Abstract
Epithelial tubes are the infrastructure for organs and tissues, and tube morphogenesis requires precise orchestration of cell signaling, shape, migration, and adhesion. Follicle cells in the Drosophila ovary form a pair of epithelial tubes whose lumens act as molds for the eggshell respiratory filaments, or dorsal appendages (DAs). DA formation is a robust and accessible model for studying the patterning, formation, and expansion of epithelial tubes. Tramtrack69 (TTK69), a transcription factor that exhibits a variable embryonic DNA-binding preference, controls DA lumen volume and shape by promoting tube expansion; the tramtrack mutation twin peaks (ttktwk) reduces TTK69 levels late in oogenesis, inhibiting this expansion. Microarray analysis of wild-type and ttktwk ovaries, followed by in situ hybridization and RNAi of candidate genes, identified the Phospholipase B-like protein Lamina ancestor (LAMA), the scaffold protein Paxillin, the endocytotic regulator Shibire (Dynamin), and the homeodomain transcription factor Mirror, as TTK69 effectors of DA-tube expansion. These genes displayed enriched expression in DA-tube cells, except lama, which was expressed in all follicle cells. All four genes showed reduced expression in ttktwk mutants and exhibited RNAi phenotypes that were enhanced in a ttktwk/+ background, indicating ttktwk genetic interactions. Although previous studies show that Mirror patterns the follicular epithelium prior to DA tubulogenesis, we show that Mirror has an independent, novel role in tube expansion, involving positive regulation of Paxillin. Thus, characterization of ttktwk-differentially expressed genes expands the network of TTK69 effectors, identifies novel epithelial tube-expansion regulators, and significantly advances our understanding of this vital developmental process.
Keywords: Tramtrack69 (TTK69), Paxillin, Dynamin (shibire), Mirror, Epithelial tube morphogenesis, TTK69-binding preference
Introduction
Epithelial tubes are essential structures in metazoan organs and tissues and thus, errors during tube morphogenesis can have profound developmental consequences. Failure of gastrulation will arrest development and defects in neural tube closure may result in spina bifida or anencephaly (Botto et al., 1999; Davidoff et al., 2002; Wallingford, 2005; Ray and Niswander, 2012). Gaining insight into the molecular and cellular requirements of tubulogenesis will augment our understanding of this developmental process and illuminate underlying causes of developmental tube defects, leading to better diagnostics and treatments.
To create an epithelial tube, cells must adopt a tube fate distinct from neighboring cells, coordinate their movements to form rudimentary tubes, and expand those tubes into a terminal morphology (Andrew and Ewald, 2010). Tube-forming events involving epithelial sheets share common molecular and cellular mechanisms (patterning, adhesion, polarity, guidance and morphogenetic movements), but the network of effectors and pathways that coordinate these mechanisms, from cell fate to terminal tube morphology, is not well defined. To advance our understanding of these networks, we identify and characterize downstream effectors of a key transcription factor that regulates tube morphogenesis in the Drosophila ovary.
Our model for epithelial tubulogenesis resembles vertebrate neural tube formation and involves the synthesis of the dorsal appendages [DAs], respiratory structures of the eggshell (Dorman et al., 2004). During oogenesis, egg chambers develop in an assembly-line fashion through 14 stages (S1–S14); a single layer of somatic, epithelial, follicle cells (FCs) envelops the developing germline (oocyte and nurse cells), undergoes coordinated morphogenesis to give shape to the egg, and secretes the eggshell (Spradling, 1993). This epithelium is polarized such that the apical surface contacts the germline and the basal surface faces outward. During stages S10B through S14, when growth and cell division have ceased, two dorsal anterior groups of FCs become patterned through EGF, BMP, and Notch signaling, form tubes through apical constriction and zippering, and elongate the tubes by migrating anteriorly, expanding apices, and intercalating through convergence and extension (Fig. 1A; Berg, 2005). The apical lumens of these tubes act as molds for the DAs of the mature eggshell, so although the FCs slough off during oviposition, the number, position, and morphology of the DA structures on laid eggs provide physical evidence for the efficacy of tube patterning and morphogenesis during oogenesis. Furthermore, since ovaries from well-nourished females contain all stages of egg chamber development, and since egg chambers can be dissected and cultured outside of the ovary, this system is ideal for investigating epithelial tube patterning, formation, and expansion (Berg, 2005).
Fig. 1.

twin peaks (ttktwk), a female sterile tramtrack69 mutation, affects gene expression during late oogenesis. (A) WT vs. ttktwk DA tubulogenesis, dorsal view. All subsequent images will be shown with the anterior facing left. S10B–S14=late stages of oogenesis, DM=Dorsal Midline. Blue=DA-roof cells. Red=DA-floor cells. The roof cells above the dorsal midline in S11–S14 are transparent to show the underlying DA-tube lumen (black). From S10B-S12, ttktwk resembles WT. During S13–S14, WT DA tubes expand but ttktwk DA tubes do not (indicated by orange). (B) Experimental pipeline. (C) Genes with significant differential expression in ttktwk (q≤0.05), plotted using mean log2-expression values for WT and ttktwk. Colors represent degree of raw fold change. ttktwk-down-regulated genes are above the dotted line (0-fold change) and ttktwk-up-regulated genes are below. (D) Subset of ttktwk-differentially expressed genes (i.e., panel C) with FIMO motif incidence ≥ 1. Colors indicate number of TTK69 motif instances in a gene’s promoter. (E) Subset of ttktwk-differentially expressed genes selected for subsequent analysis.
The female sterile tramtrack69 (ttk69) mutant twin peaks (ttktwk) produces eggs with severely stunted DAs and weak eggshells that cannot support fertilization. These defects are separable, the former resulting from a tube expansion defect (French et al., 2003). In ttktwk mutants, DA-tube cells undergo proper patterning and tube formation but fail to change shape and move during tube elongation. These cells retain some migratory ability and stretch along a correct, anterior path, but the DA tube itself fails to expand (Fig. 1A; French et al., 2003; Boyle and Berg, 2009).
What is the underlying mechanism causing this failure in tube expansion? The ttktwk mutation is an unusual, hypomorphic allele of the tramtrack69 gene that does not influence overall viability or development; the ttktwkP-element insertion disrupts a promoter required for late-oogenic ttk69 expression, inducing visible defects only in the DAs and eggshell (French et al., 2003).
TTK69 is a zinc-finger transcription factor originally identified as a repressor in embryogenesis (Harrison and Travers, 1990; Brown et al., 1991; Read et al., 1992); it is also a founding member of the BTB protein family that shares a Bric-á-brac—Tramtrack—Broad protein–protein interaction domain (Godt et al., 1993; Zollman et al., 1994). Best known for its role downstream of Notch in repressing neural cell fates (Guo et al., 1995), TTK69 also regulates diverse processes during fly development, from cell-cycle regulation to tracheal tubulogenesis (Baonza et al., 2002; Araújo et al., 2007). Genome-wide expression profiling in S2 cells (Reddy et al., 2010) identified broad classes of TTK69-regulated genes: protein folding, mRNA splicing, cell proliferation, phagocytosis, tracheal development, and axon guidance. Microarray profiling of embryonic trachea (Rotstein et al., 2011) indicated that TTK69 interacts with known pathways (e.g., Notch), and is upstream of known tracheal-tube size genes (e.g., chitin metabolism, septate junction, polarity proteins), corroborating previous observations.
To define TTK69's regulatory role during epithelial tubulogenesis and to identify novel effectors, we exploited the unique ttk69 allele, ttktwk. We compared wild type and ttktwk expression profiles via microarrays to identify direct and indirect targets of TTK69, evaluated the utility of TTK69-binding motifs as predictive tools, and ascertained which TTK69-regulated genes are required for DA-tube expansion through in situ hybridization (ISH) and tissue-specific RNAi. These studies reveal regulatory links between TTK69 and known or novel tubulogenesis effectors, show that TTK69-binding preference is highly variable, and demonstrate that ttk69's influence in late oogenesis extends beyond tubulogenesis.
Materials and methods
Drosophila strains
For microarrays and in situ analyses, we compared Canton S wild type to homozygous ry506P{PZ}07223=P{ry+t7.2, PlacZ}ttktwk mutants (French et al., 2003). For RNAi, we used UAS-RNAi transgenic strains from Drosophila stock centers ( Table S1). UAS-mirror was a gift from Helen McNeill (McNeill et al., 1997) and UAS-Paxillin (UAS-GFP::Paxillin) was a gift from Denise Montell (He et al., 2010). GAL4 driver strains were w*; CY2-GAL4; ry506ttktwk/TM3 (Queenan et al., 1997) and w*; Vm26Aa-GAL4; ry506ttktwk/TM3. To characterize GAL4 expression (Fig. 4A), we crossed GAL4 lines to UAS-GFP::moesin (Bloor and Kiehart, 2001). To visualize floor cells, we used y*w*; P{w+mC=Rho(ve)-lacZ.0.7} (Ip et al., 1992).
Fig. 4.

Expression of tramtrack69, mirror, and Paxillin mRNA during late oogenesis. Spatial and temporal comparison of ttk69, mirr, and Pax mRNA expression, visualized by FISH (A–X) and DAPI staining (A′–X′). At S10B (A–F′), all columnar FCs uniformly express ttk69 mRNA (A–A′); a dorsal saddle and ventral belt of FCs express mirr mRNA (C–C′); and the DA-floor cells are the first to express Pax mRNA (E–E′; arrowheads). ttk69 mRNA and Pax mRNA are absent in ttktwk (B–B′; F–F′), and expression of mirr mRNA is reduced (D–D′). At S11 (G–L′), leading DA-tube cells and, to a lesser degree, midline columnar FCs, express ttk69 mRNA (G–G′); several cells at the leading tip of each DA-tube express mirr mRNA (I–I′; arrowheads); and both DA-floor cells (highest levels) and DA-roof cells express Pax mRNA (K–K′). Transcripts from ttk69 and mirr are absent in ttktwk (H–H′; J–J′), and Pax mRNA expression is greatly reduced (L–L′). At S12 (M–R′), only leading DA-tube cells express ttk69 mRNA (M–M′); no FCs express mirr mRNA (O–O′); and all DA-tube cells express high levels of Pax mRNA (Q–Q′). In ttktwk, ttk69 mRNA and mirr mRNA are absent (N–N′; P–P′) and Pax mRNA expression is greatly reduced (R–R′). At S13 (S–X′), a few weak puncta of ttk69 mRNA are visible (S–S′); DA-tube cells begin to express high levels of mirr mRNA again (U–U′); and DA-tube cells continue to express Pax mRNA (W–W′). In ttktwk, a few weak puncta of ttk69 mRNA are visible; no mirr mRNA is visible (V–V′); and a low level of Pax mRNA is detectable around the dying nurse cells and stretch FCs (X–X′). Dotted lines = dorsal midlines; scale bar = 50 μm.
