins with a common glycosylated proline-threonine-serine domain. They are normally either secreted to extracellular compartments or are associated with the plasma membrane [6]. Secreted and transmembrane mucins differ mainly in the absence and presence of a single membrane-spanning region, respectively. Adenocarcinomas release high levels of mucin, which is thought to shield tumors from toxic microenvironments [6]. Membrane-bound mucin is normally expressed at the apical borders of glandular epithelial cells [7]. In carcinoma cells, mucin is overexpressed over the entire cell surface as well as in the nucleus, mitochondria, and cytoplasm [7]. Expression of mucin on the membrane may confer anti-adhesive properties, leading to the loss of cell–cell and cell–matrix interactions of cancer cells. In addition, membranous mucin confers anti-recognition properties, leading to immune surveillance evasion. Furthermore, transmembrane mucin can transduce signaling to regulate cancer cell proliferation and growth by interacting with EGFR and β-catenin [8].
The first report of nuclear localization of the MUC1 protein was made by Wen et al. in 2003 [9]. That group found that the MUC1 cytoplasmic domain (MUC1-C) was distributed on the plasma membrane, in the cytoplasm, and in the nucleus of S2-013 and Panc-1 human pancreatic cancer cells [9]. The nuclear localization of MUC1-C and its interaction with β-catenin has since been confirmed for various adenocarcinomas [9–14]. β-Catenin binds directly to the MUC1-C SAGNGGSSL motif (amino acid residues from 50–59) [15]. EGFR and c-Src phosphorylate MUC1-C at Y-46, thereby increasing the binding of MUC1 and β-catenin [16, 17]. In contrast, GSK3β binds to and phosphorylates the MUC1-C at S-44 and decreases the interaction of MUC1-C and β-catenin in the nucleus [18].
There is substantial evidence that MUC1-C contributes to the growth and metastatic properties of tumors. This contribution is at least partially mediated by nuclear MUC1-C, which regulates the functions of several important tumor regulators, including β-catenin, EGFR, and p53. β-catenin is associated with MUC1-C in both the cytoplasm and nucleus [9]. Nuclear MUC1 co-activates β-catenin–dependent gene transcription [10], whereas the Y46F mutation decreases the MUC1 association with β-catenin, anchorage-independent growth, and tumorigenicity [10]. Nuclear localization of MUC1 and its interaction with γ-catenin can be induced by heregulin [12]; furthermore, mutation of an RRK motif in MUC1-C abrogates the nuclear localization of MUC1 and γ-catenin [12]. In addition, MUC1-C regulates the trafficking and nuclear activity of EGFR, which binds to the promoter of cyclin D1 and therefore induces gene expression and cell proliferation [19]. MUC1 can also associate with estrogen receptor α (ERα) complexes on estrogen-responsive promoters, which enhances ERα promoter occupancy and recruitment of the p160 co-activators SRC-1 and GRIP1. Consequently, MUC1 stimulates ERα-mediated transcription and contributes to the E2-mediated growth and survival of breast cancer cells [20].
The nuclear localization of MUC1-C has an anti-apoptotic role in drug-resistant cancers. MUC1-C binds directly to p53 in the nucleus, which in turn increases the occupancy of p53 on the p21 promoter region while decreasing the binding of p53 to the Bax promoter [6]. Upregulation of p21 induces cell cycle arrest, which can protect cells from p53-mediated apoptosis [21]. On the other hand, Bax is a pro-apoptotic protein that mediates p53-induced apoptosis [22]. Therefore, as a consequence of MUC1-p53 interaction in the nucleus, MUC1 activates p53-dependent growth arrest and suppresses p53-dependent apoptotic response to DNA damage [6]. MUC1-C is constitutively associated with nuclear factor κB (NF-κB) p65, and tumor necrosis factor α stimulation induces occupancy and activation of these complexes on the NF-κB response element in the Bcl-xL gene promoter [23]. Bcl-xL, which acts as a pro-survival/anti-apoptotic factor, is often overexpressed in cancer cells during the development of chemoresistance [24, 25]. Therefore, nuclear MUC1 may protect cell chemoresistance by up-regulating Bcl-xL. MUC1 has been shown to protect multiple myeloma cells against apoptosis induced by melphalan and dexamethasone through activating the β-catenin and NF-κB pathways [26]. Moreover, MUC1 sequesters c-Abl in the cytoplasm, and thereby inhibiting the action of genotoxic anticancer agents [27].
Nuclear MUC1-C may also play an important role in inducing endothelial–mesenchymal transition (EMT) and cellular invasion [28]. MUC1-C forms a complex with NF-κB p65 and functions as a co-activator of p65 in the nucleus [23]. The MUC1-C–p65 complex occupies and activates the promoter of ZEB1, a crucial transcriptional factor that induces EMT [28, 29]. MUC1 in turn associates with ZEB1 and contributes to the ZEB1-mediated transcriptional suppression of miR-200c, an EMT suppressor. As a consequence of MUC1-mediated ZEB1 activation and miR-200c suppression, nuclear MUC1-C induces EMT and cellular invasion of breast cancer cells and possibly other cancers [28].
In addition to nuclear localization, localization of MUC1 in the mitochondria has been reported for a variety of cancer cell lines such as HCT116 colon carcinoma cells and ZR-75-1 breast cancer, as well as primary tumors [30–35]. These observations were confirmed using both confocal microscope imaging and western blotting of mitochondrial lysate fractions [30, 31]. Furthermore, MUC1 is localized to mitochondria in 33.33% (5 of 15) of dysplasia samples and in 47.05% (8 of 17) of adenocarcinoma samples of human gastric tissues [35]. The transport of MUC1-C to mitochondria can be induced by heregulin, a pleiotropic growth factor [30]. Heregulin induces the activation of c-Src kinase, which phosphorylates MUC1-C and stimulates the binding of MUC1 to HSP90 [32, 33]. Whereas nuclear localization of MUC1-C depends on its association with β-catenin, delivery of MUC1 to the mitochondrial outer membrane is facilitated by HSP90 [32, 33].
Mitochondrial MUC1-C plays a protective role for tumor cells by suppressing intrinsic apoptosis, which contributes to the drug-resistant phenotype of cancer cells. Mitochondrial MUC1-C attenuates cytochrome c release and caspase-3 activation and therefore suppresses apoptosis [30]. Furthermore, mitochondrial MUC1-C also binds directly to the BAX BH3 domain in the cytoplasm and mitochondria, thereby blocking the function of BAX in activating the mitochondrial death pathway [31]. Treatment of multiple myeloma cells with a MUC1-C inhibitor causes cell death associated with increased levels of reactive oxygen species, oxidation of mitochondrial cardiolipin, and loss of the mitochondrial transmembrane potential [36]. Many genotoxic anticancer drugs induce apoptosis by activating the intrinsic apoptosis pathway. Mitochondrial MUC1 may confer the drug-resistant phenotype to cancer cells by attenuating the release of mitochondrial cytochrome c and the activation of other pro-apoptotic factors. For example, overexpression of MUC1-C in HCT116 cancer cells reduces the cell apoptosis induced by cisplatin and TRAIL; this effect is abolished by Y46F mutation of MUC1, a mutation that is ineffective in blocking cisplatin-induced cytochrome c release [30]. Conversely, small interfering RNA knockdown of MUC1-C in A549 carcinoma cells sensitizes them to apoptosis induced by cisplatin in vitro [30]. Moreover, HCT116 tumor cells expressing MUC1 are resistant to cisplatin treatment in vivo compared with cells that do not express this protein [30].
Hypoxia induces cell apoptosis, mainly through the intrinsic or mitochondrial pathway [37]. It has been reported that MUC1 plays a critical role in attenuating hypoxia-induced loss of mitochondrial transmembrane potential and apoptosis [38]. This effect is mediated by inhibiting the activation of hypoxia-inducible factor 1α, the key regulator of hypoxia-induced apoptosis, as well as by suppressing the accumulation of reactive oxygen species [38]. The suppression of hypoxia-induced apoptosis can be abolished by the MUC1-C Y46F mutation [38]. Thus, localization of MUC1-C in the mitochondria may be an important mechanism for cancer cell survival under hypoxia stress, which is a general feature of the microenvironment for solid tumors.
3. Mislocalization of tumor suppressor proteins
Tumor suppressors are generally proteins that slow down the cell cycle, promote apoptosis, or both. Many of these proteins have been found to display different subcellular localization patterns between physiologically normal cells and cancer cells. These include BRCA1, p53, retinoblastoma, ING1/p33 [39], and adenomatous polyposis coli (APC) [40] (Table 1). In general, tumor suppressors possessing transcriptional functions tend to localize in the nucleus of normal cells but in the cytoplasm of cancer cells. We focus here on p53 as an example of how the localization of tumor suppressors can affect tumor development.
The p53 protein is a homotetrametric transcription factor that is well known to safeguard against cancer. It exerts its antitumor role mainly through regulating the cell cycle checkpoint, DNA damage repair, and cell apoptosis. In order to regulate the target gene transcription, p53 has to be translocated into the nucleus when DNA damages occurs or cellular stress reaches a dangerous level [41]. Thus, the nucleus is the primary site of p53 function in tumor suppression.
The nuclear–cytoplasmic shuttling of p53 is tightly controlled by nuclear import/export as well as cytoplasmic sequestration mechanisms. The nuclear localization signal (NLS) and nuclear export signal (NES) are the essential elements for a protein undergoing nucleocytoplasmic transportation. The p53 protein harbors three NLSs [42, 43] and two NESs [44, 45]. Many other proteins work directly or indirectly with these signals to regulate p53 translocation. For example, the binding of importin-α with the NLS of p53 enables the nuclear import of p53 [43, 46]. In contrast, the NES of p53 can bind to CRM1, leading to the nuclear export of p53 [44, 47]. MDM2 can bind to nuclear p53, inducing p53 ubiquitination and export from the nucleus [48]. Other modifications, such as S315 phosphorylation of p53, mediate cell cycle-dependent nuclear retention of p53 by E2F1 [49]. Other proteins, such as c-Abl and PI3K/AKT, can regulate MDM2 activity, which in turn influences the nuclear export of p53. The nuclear localization of p53 protein allows it access to its many target genes, such as p21, p53R2, MDM2, p53R2, BAX, p53AIP1, NOXA, and PUMA. The expression of these p53-targeted genes results in cell cycle arrest, DNA repair, and cell apoptosis [50].
In unstressed cells, p53 protein is present at a low level in the cytosol due to MDM2-mediated targeted degradation. In a study of neuroblastomas, 96% (30 of 31) of undifferentiated tumor samples exhibited an increased level of wild-type p53 in the cytoplasm and a lack of nuclear staining; in contrast, cytoplasmic p53 was not detected in 14 differentiated ganglioneuroblastomas [51]. Cytoplasmic accumulation and retention of p53 has also been found in about 40% of breast cancer tissues, retinoblastomas, glioblastomas, and hepatocellular carcinomas [41, 52]. The retention of cytoplasmic p53 by the heat shock protein Mortalin has been reported for several tumor cell types, including human glioblastomas, hepatocellular carcinomas, and colorectal adenocarcinomas [53]. In contrast to the retention of wild-type p53 in the cytoplasm, mutant p53 tends to accumulate in the nucleus of colorectal adenocarcinoma [54]. Cytoplasmic accumulation of p53 could be due to the excess nuclear export of p53, defective cytoplasmic degradation, retention by cytoskeleton proteins, or other mechanisms.
Constitutive cytoplasmic localization of p53 in cancer cells has been associated with poor response to chemotherapy, tumor metastasis, and short-term patient survival [41]. For example, p53 is accumulated and sequestered in the cytoplasm of estrogen-independent human breast cancer cells that are resistant to tamoxifen and methotrexate [55]. In addition, cisplatin sensitivity is greatly reduced in head and neck squamous cell carcinomas with loss of nuclear p53 signal [56].
Activation of programmed cell death, i.e., apoptosis, is an important mechanism in p53 tumor suppression. Translocation of p53 to mitochondria has been reported as a feature of cancer cells. This translocation and the subsequent inactivation of manganese superoxide dismutase explain the observed mitochondrial dysfunction, which leads to transcription-dependent mechanisms of p53-induced apoptosis [57]. Monoubiquitylation promotes mitochondrial p53 translocation, which can initiate apoptosis if DNA damage proves to be irreparable [58].
4. Mislocalization of other cancer-related proteins
Many other types of proteins are differentially located in physiologically normal cells and tumor cells. For example, sphingosine 1-phosphate is a bioactive lipid that has an important role in promoting tumor survival, growth, and invasiveness. Mislocalization of sphingosine 1-phosphate receptor regulators is associated with poor prognosis and with the development of tamoxifen resistance for ER-positive breast cancer patients [59]. This study demonstrated that sphingosine 1-phosphate receptors 1 and 3 were not only located in the cell membrane but can be also found in the cytoplasm in breast cancer; and cytoplasmic expression of these proteins was associated with disease-specific poor survival [59].
Another example is B7-H1. This protein is of particular interest because its expression on the surface of cancer cells could provide inhibitory signaling to T cells, inducing the lymphocyte apoptosis [60]. Many cancer cells express B7-H1 as a mechanism to evade immune attack and surveillance. A recent study found that doxorubicin down-regulated cell surface expression of B7-H1 in breast cancer cell lines, which is concurrently translocated into the nucleus [61]. Gene silencing of B7-H1 in breast cancer cells has been found to increase doxorubicin-induced apoptosis, suggesting that nuclear localization of this protein may play an anti-apoptotic role during chemotherapy [61]. Similarly, the inhibitory co-stimulatory membrane molecules B7-H3 and B7-H4 have been reported to redistribute into the nucleus in colon cancer and renal cell carcinoma and to be associated with more advanced disease and poor prognosis [62, 63]. Furthermore, B7-H4 confers chemoresistance to renal cell carcinoma by regulating cell cycle checkpoints [62].
5. Mechanisms of protein mislocalization
Protein synthesis occurs primarily in the cytosol. From there, proteins are transported to their functional sites, such as the nucleus, plasma membrane, mitochondria, and other organelles. Mechanisms for protein trafficking and translocation have been reviewed in depth by Wickner and Schekman [64]. Briefly, transport systems recognize proteins by their signal peptide sequences. In most cases, proteins form complexes with chaperones or ribosomes, bind to a membrane receptor, are transferred through a membrane-embedded translocator or channel, and are released into or cross the membrane [64]. In addition to this relatively universal transporter system, a protein may be redeposited by its interacting regulatory proteins. Furthermore, modification of the signal transduction and post-translocation systems is essential for triggering of the protein translocation events. In cancer cells, several mechanisms are responsible for the dysregulation of protein trafficking, which leads to abnormal subcellular localization of proteins.
5.1 Mutation of protein-targeting signals
The NLS and NES are the essential elements required for nucleocytoplasmic transportation of a protein. For example, p53 harbors three NLSs [42, 43] and two NESs [44, 45]. Complete loss of nuclear p53 signal was observed in three of nine investigated head and neck squamous cell carcinoma cell lines; this loss was due to mutations and disruption of the p53 COOH-terminal NLS [56]. In human ovarian cancer line OV-MZ-32, p53 staining occurs exclusively in the cytoplasm, which is due to a deletion mutation of the major NES [65]. Mutations of exon 12 of the nucleophosmin-1 gene are frequently identified in acute myeloid leukemia (AML) [66]. Nucleophosmin-1 mutations create a NES motif and disrupt tryptophans at the NPM1 C-terminus, resulting in cytoplasmic accumulation of this protein in leukemic cells [67, 68]. In addition, almost all solid pseudopapillary tumors of the pancreas contain a mutation in the β-catenin gene, which may account for the nuclear localization of β-catenin in these tumors [69].
5.2 Dysregulation of transporter machinery
NLS and NES are recognized by nuclear–cytoplasmic transport receptors, which belong mostly to the family of β-karyopherins. These receptors are called importins or exportins, depending on their mode of action [70]. Mutations of NLS and NES are relatively rare in cancer. In contrast, the dysregulation of importins and exportins seems to be a universal mechanism for protein mislocalization. For example, CRM1/exportin 1 mediates the nuclear export of many tumor suppressor proteins, including retinoblastoma, APC, p53, BRAC1, and FOXO. High levels of CRM1 protein expression have been reported for various cancers, including AML, ovarian cancer, pancreatic cancer, osteosarcoma, glioma, and cervical cancer [44, 47, 71, 72]. CRM1 overexpression in these cancers may account for the cytoplasmic accumulation of retinoblastoma, APC, p53, β-catenin, BRAC1, FOXO, INI1/hSNF5, galectin-3, Bok, nucleophosmin, RASSF2, Merlin, p21CIP, p27KIP1, N-WASP/FAK, estradiol receptor, Tob, topoisomerases I and IIα, BCR-ABL, and Hsp90 [71]. In addition, the expression of Karyopherin α2, a member of the karyopherin α (importin α) family, has been found to be higher in a variety of malignancies compared with physiologically normal cells and to correlate with a poor prognosis irrespective of the cancer type [73]. Karyopherin α2 promotes tumorigenesis through the translocation of cancer-associated cargo proteins, which include the cell-cycle regulator Chk2, BRCA1, E2F1, NBS1, androgen receptor, p53, and c-Myc [73].
5.3 ER retention of misfolded proteins
Protein folding occurs at the endoplasmic reticulum (ER). The folding is orchestrated by various chaperone proteins. Protein misfolding and aberrant aggregation not only lead to the diminished or altered function of the protein from its normal place of action and but also induce ER stress. The accumulation of unfolded proteins and protein aggregates triggers unfolded protein responses such as inducing expression of the Hsp70 family of chaperone. Hsp90 overexpression is observed in the majority of human tumors [74]. Many tumorigenic p53 mutants are folding defective. Hsp90 interacts with p53 mutants and MDM2, increasing the stability of and cytoplasmic and nuclear localization of p53/MDM2 in tumor cells [75,76]. Furthermore, ER stress causes the retention of wild-type p53 in cytoplasm and blocks the nuclear localization and function of p53 [77]. Cytoplasmic retention of p53 is mediated by a pathway involving ER stress-induced activation of GSK-3β, which in turn phosphorylates p53 and prevents its nuclear translocation [77]. Several in-depth reviews of targeting protein misfolding and ER stress for cancer therapy have been published elsewhere [78, 79].
5.4 Aberrant endocytosis and vesicular trafficking
Membrane receptors are internalized via endocytosis. Recycling receptors are subsequently transported from early endosomes to the endosomal recycling compartment and then returned to the plasma membrane. Receptor tyrosine kinases (RTKs) such as EGFR have important tumorigenic function. Endocytic vesicles may serve as carriers to shuttle RTKs into the cell nucleus [80, 81]. Aberrant endocytosis and vesicle trafficking of EGFR mutants have been identified in non-small cell lung cancer lines and contribute to the nuclear localization of EGFR [82]. Intracellular trafficking of MUC1 is mediated by clathrin-coated pits and regulated by dynamin and Rab5, all of which are important for endocytosis and receptor recycling [83]. Many cancers exhibit overexpression or mutations of small GTPases (e.g., Rab25 overexpression in ovarian cancer and colon cancer [84]) or their regulators (e.g., Rab-coupling protein gene amplification in breast cancer [85]). As a consequence, protein mislocalization and dysfunction may occur. More information regarding endosome trafficking of EGFR to the nucleus can be found in a recent review published by Wang et al. [81].
5.5 Dysregulation of signal transduction and protein post-translational modification
Differences in phosphorylation and signal transduction are constantly observed in various tumors and play key roles in all aspects of neoplasia, including proliferation, invasion, angiogenesis and metastasis. One of the action modes of perturbed kinase activation is the altering of protein trafficking and subcellular localization of cancer regulators. The PI3K/AKT pathway is activated in many human cancers and plays a key role in cell proliferation and survival. The AKT1 (E17K) mutation has been reported in breast, colorectal, ovarian, and lung cancers and in endometrial carcinoma [86]. The E17K mutation, which occurs in the pleckstrin homology domain of AKT1, results in its constitutive phosphorylation and membrane localization. Six non-hotspot AKT1 pleckstrin homology domain mutants have also been identified in large-scale breast cancer sequencing studies [87]. Three of these mutants cause constitutive activation of AKT1 and confer constitutive membrane localization of Akt1. In addition, these three AKT mutants showed oncogenic activity in a cellular transformation assay [87]. The increased PI3K/AKT signaling promotes the nuclear export of GSK3β, thereby restricting its access to nuclear substrates such as c-myc and β-catenin. PI3K/AKT can also regulate MDM2 activity, which in turn influences the nuclear export of p53. As another example, alterations of the O-glycan structure of MUC1 have been observed in cancer cells, which promotes MUC1 endocytosis and intracellular sequestration [6].
5.6 Alteration of protein–protein interactions
Point mutation, deletion, and hypermethylation of p14ARF have been widely documented in various cancers. A consequence of these genetic or epigenetic changes is that p14ARF is loss of interaction with MDM2 and subsequent retention of MDM2 in the nucleus [88]. The cytoplasm translocation of p53 can be mediated by the interactions of p53 with cytoskeletal proteins such as actin and vimentin and with microtubules [89]. Cytoplasmic accumulation of p53 has been correlated with the presence of vimentin in rat glioma cells, and nuclear p53 has been found in vimentin-negative rat glioma cells [90]. In another study, STAT3 mutations were found in 40% of (31 of 77) patients with large granular lymphocytic leukemia. All mutations were located in exon 21, which encodes the SH2 domain. The mutations resulted in an association between the phosphorylation of STAT3 and its localization in the nucleus [91].
5.7 Cross-regulation of cancer-related proteins
The cooperation and antagonization of oncoproteins and tumor suppressors are necessary for tumorigenesis. This effect can be mediated by changing the subcellular localization of their interacting proteins. For example, MDM2 binds to nuclear p53 and helps export it to the cytosol, where p53 is sequestered and targeted for degradation. Overexpression of MDM2 in human leukemia could explain the cytosolic accumulation of p53 in these diseases [92, 93]. Another example is EGFR nuclear localization, which is regulated by trafficking of other oncoproteins and tumor suppressors (Figure 1). EGFR is associated with MUC1 on the plasma membrane; constitutive internalization of MUC1 results in EGFR being endocytosed [19, 94]. Galectin-3 mediates EGFR and MUC1 interaction, which favors the endocytosis of MUC1-C and EGFR [94]. The EGFR/MUC1-C/Galectin-3 protein complex undergoes retrograde trafficking through the endoplasmic reticulum, where they are released to cytosol via the Sec61 translocon and further imported into the nucleus through importin β1 [80, 95]. The interaction of MUC1-C with EGFR promotes the nuclear accumulation of EGFR, which binds to chromatin and colocalizes with transcriptional initiation elements on the targeted genes, inducing gene expression [19]. The constitutive nuclear import of EGFR can occur in the absence of MUC1-C [19]. This import is assisted by the Sec61 translocon and is mediated by the direct interaction of EGFR tripartite NLS with importin β1 [80, 95, 96]. However, in the absence of MUC1-C, nuclear EGFR is unable to bind to chromatin and be ready to export to cytosol for degradation [19]. Therefore, MUC1-C is important for increasing the nuclear localization and transcriptional activity of EGFR, thereby promoting cancer cell growth and proliferation.
6. Relevance of protein subcellular localization to cancer diagnosis and prognosis
Examples of clinical application of our knowledge of protein subcellular localization to cancer diagnosis and prognosis are shown in Table 2. For some proteins, subcellular localization may be different between cancer cells and normal cells; immunohistochemical staining of these proteins can be implemented for diagnostic purposes. For example, positive nuclear staining of E-cadherin and β-catenin occurs in 100% of solid pseudopapillary tumors of the pancreas and is of diagnostic use as a biomarker [97]. In addition, a study of 102 tumor specimens from breast cancer patients revealed that breast cancer cells stain strongly for serotonin receptor 1A (5-HTR1A) on the membrane, whereas non-malignant cells stain positively in only the cytoplasm. Furthermore, 5-HTR1B is predominantly expressed in the cytoplasm of breast cancer cells but stains weakly in the cytoplasm of nonmalignant epithelial cells. Thus, 5-HTR1A and 5-HTR1B could be used as diagnostic markers for breast cancer [98].
Table 2.