Vm26Aa-GAL4
To facilitate transcript production of GAL4 (Furger et al., 2002), we created pF-GAL4 by modifying the pGaTB GAL4 expression vector (Brand and Perrimon, 1993); we inserted a 150-bp fragment, containing splice donor, intron, and splice acceptor sequences from the Cp36 gene, into the hsp70 sequences that reside upstream of the Gal4 translational start site. To create pVm26Aa-F-GAL4, we inserted a 1047-bp PCR product from the Vm26Aa regulatory region, including ~950 bp of upstream sequences, the transcription start site, and the 5′ UTR, upstream of the Cp36 sequences. After verifying the construct by Sanger sequencing, we sent plasmid DNA to Rainbow Transgenics, who created > 100 random insertion lines. We selected the best expressing lines on X, 2 (used here), and 3. The Vm26Aa fragment is sufficient to direct gene expression in columnar FCs from S10 onward (Jin and Petri, 1993). The complete pVm26Aa-F-GAL4 plasmid sequence is available at NCBI (BankIt1606792 Seq1 KC664779).
Microarrays
Canton S and ttktwk ovaries came from 36-hr-old females; for each sample (3 biological replicates for each condition), 50 ovary pairs were dissected into RNAlater® (Life Technologies) and processed in < 20 min to minimize experimental effects on gene expression. Total RNA was extracted via the RNAqueous® Kit (Ambion) and processed at the UW Center for Array Technologies: RNA quality was evaluated via BioAnalyzer (Agilent); 100 ηg of total RNA was used to generate labeled probes via the Affymetrix GeneChip®3′ IVT Express Kit (2008), and following fragmentation, 10 μg labeled antisense RNA was hybridized for 16 h at 45°C to GeneChip®Drosophila Genome 2.0 Arrays (> 18,500 transcripts). The arrays were washed and stained in an Affymetrix Fluidics Station FS450 and scanned using an Affymetrix GeneChip® Scanner 3000 7G. The data were analyzed using GeneChip® Operating Software (GCOS) and the Microarray Suite version 5.0 (MAS 5.0) algorithm. Expression Console (Affymetrix) was used for robust multi-chip analysis (RMA) normalization. ttktwk-differentially expressed genes were then identified via SAM (Significance Analysis of Microarrays), q≤0.05.
TTK69 binding preference
Using modENCODE data from 0 to 12-h embryos (the mod-ENCODE consortium et al., 2012), ChIP-Seq sequences from 384 TTK69-bound sites were used to generate a 21-bp TTK69-preferred motif (E-value=2.2e−8) by employing the MEME-ChIP algorithm (MEME-Suite; Bailey et al., 2009). TOMTOM (MEME-Suite) was used to search for similarities with published motifs, and FIMO (MEME-Suite) was used to examine motif-incidence within ttktwk-differentially-expressed-gene promoters (including 2 kb upstream and 1 kb downstream).
In situ hybridization; immunostaining
We optimized colorimetric and fluorescence, tyramide-amplified (Lécuyer et al., 2007) in situ hybridization (ISH) protocols for the ovary and formulated a protocol for dual immunostaining:FISH in the ovary (Zimmerman et al., in preparation). For a complete protocol, contact Celeste Berg. We generated probes from sequenced cDNA clones ( Table S1; DGRC) with three exceptions: ttk69, mirr, and Rac2. For these genes, we amplified sequences unique to each gene from w1118 genomic DNA, cloned the sequences into TOPO-TA vectors (Invitrogen), and verified the clones by sequencing. For ttk69, we amplified 792 bp from the 3′-UTR of ttk, using sequence specific to the ttk69 isoform (ttk69-FW: 5′-CGCTCTTCGGGATTTAGTTG-3′; ttk69-RV: 5′-GTTGGTTTTTGAGGGTGTGG-3′). For mirr, we amplified 419 bp from the mirr 3′-UTR, a region that is mirr-specific and shared by all mirr transcripts (mirr-FW: 5′-GCCGTAGTCACTCCCAGTTT-3′; mirr-RV: 5′-GCGTCGAATTGTTTGCATCT-3′). For Rac2, we amplified 389 bp (unique) from the Rac2 5′-UTR to corroborate full-length Rac2 probe results that could reflect non-specific Rho-GTPase expression (Rac2-FW: 5′-TCTCTGTACGCGATTGCTTG-3′; Rac2-RV: 5′-GCAGAGGGTTTTTCAGTGGA-3′). To generate plots of late oogenesis expression (Figs. 2, S1), we scored relative germline and FC expression in 7–50 egg chambers for each stage using a 0–2 scoring system (0=weak/no expression, 1 = moderate expression, 2 = strong expression). Each point represents the average score for a given stage, tissue type, and background. Asterisks indicate significant differences (* = p < 0.05, **= p < 0.001) calculated by Chi-square analysis. Table S2 includes qualitative descriptions of spatial expression. Ovary immunostaining was as described (Ward and Berg, 2005; Zimmerman et al., in preparation), using rabbit anti-β-galactosidase (preabsorbed-1:2000; Cappel), mouse anti-GFP (1:200, Molecular Probes/Invitrogen), mouse anti-BR-core (1:500, 25E9.D7-concentrate, DSHB; Emery et al., 1994), mouse anti-α-Spectrin (1:50, 3A9-concentrate, DSHB; Dubreuil et al., 1987), and goat anti-mouse Alexafluor 488-, 555-, and 647-conjugated antibodies (1:500, Molecular Probes/Invitrogen). Imaging was performed on a Zeiss 510 scanning confocal microscope or a Nikon Microphot FXA. Images were processed using Photoshop CS (Adobe), Helicon Focus (Helicon Soft Ltd.) and FIJI (ImageJ-based, NIH).
Fig. 2.

FISH demonstrates FC ttktwk-differential-gene expression and reveals patterns relevant to DA-tube expansion. WT (A,D,G,J) vs. ttktwk (B, E, H, K) mRNA expression for lamina ancestor (lama), shibire (shi), Paxillin (Pax), and mirror (mirr). (A–K′) All images are oriented with anterior to the left. Dotted lines indicate the dorsal midline and insets indicate egg chamber orientation and stage. Scale bar = 50 μm. (C, F, I and L) Plots generated using a 0–2 scoring system (0=weak/no expression, 1 = moderate expression and 2 = strong expression); asterisks indicate significant differences (see Materials and methods); blue solid lines = WT, red dashed lines = ttktwk. (A–C) From S10A-S12, lamina ancestor mRNA is visible in all FCs and is reduced in ttktwk. (D–F) From S11–13, shibire (dynamin) mRNA is visible in anterior FCs and nurse cells and is reduced in ttktwk. (G–I) From S10B-S13, Paxillin mRNA is enriched in DA-tube cells, and expression is almost absent in ttktwk. (J–L) Previous studies show that mirror is expressed in a dorsal anterior saddle at S10B. We find new expression at S13 in DA-tube cells. In ttktwk, mirr mRNA expression is significantly reduced at S10B and is absent at S13–14.
Tissue-specific expression: GAL4-UAS
For RNAi assays, we crossed GAL4-bearing virgin females to UAS-RNAi-bearing (Table S1) males at 25 °C; we mated female progeny of the desired genotype to w1118 males at 30 °C in the presence of wet yeast, to optimize GAL4 expression and egg production. After > 24 h at 30 °C, we collected eggs over successive 8–12-h periods on grape juice/agar plates, then rinsed, pooled, and mounted the eggs in Hoyer's medium. For over-expression assays (UAS-mirr and UAS-Pax), we performed assays at 30 °C and 25 °C, to compare the effects of stronger and weaker GAL4 expression, respectively. We evaluated DAs using a 0–2 scoring system (0 = no DA defect, 1 = moderate DA defect and 2 = severe DA defect) and calculated an average score to facilitate comparison between strains. Moderate defects included rough/feathered DA shape, difference in DA length within the DA pair, wide DA paddles, wide DA shafts, and short DAs that extended past the micropyle. Severe DA defects included short and/or wide DAs not extending past the micropyle, fused DAs, or a combination of 2 or more category-1 criteria. Table 1 includes qualitative descriptions of DA morphology. Viability assays were performed at 30 °C as follows: after 8 h of egg laying, laid eggs were counted; plates were incubated at 25 °C for an additional 30 h, after which unhatched eggs were counted.
Table I.
| Genetic Background | Expressed UAS Construct |
Vm26Aa-GAL4
|
CY2-GAL4
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| % Normal |
% Moderate |
% Severe |
n Weighted Score (0-2) | Description of DA defect | % Normal |
% Moderate |
% Severe |
n Weighted Score (0-2) | Description of DA Defect | ||
| +/+ | GFP | 91 | 9 | 0 | 1060.09 | rough DAs | 81 | 19 | 1 | 1770.2 | rough DAs |
| lamina ancestor RNAi | 76 | 23 | 1 | 2050.25 | rough paddles | 50 | 49 | 1 | 1010.5 | broad DAs | |
| CG31918 RNAi | 63 | 36 | 1 | 1030.38 | broad DAs | 55 | 43 | 2 | 1300.48 | broad DAs | |
| katanin 80 RNAi | 60 | 31 | 9 | 1670.49 | rough, broad paddles | 43 | 52 | 5 | 1220.62 | rough, broad paddles | |
| Cp16 RNAi | 68 | 31 | 1 | 90 0.33 | narrow DA paddles | 66 | 32 | 2 | 1050.36 | narrow DA paddles | |
| Paxillin RNAi | 79 | 15 | 6 | 96 0.27 | rough DAs | 52 | 48 | 0 | 1450.48 | short DAs | |
| Rac2 RNAi | 36 | 56 | 8 | 1460.73 | rough DAs | 52 | 36 | 11 | 1050.59 | wide-based DAs | |
| shibire RNAi | 72 | 27 | 1 | 1010.29 | broad DAs | 61 | 37 | 2 | 110 0.41 | short, rough DAs | |
| mirror RNAi | 7 | 31 | 62 | 1431.24 | short, smooth DAs | 0 | 0 | 100 | 1852.00 | no DAs present | |
| tramtrack RNAi | 0 | 0 | 100 | 2242.00 | stunted, rough DAs | 0 | 0 | 100 | 1782.00 | stunted, rough DAs | |
| ttktwk/+ | GFP | 73 | 27 | 0 | 1170.27 | rough DAs | 68 | 30 | 2 | 210 0.34 | rough DAs |
| lamina ancestor RNAi | 71 | 29 | 0 | 116 0.29 | broad, rough paddles | 29 | 71 | 0 | 1900.71 | broad, short DAs | |
| CG31918 RNAi | 42 | 57 | 2 | 65 0.60 | broad DAs | 29 | 68 | 3 | 1540.74 | broad DAs | |
| katanin 80 RNAi | 44 | 53 | 3 | 1820.59 | rough, broad paddles | 28 | 71 | 2 | 1890.74 | rough, broad paddles | |
| Cp16 RNAi | 36 | 56 | 8 | 95 0.73 | narrow DA paddles | 31 | 66 | 2 | 2170.71 | narrow DAs | |
| Paxillin RNAi | 44 | 51 | 4 | 90 0.60 | rough DAs | 24 | 70 | 5 | 115 0.81 | short, broad, DAs | |
| Rac2 RNAi | 16 | 79 | 5 | 1540.88 | wide-based DAs | 25 | 66 | 9 | 1360.84 | wide-based DAs | |
| shibire RNAi | 6 | 74 | 19 | 1241.13 | broad DAs | 0 | 70 | 30 | 1381.30 | short, broad DAs | |
| mirror RNAi | 1 | 9 | 90 | 1611.89 | short, smooth DAs | 0 | 0 | 100 | 118 2.00 | no DAs present | |
| tramtrack RNAi | 0 | 0 | 100 | 1972.00 | stunted, rough DAs | 0 | 0 | 100 | 2012.00 | stunted, rough DAs | |
Results
Microarray analysis of twin peaks (ttktwk), a female sterile tramtrack69 mutant, reveals downstream genes during oogenesis
To identify TTK69 effectors in the ovary, we compared expression profiles between wild type and ttktwk using microarrays (Fig. 1B). This approach has been effective with whole ovaries (Jordan et al., 2005), staged egg chambers (Yakoby et al., 2008; Tootle et al., 2011), and purified follicle cells (Bryant et al., 1999; Wang et al., 2006). Although only a subset of FCs require TTK69 for DA-tube expansion (Boyle et al., 2010), we chose to analyze whole ovaries because this approach can identify differential gene expression associated with restricted FC-patterns (Jordan et al., 2005), the ttktwk mutation is specific, affecting TTK69 production only during late oogenesis (French et al., 2003), and germline TTK69 expression is not required for DA-tube expansion (Boyle and Berg, 2009). To increase the likelihood of identifying gene-expression differences relevant to DA-tube expansion, we dissected wild-type and ttktwk ovaries from ∼36-hr-old females to ensure comparable distributions of late-stage (S10–S14) egg chambers. Using Affymetrix Drosophila 2.0 whole-genome arrays and Significance Analysis of Microarrays (SAM), we identified ∼1000 differentially expressed transcripts in ttktwk ovaries (q≤0.05), 251 of which displayed more than a 2-fold change in expression (Fig. 1C; Gene Expression Omnibus: GSE42758).
As preliminary validation of array efficacy, we examined the expression data for expected features. We observed down-regulation of ttk69 transcripts in ttktwk, but transcripts from ttk88, a tramtrack isoform with no apparent ovarian function (French et al., 2003), remained low. Chorion protein 16 (Cp16) transcripts were also reduced in ttktwk (Table S1), corroborating previous northern blot analysis (French et al., 2003). Consistent with ttktwk's fragile eggshell phenotype, we noted decreased transcript levels for other eggshell genes (Cp18, Cp19, Cp7Fa, CG15570, and CG15571) and for an upstream regulator of eggshell gene expression, Cytochrome P450-18a1 (Cyp18a1; Tootle et al., 2011). These data validate our whole-ovary microarray approach.
In addition to eggshell genes, ttktwk altered expression of genes involved in metabolism, response to stress, DNA repair, ion transport, cell-cycle regulation, or phagocytosis; ∼42% of differentially expressed genes had no known function. The most enriched gene ontology terms were associated with mitochondrial functions (e.g., oxidoreductase, electron acceptor and heme), eggshell synthesis (chorion), or the cytoskeleton (actin). These results are consistent with genome-wide proteomic analyses in S2 cells that show that TTK69 is present in a complex with the mitochondrial transcription factor TFAM (Guruharsha et al., 2011) and substantiate TTK69's role in eggshell and cytoskeletal regulation.
modENCODE data provide insight into TTK69-binding preference
To understand the relationship between TTK69 binding and the ∼1000 genes that were differentially expressed in ttktwk, and to potentially make predictions about direct vs. indirect TTK69 interactions, we utilized MEME SUITE software components MEME-ChIP, TOMTOM, and FIMO (Bailey et al., 2009) to analyze modENCODE ChIP-Seq TTK69-binding data from 0 to 12 h embryos (384 bound sites; The modENCODE Consortium et al., 2010). We generated a preferred binding motif for TTK69 with MEME-ChIP (Multiple Em for Motif Elicitation, ChIP-optimized; Fig. S1A). TOMTOM, a motif-database scanning algorithm, showed that this motif partially matched another proposed Tramtrack motif generated by meta-analysis of experimental data (Kulakovskiy and Makeev, 2009). We then searched for the modENCODE motif within promoters of ttktwk-differentially expressed genes using FIMO (Find Individual Motif Occurrences). 54% of ttktwk-differentially expressed genes possessed ≥1 FIMO promoter hit, with 120 genes possessing ≥5 hits and 16 genes possessing ≥ 10 hits (Fig. 1D; Table S1). These data suggest that TTK69 both directly and indirectly regulates the expression of target genes. With the caveats that our modENCODE motif was derived from sites identified in embryos, and the MEME analysis indicates that TTK69-binding preference is variable, we considered the FIMO results when selecting candidates for further analysis.
TTK69 positively and negatively regulates FC gene-expression
To confirm that our array data reflected actual gene-expression differences during oogenesis, we selected candidate genes and used in situ hybridization (ISH) to compare transcript levels between wild-type and ttktwk ovaries. Using gene ontologies and the literature, we identified genes with functions relevant to tube expansion (cytoskeletal regulation, cell adhesion, migration and vesicle trafficking; Lubarsky and Krasnow, 2003). We selected genes with a wide range of expression changes, reasoning that large differences would be readily discernable by ISH, and that small differences were also relevant, since mRNA differences in DA-tube-forming cells might be subtle relative to total mRNA from whole ovaries. To provide additional diversity to our candidate pool, we selected both up- and down-regulated genes, genes with a range of FIMO-motif hits, and un-annotated genes (Fig. 1E; Table S1).
We compared transcript abundance and localization for 29 genes in wild-type and ttktwk ovaries by colorimetric ISH, and for FC-expressing genes, also by fluorescent ISH (FISH), noting the expressing tissue (germline, FC, or both) and any spatially restricted expression (Figs. 1E, 2 and S2; Tables S1 and S2). We generated expression timelines by assessing relative expression at each late-oogenic stage (Fig. 2C, F, I, L and Fig. S2A″–L″). For most transcripts (24/29), we observed ttktwk-differential gene expression as predicted by the arrays. Some transcripts exhibited subtle changes, but none contradicted the array results (Table S2; Figs. 2 and S2). In addition to FC expression, we discovered that TTK69 influences gene expression in the germ-line. Indeed, more than half (18/29) of the tested genes displayed germline expression, with 12 appearing to express strictly in the germline (Table S2; Fig. S2). Since germline ttktwk clones do not result in DA defects (Boyle and Berg, 2009), however, and since TTK69 is required specifically in FCs for DA-tube expansion (Boyle et al., 2010), we focused our attention on the most interesting genes, those with FC expression (15/29; Figs. 2 and S2; Table S2).
Of the 24 genes for which we observed ttktwk-differential gene expression, 8 genes displayed uniform FC expression and/or mixed FC-germline expression. We observed expression in all FCs for three genes: lamina ancestor (lama), Cyp18a1, and Cyp9b2. lama and Cyp18a1 transcripts were substantially reduced in ttktwk (Fig. 2A –C; Fig. S2D–D″), whereas Cyp9b2 expression increased in ttktwk (Fig. S2B–B″). Interestingly, late wild-type Cyp9b2 transcripts were enriched in anterior FCs relative to other FCs, while in ttktwk this pattern was reversed. One gene, rolled (rl; MAPK) was detected in all cell types, FC and germline, and expression increased slightly in ttktwk (Table S2). Four genes displayed late FC-expression in anterior FCs (including DA-tube cells) and germline expression at earlier stages: shibire (shi; dynamin); huckebein (hkb), Rab40, and (Centaurin beta 1A (CenB1A)). The level of shi transcripts was reduced in ttktwk, particularly in anterior FCs (Fig. 2D –F), whereas levels increased uniformly for hkb, Rab40, and CenB1A (Table S2).
Notably, 7 genes displayed expression in specific FC-types: CG7997, nervana 3 (nrv3), Cytochrome P450-12d1 (Cyp12d1), Glutathione S transferase E1 (GstE1), Rac2, Paxillin (Pax), and mirror (mirr). CG7997 transcripts were restricted to the squamous stretch FCs that envelop the nurse cells, while nrv3 transcripts were restricted to the columnar FCs covering the oocyte; in ttktwk, both CG7997 and nrv3 mRNA levels were higher than in wild type ( Fig. S2A–A″; Table S2). Cyp12d1 transcripts were present in anterior FCs during late oogenesis (S13+), and were absent in ttktwk ( Fig. S2C–C″). GstE1 expression began in all columnar FCs at S10, later appeared in small patches of anterior FCs ( Fig. S2E), and was absent in ttktwk ( Fig. S2E′–E″). Rac2 transcripts were visible in stretch FCs from S10 to 12, and in DA-tube cells at S12 ( Fig. S2K; inset, white arrowheads); levels were slightly reduced in ttktwk ( Fig. S2K–K″; black arrowhead). Both Pax and mirr transcripts localized primarily to DA-tube cells and therefore merited special attention, as discussed below.
In addition to the previously reported expression in migrating border cells (Chen et al., 2005), Pax transcripts were highly enriched in DA-tube cells (Fig. 2G–G′) and substantially reduced in ttktwk (Fig. 2H–I). Dual immunostaining:FISH revealed that Pax expression is dynamic during late oogenesis specifically in DA-tube cells (Fig. 3). Visualization of nuclear morphology (DAPI; Fig. 3A–F) and cell shape (spectrin protein; Fig. 3A′–F′) along with Pax mRNA (Fig. 3A ″–F″) indicated that peak transcript levels corresponded precisely to the initiation of DA-tube expansion (S12–13; Fig. 3C″–D″). Co-staining for a DA-tube-cell reporter (rhomboid-lacZ; Fig. 3A′–F′) showed conclusively that high Pax expression is contained within rhomboid-expressing floor cells (Fig. 3A‴–F‴) and the adjacent roof cells (see below). Pax encodes a focal adhesion scaffold protein with important roles in cell migration, adhesion, and signaling (Turner et al., 1990; Brown and Turner, 2004), but a role during tubulogenesis has not been described previously.
Fig. 3.