Implications of protein subcellular localization in cancer diagnosis and prognosis
| Protein | Relevance to cancer diagnosis and prognosis | Reference |
|---|---|---|
| E-cadherin | Nuclear expression, loss of plasma membrane expression, or both was present in 97% of Merkel cell carcinomas, 100% of solid pseudopapillary tumors of the pancreas, and 86% of pituitary adenomas, pancreatic neuroendocrine tumors, clear cell renal cell carcinoma, esophageal squamous carcinoma, colorectal and gastric cancer, or synovial sarcoma. | [69, 176–180] |
| Epithelial Cell Adhesion Molecule (EPCAM) | Extracellular domain was lost on the plasma membrane but the intracellular domain increased in cytoplasm and nucleus for all epithelial cancers, including thyroid, breast, prostate, head and neck, esophagus, lung, colon, liver, bladder, pancreatic, and ovarian. | [170, 181–183] |
| Vascular Endothelial Growth Factor Receptor (VEGFR) 2 | Positive nuclear staining in invasive lobular breast carcinomas but negative nuclear staining in invasive ductal carcinomas; cytoplasmic localization was more pronounced in patients with melanoma and diffuse large B-cell lymphoma who had poor prognoses. | [99] |
| Serotonin receptor 1A (5-HTR1A) | Moderate to strong expression, predominantly in the plasma membrane, in all 102 human breast cancer samples; cytoplasmic expression detected in non-malignant cells. | [98] |
| Epidermal growth factor receptor (EGFR) | Nuclear level was associated with higher local recurrence rate in oropharyngeal squamous cell cancer and with worse disease-free survival in gallbladder carcinoma; cytosolic phosphorylated EGFR was strongly related to increased risk of recurrence and shorter overall survival in penile cancer (36 patients). | [184–187] |
| Mucin 1 (MUC1) | Expressed on the reversed apical plasma membrane of neoplastic cell clusters in pure invasive micropapillary carcinoma cases but in the whole cytoplasmic membrane or cytoplasm in pseudoinvasive micropapillary carcinomas cases. | [100] |
| Human Epidermal Growth Factor Receptor 2 (HER2) | Cytoplasmic staining was correlated with neuroendocrine differentiation in breast carcinoma (1053 patients). | [188] |
| β-Catenin | Cytoplasmic or nuclear expression was strongly associated with poor prognosis and was an independent prognosticator for overall survival in non-small cell lung cancer 309 patients; MTC score (membrane minus cytoplasmic expression) was associated with a worse outcome in invasive ductal breast cancers (292 patients). | [189, 190] |
| Homeobox Transcription Factor Nanog | Nuclear expression was significantly correlated with nuclear β-catenin expression and poor prognosis in non-small cell lung cancer (309 patients). | [190] |
| Mammary serine protease inhibitor (MASPIN) | Nuclear expression in 108 tissue samples of laryngeal squamous cell carcinoma was associated with longer disease-free survival. | [191] |
| Transcriptional Adaptor 3 (ADA3) | Nuclear staining in breast cancer tissues served as a marker of good prognosis; predominant cytoplasmic expression was a marker of poor prognosis in breast cancer (900 cases). | [192] |
| Cytokine-induced apoptosis inhibitor 1 (CIPIN1) | Nuclear localization was an unfavorable prognostic factor in epithelial ovarian cancer (108 patients). | [193] |
| NME/NM23 nucleoside diphosphate kinase 1 (NME 1), NME23) | High nuclear expression was associated with a better prognosis in elderly patients with laryngeal carcinoma. | [194] |
| Cyclin B2 | Elevated cytoplasmic level was strongly associated with short-term disease-specific survival of breast cancer patients and with histological tumor type (80 patients). | [195] |
On the other hand, the localization of some proteins differs by cancer type and thus might be used for differential diagnosis. For example, staining of nuclear vascular epithelial growth factor 2 is positive in invasive lobular breast carcinomas but negative in invasive ductal breast carcinomas [99]. As another example, the detection of subcellular localization of MUC1 is useful in differentiating between invasive micropapillary carcinoma (IMPC) of the breast and conventional invasive ductal carcinoma showing an IMPC-like pattern due to artifact (pseudo-IMPC) [100].
Although the expression level and subcellular localization of a specific protein may be highly associated with some tumor types, they may not necessarily indicate cancer. Rather, a combination of markers can provide values for difficult-to-diagnose cancers. For example, the results of immunohistochemical staining of β-catenin and E-cadherin are useful for differentiating a solid pseudopapillary neoplasm from a pancreatic endocrine neoplasm or adenocarcinoma [101]. Kim et al. found that 94% of solid pseudopapillary neoplasm cases were positive for nuclear β-catenin and none was positive for nuclear E-cadherin, whereas 96% of pancreatic adenocarcinoma or endocrine neoplasm cases were positive for nuclear E-cadherin and none was positive for nuclear expression of β-catenin [101]. Molecular morphologic techniques may achieve a more objective and reproducible diagnosis of difficult-to-diagnose cancers. For example, three experts who evaluated results from immunohistochemical analysis of HMB-45, Ki67, cyclin D1, E-cadherin, and p16 and fluorescence in situ hybridization for melanoma developed a “consensus diagnosis” for 13 melanocytic skin neoplasms that provided an objective and reproducible result [102].
For many cancers, clinical staging and histological grading of biopsy or surgical tumor samples are recognized as “gold standards” for predicting the prognosis and planning adjuvant therapy. Immunohistochemical staining of the expression and subcellular localization of some biomarkers alone or in combination provides additional prognostic information. In a study of E-cadherin and EGFR expression in lung adenocarcinoma or squamous cell carcinoma tissues from 131 patients, both negative and cytoplasmic staining of E-cadherin correlated with shorter patient survival [84]. Patients with lung adenocarcinoma or squamous cell carcinoma with negative E-cadherin expression and positive EGFR expression had a worse disease outcome [103]. Similarly, immunohistochemical analysis of E-cadherin and EGFR in 143 surgical specimens of head and neck squamous cell carcinoma revealed that the expression or membrane localization (or both) of these proteins was useful in predicting lymph node metastasis, patient survival, and response to EGFR-targeted therapy [104]. Furthermore, double staining of MUC1 and β-catenin in colorectal carcinoma tissues can provide prognostic value: the combination of MUC1 expression and low membranous β-catenin expression distinguishes a subgroup of patients with a worse prognosis [105].
7. Targeting protein subcellular localization for cancer therapy
The translocation of many potential oncoproteins, tumor suppressors, and other cancer-related proteins plays critical roles in cancer progression, metastasis, and drug resistance. Thus, blocking protein translocation is a strategy for cancer treatment. Depending on the cancer type and the translocation mechanism, small molecule inhibitors or analogs that directly interfere with the protein trafficking, interactions, or signaling required for protein translocation could be used.
7.1 Targeting nuclear exporters or importers
Overexpressed in many cancers, the protein CRM1 is responsible for the nuclear export of many tumor suppressor proteins, such as p53, APC, BRAC1, and FOXO [44, 47, 71, 72, 106]. Blocking the CRM1-mediated nuclear export of such proteins may restore their tumor suppressor function. Orally bioavailable small-molecule selective inhibitors of nuclear exporter (SINE) that can irreversibly bind to CRM1 and block its function have been developed [107]. In in vitro studies, SINE compounds (KPT-185 and KPT-276) can induce apoptosis and inhibit proliferation in many cancer cell lines, including renal cell carcinoma, colon cancer, pancreatic cancer, breast cancer, chronic myeloid leukemia, lymphocytic leukemia, and AML[107, 108] [109] [109–112]. KPT-185 caused nuclear retention of p53 and p21, suggesting that the mechanism of action of these compounds depends on tumor suppressor protein localization [108]. KPT-276 has the same CRM1 binding warhead and specificity as KPT-185 but appears to have superior oral bioavailability and pharmacokinetics. Thus, KPT-276 was selected for in vivo testing. Oral administration of the KPT-276 in in various preclinical models showed remarkable anti-tumor efficacy without any sign of toxicity effect [107–112] (Table 3). These promising results warrant testing in a clinical trial.
Table 3.
Recent preclinical studies of targeting protein translocalization for cancer treatment*
| Compounds | Mechanisms | In vivo tumor models | Effects | Reference |
|---|---|---|---|---|
| Selective Inhibitors of Nuclear Export (SINE) (e.g., KPT251, KPT-276 and KPT-330) | Orally bioavailable small-molecule selective inhibitors of nuclear exporter CRM1 cause nuclear retention of p53 and p21, increase ERK phosphorylation, and reduce retinoblastoma protein (pRb) phosphorylation. They can also block RANKL-induced NF-κ B and NFATc1. | NSG immune deficient mice transplanted with acute myeloid leukemia | KPT-276 treatment prolonged mice survival (median survival drug-treated vs vehicle, 39.5 vs 27 days, respectively). | [107] |
| SCID mice engrafted with active CLL-like leukemia from Eμ-TCL1 transgenic mice | KPT-276 treatment slowed disease progression, improved overall survival with minimal weight loss or other toxicities. | [109] | ||
| NSG immunodeficient mice engrafted with human T-ALL MOLT-4 cells | KPT-330 showed striking in vivo activity against T-ALL and AML cells, with little toxicity to normal murine hematopoietic cells. | [111] | ||
| SCID mice engrafted with diffuse human multiple myeloma bone lesions | KPT-251 or KPT-276 treatment showed strong anti-MM activity, inhibit MM-induced bone lysis and prolong survival. Moreover, they directly impaired osteoclastogenesis and bone resorption with minimal impact on osteoblasts and BMSCs. | [112] | ||
| Nude mice engrafted with human renal cell carcinoma | KPT-276 treatment inhibited tumor growth without adverse effects. | [108] | ||
| Peptide inhibitor of MUC1-C(e.g., GO-201 and GO-203) | They are cell-penetrating peptide inhibitors of oncogenic protein MUC1-C; they interact directly with the MUC1-C subunit at its oligomerization domain; they can attenuate MUC1-C nuclear translocation, increase the association of MUC1-C and HSP70 in the cytosol, induce MUC1 transport into lysosomes, cause mitochondrial dysfunction, and activate the DNA damage response. | Nude mice engrafted with human prostate cancer cells | GO-201 treatment resulted in complete tumor regression and prolonged lack of recurrence. | [117] |
| Nude mice bearing human breast tumor xenografts | GO-201 treatment resulted in loss of tumorigenicity and extensive necrosis of tumor cells, and prolonged regression of tumor growth of breast cancer. | [196] | ||
| Nude mice engrafted with orthotopic human pancreatic cancer | GO-201 treatment reduced tumor incidence, tumor volume, and metastasis in pancreatic cancer. | [197] | ||
| Nude mice engrafted with human lung cancer cells | GO-203 treatment resulted in complete tumor regressions of lung cancer. | [120] | ||
| NSG immunodeficient mice engrafted with AML stem cells | GO-203 treatment depleted established AML in vivo, but did not affect engraftment of normal hematopoietic cells. | [198] |
The preclinical studies that target kinases and signaling pathways involved in protein translocation are not included in this table.
Two peptide inhibitors, bimax1 and bimax2, have been identified as specific blockers of the classic importin α/β pathway [113]. Bimax1 treatment of HeLa cells impaired nuclear localization of NAC1, an oncogenic nuclear protein involved in ovarian cancers and cervical carcinomas [114, 115]. Another cell-permeable SN52 peptide was shown to block nuclear import of NF-κB family members, p52 and RelB, through competing with p52/RelB for nuclear import proteins [116]. Importantly, SN52 can sensitize prostate cancer cells to ionizing radiation at clinically relevant radiation doses with little cytotoxicity to normal prostate epithelial cells [116]. The usage of these peptide inhibitors in in vivo tumor models has not been reported yet.
7.2 Targeting protein–protein interactions
The synthesized cell-penetrating peptide inhibitor GO-201 interacts directly with the MUC1-C subunit at its oligomerization domain. This peptide inhibitor has a potent antitumor role in various cancer cell lines [8, 117, 118]. Inhibition of the MUC1-C oligomerization domain by GO-201 subsequently affects the interactions of MUC1-C with multiple effectors at the cell membrane, in the cytoplasm, and in the nucleus as well as the localization of MUC1-C in the nucleus. Treatment with GO-201 suppresses interaction between MUC1-C and TCF7L2, resulting in the inhibition of Wnt/β-catenin pathway in breast cancer cells. Furthermore, inhibition of MUC1-C blocks its interaction with STAT3 and RelA, resulting in STAT3 inhibition and NF-κB activation, respectively. On the basis of these findings and activity identified in other carcinoma models, a second-generation MUC1-C inhibitor, GO-203, has been developed and is undergoing phase I/II evaluation for patients with refractory solid tumors and AML [119]. Treatment of chronic myelogenous leukemia cells and non-small cell lung cancer cells with GO-203 increases these cells' expression of reactive oxygen species (ROS), resulting in cell cycle arrest and apoptosis [118, 120]. The induction of ROS by GO-203 decreases protein levels of Bcr-Abl and β-catenin, which in turn induce terminal myeloid differentiation [118]. How ROS downregulates Bcr-Abl and β-catenin protein levels remains unclear. ROS may induce the cleavage and hence dysfunction of Hsp90, an essential chaperone that regulates the folding and stability of Bcr-Abl and β-catenin [121]. In addition, GO-203 can directly disrupt the interaction of MUC1-C with Bcr-Abl and β-catenin, which in turn destabilize these oncogenic proteins [122, 123].Preclinical studies of the anti-tumor efficacy and safety of MUC1–C inhibitors are summarized in Table 3.