Paxillin expression in DA-tube cells peaks immediately prior to tube expansion. Timeline of Pax expression during late oogenesis (S10B–S14), visualized by dual immunostaining:FISH. (A–F) DAPI nuclear stains reveal stage. (A′–F′) rhomboid-lacZ, visualized by anti-βGal immunostaining, marks DA floor cells; α-Spectrin protein immunostain distinguishes cell membranes. (A″–F″) Pax mRNA levels in the DA-tube cells are dynamic, peaking at S12 prior to DA tube expansion. (A‴–F‴) Pax mRNA expresses in floor cells (rhomboid-lacZ) and overlying roof cells. Dotted lines = dorsal midlines; insets = egg chamber orientation and stage; scale bar = 50 μm.
mirr encodes a homeodomain transcription factor with dorsoventral (DV) patterning roles in the eye and ovary (McNeill et al., 1997; Jordan et al., 2000; Zhao et al., 2000; Atkey et al., 2006). Expression of mirr at S10 in a dorsal anterior FC ‘saddle’ helps define the DV axis of the eggshell and embryo and determines the placement of the DAs (Nakamura et al., 2007; Lachance et al., 2009; Fuchs et al., 2012). We confirmed this reported S10 mirr mRNA expression (Figs. 2L and 4C– C′), and noted a previously unreported, second wave of mirr mRNA expression in DA-tube cells during S13–14 (Fig. 2J–J′). This observation was intriguing since Mirror's known DV patterning function is accomplished by the conclusion of S10. Analysis of mirr mRNA expression in ttktwk revealed two important differences from wild type: the S10B expression of mirr mRNA in ttktwk was reduced (Figs. 2L and 4D–D′), and the late wave of mirr mRNA expression was absent (Figs. 2K–Land 4V–V′). We showed previously that patterning is normal in ttktwk (French et al., 2003), so we reasoned that the reduced mirr mRNA expression at S10B must still be sufficient for patterning in ttktwk. We hypothesized, however, that this reduced mirr mRNA expression at S10B and the lack of mirr mRNA expression at S13–14 could contribute to the defect in DA-tube expansion in ttktwk.
To understand how the patterns of ttk69, mirr, and Pax mRNA expression relate to each other and to DA-tube expansion, and how this mRNA expression is affected in ttktwk, we compared the spatial and temporal patterns of mRNA expression for these genes in wild type and ttktwk during DV patterning (S10B), DA-tube formation (S11), and at the initiation (S12) and conclusion (S13) of DA-tube expansion (Fig. 4). At S10B, ttk69 transcripts were present at high levels in all wild-type columnar FCs and were absent in ttktwk at this stage (Fig. 4A –B′) and all subsequent stages (Fig. 4H–H′, N–N′, T–T′). At this time, high levels of mirr transcripts were present in a dorsal saddle (Fig. 4C –C′); both the level and posterior extent of mirr expression were reduced in ttktwk (Fig. 4D –D′). Pax transcripts were present throughout the tissue at low levels and punctate expression was just becoming visible within the nuclei of the DA-floor cells (Fig. 4E –E′, arrowheads); this expression was not detectable in ttktwk (Fig. 4F –F').
At S11, ttk69 transcripts were still present in anterior and midline FCs but were reduced in more posterior FCs (Fig. 4G–G′). Transcripts of mirr were detectable only in a few cells at the leading tip of each DA tube (Fig. 4I–I'; arrowheads) and this expression was variably present in ttktwk (Fig. 4J –J′). Pax transcript levels increased notably during S11 in wild-type DA-tube cells (Fig. 4K –K'), while in ttktwk, relatively little Pax mRNA expression was visible in these cells (Fig. 4L –L′). We noted that at S11, Pax mRNA expression was increasing in cells that had been expressing high levels of mirr mRNA at S10B, suggesting that Mirror, as well as TTK69, could play a role in regulating Pax mRNA expression.
At S12, when DA-tube expansion initiates, ttk69 transcript expression was restricted to the anterior-most, leading cells of the DA tube (Fig. 4M –M′), consistent with a previously demonstrated requirement for TTK69 within those cells (Boyle et al., 2010). Transcripts of mirr were not detectable at S12 in either wild-type or ttktwk FCs (Fig. 4O –P'). Pax transcript levels were dramatically elevated during S12 specifically in DA-tube cells, and this expression was greatly reduced in ttktwk (Fig. 4Q -R').
At the end of S13, ttk69 transcripts were barely detectable in leading DA-tube FCs (Fig. 4S –S′). By S13 mirr transcription had re-initiated within the DA-tube FCs (Fig. 4U –U′), and this late wave of mirr expression was dependent on TTK69 (Fig. 4V –V′). Pax transcripts were present at high levels during this stage in DA-tube FCs, although not to the degree observed at S12 (Fig. 4W –W′); this expression was barely detectable in ttktwk (Fig. 4X –X′). These mRNA expression data are consistent with the reduced TTK69 protein expression observed in ttktwk (French et al., 2003; Boyle and Berg, 2009), confirm that TTK69 regulates both mirr and Pax expression, and demonstrate that ttk69, mirr, and Pax could interact with one another because their mRNAs are expressed in overlapping domains. Furthermore, this analysis confirms that the late expression of mirr in DA-tube cells is distinct from the earlier mirr expression required for DV patterning.
Following this ISH-based expression analysis, which effectively validated our arrays and distinguished genes with spatially and temporally relevant FC expression, we sought to address gene function within the FCs. To this end, we employed a FC-specific, RNAi-based analysis of candidate-gene function during DA tubulogenesis.
FC-specific RNAi against ttktwk-differentially-expressed transcripts identifies genes with functional roles in tubulogenesis and genetic interactions with ttktwk
To assess potential function in DA tubulogenesis (S10–14), we expressed RNAi constructs specifically in the FCs through the GAL4-UAS system (Brand and Perrimon, 1993; Dietzl et al., 2007), assaying DA morphology in laid eggs. Defects specific to DA-tube expansion manifest as properly located and formed, but abnormally shortened or misshapen, DAs, as in ttktwk (Fig. 1A). To create a spatiotemporally specific GAL4 driver, we cloned the Vitelline membrane 26Aa (Vm26Aa) regulatory region upstream of GAL4; this construct expresses at moderate levels from S10B–S14 in columnar FCs and in no other developmental context (Fig. 5A; Popodi et al., 1988). We also employed an enhancer trap line, CY2-GAL4, which drives strong expression in all FCs from S8 onward, but infrequently causes lethality due to expression in other tissues (Fig. 5A; Queenan et al., 1997). FC-specific expression of ttk-RNAi by either driver results in fully penetrant, DA and eggshell defects similar to ttktwk, providing a robust positive control (Fig. 5B; Table 1). As negative controls for each driver we expressed GFP (Fig. 5C –C″; Table 1).
Fig. 5.

RNAi demonstrates DA-tubulogenesis function and ttktwk interactions. At 30 °C, FC-RNAi against lama, shi, Pax, or mirr produces DA-tube defects. (A) GAL4-driven UAS–GFP::moesin expression. Left panel: Vm26Aa–GAL4 expresses only in columnar FCs from S10–S14. Right panel: CY2–GAL4 expresses in all FCs from S8-S14. (B–G'') Laid eggs from GAL4-UAS females in +/+ or ttktwk/+ genetic backgrounds. Numbers indicate percentages of normal (N), moderately defective (M), and severely defective (S) DAs; yellow numbers indicate the category of egg being shown. White arrowheads indicate the 8 abdominal ventral denticle belts visible on near-hatching larvae. (B) Vm26Aa-driven ttk-RNAi (positive control) disrupts DA-tube expansion and eggshell secretion. (C–C′′) GFP expression (negative control) indicates baseline DA-defect frequency for each genetic background. (D–G) Vm26Aa-driven RNAi causes subtle DA defects, with the exception of mirr-RNAi. (D′–G′) CY2-driven RNAi causes more frequent and severe DA defects for all conditions shown. (D″–G″) DA defects were enhanced in ttktwk/+, and, for shi-RNAi and Pax-RNAi, were frequently accompanied by short, round eggs. Scale bars = 50 μm.
In addition to Pax and mirr, which showed particularly striking expression patterns, we concentrated our functional analysis on the 11 other down-regulated candidates from the in situ analysis that displayed relevant FC expression (e.g., shibire and Rac2). We chose genes with reduced expression in ttktwk because mimicking ttktwk-down-regulation is simpler than expressing RNAi in a ttktwk homozygous background to suppress the ttktwk phenotype. Additionally, this approach let us compare the effects of FC-RNAi in both wild type (+/+) and ttktwk heterozygous (ttktwk/+) backgrounds, providing a sensitized background and a tool for identifying ttktwk genetic interactions. We also examined the effects of Cp16 RNAi, since Cp16 is expressed in dorsal anterior FCs (Parks and Spradling, 1987) and expression is reduced in ttktwk (French et al., 2003).
RNAi against 8 ttktwk-down-regulated genes resulted in DA defects (Table 1; Fig. 5D –G″). In general, these defects were more severe with the stronger CY2–GAL4 driver (Table 1; Fig. 5D ′–G′) and were exacerbated in the ttktwk/+ background (Table 1; Fig. 5D ″–G″). For genes such as lama, CG31918, Cp16, and kat80, RNAi knockdown primarily affected DA shape as opposed to length (Fig. 5D –D″; Table 1). RNAi against shi resulted in short, wide DAs, a defect that was greatly enhanced in ttktwk/+, indicating a strong ttktwk interaction (Fig. 5E –E″; Table 1). Similar but less pronounced DA defects occurred following Pax-RNAi; the phenotype was significantly enhanced in ttktwk/+ (Fig. 5F–F″; Table 1). In contrast, Rac2-RNAi produced DAs with wide bases, a defect that was not significantly enhanced in ttktwk/+ (Table 1). Finally, mirr-RNAi caused some unexpected phenotypes, described in detail below. These results reveal potential roles for ttktwk-differentially-expressed genes in DA tubulogenesis, and provide evidence for ttk69 genetic interactions.
Mirror regulates DA tubulogenesis and Paxillin expression separately from DV patterning
RNAi against mirr produced a profound and penetrant DA defect with both GAL4 drivers (Fig. 5G –G″; Table 1). Four additional mirr-RNAi constructs produced similar effects (not shown) and these effects could be ameliorated by ectopic mirr expression (Fig. 6; Table S3), indicating that the effects of mirr-RNAi are specific. Fine-tuning mirr-RNAi activity by reducing the assay temperature reduced the severity of the observed DA defects compared with controls, indicating that the level, not just the presence, of mirr mRNA expression influences DA-tube expansion (30–25 °C; Fig. 6A and B′). Ectopic mirr expression, representing a gain of function due to expression throughout the follicular epithelium, had a profound effect on eggshell morphology, making it difficult to interpret the DA-tube expansion defects (Fig. 6C –C′). Nevertheless, co-expression of mirr-RNAi and ectopic mirr produced DAs of an intermediate length (Fig. 6D –D'), occasionally producing DAs of a length comparable to controls. These results confirm that mirr-RNAi does indeed disrupt mirr expression and indicate that the precise level of mirr expression dictates the degree of DA-tube expansion.