7.3 Targeting signaling pathways that regulate protein localization
Cetuximab is an anti-EGFR antibody that has been used successfully to treat EGFR-expressing head and neck cancer and colorectal cancer. However, cetuximab resistance has been observed in patients with various types of cancer [124, 125]. Nuclear localization of EGFR may be a critical mechanism for cetuximab resistance [126, 127]. The translocation of EGFR to the nucleus involves different mechanisms, but the phosphorylation and activation of EGFR by Src family kinases in particular seems to be critical for nuclear entry of EGFR [126]. Dasatinib, an inhibitor of c-Src kinases, can interfere with the nuclear localization of EGFR in vitro, thereby enhancing the radiosensitivity of human head and neck squamous cell carcinomas in vitro [128]. When cetuximab fails, this approach may provide an alternative therapeutic strategy by targeting EGFR nuclear translocation. This is supported by the observation that combination of Dasatinib and Cetuximab sensitized KRAS mutant colorectal tumor, which exhibited minimal response to dasatinib or cetuximab monotherapy in vivo [129].
The overexpression and hyperactivation of PI3K/AKT/mTOR signaling are consistently observed in various tumors, such as hepatocellular carcinoma and cancers of the ovaries, pancreas, stomach, colorectum, prostate, and breast [130, 131]. PI3K/AKT/mTOR signaling promotes tumor survival, proliferation, and drug resistance via different mechanisms, such as by regulating the nuclear export and mitochondrial translocation of p53 [132]. Inhibitors of the PI3K/AKT/mTOR pathway have been intensively evaluated in various preclinical cancer models, showing promising anti-tumor activity and restoration of drug sensitivity [133–135]. The evaluation of these agents in early stage clinical trials has reported to be well tolerated and show moderate clinical benefits in multiple tumor types, which are reviewed in recent paper published by Rodon et al [136].
8. Concluding remarks
We have described evidence from the literature that deregulation of protein translocation plays critical roles in inducing cell transformation, survival, proliferation, apoptosis, and drug resistance. The differential subcellular localization pattern of potential oncoproteins, tumor-suppressor proteins, and other cancer-related proteins compared with physiologically normal proteins provides clues for the clinical diagnosis and prognosis of a variety of human cancers. Drugs that have been developed to target protein translocation mechanisms have shown great promise in preclinical and clinical studies.
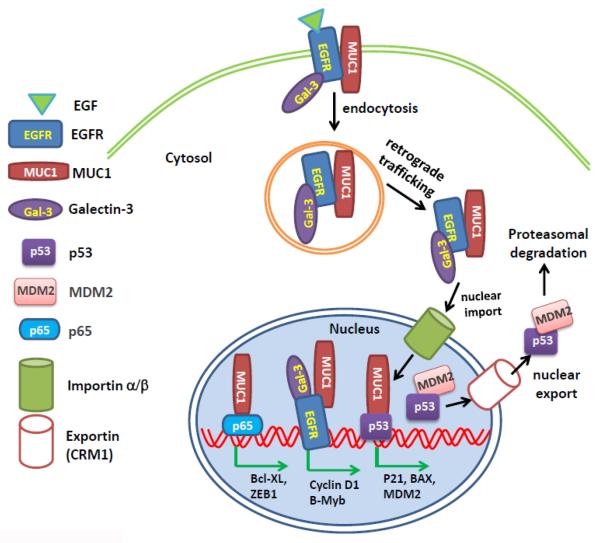
Figure 2. Schematic model of cross-regulation of protein subcellular localization of oncoproteins and tumor suppressor proteins.
EGFR is constitutively associated with MUC1 on the plasma membrane. On binding of epidermal growth factor, EGFR and MUC1 are endocytosed. Both proteins undergo retrograde trafficking through the endoplasmic reticulum and are imported into the nucleus via importin β1. Galectin-3 associates with EGFR to facilitate the nuclear import of MUC1 and EGFR [94]. The interaction of MUC1 and EGFR in the nucleus promotes nuclear EGFR accumulation and transcriptional activation, which induces the gene expression of cyclin D and B-Myb and promotes cell proliferation. Nuclear MUC1 also binds to NF-κB p65 and induces p65-mediated gene expression of Bcl-xL and ZEB1. The nuclear association between MUC1 and p53 induces transcription of the p53 promoter and expression of p21, Bax, and MDM2. MDM2 is a negative regulator of p53. It binds to p53 in the nucleus and assists the nuclear export of MDM2 to the cytosol, where p53 is sequestered and targeted for degradation. The coordination and antagonization between these oncoproteins and tumor suppressor proteins result in tumor cell survival, proliferation, and progression.
Acknowledgement
we would like to thank Ms. Elizabeth L Hess from Department of Scientific Publications, The University of Texas MD Anderson Cancer Center, for her critical review and editing for this manuscript. This study was supported by National Cancer Institute grants RO1CA098928 and RO1CA142855.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- [3].Cohen S. The epidermal growth factor (EGF) Cancer. 1983;51:1787–1791. doi: 10.1002/1097-0142(19830515)51:10<1787::aid-cncr2820511004>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- [4].Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nature cell biology. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- [5].Dittmann K, Mayer C, Rodemann HP. Nuclear EGFR as novel therapeutic target: insights into nuclear translocation and function. Strahlentherapie und Onkologie : Organ der Deutschen Rontgengesellschaft … [et al] 2010;186:1–6. doi: 10.1007/s00066-009-2026-4. [DOI] [PubMed] [Google Scholar]
- [6].Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface, Nature reviews. Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- [7].Kufe D, Inghirami G, Abe M, Hayes D, Justi-Wheeler H, Schlom J. Differential reactivity of a novel monoclonal antibody (DF3) with human malignant versus benign breast tumors. Hybridoma. 1984;3:223–232. doi: 10.1089/hyb.1984.3.223. [DOI] [PubMed] [Google Scholar]
- [8].Kufe DW. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32:1073–1081. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wen Y, Caffrey TC, Wheelock MJ, Johnson KR, Hollingsworth MA. Nuclear association of the cytoplasmic tail of MUC1 and beta-catenin. The Journal of biological chemistry. 2003;278:38029–38039. doi: 10.1074/jbc.M304333200. [DOI] [PubMed] [Google Scholar]
- [10].Huang L, Ren J, Chen D, Li Y, Kharbanda S, Kufe D. MUC1 cytoplasmic domain coactivates Wnt target gene transcription and confers transformation. Cancer biology & therapy. 2003;2:702–706. [PubMed] [Google Scholar]
- [11].Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003;22:6107–6110. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li Y, Yu WH, Ren J, Chen W, Huang L, Kharbanda S, Loda M, Kufe D. Heregulin targets gamma-catenin to the nucleolus by a mechanism dependent on the DF3/MUC1 oncoprotein. Molecular cancer research : MCR. 2003;1:765–775. [PubMed] [Google Scholar]
- [13].Baldus SE, Monig SP, Huxel S, Landsberg S, Hanisch FG, Engelmann K, Schneider PM, Thiele J, Holscher AH, Dienes HP. MUC1 and nuclear beta-catenin are coexpressed at the invasion front of colorectal carcinomas and are both correlated with tumor prognosis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:2790–2796. doi: 10.1158/1078-0432.ccr-03-0163. [DOI] [PubMed] [Google Scholar]
- [14].Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer cell. 2005;7:167–178. doi: 10.1016/j.ccr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- [15].Yamamoto M, Bharti A, Li Y, Kufe D. Interaction of the DF3/MUC1 breast carcinoma-associated antigen and beta-catenin in cell adhesion. The Journal of biological chemistry. 1997;272:12492–12494. doi: 10.1074/jbc.272.19.12492. [DOI] [PubMed] [Google Scholar]
- [16].Li Y, Ren J, Yu W, Li Q, Kuwahara H, Yin L, Carraway KL, 3rd, Kufe D. The epidermal growth factor receptor regulates interaction of the human DF3/MUC1 carcinoma antigen with c-Src and beta-catenin. The Journal of biological chemistry. 2001;276:35239–35242. doi: 10.1074/jbc.C100359200. [DOI] [PubMed] [Google Scholar]
- [17].Li Y, Kuwahara H, Ren J, Wen G, Kufe D. The c-Src tyrosine kinase regulates signaling of the human DF3/MUC1 carcinoma-associated antigen with GSK3 beta and beta-catenin. The Journal of biological chemistry. 2001;276:6061–6064. doi: 10.1074/jbc.C000754200. [DOI] [PubMed] [Google Scholar]
- [18].Li Y, Bharti A, Chen D, Gong J, Kufe D. Interaction of glycogen synthase kinase 3beta with the DF3/MUC1 carcinoma-associated antigen and beta-catenin. Molecular and cellular biology. 1998;18:7216–7224. doi: 10.1128/mcb.18.12.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bitler BG, Goverdhan A, Schroeder JA. MUC1 regulates nuclear localization and function of the epidermal growth factor receptor. Journal of cell science. 2010;123:1716–1723. doi: 10.1242/jcs.062661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wei X, Xu H, Kufe D. MUC1 oncoprotein stabilizes and activates estrogen receptor alpha. Molecular cell. 2006;21:295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- [21].Warfel NA, El-Deiry WS. p21WAF1 and tumourigenesis: 20 years after. Current opinion in oncology. 2013;25:52–58. doi: 10.1097/CCO.0b013e32835b639e. [DOI] [PubMed] [Google Scholar]
- [22].Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- [23].Ahmad R, Raina D, Joshi MD, Kawano T, Ren J, Kharbanda S, Kufe D. MUC1-C oncoprotein functions as a direct activator of the nuclear factor-kappaB p65 transcription factor. Cancer research. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Heere-Ress E, Thallinger C, Lucas T, Schlagbauer-Wadl H, Wacheck V, Monia BP, Wolff K, Pehamberger H, Jansen B. Bcl-X(L) is a chemoresistance factor in human melanoma cells that can be inhibited by antisense therapy, International journal of cancer. Journal international du cancer. 2002;99:29–34. doi: 10.1002/ijc.10248. [DOI] [PubMed] [Google Scholar]
- [25].Karnak D, Xu L. Chemosensitization of prostate cancer by modulating Bcl-2 family proteins. Current drug targets. 2010;11:699–707. doi: 10.2174/138945010791170888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kawano T, Ahmad R, Nogi H, Agata N, Anderson K, Kufe D. MUC1 oncoprotein promotes growth and survival of human multiple myeloma cells. International journal of oncology. 33(2008):153–159. [PMC free article] [PubMed] [Google Scholar]
- [27].Raina D, Ahmad R, Kumar S, Ren J, Yoshida K, Kharbanda S, Kufe D. MUC1 oncoprotein blocks nuclear targeting of c-Abl in the apoptotic response to DNA damage. The EMBO journal. 2006;25:3774–3783. doi: 10.1038/sj.emboj.7601263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rajabi H, Alam M, Takahashi H, Kharbanda A, Guha M, Ahmad R, Kufe D. MUC1-C oncoprotein activates the ZEB1/miR-200c regulatory loop and epithelial-mesenchymal transition. Oncogene. 2013 doi: 10.1038/onc.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nature cell biology. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- [30].Ren J, Agata N, Chen D, Li Y, Yu WH, Huang L, Raina D, Chen W, Kharbanda S, Kufe D. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer cell. 2004;5:163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ahmad R, Alam M, Rajabi H, Kufe D. The MUC1-C oncoprotein binds to the BH3 domain of the pro-apoptotic BAX protein and blocks BAX function. The Journal of biological chemistry. 2012;287:20866–20875. doi: 10.1074/jbc.M112.357293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ren J, Bharti A, Raina D, Chen W, Ahmad R, Kufe D. MUC1 oncoprotein is targeted to mitochondria by heregulin-induced activation of c-Src and the molecular chaperone HSP90. Oncogene. 2006;25:20–31. doi: 10.1038/sj.onc.1209012. [DOI] [PubMed] [Google Scholar]
- [33].Ren J, Raina D, Chen W, Li G, Huang L, Kufe D. MUC1 oncoprotein functions in activation of fibroblast growth factor receptor signaling. Molecular cancer research : MCR. 2006;4:873–883. doi: 10.1158/1541-7786.MCR-06-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Siragusa M, Zerilli M, Iovino F, Francipane MG, Lombardo Y, Ricci-Vitiani L, Di Gesu G, Todaro M, De Maria R, Stassi G. MUC1 oncoprotein promotes refractoriness to chemotherapy in thyroid cancer cells. Cancer research. 2007;67:5522–5530. doi: 10.1158/0008-5472.CAN-06-4197. [DOI] [PubMed] [Google Scholar]