Fig. 6.

The degree of DA-tube expansion depends on the levels of Mirror. The effects of Vm26Aa-GAL4-driven mirr-RNAi are reversible through ectopic expression of UAS-mirr. At 30 °C (A–D), the GAL4-UAS system is stronger than at 25 °C (A′–D′), but causes more background defects. (A–A′) Control eggs (GAL4 driver only) have no DA defects. (B–B′) Vm26Aa-GAL4-driven mirr-RNAi causes severe, highly penetrant DA defects. (C–C′) Vm26Aa-GAL4-driven ectopic mirr expression causes severe defects throughout the egg. (D–D′) Vm26Aa-GAL4-driven mirr expression can suppress the DA defects of Vm26Aa-GAL4-driven mirr-RNAi. Scale bar = 50 μm.
DA defects following mirr-RNAi resembled phenotypes produced by mirr[null] clones (Jordan et al., 2000; Zhao et al., 2000; Lachance et al., 2009; Fuchs et al., 2012), yet other features of these eggs differed significantly. mirr-RNAi eggs lacked DAs, yet DV polarity was unaffected: the dorsal surface of the egg was flat and the ventral surface was curved (Fig. 5G –G″; Lachance et al., 2009), and the embryos developing within these eggshells exhibited the normal pattern of ventral denticle belts (Fig. 5G–G″, arrowheads). Embryonic viability assays revealed marginal differences in hatch rates compared to Vm26Aa-GAL4 controls; the small increase in lethality with CY2-GAL4 was likely due to earlier RNAi expression ( Table S4). These data suggested that late-oogenic mirr-RNAi does not significantly affect DV patterning, and that mirr has a novel role in DA-tube expansion.
To test this hypothesis, we examined the expression of Broad, a DA-tube cell-fate marker. DV patterning begins by localized Gurken (EGF) signaling (Schüpbach, 1987), which activates mirr transcription in a dorsal ‘saddle’ at S10 (Fig. 4C –C′; Jordan et al., 2000; Zhao et al., 2000). Mirror helps establish DA-tube cell fate by dorsally activating broad and repressing pipe (Fuchs et al., 2012; Andreu et al., 2012). Broad protein remains high in DA-roof cells and is a terminal readout for dorsal FC fate (Tzolovsky et al., 1999; Dorman et al., 2004). Reducing mirr levels via Vm26Aa-GAL4-driven RNAi resulted in normal Broad protein levels at S11–S12 (N = 10/10; Fig. 7A ′–D′), yet laid eggs exhibited short, stubby DAs (Figs. 5G; 6B).
Fig. 7.
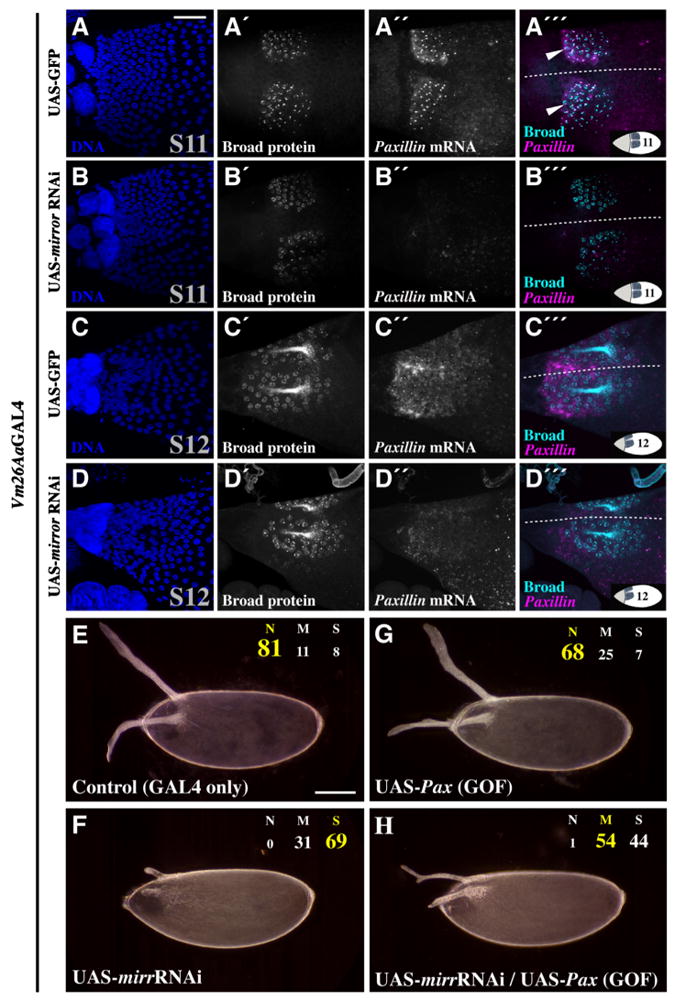
Mirror regulates DA tubulogenesis and Paxillin expression, independently of DV patterning. Dual immunostaining:FISH indicates that Vm26Aa-GAL4-driven mirr-RNAi disrupts DA tubulogenesis without affecting DV patterning (A–D″′), and over-expression of Pax can suppress the mirr-RNAi phenotype (E–H). At S11 (A–B″′) and S12 (C–D″′), egg chambers expressing GFP (A–A″′, C–C″′) and mirr-RNAi (B–B″′, D–D″′) display relatively normal Broad protein expression. Pax mRNA expression in DA-tube cells is reduced following mirr-RNAi (compare A″–B″ and C″–D″). (A″′) Arrowheads indicate co-localization of Broad protein and nascent Pax transcripts. At 30 °C, eggs from females expressing the Vm26Aa-GAL4 driver alone have normal DAs (E) and Vm26Aa-GAL4-driven mirr-RNAi causes severe, highly penetrant DA defects (F). Vm26Aa-GAL4-driven, ectopic Pax expression alone causes only minor DA defects (G), but can partially suppress the DA defects of Vm26Aa-GAL4-driven mirr-RNAi when co-expressed (H). Dotted lines = dorsal midlines; insets = egg chamber orientation and stage. Scale bars = 50 μm.
Since mirr and Pax share spatial expression domains (Fig. 4) and high mirr mRNA expression at S10B precedes high Pax mRNA expression at S11–12, we asked whether mirr acts upstream of Pax in DA-tube cells. Vm26Aa-GAL4-driven mirr-RNAi caused a reduction in Pax mRNA expression (N = 7/10; Fig. 7A ″–D″). Simultaneous assessment of Broad protein and Pax mRNA using dual immunostaining:FISH confirmed that Pax mRNA expression is high in DA-roof cells (Fig. 7A ‴ and C‴) and indicated that Vm26Aa-GAL4-driven mirr-RNAi affects Pax expression without disrupting DV patterning (Broad protein; Fig. 7B ‴ and D‴). Interestingly, puncta of Broad protein, indicating sites of high DNA occupancy, co-localized with puncta of Pax mRNA, indicating nascent Pax transcripts (Fig. 7A‴, arrowheads).
If Pax is indeed downstream of mirr, as implicated by the reduced Pax mRNA expression following mirr-RNAi, then over-expression of Pax might be able to reverse the effects of mirr-RNAi. To test this hypothesis, we compared DA morphology at 30 °C (Fig. 7E–H; Table S3) following RNAi against mirr alone (Fig. 7F), Pax alone (Fig. 7G), and co-expression of mirr-RNAi and Pax-RNAi (Fig. 7H). As observed previously (Figs. 5G and 6B), mirr-RNAi produced short, stubby DAs compared to driver controls (Fig. 7E and F). Over-expression of Pax resulted in some moderate DA defects, but the majority of eggs bore normal DAs (Fig. 7G). Importantly, expression of Pax in a mirr-RNAi background partially suppressed the mirr-RNAi DA defects, producing eggs with narrow DAs that often extended well beyond the anterior of the egg (Fig. 7H). We observed this effect to a lesser degree at 25 °C (Table S3). These results suggest that Mirror regulates DA-tube expansion at least in part through positive regulation of Pax. Given the co-localization of Broad protein with nascent Pax transcripts, Broad may contribute to this regulation. These data support our hypothesis that Mirror has an additional function in tubulogenesis separate from its role in DV patterning and suggest that Mirror and Paxillin function in a similar pathway downstream of TTK69 to promote DA-tube expansion.
Discussion
Here we significantly advance our understanding of TTK69 by characterizing its regulatory role during late oogenesis (Fig. 8) through analysis of the unique ttk69 allele, ttktwk. We identify differentially expressed genes in ttktwk, demonstrate that TTK69 effectors facilitate tube expansion and interact with one another, and elucidate novel roles for known proteins such as the homeobox transcription factor Mirror. By analyzing data for TTK69 binding during embryogenesis, we detect a variable sequence preference that lets us speculate about direct binding of TTK69 to ttktwk-differentially expressed genes, but also raises concerns about using a TTK69 motif as a predictive tool. Finally, our data substantiate TTK69's role in eggshell synthesis and indicate that TTK69's regulatory influence during oogenesis extends into the germline.
Fig. 8.

Model for TTK69 function during late oogenesis. Mirror's regulation of DV patterning (left) is distinct from its TTK69-influenced regulation of DA-tube expansion (middle). Previously known interactions are indicated by black arrows; in situ-verified regulation of expression is indicated by purple arrows; RNAi-verified, functional interactions are indicated by red arrows; potential interactions are indicated by gray dashed arrows. Line weight indicates relative strength of an interaction, based on our observations.
TTK69 regulates FC gene expression required for epithelial tube expansion
The ttktwk mutation disrupts TTK69 expression late in oogenesis, resulting in eggshell and DA defects (French et al., 2003; Boyle and Berg, 2009). Our microarray analyses corroborated these results by identifying differentially expressed eggshell and tube expansion genes in ttktwk. Analysis of expression by in situ hybridization distinguished genes whose FC expression was consistent with roles in DA-tube expansion, and tissue-specific RNAi analysis revealed functional roles in this process as well as ttktwk interactions. Of the 14 ttktwk-down-regulated genes knocked down by FC-specific RNAi, 8 produced measureable DA defects ranging widely in severity and penetrance.
For lama, CG31918, Cp16, and kat80, the RNAi defects were subtle or moderately penetrant, affecting DA shape. lama encodes a Phospholipase B-like protein that is conserved from human to Dictyostelium, with roles in Drosophila neural and imaginal-disc differentiation (Perez and Steller, 1996; Klebes et al., 2005). In Drosophila, lama has not previously been implicated in morphogenesis, but rather, is associated with undifferentiated cell types. In yeast, however, phospholipase B-like proteins regulate membrane bending during spore formation (Maier et al., 2008), and a role in regulating membrane dynamics would fit with the apical expansion required for DA-tube elongation. Since lama mRNA is expressed in all FCs, however, its precise role in DA tubulogenesis is unclear. CP16 is an extracellular matrix protein, a minor constituent of the eggshell that is synthesized late in oogenesis (Griffin-Shea et al., 1982). Consistent with this structural role, Cp16-RNAi produced narrow DA paddles. CG31918 is a predicted metalloendopeptidase, which could regulate the extracellular matrix during tube elongation, and kat80 encodes a predicted microtubule-severing protein, suggesting a regulatory role in micro-tubule dynamics/transport. Although these proteins could affect tube morphology by controlling DA-tube cell behaviors, the majority of CG31918 and kat80 mRNA expression is in the germline and/or overlying stretch FCs, suggesting that the products of these genes may function outside of DA-tube cells. Indeed, our lab has shown that germline and stretch FC-expressed genes also regulate DA-tube expansion (Rittenhouse and Berg, 1995; Tran and Berg, 2003). Nevertheless, RNAi against CG31918 and kat80 showed enhanced defects in ttktwk heterozygotes, suggesting they do play a role in DA-tube expansion, but their specific mechanisms of action are unclear.
For the other genes identified and characterized as tube-expansion effectors in this study, we can make more educated predictions about mechanism. Rac2, and Rho-GTPases in general, regulate the actin cytoskeleton, cell adhesion, and cell migration (Hall 2005). Although the difference between wild-type and ttktwkRac2 expression was small, Rac2 is noteworthy. Rac2 expression was enriched in DA-tube cells relative to other columnar FCs and Rac2 RNAi produced wide, often short, DAs. Also, Rho-GTPases such as Rac2 interact with two other tube-expansion genes identified here: Pax and shi. Paxillin's role in Drosophila development was first characterized through regulation of Rho and Rac signaling (Chen et al., 2005), and shi acts downstream of Rac during salivary gland morphogenesis, regulating E-cadherin levels via endocytosis (Pirraglia et al., 2006). These functions in other tissues support a role for Rac2 in DA tubulogenesis.
shi, which encodes Drosophila Dynamin, regulates endocytosis throughout development. Dynamin is required during cell migration and morphogenesis events, including border cell migration, salivary gland morphogenesis, and tracheal morphogenesis (Medioni and Noselli, 2005; Pirraglia et al., 2006; Hsouna et al., 2007). Dynamin may promote DA-tube expansion by facilitating apical cell-membrane turnover following DA-tube formation, as in the salivary gland, or by regulating basement membrane features, as in the border cells.
Pax encodes a molecular scaffold that localizes to focal adhesions, regulates Rho and Rac activity, modulates cell adhesion and migration, and facilitates epithelial tissue elongation (Chen et al., 2005; Deakin and Turner, 2008; Llense and Martín-Blanco, 2008; He et al., 2010). RNAi against Pax in migrating border cells delays migration and produces elongated cell morphologies (Llense and Martín-Blanco, 2008), similar to the shapes of ttktwk DA-tube cells (Boyle and Berg, 2009). The exquisite spatiotemporal pattern of Pax mRNA expression in DA-tube cells, the reduction of Pax mRNA expression in ttktwk, and the enhancement of Pax-RNAi DA defects in a ttktwk/+ background support a role for Paxillin in DA-tube expansion. Furthermore, the reduced mRNA expression following mirr-RNAi and the partial suppression of mirr-RNAi DA defects following Pax over-expression indicate that Pax is downstream of mirr as well as ttk69. Co-localization of Broad with nascent Pax transcripts also raises the possibility that a BTB-mediated interaction between Broad and TTK69 might contribute to Pax regulation as well.
How might these TTK69-regulated effectors interact with one another to facilitate DA-tube expansion? Our in situ hybridization data support a temporal and spatial program of gene regulatory interactions. Although we lack information about protein stability for some of these factors (e.g., Mirror), our genetic studies show that ttk69 regulates mirr mRNA expression, that mirr regulates Pax mRNA expression, and that ttk69 also regulates Rac2 and shi mRNA expression. In principle, TTK69 and Mirror could work together to regulate transcription of Pax, Rac2, and shi, so that sufficient levels of these proteins exist at the initiation of, and during, DA-tube expansion. During DA-tube expansion, Paxillin protein may regulate the activity of Rac2, and Rac-related signaling, as it has been shown to do in the Drosophila eye and wing (Chen et al., 2005). Rac-related signaling could then promote Shibire-mediated recycling of adhesion machinery as it does in the Drosophila salivary gland (Pirraglia et al., 2006), relaxing the apical constriction in the DA-tube cells and allowing the DA-tube to expand. Notably, this model could help explain the prominent apical-expansion defect that we have previously documented in ttktwk (Boyle and Berg, 2009).
1.1. Mirror has distinct oogenic roles in DV patterning and epithelial tube expansion
Mosaic analyses with mirr[null] alleles demonstrate that Mirror provides dorsal cues to the egg chamber and embryo (Jordan et al., 2000; Zhao et al., 2000; Lachance et al., 2009), in part by dorsally activating broad and repressing pipe (Fig. 8; Fuchs et al., 2012; Andreu et al., 2012). Since no cell division occurs after S6 (King, 1970), clonal analyses necessarily disrupt S10 patterning events. Using our new Vm26Aa–GAL4 driver, however, we knocked down mirr beginning in S10 and revealed a role for mirr beyond DV patterning, a role in tube expansion.
Many lines of evidence support our assertion that Mirror regulates DA-tube expansion downstream of TTK69. First, we show that mirr mRNA expression at S10B is significantly reduced in ttktwk and that the degree of DA-tube expansion depends on the precise level of mirr mRNA; we conclude that the S10-expression of mirr mRNA in ttktwk is sufficient to pattern the DA-tube cells but is insufficient to activate a tube-expansion program within these cells. Also, a distinct second wave of mirr expression, absent in ttktwk, occurs during later DA-tube expansion (S13–S14). This expression may be important for the terminal widening of the DA paddles at the end of oogenesis. Second, post-patterning, Vm26Aa-driven mirr-RNAi produces severe, fully penetrant DA defects resembling ttktwk, yet unlike ttktwk, eggshell integrity is normal. This result is consistent with the observation that TTK69 separately regulates DA-tube expansion and eggshell secretion (French et al., 2003) and suggests that Mirror could be important for mediating some TTK69 functions during late oogenesis. Third, under post-patterning RNAi conditions, rates of egg laying and hatching are unaffected: larvae are viable, displaying normal, ventral-restricted, pipe-induced denticle bands. Fourth, distinct mirr-RNAi constructs have similar effects, which can be reversed by over-expressing mirr. Fifth, the fate of DA-tube cells, assessed by the expression of Broad protein, is unaffected by post-patterning mirr-RNAi, as is the case in ttktwk (French et al., 2003). Finally, Pax mRNA expression is reduced by mirr-RNAi and Pax over-expression can suppress these defects. Therefore, we assert that Mirror is an important, TTK69-regulated effector of DA-tube expansion that acts upstream of Pax.
Towards a better understanding of TTK69 binding preference
TTK69's DNA-binding preference has been a topic of debate since its discovery. The first proposed TTK69 motifs were based on binding interactions with the fushi tarazu (ftz) promoter (TTATCCG, Harrison and Travers, 1990; TGCNAGGACNT, Brown et al., 1991); or on interactions with both ftz and even-skipped (eve). This knowledge facilitated analysis of TTK69 binding sites in the tailless (tll) promoter (TCCT; Chen et al., 2002). A weighted TTK69 motif, ascertained through bacterial one-hybrid and DNAseI-footprinting assays, expanded upon the previous TCCT motif (∼ttaTCCTg; Kulakovskiy and Makeev, 2009).
The modENCODE ChIP analysis of TTK69 binding in embryos was the first genome-wide, in vivo study examining TTK69 binding. We used these data to generate a longer motif; its loose nature and slight similarity to previous motifs indicate that DNA-binding by TTK69 is complex, potentially due to variable BTB-mediated protein–protein interactions (Bonchuk et al., 2011).
Since TTK69 could interact with different binding partners in oogenesis and embryogenesis, we view our FIMO results conservatively. Nevertheless, with the exception of lama, all genes exhibiting RNAi phenotypes possessed at least one FIMO hit in their promoter. Several genes with pronounced reduction in ttktwk expression also possessed promoter enrichment for the TTK69 binding motif: Arc1 (9 instances), scramb2 (11), kat80 (12), and Cyp18a1 (5), suggesting that TTK69 might have an activating function (Reddy et al., 2010; Rotstein et al., 2011).
TTK69 regulates eggshell and germline gene expression
Consistent with ttktwk’s fragile eggshell phenotype, microarray analysis revealed reduced expression of numerous chorion genes and Cyp18a1, an upstream regulator of eggshell gene expression (Fig. 8; Tootle et al., 2011). CYP18A1 processes the steroid hormone Ecdysone (Guittard et al., 2011), a signaling molecule that initiates metamorphosis (reviewed by Andres and Thummel, 1992), the FC endocycle-to-gene-amplification switch, and FC patterning (Sun et al., 2008; Boyle and Berg, 2009). Ecdysone is required for anterior FC migration, including centripetal migration and DA-tube formation (Hackney et al., 2007); by extension, Cyp18a1 could function during both migratory events. Alternatively, CYP18A1's role in late oogenesis may be strictly eggshell-related. RNAi against Cyp18a1 did not affect the DAs or eggshell, but RNAi may not sufficiently reduce Cyp18a1 transcripts to cause a visible defect. Nevertheless, ttktwk affects expression of many eggshell genes and an upstream eggshell-gene regulator; we hypothesize that reduced expression of these genes contributes to the fragile eggshell phenotype of ttktwk.
Finally, TTK69 regulates gene expression in the germline. Since eggs from ttktwk homozygous females are not fertilized, there are no visible ttktwk-associated defects during embryogenesis (French et al., 2003). Nevertheless, TTK69 could function in the germline to regulate maternally loaded transcripts.
Where do the ‘tracks’ lead?
We have identified and characterized downstream effectors of TTK69 required for epithelial tube expansion; we show that mirr plays a pivotal role in this process, acting upstream of Pax, and we provide corroborating evidence that TTK69 regulates eggshell gene expression. Nevertheless, there is much to learn about TTK69 and oogenesis. Not only does TTK69 regulate germline gene expression, it also regulates expression of stretch-FC-specific genes (e.g., CG7997) and phagocytosis-associated genes (e.g., Bx, CenB1A, and Rac2). Since stretch-FCs engulf dying nurse cells (Tran and Berg, 2003), it is possible that TTK69 has a regulatory role in nurse-cell engulfment. Furthermore, the altered expression of genes involved in oxidation-reduction (e.g., Cyp9b2, Cyp12d1, and CG31673) reveals an as yet unexplored protective function for TTK69 late in oogenesis. Clearly we have much to learn about TTK69's role in all ovarian cell types.