- [35].Benjamin JB, Jayanthi V, Devaraj H. MUC1 expression and its association with other aetiological factors and localization to mitochondria in preneoplastic and neoplastic gastric tissues. Clinica chimica acta; international journal of clinical chemistry. 2010;411:2067–2072. doi: 10.1016/j.cca.2010.09.003. [DOI] [PubMed] [Google Scholar]
- [36].Yin L, Kosugi M, Kufe D. Inhibition of the MUC1-C oncoprotein induces multiple myeloma cell death by down-regulating TIGAR expression and depleting NADPH. Blood. 2012;119:810–816. doi: 10.1182/blood-2011-07-369686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Weinmann M, Jendrossek V, Handrick R, Guner D, Goecke B, Belka C. Molecular ordering of hypoxia-induced apoptosis: critical involvement of the mitochondrial death pathway in a FADD/caspase-8 independent manner. Oncogene. 2004;23:3757–3769. doi: 10.1038/sj.onc.1207481. [DOI] [PubMed] [Google Scholar]
- [38].Yin L, Kharbanda S, Kufe D. Mucin 1 oncoprotein blocks hypoxia-inducible factor 1alpha activation in a survival response to hypoxia. The Journal of biological chemistry. 2007;282:257–266. doi: 10.1074/jbc.M610156200. [DOI] [PubMed] [Google Scholar]
- [39].Nouman GS, Anderson JJ, Lunec J, Angus B. The role of the tumour suppressor p33 ING1b in human neoplasia. Journal of clinical pathology. 2003;56:491–496. doi: 10.1136/jcp.56.7.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Aoki K, Taketo MM. Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. Journal of cell science. 2007;120:3327–3335. doi: 10.1242/jcs.03485. [DOI] [PubMed] [Google Scholar]
- [41].O'Brate A, Giannakakou P. The importance of p53 location: nuclear or cytoplasmic zip code? Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 2003;6:313–322. doi: 10.1016/j.drup.2003.10.004. [DOI] [PubMed] [Google Scholar]
- [42].Liang SH, Clarke MF. The nuclear import of p53 is determined by the presence of a basic domain and its relative position to the nuclear localization signal. Oncogene. 1999;18:2163–2166. doi: 10.1038/sj.onc.1202350. [DOI] [PubMed] [Google Scholar]
- [43].Liang SH, Clarke MF. A bipartite nuclear localization signal is required for p53 nuclear import regulated by a carboxyl-terminal domain. The Journal of biological chemistry. 1999;274:32699–32703. doi: 10.1074/jbc.274.46.32699. [DOI] [PubMed] [Google Scholar]
- [44].Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. The EMBO journal. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang Y, Xiong Y. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science. 2001;292:1910–1915. doi: 10.1126/science.1058637. [DOI] [PubMed] [Google Scholar]
- [46].Kim IS, Kim DH, Han SM, Chin MU, Nam HJ, Cho HP, Choi SY, Song BJ, Kim ER, Bae YS, Moon YH. Truncated form of importin alpha identified in breast cancer cell inhibits nuclear import of p53. The Journal of biological chemistry. 2000;275:23139–23145. doi: 10.1074/jbc.M909256199. [DOI] [PubMed] [Google Scholar]
- [47].Chen L, Moore JE, Samathanam C, Shao C, Cobos E, Miller MS, Gao W. CRM1-dependent p53 nuclear accumulation in lung lesions of a bitransgenic mouse lung tumor model. Oncology reports. 2011;26:223–228. doi: 10.3892/or.2011.1279. [DOI] [PubMed] [Google Scholar]
- [48].Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nature cell biology. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- [49].Fogal V, Hsieh JK, Royer C, Zhong S, Lu X. Cell cycle-dependent nuclear retention of p53 by E2F1 requires phosphorylation of p53 at Ser315. The EMBO journal. 2005;24:2768–2782. doi: 10.1038/sj.emboj.7600735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ozaki T, Nakagawara A. p53: the attractive tumor suppressor in the cancer research field. Journal of biomedicine & biotechnology. 2011;2011:603925. doi: 10.1155/2011/603925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Moll UM, LaQuaglia M, Benard J, Riou G. Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:4407–4411. doi: 10.1073/pnas.92.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Moll UM, Riou G, Levine AJ. Two distinct mechanisms alter p53 in breast cancer: mutation and nuclear exclusion. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:7262–7266. doi: 10.1073/pnas.89.15.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gestl EE, Anne Bottger S. Cytoplasmic sequestration of the tumor suppressor p53 by a heat shock protein 70 family member, mortalin, in human colorectal adenocarcinoma cell lines. Biochemical and biophysical research communications. 2012;423:411–416. doi: 10.1016/j.bbrc.2012.05.139. [DOI] [PubMed] [Google Scholar]
- [54].Bosari S, Viale G, Roncalli M, Graziani D, Borsani G, Lee AK, Coggi G. p53 gene mutations, p53 protein accumulation and compartmentalization in colorectal adenocarcinoma. The American journal of pathology. 1995;147:790–798. [PMC free article] [PubMed] [Google Scholar]
- [55].Lilling G, Nordenberg J, Rotter V, Goldfinger N, Peller S, Sidi Y. Altered subcellular localization of p53 in estrogen-dependent and estrogen-independent breast cancer cells. Cancer investigation. 2002;20:509–517. doi: 10.1081/cnv-120002151. [DOI] [PubMed] [Google Scholar]
- [56].Mandic R, Schamberger CJ, Muller JF, Geyer M, Zhu L, Carey TE, Grenman R, Dunne AA, Werner JA. Reduced cisplatin sensitivity of head and neck squamous cell carcinoma cell lines correlates with mutations affecting the COOH-terminal nuclear localization signal of p53. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:6845–6852. doi: 10.1158/1078-0432.CCR-05-0378. [DOI] [PubMed] [Google Scholar]
- [57].Zhao Y, Chaiswing L, Velez JM, Batinic-Haberle I, Colburn NH, Oberley TD, St Clair DK. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer research. 2005;65:3745–3750. doi: 10.1158/0008-5472.CAN-04-3835. [DOI] [PubMed] [Google Scholar]
- [58].Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. The EMBO journal. 2007;26:923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Watson C, Long JS, Orange C, Tannahill CL, Mallon E, McGlynn LM, Pyne S, Pyne NJ, Edwards J. High expression of sphingosine 1-phosphate receptors, S1P1 and S1P3, sphingosine kinase 1, and extracellular signal-regulated kinase-1/2 is associated with development of tamoxifen resistance in estrogen receptor-positive breast cancer patients. The American journal of pathology. 2010;177:2205–2215. doi: 10.2353/ajpath.2010.100220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, Horton HF, Fouser L, Carter L, Ling V, Bowman MR, Carreno BM, Collins M, Wood CR, Honjo T. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. The Journal of experimental medicine. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ghebeh H, Lehe C, Barhoush E, Al-Romaih K, Tulbah A, Al-Alwan M, Hendrayani SF, Manogaran P, Alaiya A, Al-Tweigeri T, Aboussekhra A, Dermime S. Doxorubicin downregulates cell surface B7-H1 expression and upregulates its nuclear expression in breast cancer cells: role of B7-H1 as an anti-apoptotic molecule. Breast cancer research : BCR. 2010;12:R48. doi: 10.1186/bcr2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zhang L, Wu H, Lu D, Li G, Sun C, Song H, Li J, Zhai T, Huang L, Hou C, Wang W, Zhou B, Chen S, Lu B, Zhang X. The costimulatory molecule B7-H4 promote tumor progression and cell proliferation through translocating into nucleus. Oncogene. 2013 doi: 10.1038/onc.2012.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ingebrigtsen VA, Boye K, Tekle C, Nesland JM, Flatmark K, Fodstad O. B7-H3 expression in colorectal cancer: nuclear localization strongly predicts poor outcome in colon cancer. International journal of cancer. Journal international du cancer. 2012;131:2528–2536. doi: 10.1002/ijc.27566. [DOI] [PubMed] [Google Scholar]
- [64].Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310:1452–1456. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
- [65].Runnebaum IB, Kieback DG, Mobus VJ, Tong XW, Kreienberg R. Subcellular localization of accumulated p53 in ovarian cancer cells. Gynecologic oncology. 1996;61:266–271. doi: 10.1006/gyno.1996.0137. [DOI] [PubMed] [Google Scholar]
- [66].Falini B, Martelli MP, Bolli N, Bonasso R, Ghia E, Pallotta MT, Diverio D, Nicoletti I, Pacini R, Tabarrini A, Galletti BV, Mannucci R, Roti G, Rosati R, Specchia G, Liso A, Tiacci E, Alcalay M, Luzi L, Volorio S, Bernard L, Guarini A, Amadori S, Mandelli F, Pane F, Lo-Coco F, Saglio G, Pelicci PG, Martelli MF, Mecucci C. Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood. 2006;108:1999–2005. doi: 10.1182/blood-2006-03-007013. [DOI] [PubMed] [Google Scholar]
- [67].Bedekovics J, Rejto L, Telek B, Udvardy M, Ujfalusi A, Olah E, Hevessy Z, Kappelmayer J, Kajtar B, Mehes G. Immunohistochemical demonstration of mutant nucleophosmin in acute myeloid leukemia: biological and clinical features related to NPMc expression. Orvosi hetilap. 2009;150:1031–1035. doi: 10.1556/OH.2009.28623. [DOI] [PubMed] [Google Scholar]
- [68].Bolli N, De Marco MF, Martelli MP, Bigerna B, Pucciarini A, Rossi R, Mannucci R, Manes N, Pettirossi V, Pileri SA, Nicoletti I, Falini B. A dose-dependent tug of war involving the NPM1 leukaemic mutant, nucleophosmin, and ARF. Leukemia. 2009;23:501–509. doi: 10.1038/leu.2008.326. [DOI] [PubMed] [Google Scholar]
- [69].Tang WW, Stelter AA, French S, Shen S, Qiu S, Venegas R, Wen J, Wang HQ, Xie J. Loss of cell-adhesion molecule complexes in solid pseudopapillary tumor of pancreas. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2007;20:509–513. doi: 10.1038/modpathol.3800764. [DOI] [PubMed] [Google Scholar]
- [70].Grossman E, Medalia O, Zwerger M. Functional architecture of the nuclear pore complex. Annual review of biophysics. 2012;41:557–584. doi: 10.1146/annurev-biophys-050511-102328. [DOI] [PubMed] [Google Scholar]
- [71].Turner JG, Dawson J, Sullivan DM. Nuclear export of proteins and drug resistance in cancer. Biochemical pharmacology. 2012;83:1021–1032. doi: 10.1016/j.bcp.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kojima K, Kornblau SM, Ruvolo V, Dilip A, Duvvuri S, Davis RE, Zhang M, Wang Z, Coombes KR, Zhang N, Qiu YH, Burks JK, Kantarjian H, Shacham S, Kauffman M, Andreeff M. Prognostic impact and targeting of CRM1 in acute myeloid leukemia. Blood. 2013;121:4166–4174. doi: 10.1182/blood-2012-08-447581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Christiansen A, Dyrskjot L. The functional role of the novel biomarker karyopherin alpha 2 (KPNA2) in cancer. Cancer letters. 2013;331:18–23. doi: 10.1016/j.canlet.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Murphy ME. The HSP70 family and cancer. Carcinogenesis. 2013;34:1181–1188. doi: 10.1093/carcin/bgt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Muller P, Hrstka R, Coomber D, Lane DP, Vojtesek B. Chaperone-dependent stabilization and degradation of p53 mutants. Oncogene. 2008;27:3371–3383. doi: 10.1038/sj.onc.1211010. [DOI] [PubMed] [Google Scholar]
- [76].Peng Y, Chen L, Li C, Lu W, Chen J. Inhibition of MDM2 by hsp90 contributes to mutant p53 stabilization. The Journal of biological chemistry. 2001;276:40583–40590. doi: 10.1074/jbc.M102817200. [DOI] [PubMed] [Google Scholar]
- [77].Qu L, Huang S, Baltzis D, Rivas-Estilla AM, Pluquet O, Hatzoglou M, Koumenis C, Taya Y, Yoshimura A, Koromilas AE. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3beta. Genes & development. 2004;18:261–277. doi: 10.1101/gad.1165804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Liu Y, Ye Y. Proteostasis regulation at the endoplasmic reticulum: a new perturbation site for targeted cancer therapy. Cell research. 2011;21:867–883. doi: 10.1038/cr.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Nagaraj NS, Singh OV, Merchant NB. Proteomics: a strategy to understand the novel targets in protein misfolding and cancer therapy. Expert review of proteomics. 2010;7:613–623. doi: 10.1586/epr.10.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lo HW, Ali-Seyed M, Wu Y, Bartholomeusz G, Hsu SC, Hung MC. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. Journal of cellular biochemistry. 2006;98:1570–1583. doi: 10.1002/jcb.20876. [DOI] [PubMed] [Google Scholar]