TTK69's regulatory function may also be conserved outside of the ovary, and perhaps in other organisms. TTK69 regulates tracheal tube morphogenesis, and, although this process occurs by branching morphogenesis rather than wrapping, as in DA tubulo-genesis, TTK69 might regulate similar tube expansion effectors in both contexts. The BTB domain-containing protein, Ribbon, has roles in Drosophila tracheal and salivary gland tubulogenesis, and disruption of ribbon causes cell defects similar to ttktwk (Shim et al., 2001; Bradley and Andrew, 2001). Since BTB-domain-containing proteins often dimerize or multimerize (Bonchuk et al., 2011), it is possible that TTK69 could interact with Ribbon through its BTB domain, or play a functionally equivalent role to Ribbon, in the ovary. Both TTK69's BTB and DNA-binding domains are conserved (e.g., 218 BLAST hits to the human genome). Indeed, the BTB domain from human Bcl-6 can functionally replace the BTB domain of TTK69 during eye development (Wen et al., 2000). While known functions of ttk69-related human genes include B-cell development and immunity, which share features with Drosophila innate immunity, functional roles for the vast majority of these TTK69-like proteins are unknown but could include tube morphogenesis. The conservation of TTK69's BTB and DNA-binding domains reveals constraints on TTK69's molecular activity during evolution; soon we may understand whether this conservation extends into TTK69's regulatory relationships, such as those we have studied in Drosophila.
Supplementary Material
Acknowledgments
We thank Rob Hall and Kurt Hardesty at the UW Center for Array Technologies; Brigham Mecham for statistical analysis; Greg Martin at the UW Keck Microscopy Center for imaging; Sandra Zimmerman for protocol optimization and statistical consultation; Faith Hassinger for construct analysis, Robert Matlock for egg-processing; Sandra Zimmerman and Kelsey Kaeding for in situ contributions; Hannele Ruohola-Baker, Dan Kiehart, Denise Mon-tell, Helen McNeil, Susan Parkhurst, Trudi Schüpbach, the Bloomington Stock Center, NIG-Fly, and the VDRC for fly strains; the DGRC for cDNA clones; Barbara Wakimoto and Sandra Zimmerman for critical reading of this manuscript; NIH R01-GM079433 for funding to CAB; and NSF Graduate Research Fellowship DGE-0718124 for supporting NCP.
Footnotes
Accession numbers
The microarray data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE42758 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42758).
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2013.03.017.
References
- Andres AJ, Thummel CS. Hormones, puffs and flies: the molecular control of metamorphosis by ecdysone. Trends Genet. 1992;8:132–138. doi: 10.1016/0168-9525(92)90371-A. [DOI] [PubMed] [Google Scholar]
- Andreu MJ, González-Pérez E, Ajuria L, Samper N, González-Crespo S, Campuzano S, Jiménez G. Mirror represses pipe expression in follicle cells to initiate dorsoventral axis formation in Drosophila. Development. 2012;139:1110–1114. doi: 10.1242/dev.076562. [DOI] [PubMed] [Google Scholar]
- Andrew DJ, Ewald AJ. Morphogenesis of epithelial tubes: Insights into tube formation, elongation, and elaboration. Dev Biol. 2010;341:34–55. doi: 10.1016/j.ydbio.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo SJ, Cela C, Llimargas M. Tramtrack regulates different morphogenetic events during Drosophila tracheal development. Development. 2007;134:3665–3676. doi: 10.1242/dev.007328. [DOI] [PubMed] [Google Scholar]
- Atkey MR, Lachance JB, Walczak M, Rebellow T, Nilson LA. Capicua regulates follicle cell fate in the Drosophila ovary through repression of mirror. Development. 2006;133:2115–2123. doi: 10.1242/dev.02369. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baonza A, Murawsky CM, Travers AA, Freeman M. Pointed and Tram-track69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat Cell Biol. 2002;4:976–980. doi: 10.1038/ncb887. [DOI] [PubMed] [Google Scholar]
- Berg CA. The Drosophila shell game: patterning genes and morphological change. Trends Genet. 2005;21:346–355. doi: 10.1016/j.tig.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Bloor JW, Kiehart DP. Zipper non-muscle myosin-II functions downstream of PS2 Integrin in Drosophila myogenesis and is necessary for myofibril formation. Dev Biol. 2001;239:215–228. doi: 10.1006/dbio.2001.0452. [DOI] [PubMed] [Google Scholar]
- Bonchuk A, Denisov S, Georgiev P, Maksimenko O. Drosophila BTB/POZ domains of ‘ttk group’ can form multimers and selectively interact with each other. J Mol Biol. 2011;412:423–436. doi: 10.1016/j.jmb.2011.07.052. [DOI] [PubMed] [Google Scholar]
- Botto LD, Moore CA, Khoury MJ, Erickson JD. Neural-tube defects. New Engl J Med. 1999;341:1509–1519. doi: 10.1056/NEJM199911113412006. [DOI] [PubMed] [Google Scholar]
- Boyle MJ, Berg CA. Control in time and space: Tramtrack69 cooperates with Notch and Ecdysone to repress ectopic fate and shape changes during Drosophila egg chamber maturation. Development. 2009;136:4187–4197. doi: 10.1242/dev.042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle MJ, French RL, Cosand AK, Dorman JB, Kiehart DP, Berg CA. Division of labor: Subsets of dorsal-appendage-forming cells control the shape of the entire tube. Dev Biol. 2010;346:68–79. doi: 10.1016/j.ydbio.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley PL, Andrew DJ. ribbon encodes a novel BTB/POZ protein required for directed cell migration in Drosophila melanogaster. Development. 2001;128:3001–3015. doi: 10.1242/dev.128.15.3001. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brown JL, Sonoda S, Ueda H, Scott MP, Wu C. Repression of the Drosophila fushi tarazu (ftz) segmentation gene. EMBO J. 1991;10:665–674. doi: 10.1002/j.1460-2075.1991.tb07995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MC, Turner CE. Paxillin: adapting to change. Physiol Rev. 2004;84:1315–1339. doi: 10.1152/physrev.00002.2004. [DOI] [PubMed] [Google Scholar]
- Bryant Z, Subrahmanyan L, Tworoger M, LaTray L, Liu C, Li M, van denEngh G, Ruohola-Baker H. Characterization of differentially expressed genes in purified Drosophila follicle cells: toward a general strategy for cell type-specific developmental analysis. Proc Natl Acad Sci USA. 1999;96:5559–5564. doi: 10.1073/pnas.96.10.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GC, Turano B, Ruest PJ, Hagle M, Settleman J, Thomas SM. Regulation of Rho and Rac signaling to the actin cytoskeleton by Paxillin during Drosophila development. Mol Cell Biol. 2005;25:979–987. doi: 10.1128/MCB.25.3.979-987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Chiang CS, Weng LC, Lengyel JA, Liaw GJ. Tramtrack69 is required for the early repression of tailless expression. Mech Dev. 2002;116:75–83. doi: 10.1016/s0925-4773(02)00143-0. [DOI] [PubMed] [Google Scholar]
- Davidoff MJ, Petrini J, Damus K, Russell RB, Mattison D. Neural tube defect-specific infant mortality in the United States. Teratology. 2002;66(Suppl. 1):S17–22. doi: 10.1002/tera.90005. [DOI] [PubMed] [Google Scholar]
- Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci. 2008;121:2435–2444. doi: 10.1242/jcs.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su K, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dorman JB, James KE, Fraser SE, Kiehart DP, Berg CA. bullwinkle is required for epithelial morphogenesis during Drosophila oogenesis. Dev Biol. 2004;267:320–341. doi: 10.1016/j.ydbio.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Dubreuil R, Byers TJ, Branton D, Goldstein LS, Kiehart DP. Drosophila spectrin I. Characterization of the purified protein. J Cell Biol. 1987;105:2095–2102. doi: 10.1083/jcb.105.5.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery IF, Bedian V, Guild GM. Differential expression of Broad-Complex transcription factors may forecast tissue-specific developmental fates during Drosophila metamorphosis. Development. 1994;120:3275–3287. doi: 10.1242/dev.120.11.3275. [DOI] [PubMed] [Google Scholar]
- French RL, Cosand AK, Berg CA. The Drosophila female sterile mutation twin peaks is a novel allele of tramtrack and reveals a requirement for TTK69 in epithelial morphogenesis. Dev Biol. 2003;253:18–35. doi: 10.1006/dbio.2002.0856. [DOI] [PubMed] [Google Scholar]
- Fuchs A, Cheung LS, Charbonnier E, Shvartsman SY, Pyrowolakis G. Transcriptional interpretation of the EGF receptor signaling gradient. Proc Natl Acad Sci USA. 2012;109:1572–1577. doi: 10.1073/pnas.1115190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furger A, O’Sullivan JM, Binnie A, Lee BA, Proudfoot NJ. Promoter proximal splice sites enhance transcription. Genes Dev. 2002;16:2792–2799. doi: 10.1101/gad.983602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godt D, Couderc JL, Cramton SE, Laski FA. Pattern formation in the limbs of Drosophila: bric à brac is expressed in both a gradient and a wave-like pattern and is required for specification and proper segmentation of the tarsus. Development. 1993;119:799–812. doi: 10.1242/dev.119.3.799. [DOI] [PubMed] [Google Scholar]
- Griffin-Shea R, Thireos G, Kafatos FC. Organization of a cluster of four chorion genes in Drosophila and its relationship to developmental expression and amplification. Dev Biol. 1982;91:325–336. doi: 10.1016/0012-1606(82)90039-2. [DOI] [PubMed] [Google Scholar]
- Guittard E, Blais C, Maria A, Parvy J, Pasricha S, Lumb C, Lafont R, Daborn P, Dauphin-Villemant C. CYP18A1, a key enzyme of Drosophila steroid hormone inactivation, is essential for metamorphosis. Dev Biol. 2011;349:35–45. doi: 10.1016/j.ydbio.2010.09.023. [DOI] [PubMed] [Google Scholar]
- Guo M, Bier E, Jan LY, Jan YN. tramtrack acts downstream of numb to specify distinct daughter cell fates during asymmetric cell divisions in the Drosophila PNS. Neuron. 1995;14:913–925. doi: 10.1016/0896-6273(95)90330-5. [DOI] [PubMed] [Google Scholar]