- [81].Wang YN, Yamaguchi H, Hsu JM, Hung MC. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene. 2010;29:3997–4006. doi: 10.1038/onc.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chung BM, Raja SM, Clubb RJ, Tu C, George M, Band V, Band H. Aberrant trafficking of NSCLC-associated EGFR mutants through the endocytic recycling pathway promotes interaction with Src. BMC cell biology. 2009;10:84. doi: 10.1186/1471-2121-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Liu X, Yuan Z, Chung M. MUC1 intra-cellular trafficking is clathrin, dynamin, and rab5 dependent. Biochemical and biophysical research communications. 2008;376:688–693. doi: 10.1016/j.bbrc.2008.09.065. [DOI] [PubMed] [Google Scholar]
- [84].Goldenring JR, Nam KT. Rab25 as a tumour suppressor in colon carcinogenesis. British journal of cancer. 2011;104:33–36. doi: 10.1038/sj.bjc.6605983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zhang J, Liu X, Datta A, Govindarajan K, Tam WL, Han J, George J, Wong C, Ramnarayanan K, Phua TY, Leong WY, Chan YS, Palanisamy N, Liu ET, Karuturi KM, Lim B, Miller LD. RCP is a human breast cancer-promoting gene with Ras-activating function. The Journal of clinical investigation. 2009;119:2171–2183. doi: 10.1172/JCI37622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Shoji K, Oda K, Nakagawa S, Hosokawa S, Nagae G, Uehara Y, Sone K, Miyamoto Y, Hiraike H, Hiraike-Wada O, Nei T, Kawana K, Kuramoto H, Aburatani H, Yano T, Taketani Y. The oncogenic mutation in the pleckstrin homology domain of AKT1 in endometrial carcinomas. British journal of cancer. 2009;101:145–148. doi: 10.1038/sj.bjc.6605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Yi KH, Axtmayer J, Gustin JP, Rajpurohit A, Lauring J. Functional analysis of non-hotspot AKT1 mutants found in human breast cancers identifies novel driver mutations: implications for personalized medicine. Oncotarget. 2013;4:29–34. doi: 10.18632/oncotarget.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Esteller M, Cordon-Cardo C, Corn PG, Meltzer SJ, Pohar KS, Watkins DN, Capella G, Peinado MA, Matias-Guiu X, Prat J, Baylin SB, Herman JG. p14ARF silencing by promoter hypermethylation mediates abnormal intracellular localization of MDM2. Cancer research. 2001;61:2816–2821. [PubMed] [Google Scholar]
- [89].Yang X, Wang J, Liu C, Grizzle WE, Yu S, Zhang S, Barnes S, Koopman WJ, Mountz JD, Kimberly RP, Zhang HG. Cleavage of p53-vimentin complex enhances tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis of rheumatoid arthritis synovial fibroblasts. The American journal of pathology. 2005;167:705–719. doi: 10.1016/S0002-9440(10)62045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Klotzsche O, Etzrodt D, Hohenberg H, Bohn W, Deppert W. Cytoplasmic retention of mutant tsp53 is dependent on an intermediate filament protein (vimentin) scaffold. Oncogene. 1998;16:3423–3434. doi: 10.1038/sj.onc.1202155. [DOI] [PubMed] [Google Scholar]
- [91].Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI, Lagstrom S, Clemente MJ, Olson T, Jalkanen SE, Majumder MM, Almusa H, Edgren H, Lepisto M, Mattila P, Guinta K, Koistinen P, Kuittinen T, Penttinen K, Parsons A, Knowles J, Saarela J, Wennerberg K, Kallioniemi O, Porkka K, Loughran TP, Jr., Heckman CA, Maciejewski JP, Mustjoki S. Somatic STAT3 mutations in large granular lymphocytic leukemia. The New England journal of medicine. 2012;366:1905–1913. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kojima K, Konopleva M, Samudio IJ, Shikami M, Cabreira-Hansen M, McQueen T, Ruvolo V, Tsao T, Zeng Z, Vassilev LT, Andreeff M. MDM2 antagonists induce p53-dependent apoptosis in AML: implications for leukemia therapy. Blood. 2005;106:3150–3159. doi: 10.1182/blood-2005-02-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Konikova E, Kusenda J. Altered expression of p53 and MDM2 proteins in hematological malignancies. Neoplasma. 2003;50:31–40. [PubMed] [Google Scholar]
- [94].Merlin J, Stechly L, de Beauce S, Monte D, Leteurtre E, van Seuningen I, Huet G, Pigny P. Galectin-3 regulates MUC1 and EGFR cellular distribution and EGFR downstream pathways in pancreatic cancer cells. Oncogene. 2011;30:2514–2525. doi: 10.1038/onc.2010.631. [DOI] [PubMed] [Google Scholar]
- [95].Leng Y, Cao C, Ren J, Huang L, Chen D, Ito M, Kufe D. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. The Journal of biological chemistry. 2007;282:19321–19330. doi: 10.1074/jbc.M703222200. [DOI] [PubMed] [Google Scholar]
- [96].Liao HJ, Carpenter G. Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Molecular biology of the cell. 2007;18:1064–1072. doi: 10.1091/mbc.E06-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Serra S, Chetty R. Revision 2: an immunohistochemical approach and evaluation of solid pseudopapillary tumour of the pancreas. Journal of clinical pathology. 2008;61:1153–1159. doi: 10.1136/jcp.2008.057828. [DOI] [PubMed] [Google Scholar]
- [98].Kopparapu PK, Tinzl M, Anagnostaki L, Persson JL, Dizeyi N. Expression and localization of serotonin receptors in human breast cancer. Anticancer research. 2013;33:363–370. [PubMed] [Google Scholar]
- [99].Kampen KR. The mechanisms that regulate the localization and overexpression of VEGF receptor-2 are promising therapeutic targets in cancer biology. Anti-cancer drugs. 2012;23:347–354. doi: 10.1097/CAD.0b013e32835004ac. [DOI] [PubMed] [Google Scholar]
- [100].Li YS, Kaneko M, Sakamoto DG, Takeshima Y, Inai K. The reversed apical pattern of MUC1 expression is characteristics of invasive micropapillary carcinoma of the breast. Breast Cancer. 2006;13:58–63. doi: 10.2325/jbcs.13.58. [DOI] [PubMed] [Google Scholar]
- [101].Kim MJ, Jang SJ, Yu E. Loss of E-cadherin and cytoplasmic-nuclear expression of beta-catenin are the most useful immunoprofiles in the diagnosis of solid-pseudopapillary neoplasm of the pancreas. Human pathology. 2008;39:251–258. doi: 10.1016/j.humpath.2007.06.014. [DOI] [PubMed] [Google Scholar]
- [102].Ferrara G, Misciali C, Brenn T, Cerroni L, Kazakov DW, Perasole A, Russo R, Ricci R, Crisman G, Fanti PA, Passarini B, Patrizi A. The Impact of Molecular Morphology Techniques on the Expert Diagnosis in Melanocytic Skin Neoplasms. International journal of surgical pathology. 2013 doi: 10.1177/1066896913491323. [DOI] [PubMed] [Google Scholar]
- [103].Deeb G, Wang J, Ramnath N, Slocum HK, Wiseman S, Beck A, Tan D. Altered E-cadherin and epidermal growth factor receptor expressions are associated with patient survival in lung cancer: a study utilizing high-density tissue microarray and immunohistochemistry. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2004;17:430–439. doi: 10.1038/modpathol.3800041. [DOI] [PubMed] [Google Scholar]
- [104].Muller S, Su L, Tighiouart M, Saba N, Zhang H, Shin DM, Chen Z. Distinctive E-cadherin and epidermal growth factor receptor expression in metastatic and nonmetastatic head and neck squamous cell carcinoma: predictive and prognostic correlation. Cancer. 2008;113:97–107. doi: 10.1002/cncr.23557. [DOI] [PubMed] [Google Scholar]
- [105].Zhang W, Tang W, Inagaki Y, Qiu M, Xu HL, Li X, Sugawara Y, Nagawa H, Nakata M, Kokudo N. Positive KL-6 mucin expression combined with decreased membranous beta-catenin expression indicates worse prognosis in colorectal carcinoma. Oncology reports. 2008;20:1013–1019. [PubMed] [Google Scholar]
- [106].Mao L, Yang Y. Targeting the nuclear transport machinery by rational drug design. Current pharmaceutical design. 2013;19:2318–2325. doi: 10.2174/1381612811319120018. [DOI] [PubMed] [Google Scholar]
- [107].Ranganathan P, Yu X, Na C, Santhanam R, Shacham S, Kauffman M, Walker A, Klisovic R, Blum W, Caligiuri M, Croce CM, Marcucci G, Garzon R. Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood. 2012;120:1765–1773. doi: 10.1182/blood-2012-04-423160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Inoue H, Kauffman M, Shacham S, Landesman Y, Yang J, Evans CP, Weiss RH. CRM1 Blockade by Selective Inhibitors of Nuclear Export Attenuates Kidney Cancer Growth. The Journal of urology. 2013;189:2317–2326. doi: 10.1016/j.juro.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Lapalombella R, Sun Q, Williams K, Tangeman L, Jha S, Zhong Y, Goettl V, Mahoney E, Berglund C, Gupta S, Farmer A, Mani R, Johnson AJ, Lucas D, Mo X, Daelemans D, Sandanayaka V, Shechter S, McCauley D, Shacham S, Kauffman M, Chook YM, Byrd JC. Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood. 2012;120:4621–4634. doi: 10.1182/blood-2012-05-429506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Salas Fragomeni RA, Chung HW, Landesman Y, Senapedis W, Saint-Martin JR, Tsao H, Flaherty KT, Shacham S, Kauffman M, Cusack JC. CRM1 and BRAF Inhibition Synergize and Induce Tumor Regression in BRAF-Mutant Melanoma. Molecular cancer therapeutics. 2013;12:1171–1179. doi: 10.1158/1535-7163.MCT-12-1171. [DOI] [PubMed] [Google Scholar]
- [111].Etchin J, Sanda T, Mansour MR, Kentsis A, Montero J, Le BT, Christie AL, McCauley D, Rodig SJ, Kauffman M, Shacham S, Stone R, Letai A, Kung AL, Thomas Look A. KPT-330 inhibitor of CRM1 (XPO1)-mediated nuclear export has selective anti-leukaemic activity in preclinical models of T-cell acute lymphoblastic leukaemia and acute myeloid leukaemia. British journal of haematology. 2013;161:117–127. doi: 10.1111/bjh.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Tai YT, Landesman Y, Acharya C, Calle Y, Zhong MY, Cea M, Tannenbaum D, Cagnetta A, Reagan M, Munshi AA, Senapedis W, Saint-Martin JR, Kashyap T, Shacham S, Kauffman M, Gu Y, Wu L, Ghobrial I, Zhan F, Kung AL, Schey SA, Richardson P, Munshi NC, Anderson KC. CRM1 inhibition induces tumor cell cytotoxicity and impairs osteoclastogenesis in multiple myeloma: molecular mechanisms and therapeutic implications. Leukemia. 2013 doi: 10.1038/leu.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Kosugi S, Hasebe M, Entani T, Takayama S, Tomita M, Yanagawa H. Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. Chemistry & biology. 2008;15:940–949. doi: 10.1016/j.chembiol.2008.07.019. [DOI] [PubMed] [Google Scholar]
- [114].Zhang Y, Cheng Y, Ren X, Zhang L, Yap KL, Wu H, Patel R, Liu D, Qin ZH, Shih IM, Yang JM. NAC1 modulates sensitivity of ovarian cancer cells to cisplatin by altering the HMGB1-mediated autophagic response. Oncogene. 2012;31:1055–1064. doi: 10.1038/onc.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Yeasmin S, Nakayama K, Rahman MT, Rahman M, Ishikawa M, Katagiri A, Iida K, Nakayama N, Otuski Y, Kobayashi H, Nakayama S, Miyazaki K. Biological and clinical significance of NAC1 expression in cervical carcinomas: a comparative study between squamous cell carcinomas and adenocarcinomas/adenosquamous carcinomas. Human pathology. 2012;43:506–519. doi: 10.1016/j.humpath.2011.05.021. [DOI] [PubMed] [Google Scholar]