- Guruharsha KG, Rual J, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, McKillip E, Shah S, Stapleton M, Wan KH, Yu C, Parsa B, Carlson JW, Chen X, Kapadia B, VijayRaghavan K, Gygi SP, Celniker SE, Obar RA, Artavanis-Tsakonas S. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney JF, Pucci C, Naes E, Dobens L. Ras signaling modulates activity of the Ecdysone receptor EcR during cell migration in the Drosophila ovary. Dev Dyn. 2007;236:1213–1226. doi: 10.1002/dvdy.21140. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the control of cell behaviour. Biochem Soc Trans. 2005;33:891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- Harrison SD, Travers AA. The tramtrack gene encodes a Drosophila finger protein that interacts with the ftz transcriptional regulatory region and shows a novel embryonic expression pattern. EMBO J. 1990;9:207–216. doi: 10.1002/j.1460-2075.1990.tb08097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Wang X, Tang HL, Montell DJ. Tissue elongation requires oscillating contractions of a basal actomyosin network. Nat Cell Biol. 2010;12:1133–1142. doi: 10.1038/ncb2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsouna A, Lawal HO, Izevbaye I, Hsu T, O’Donnell JM. Drosophila dopamine synthesis pathway genes regulate tracheal morphogenesis. Dev Biol. 2007;308:30–43. doi: 10.1016/j.ydbio.2007.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- Jin J, Petri WH. Developmental control elements in the promoter of a Drosophila vitelline membrane gene. Dev Biol. 1993;156:557–565. doi: 10.1006/dbio.1993.1102. [DOI] [PubMed] [Google Scholar]
- Jordan KC, Clegg NJ, Blasi JA, Morimoto AM, Sen J, Stein D, McNeill H, Deng WM, Tworoger M, Ruohola-Baker H. The homeobox gene mirror links EGF signalling to embryonic dorso–ventral axis formation through Notch activation. Nat Genet. 2000;24:429–433. doi: 10.1038/74294. [DOI] [PubMed] [Google Scholar]
- Jordan KC, Hatfield SD, Tworoger M, Ward EJ, Fischer KA, Bowers S, Ruohola-Baker H. Genome wide analysis of transcript levels after perturbation of the EGFR pathway in the Drosophila ovary. Dev Dyn. 2005;232:709–724. doi: 10.1002/dvdy.20318. [DOI] [PubMed] [Google Scholar]
- King RC. Ovarian development in Drosophila melanogaster. Academic Press, Inc.; NY: 1970. [Google Scholar]
- Klebes A, Sustar A, Kehris K, Li H, Schubiger G, Kornberg TB. Regulation of cellular plasticity in Drosophila imaginal disc cells by the Polycomb group, trithorax group and lama genes. Development. 2005;132:3753–3765. doi: 10.1242/dev.01927. [DOI] [PubMed] [Google Scholar]
- Kulakovskiy IV, Makeev VJ. Discovery of DNA motifs recognized by transcription factors through integration of different experimental sources. Mol Biophys. 2009;54:667–674. [Google Scholar]
- Lachance JB, Lomas MF, Eleiche A, Kerr PB, Nilson LA. Graded Egfr activity patterns the Drosophila eggshell independently of autocrine feedback. Development. 2009;136:2893–2902. doi: 10.1242/dev.036103. [DOI] [PubMed] [Google Scholar]
- Lécuyer E, Parthasarathy N, Krause HM. Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. In: Dahmann C, editor. Methods in Molecular Biology. Humana Press, Inc.; Totowa, NJ: 2007. pp. 289–302. [DOI] [PubMed] [Google Scholar]
- Llense F, Martín-Blanco E. JNK signaling controls border cell cluster integrity and collective cell migration. Curr Biol. 2008;18:538–544. doi: 10.1016/j.cub.2008.03.029. [DOI] [PubMed] [Google Scholar]
- Lubarsky B, Krasnow MA. Tube morphogenesis: making and shaping biological tubes. Cell. 2003;112:19–28. doi: 10.1016/s0092-8674(02)01283-7. [DOI] [PubMed] [Google Scholar]
- Maier P, Rathfelder N, Maeder CI, Colombelli J, Stelzer EHK, Knop M. The SpoMBe pathway drives membrane bending necessary for cytokinesis and spore formation in yeast meiosis. EMBO J. 2008;27:2363–2374. doi: 10.1038/emboj.2008.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill H, Yang CH, Brodsky M, Ungos J, Simon MA. mirror encodes a novel PBX-class homeoprotein that functions in the definition of the dorsal–ventral border in the Drosophila eye. Genes Dev. 1997;11:1073–1082. doi: 10.1101/gad.11.8.1073. [DOI] [PubMed] [Google Scholar]
- Medioni C, Noselli S. Dynamics of the basement membrane in invasive epithelial clusters in Drosophila. Development. 2005;132:3069–3077. doi: 10.1242/dev.01886. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kagesawa T, Nishikawa M, Hayashi Y, Kobayashi S, Niimi T, Matsuno K. Soma-dependent modulations contribute to divergence of rhomboid expression during evolution of Drosophila eggshell morphology. Development. 2007;134:1529–1537. doi: 10.1242/dev.001578. [DOI] [PubMed] [Google Scholar]
- Parks S, Spradling A. Spatially regulated expression of chorion genes during Drosophila oogenesis. Genes Dev. 1987;1:497–509. [Google Scholar]
- Perez SE, Steller H. Molecular and genetic analyses of lama, an evolutionarily conserved gene expressed in the precursors of the Drosophila first optic ganglion. Mech Dev. 1996;59:11–27. doi: 10.1016/0925-4773(96)00556-4. [DOI] [PubMed] [Google Scholar]
- Pirraglia C, Jattani R, Myat MM. Rac function in epithelial tube morphogenesis. Dev Biol. 2006;290:435–446. doi: 10.1016/j.ydbio.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Popodi E, Minoo P, Burke T, Waring GL. Organization and expression of a second chromosome follicle cell gene cluster in Drosophila. Dev Biol. 1988;127:248–256. doi: 10.1016/0012-1606(88)90312-0. [DOI] [PubMed] [Google Scholar]
- Queenan AM, Ghabrial A, Schüpbach T. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development. 1997;124:3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- Ray HJ, Niswander L. Mechanisms of tissue fusion during development. Development. 2012;139:1701–1711. doi: 10.1242/dev.068338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read D, Levine M, Manley JL. Ectopic expression of the Drosophila tramtrack gene results in multiple embryonic defects, including repression of even-skipped and fushi tarazu. Mech Dev. 1992;38:183–195. doi: 10.1016/0925-4773(92)90052-l. [DOI] [PubMed] [Google Scholar]
- Reddy BA, Bajpe PK, Bassett A, Moshkin YM, Kozhevnikova E, Bezstarosti K, Demmers JAA, Travers AA, Verrijzer CP. Drosophila transcription factor Tramtrack69 binds MEP1 to recruit the chromatin remodeler NuRD. Mol Cell Biol. 2010;30:5234–5244. doi: 10.1128/MCB.00266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittenhouse KR, Berg CA. Mutations in the Drosophila gene bullwinkle cause the formation of abnormal eggshell structures and bicaudal embryos. Development. 1995;121:3023–3033. doi: 10.1242/dev.121.9.3023. [DOI] [PubMed] [Google Scholar]
- Rotstein B, Molnar D, Adryan B, Llimargas M. Tramtrack is genetically upstream of genes controlling tracheal tube size in Drosophila. PLoS One. 2011;6:e28985. doi: 10.1371/journal.pone.0028985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T. Germ line and soma cooperate during oogenesis to establish the dorsoventral pattern of egg shell and embryo in Drosophila melanogaster. Cell. 1987;49:699–707. doi: 10.1016/0092-8674(87)90546-0. [DOI] [PubMed] [Google Scholar]
- Shim K, Blake KJ, Jack J, Krasnow MA. The Drosophila ribbon gene encodes a nuclear BTB domain protein that promotes epithelial migration and morphogenesis. Development. 2001;128:4923–4933. doi: 10.1242/dev.128.23.4923. [DOI] [PubMed] [Google Scholar]
- Spradling AC. Developmental genetics of oogenesis. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Press, Cold Spring Harbor; NY: 1993. pp. 1–70. [Google Scholar]
- Sun J, Smith L, Armento A, Deng WM. Regulation of the endocycle/gene amplification switch by Notch and ecdysone signaling. J Cell Biol. 2008;182:885–896. doi: 10.1083/jcb.200802084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The modENCODE Consortium. Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootle TL, Williams D, Hubb A, Frederick R, Spradling A. Drosophila eggshell production: Identification of new genes and coordination by Pxt. PLoS One. 2011;6:e19943. doi: 10.1371/journal.pone.0019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DH, Berg CA. bullwinkle and shark regulate dorsal-appendage morphogenesis in Drosophila oogenesis. Development. 2003;130:6273–6282. doi: 10.1242/dev.00854. [DOI] [PubMed] [Google Scholar]
- Turner CE, Glenney JR, Burridge K. Paxillin: A new Vinculin-binding protein present in focal adhesions. J Cell Biol. 1990;111:1059–1068. doi: 10.1083/jcb.111.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzolovsky G, Deng WM, Schlitt T, Bownes M. The function of the broad-complex during Drosophila melanogaster oogenesis. Genetics. 1999;153:1371–1383. doi: 10.1093/genetics/153.3.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB. Neural tube closure and neural tube defects: studies in animal models reveal known knowns and known unknowns. Am J Med Genet. 2005;135C:59–68. doi: 10.1002/ajmg.c.30054. [DOI] [PubMed] [Google Scholar]
- Wang X, Bo J, Bridges T, Dugan KD, Pan T, Chodosh LA, Montell DJ. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev Cell. 2006;10:483–495. doi: 10.1016/j.devcel.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Ward EJ, Berg CA. Juxtaposition between two cell types is necessary for dorsal appendage tube formation. Mech Dev. 2005;122:241–255. doi: 10.1016/j.mod.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Wen Y, Nguyen D, Li Y, Lai Z. The N-terminal BTB/POZ domain and C-Terminal sequences are essential for Tramtrack69 to specify cell fate in the developing Drosophila eye. Genetics. 2000;156:195–203. doi: 10.1093/genetics/156.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakoby N, Bristow CA, Gong D, Schafer X, Lembong J, Zartman JJ, Halfon MS, Schüpbach T, Shvartsman SY. A combinatorial code for pattern formation in Drosophila oogenesis. Dev Cell. 2008;15:725–737. doi: 10.1016/j.devcel.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Woolner S, Bownes M. The Mirror transcription factor links signalling pathways in Drosophila oogenesis. Dev Genes Evol. 2000;210:449–457. doi: 10.1007/s004270000081. [DOI] [PubMed] [Google Scholar]
- Zimmerman SG, Peters NC, Altaras A, Berg CA. Optimized ISH, FISH, and protein-RNA double labeling in Drosophila ovaries. Manuscript in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollman S, Godt D, Privé GG, Couderc JL, Laski FA. The BTB domain, found primarily in zinc finger proteins defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci USA. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