- [116].Xu Y, Fang F, St Clair DK, Sompol P, Josson S, St Clair WH. SN52, a novel nuclear factor-kappaB inhibitor, blocks nuclear import of RelB:p52 dimer and sensitizes prostate cancer cells to ionizing radiation. Molecular cancer therapeutics. 2008;7:2367–2376. doi: 10.1158/1535-7163.MCT-08-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Joshi MD, Ahmad R, Yin L, Raina D, Rajabi H, Bubley G, Kharbanda S, Kufe D. MUC1 oncoprotein is a druggable target in human prostate cancer cells. Molecular cancer therapeutics. 2009;8:3056–3065. doi: 10.1158/1535-7163.MCT-09-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Yin L, Kufe D. MUC1-C Oncoprotein Blocks Terminal Differentiation of Chronic Myelogenous Leukemia Cells by a ROS-Mediated Mechanism. Genes & cancer. 2011;2:56–64. doi: 10.1177/1947601911405044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Yin L, Wu Z, Avigan D, Rosenblatt J, Stone R, Kharbanda S, Kufe D. MUC1-C oncoprotein suppresses reactive oxygen species-induced terminal differentiation of acute myelogenous leukemia cells. Blood. 2011;117:4863–4870. doi: 10.1182/blood-2010-10-296632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Raina D, Kosugi M, Ahmad R, Panchamoorthy G, Rajabi H, Alam M, Shimamura T, Shapiro GI, Supko J, Kharbanda S, Kufe D. Dependence on the MUC1-C oncoprotein in non-small cell lung cancer cells. Molecular cancer therapeutics. 2011;10:806–816. doi: 10.1158/1535-7163.MCT-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Beck R, Dejeans N, Glorieux C, Creton M, Delaive E, Dieu M, Raes M, Leveque P, Gallez B, Depuydt M, Collet JF, Calderon PB, Verrax J. Hsp90 is cleaved by reactive oxygen species at a highly conserved N-terminal amino acid motif. PloS one. 2012;7:e40795. doi: 10.1371/journal.pone.0040795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Kawano T, Ito M, Raina D, Wu Z, Rosenblatt J, Avigan D, Stone R, Kufe D. MUC1 oncoprotein regulates Bcr-Abl stability and pathogenesis in chronic myelogenous leukemia cells. Cancer research. 2007;67:11576–11584. doi: 10.1158/0008-5472.CAN-07-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks glycogen synthase kinase 3beta-mediated phosphorylation and degradation of beta-catenin. Cancer research. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- [124].Saki M, Toulany M, Rodemann HP. Acquired resistance to cetuximab is associated with the overexpression of Ras family members and the loss of radiosensitization in head and neck cancer cells. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2013 doi: 10.1016/j.radonc.2013.06.023. [DOI] [PubMed] [Google Scholar]
- [125].Rosa R, Marciano R, Malapelle U, Formisano L, Nappi L, D'Amato C, D'Amato V, Damiano V, Marfe G, Del Vecchio S, Zannetti A, Greco A, De Stefano A, Carlomagno C, Veneziani BM, Troncone G, De Placido S, Bianco R. Sphingosine kinase 1 overexpression contributes to cetuximab resistance in human colorectal cancer models. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:138–147. doi: 10.1158/1078-0432.CCR-12-1050. [DOI] [PubMed] [Google Scholar]
- [126].Iida M, Brand TM, Campbell DA, Li C, Wheeler DL. Yes and Lyn play a role in nuclear translocation of the epidermal growth factor receptor. Oncogene. 2013;32:759–767. doi: 10.1038/onc.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Li C, Iida M, Dunn EF, Ghia AJ, Wheeler DL. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009;28:3801–3813. doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Raju U, Riesterer O, Wang ZQ, Molkentine DP, Molkentine JM, Johnson FM, Glisson B, Milas L, Ang KK. Dasatinib, a multi-kinase inhibitor increased radiation sensitivity by interfering with nuclear localization of epidermal growth factor receptor and by blocking DNA repair pathways. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2012;105:241–249. doi: 10.1016/j.radonc.2012.08.010. [DOI] [PubMed] [Google Scholar]
- [129].Dunn EF, Iida M, Myers RA, Campbell DA, Hintz KA, Armstrong EA, Li C, Wheeler DL. Dasatinib sensitizes KRAS mutant colorectal tumors to cetuximab. Oncogene. 2011;30:561–574. doi: 10.1038/onc.2010.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Mikalsen T, Gerits N, Moens U. Inhibitors of signal transduction protein kinases as targets for cancer therapy. Biotechnology annual review. 2006;12:153–223. doi: 10.1016/S1387-2656(06)12006-2. [DOI] [PubMed] [Google Scholar]
- [131].Nair P, Somasundaram K, Krishna S. Activated Notch1 inhibits p53-induced apoptosis and sustains transformation by human papillomavirus type 16 E6 and E7 oncogenes through a PI3K-PKB/Akt-dependent pathway. Journal of virology. 2003;77:7106–7112. doi: 10.1128/JVI.77.12.7106-7112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Shankar S, Srivastava RK. Involvement of Bcl-2 family members, phosphatidylinositol 3'-kinase/AKT and mitochondrial p53 in curcumin (diferulolylmethane)-induced apoptosis in prostate cancer. International journal of oncology. 2007;30:905–918. [PubMed] [Google Scholar]
- [133].Patel S. Exploring novel therapeutic targets in GIST: focus on the PI3K/Akt/mTOR pathway. Current oncology reports. 2013;15:386–395. doi: 10.1007/s11912-013-0316-6. [DOI] [PubMed] [Google Scholar]
- [134].Meuillet EJ. Novel inhibitors of AKT: assessment of a different approach targeting the pleckstrin homology domain. Current medicinal chemistry. 2011;18:2727–2742. doi: 10.2174/092986711796011292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Lauring J, Park BH, Wolff AC. The phosphoinositide-3-kinase-Akt-mTOR pathway as a therapeutic target in breast cancer. Journal of the National Comprehensive Cancer Network : JNCCN. 2013;11:670–678. doi: 10.6004/jnccn.2013.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials, Nature reviews. Clinical oncology. 2013;10:143–153. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- [137].Chen Y, Chen CF, Riley DJ, Allred DC, Chen PL, Von Hoff D, Osborne CK, Lee WH. Aberrant subcellular localization of BRCA1 in breast cancer. Science. 1995;270:789–791. doi: 10.1126/science.270.5237.789. [DOI] [PubMed] [Google Scholar]
- [138].Wilson CA, Ramos L, Villasenor MR, Anders KH, Press MF, Clarke K, Karlan B, Chen JJ, Scully R, Livingston D, Zuch RH, Kanter MH, Cohen S, Calzone FJ, Slamon DJ. Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nature genetics. 1999;21:236–240. doi: 10.1038/6029. [DOI] [PubMed] [Google Scholar]
- [139].Thomas JE, Smith M, Rubinfeld B, Gutowski M, Beckmann RP, Polakis P. Subcellular localization and analysis of apparent 180-kDa and 220-kDa proteins of the breast cancer susceptibility gene, BRCA1. The Journal of biological chemistry. 1996;271:28630–28635. doi: 10.1074/jbc.271.45.28630. [DOI] [PubMed] [Google Scholar]
- [140].Jiang J, Yang ES, Jiang G, Nowsheen S, Wang H, Wang T, Wang Y, Billheimer D, Chakravarthy AB, Brown M, Haffty B, Xia F. p53-dependent BRCA1 nuclear export controls cellular susceptibility to DNA damage. Cancer research. 2011;71:5546–5557. doi: 10.1158/0008-5472.CAN-10-3423. [DOI] [PubMed] [Google Scholar]
- [141].Thakur S, Zhang HB, Peng Y, Le H, Carroll B, Ward T, Yao J, Farid LM, Couch FJ, Wilson RB, Weber BL. Localization of BRCA1 and a splice variant identifies the nuclear localization signal. Molecular and cellular biology. 1997;17:444–452. doi: 10.1128/mcb.17.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Kashima K, Oite T, Aoki Y, Takakuwa K, Aida H, Nagata H, Sekine M, Wu HJ, Hirai Y, Wada Y, Yamamoto K, Hasegawa K, Sonoda T, Maruo T, Nagata I, Ohno M, Suzuki M, Kobayashi I, Kuzuya K, Takahashi T, Torii Y, Tanaka K. Screening of BRCA1 mutation using immunohistochemical staining with C-terminal and N-terminal antibodies in familial ovarian cancers. Japanese journal of cancer research : Gann. 2000;91:399–409. doi: 10.1111/j.1349-7006.2000.tb00959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Nair R, Rost B. Protein subcellular localization prediction using artificial intelligence technology. Methods Mol Biol. 2008;484:435–463. doi: 10.1007/978-1-59745-398-1_27. [DOI] [PubMed] [Google Scholar]
- [144].Jiao W, Lin HM, Datta J, Braunschweig T, Chung JY, Hewitt SM, Rane SG. Aberrant nucleocytoplasmic localization of the retinoblastoma tumor suppressor protein in human cancer correlates with moderate/poor tumor differentiation. Oncogene. 2008;27:3156–3164. doi: 10.1038/sj.onc.1210970. [DOI] [PubMed] [Google Scholar]
- [145].Jiao W, Datta J, Lin HM, Dundr M, Rane SG. Nucleocytoplasmic shuttling of the retinoblastoma tumor suppressor protein via Cdk phosphorylation-dependent nuclear export. The Journal of biological chemistry. 2006;281:38098–38108. doi: 10.1074/jbc.M605271200. [DOI] [PubMed] [Google Scholar]
- [146].Byles V, Chmilewski LK, Wang J, Zhu L, Forman LW, Faller DV, Dai Y. Aberrant cytoplasm localization and protein stability of SIRT1 is regulated by PI3K/IGF-1R signaling in human cancer cells. International journal of biological sciences. 2010;6:599–612. doi: 10.7150/ijbs.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? International journal of biological sciences. 2009;5:147–152. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Delk NA, Hunt KK, Keyomarsi K. Altered subcellular localization of tumor-specific cyclin E isoforms affects cyclin-dependent kinase 2 complex formation and proteasomal regulation. Cancer research. 2009;69:2817–2825. doi: 10.1158/0008-5472.CAN-08-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Cheng CJ, Ye XC, Vakar-Lopez F, Kim J, Tu SM, Chen DT, Navone NM, Yu-Lee LY, Lin SH, Hu MC. Bone microenvironment and androgen status modulate subcellular localization of ErbB3 in prostate cancer cells. Molecular cancer research : MCR. 2007;5:675–684. doi: 10.1158/1541-7786.MCR-06-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Gupta BK, Maher DM, Ebeling MC, Sundram V, Koch MD, Lynch DW, Bohlmeyer T, Watanabe A, Aburatani H, Puumala SE, Jaggi M, Chauhan SC. Increased expression and aberrant localization of mucin 13 in metastatic colon cancer. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2012;60:822–831. doi: 10.1369/0022155412460678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Vieyra D, Senger DL, Toyama T, Muzik H, Brasher PM, Johnston RN, Riabowol K, Forsyth PA. Altered subcellular localization and low frequency of mutations of ING1 in human brain tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9:5952–5961. [PubMed] [Google Scholar]
- [152].Saito S, Miyaji-Yamaguchi M, Nagata K. Aberrant intracellular localization of SET-CAN fusion protein, associated with a leukemia, disorganizes nuclear export, International journal of cancer. Journal international du cancer. 2004;111:501–507. doi: 10.1002/ijc.20296. [DOI] [PubMed] [Google Scholar]
- [153].Rakha EA, Boyce RW, Abd El-Rehim D, Kurien T, Green AR, Paish EC, Robertson JF, Ellis IO. Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2005;18:1295–1304. doi: 10.1038/modpathol.3800445. [DOI] [PubMed] [Google Scholar]
- [154].Inagaki Y, Tang W, Xu H, Nakata M, Mafune K, Konishi T, Seto Y, Kokudo N. Sustained aberrant localization of KL-6 mucin and beta-catenin at the invasion front of human gastric cancer cells. Anticancer research. 2011;31:535–542. [PubMed] [Google Scholar]
- [155].Ozanne DM, Brady ME, Cook S, Gaughan L, Neal DE, Robson CN. Androgen receptor nuclear translocation is facilitated by the f-actin cross-linking protein filamin. Mol Endocrinol. 2000;14:1618–1626. doi: 10.1210/mend.14.10.0541. [DOI] [PubMed] [Google Scholar]
- [156].Fernandez-Majada V, Pujadas J, Vilardell F, Capella G, Mayo MW, Bigas A, Espinosa L. Aberrant cytoplasmic localization of N-CoR in colorectal tumors. Cell Cycle. 2007;6:1748–1752. doi: 10.4161/cc.6.14.4429. [DOI] [PubMed] [Google Scholar]
- [157].Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer, Nature reviews. Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- [158].Geyer FC, Lacroix-Triki M, Savage K, Arnedos M, Lambros MB, MacKay A, Natrajan R, Reis-Filho JS. beta-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24:209–231. doi: 10.1038/modpathol.2010.205. [DOI] [PubMed] [Google Scholar]
- [159].Orsulic S, Huber O, Aberle H, Arnold S, Kemler R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. Journal of cell science. 1999;112(Pt 8):1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- [160].Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- [161].Zurawel RH, Chiappa SA, Allen C, Raffel C. Sporadic medulloblastomas contain oncogenic beta-catenin mutations. Cancer research. 1998;58:896–899. [PubMed] [Google Scholar]
- [162].Sena P, Saviano M, Monni S, Losi L, Roncucci L, Marzona L, De Pol A. Subcellular localization of beta-catenin and APC proteins in colorectal preneoplastic and neoplastic lesions. Cancer letters. 2006;241:203–212. doi: 10.1016/j.canlet.2005.10.011. [DOI] [PubMed] [Google Scholar]
- [163].Petkovic S, Mutavdzic M. The late results of conservative surgery for ureteral tumours. British journal of urology. 1968;40:412–414. doi: 10.1111/j.1464-410x.1968.tb11823.x. [DOI] [PubMed] [Google Scholar]
- [164].Singh SP, Lipman J, Goldman H, Ellis FH, Jr., Aizenman L, Cangi MG, Signoretti S, Chiaur DS, Pagano M, Loda M. Loss or altered subcellular localization of p27 in Barrett's associated adenocarcinoma. Cancer research. 1998;58:1730–1735. [PubMed] [Google Scholar]
- [165].Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy, Nature reviews. Cancer. 2008;8:253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- [166].Gu Z, Zhou L, Gao S, Wang Z. Nuclear transport signals control cellular localization and function of androgen receptor cofactor p44/WDR77. PloS one. 2011;6:e22395. doi: 10.1371/journal.pone.0022395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Nentwig A, Higgins RJ, Francey T, Doherr M, Zurbriggen A, Oevermann A. Aberrant E-cadherin, beta-catenin, and glial fibrillary acidic protein (GFAP) expression in canine choroid plexus tumors. Journal of veterinary diagnostic investigation : official publication of the American Association of Veterinary Laboratory Diagnosticians, Inc. 2012;24:14–22. doi: 10.1177/1040638711425940. [DOI] [PubMed] [Google Scholar]
- [168].Ferber EC, Kajita M, Wadlow A, Tobiansky L, Niessen C, Ariga H, Daniel J, Fujita Y. A role for the cleaved cytoplasmic domain of E-cadherin in the nucleus. The Journal of biological chemistry. 2008;283:12691–12700. doi: 10.1074/jbc.M708887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Lin YY, Hsu YH, Huang HY, Shann YJ, Huang CY, Wei SC, Chen CL, Jou TS. Aberrant nuclear localization of EBP50 promotes colorectal carcinogenesis in xenotransplanted mice by modulating TCF-1 and beta-catenin interactions. The Journal of clinical investigation. 2012;122:1881–1894. doi: 10.1172/JCI45661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Ralhan R, Cao J, Lim T, Macmillan C, Freeman JL, Walfish PG. EpCAM nuclear localization identifies aggressive thyroid cancer and is a marker for poor prognosis. BMC cancer. 2010;10:331. doi: 10.1186/1471-2407-10-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Abdelsalam M. Doxorubicin translocation B7-H1 from the cell surface to the nucleus of breast cancer cells: a novel role for B7-H1 as an anti-apoptotic molecule. Hematology/oncology and stem cell therapy. 2010;3:163. doi: 10.1016/s1658-3876(10)50028-1. [DOI] [PubMed] [Google Scholar]
- [172].Comstock CE, Revelo MP, Buncher CR, Knudsen KE. Impact of differential cyclin D1 expression and localisation in prostate cancer. British journal of cancer. 2007;96:970–979. doi: 10.1038/sj.bjc.6603615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Chung R, Peters AC, Armanious H, Anand M, Gelebart P, Lai R. Biological and clinical significance of GSK-3beta in mantle cell lymphoma--an immunohistochemical study. International journal of clinical and experimental pathology. 2010;3:244–253. [PMC free article] [PubMed] [Google Scholar]
- [174].Moore JD. In the wrong place at the wrong time: does cyclin mislocalization drive oncogenic transformation?, Nature reviews. Cancer. 2013;13:201–208. doi: 10.1038/nrc3468. [DOI] [PubMed] [Google Scholar]
- [175].Domingues I, Rino J, Demmers JA, de Lanerolle P, Santos SC. VEGFR2 translocates to the nucleus to regulate its own transcription. PloS one. 2011;6:e25668. doi: 10.1371/journal.pone.0025668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Han AC, Soler AP, Tang CK, Knudsen KA, Salazar H. Nuclear localization of E-cadherin expression in Merkel cell carcinoma. Archives of pathology & laboratory medicine. 2000;124:1147–1151. doi: 10.5858/2000-124-1147-NLOECE. [DOI] [PubMed] [Google Scholar]
- [177].Serra S, Salahshor S, Fagih M, Niakosari F, Radhi JM, Chetty R. Nuclear expression of E-cadherin in solid pseudopapillary tumors of the pancreas. JOP : Journal of the pancreas. 2007;8:296–303. [PubMed] [Google Scholar]
- [178].El-Bahrawy MA, Rowan A, Horncastle D, Tomlinson I, Theis BA, Russell RC, Stamp G. E-cadherin/catenin complex status in solid pseudopapillary tumor of the pancreas. The American journal of surgical pathology. 2008;32:1–7. doi: 10.1097/PAS.0b013e31813e0676. [DOI] [PubMed] [Google Scholar]
- [179].Elston MS, Gill AJ, Conaglen JV, Clarkson A, Cook RJ, Little NS, Robinson BG, Clifton-Bligh RJ, McDonald KL. Nuclear accumulation of e-cadherin correlates with loss of cytoplasmic membrane staining and invasion in pituitary adenomas. The Journal of clinical endocrinology and metabolism. 2009;94:1436–1442. doi: 10.1210/jc.2008-2075. [DOI] [PubMed] [Google Scholar]
- [180].Chetty R, Serra S. Nuclear E-cadherin immunoexpression: from biology to potential applications in diagnostic pathology. Advances in anatomic pathology. 2008;15:234–240. doi: 10.1097/PAP.0b013e31817bf566. [DOI] [PubMed] [Google Scholar]
- [181].Kunavisarut T, Kak I, Macmillan C, Ralhan R, Walfish PG. Immunohistochemical analysis based Ep-ICD subcellular localization index (ESLI) is a novel marker for metastatic papillary thyroid microcarcinoma. BMC cancer. 2012;12:523. doi: 10.1186/1471-2407-12-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Ralhan R, He HC, So AK, Tripathi SC, Kumar M, Hasan MR, Kaur J, Kashat L, MacMillan C, Chauhan SS, Freeman JL, Walfish PG. Nuclear and cytoplasmic accumulation of Ep-ICD is frequently detected in human epithelial cancers. PloS one. 2010;5:e14130. doi: 10.1371/journal.pone.0014130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, Kieu C, Papior P, Baeuerle PA, Munz M, Gires O. Nuclear signalling by tumour-associated antigen EpCAM. Nature cell biology. 2009;11:162–171. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- [184].Psyrri A, Yu Z, Weinberger PM, Sasaki C, Haffty B, Camp R, Rimm D, Burtness BA. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:5856–5862. doi: 10.1158/1078-0432.CCR-05-0420. [DOI] [PubMed] [Google Scholar]
- [185].Psyrri A, Egleston B, Weinberger P, Yu Z, Kowalski D, Sasaki C, Haffty B, Rimm D, Burtness B. Correlates and determinants of nuclear epidermal growth factor receptor content in an oropharyngeal cancer tissue microarray. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17:1486–1492. doi: 10.1158/1055-9965.EPI-07-2684. [DOI] [PubMed] [Google Scholar]
- [186].Li CF, Fang FM, Wang JM, Tzeng CC, Tai HC, Wei YC, Li SH, Lee YT, Wang YH, Yu SC, Shiue YL, Chu PY, Wang WL, Chen LT, Huang HY. EGFR nuclear import in gallbladder carcinoma: nuclear phosphorylated EGFR upregulates iNOS expression and confers independent prognostic impact. Annals of surgical oncology. 2012;19:443–454. doi: 10.1245/s10434-011-1942-6. [DOI] [PubMed] [Google Scholar]
- [187].Di Lorenzo G, Perdona S, Buonerba C, Sonpavde G, Gigantino V, Pannone G, Quarto G, Ferro M, Gaudioso G, Terracciano D, Di Trolio R, Rescigno P, Botti G, De Placido S, Facchini G, Ascierto PA, Franco R. Cytosolic phosphorylated EGFR is predictive of recurrence in early stage penile cancer patients: a retropective study. Journal of translational medicine. 2013;11:161. doi: 10.1186/1479-5876-11-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Horiguchi S, Hishima T, Hayashi Y, Shiozawa Y, Horiguchi K, Kuroi K, Toi M, Funata N, Eishi Y. HER-2/neu cytoplasmic staining is correlated with neuroendocrine differentiation in breast carcinoma. Journal of medical and dental sciences. 2010;57:155–163. [PubMed] [Google Scholar]
- [189].Lopez-Knowles E, Zardawi SJ, McNeil CM, Millar EK, Crea P, Musgrove EA, Sutherland RL, O'Toole SA. Cytoplasmic localization of beta-catenin is a marker of poor outcome in breast cancer patients. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19:301–309. doi: 10.1158/1055-9965.EPI-09-0741. [DOI] [PubMed] [Google Scholar]
- [190].Li XQ, Yang XL, Zhang G, Wu SP, Deng XB, Xiao SJ, Liu QZ, Yao KT, Xiao GH. Nuclear beta-catenin accumulation is associated with increased expression of Nanog protein and predicts poor prognosis of non-small cell lung cancer. Journal of translational medicine. 2013;11:114. doi: 10.1186/1479-5876-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Marioni G, Staffieri A, Bertolin A, Giacomelli L, D'Alessandro E, Ottaviano G, Accordi D, Stramare R, de Filippis C, Blandamura S. Laryngeal carcinoma lymph node metastasis and disease-free survival correlate with MASPIN nuclear expression but not with EGFR expression: a series of 108 cases. Eur Arch Otorhinolaryngol. 2010;267:1103–1110. doi: 10.1007/s00405-009-1186-2. [DOI] [PubMed] [Google Scholar]
- [192].Mirza S, Rakha EA, Alshareeda A, Mohibi S, Zhao X, Katafiasz BJ, Wang J, Gurumurthy CB, Bele A, Ellis IO, Green AR, Band H, Band V. Cytoplasmic localization of alteration/deficiency in activation 3 (ADA3) predicts poor clinical outcome in breast cancer patients. Breast cancer research and treatment. 2013;137:721–731. doi: 10.1007/s10549-012-2363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Cai X, Wang J, Xin X. CIAPIN1 nuclear accumulation predicts poor clinical outcome in epithelial ovarian cancer. World journal of surgical oncology. 2012;10:112. doi: 10.1186/1477-7819-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Lionello M, Blandamura S, Lovato A, Franchella S, Giacomelli L, Ottaviano G, Stellini E, Staffieri A, Marioni G. A high nuclear nm23-H1 expression is associated with a better prognosis in elderly patients with laryngeal carcinoma. Acta oto-laryngologica. 2013 doi: 10.3109/00016489.2013.777159. [DOI] [PubMed] [Google Scholar]
- [195].Shubbar E, Kovacs A, Hajizadeh S, Parris TZ, Nemes S, Gunnarsdottir K, Einbeigi Z, Karlsson P, Helou K. Elevated cyclin B2 expression in invasive breast carcinoma is associated with unfavorable clinical outcome. BMC cancer. 2013;13:1. doi: 10.1186/1471-2407-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Raina D, Ahmad R, Joshi MD, Yin L, Wu Z, Kawano T, Vasir B, Avigan D, Kharbanda S, Kufe D. Direct targeting of the mucin 1 oncoprotein blocks survival and tumorigenicity of human breast carcinoma cells. Cancer research. 2009;69:5133–5141. doi: 10.1158/0008-5472.CAN-09-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Banerjee S, Mujumdar N, Dudeja V, Mackenzie T, Krosch TK, Sangwan V, Vickers SM, Saluja AK. MUC1c regulates cell survival in pancreatic cancer by preventing lysosomal permeabilization. PloS one. 2012;7:e43020. doi: 10.1371/journal.pone.0043020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Stroopinsky D, Rosenblatt J, Ito K, Mills H, Yin L, Rajabi H, Vasir B, Kufe T, Luptakova K, Arnason J, Nardella C, Levine JD, Joyce RM, Galinsky I, Reiter Y, Stone RM, Pandolfi PP, Kufe D, Avigan D. MUC1 is a potential target for the treatment of acute myeloid leukemia stem cells. Cancer research. 2013;73:5569–5579. doi: 10.1158/0008-5472.CAN-13-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]