Abstract
Rodent self-grooming is an important, evolutionarily conserved behavior, highly sensitive to pharmacological and genetic manipulations. Mice with aberrant grooming phenotypes are currently used to model various human disorders. Therefore, it is critical to understand the biology of grooming behavior, and to assess its translational validity to humans. The present in-silico study used publicly available gene expression and behavioral data obtained from several inbred mouse strains in the open-field, light-dark box, elevated plus- and elevated zero-maze tests. As grooming duration differed between strains, our analysis revealed several candidate genes with significant correlations between gene expression in the brain and grooming duration. The Allen Brain Atlas, STRING, GoMiner and Mouse Genome Informatics databases were used to functionally map and analyze these candidate mouse genes against their human orthologs, assessing the strain ranking of their expression and the regional distribution of expression in the mouse brain. This allowed us to identify an interconnected network of candidate genes (which have expression levels that correlate with grooming behavior), display altered patterns of expression in key brain areas related to grooming, and underlie important functions in the brain. Collectively, our results demonstrate the utility of large-scale, high-throughput data-mining and in-silico modeling for linking genomic and behavioral data, as well as their potential to identify novel neural targets for complex neurobehavioral phenotypes, including grooming.
Keywords: Grooming behavior, Anxiety in Mice, Neurophenotypes, Behavioral domains, Gene expression and omics
1. Introduction
Large scale, high-throughput data-mining and data integration are rapidly becoming key methods for scientific discovery (Tabakoff et al., 2009, Xuan et al., 2010), emphasizing the importance of sharing biological data (Akil et al., 2011, Sears et al., 2006). Integration of behavioral phenotypes with neural and genomic data, such as phenomics and ‘genetical genomics’, is emerging as a promising strategy for the dissection of complex gene-behavior interactions (Bennett et al., 2011, Bhave et al., 2007, Tabakoff et al., 2007). Among the behavioral phenotypes, self-grooming is especially important, because it represents an evolutionarily ancient behavior with multiple biological functions (from hygiene to stress reduction) and a complex, patterned nature (Chen et al., 2010, Fentress, 1988, File et al., 1988, Sachs, 1988, Spruijt et al., 1992). In rodents, grooming is one of the most frequently occurring behaviors, often correlating with the levels of arousal (Fentress, 1968, 1977, 1988) and various anxiety-like behaviors (Denmark et al., 2010, Kalueff and Tuohimaa, 2005a, c, Kyzar et al., 2011). Mounting evidence shows the value of analyzing grooming as a behavioral endpoint following genetic or pharmacological manipulations in experimental models of various brain disorders (Audet et al., 2006, Chen, Tvrdik, 2010, Estanislau, 2012, Greer and Capecchi, 2002, Kalueff et al., 2004, Kalueff and Tuohimaa, 2005c).
While mouse self-grooming is an important behavioral domain, little is known about its genetic architectonics or genomic correlates (Bergner et al., 2010). Established in 2000, the Mouse Phenome Database (MPD) is a publicly available platform, providing phenotypic data on different mouse strains (Grubb et al., 2009). While the MPD initially lacked mouse grooming data, it now contains reports on grooming frequency in A/J, C57BL/6J, consomic (Lake et al., 2005) and wild-derived strains (Koide and Takahashi, 2006), as well as grooming duration from multiple inbred strains in several anxiety tests (Brown et al., 2004). Comparison of these data with other behaviors using the MPD online tools has revealed correlations with anxiety-sensitive behaviors, reflecting the importance of measuring grooming in animal anxiety paradigms (Crawley, 2007, Hart et al., 2010, Kalueff and Tuohimaa, 2005b).
The present study aimed to examine the potential link between mouse grooming behavior and the expression of selected genes within the brain. This study also demonstrates the utility of large-scale data-mining and in-silico (computer-based) modeling for linking genomic and behavioral data, and its potential to identify new neural targets for specific phenotypes of interest. Using mouse grooming as a representative phenotype, this proof-of-concept study can be applied in future research to other mammalian behavioral and physiological phenotypes.
2. Methods and Results
2.1. General overview
To achieve the goals of this study, we utilized the MPD data containing behavioral phenotypes (Brown, Gunn, 2004) and whole-brain genomic microarray results (Tabakoff, Bhave, 2007); see (Bennett, Saba, 2011, Stewart et al., 2011) for the conceptual framework. We first used the MPD tools to identify significant correlations for grooming and gene expression data across four widely used behavioral paradigms (open-field test, elevated plus-maze, elevated zero-maze and light-dark box); Fig. 1. We next ranked these candidate genes in terms of the strength of their correlations with grooming behaviors, identifying a sub-group of genes whose expression within the brain most strongly correlated with grooming phenotypes. We then used the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (Szklarczyk et al., 2011) and Cytoscape tools to create interactome networks for both the selected genes and their human orthologs. Based on the overlap between these networks, we identified 31 candidate genes with translational potential, and determined their associated biological roles within the brain using GoMiner and Mouse Genome Informatics tools (Shaw, 2009). Following identification of candidate genes, we completed in-silico validation of this approach by comparing patterns of expression in the candidate genes to neutral ‘control’ genes (chosen by selecting probes at random). Compared to the control genes, the candidate genes were more strongly correlated with grooming behavior, and showed altered expression levels in key brain regions involved in grooming using the Allen Brain Atlas (ABA) (Lein et al., 2007, Ng et al., 2009). These candidate genes also produced more nodes per gene within interactome networks, thus demonstrating the usefulness of this in-silico phenotype-genomic methodology.
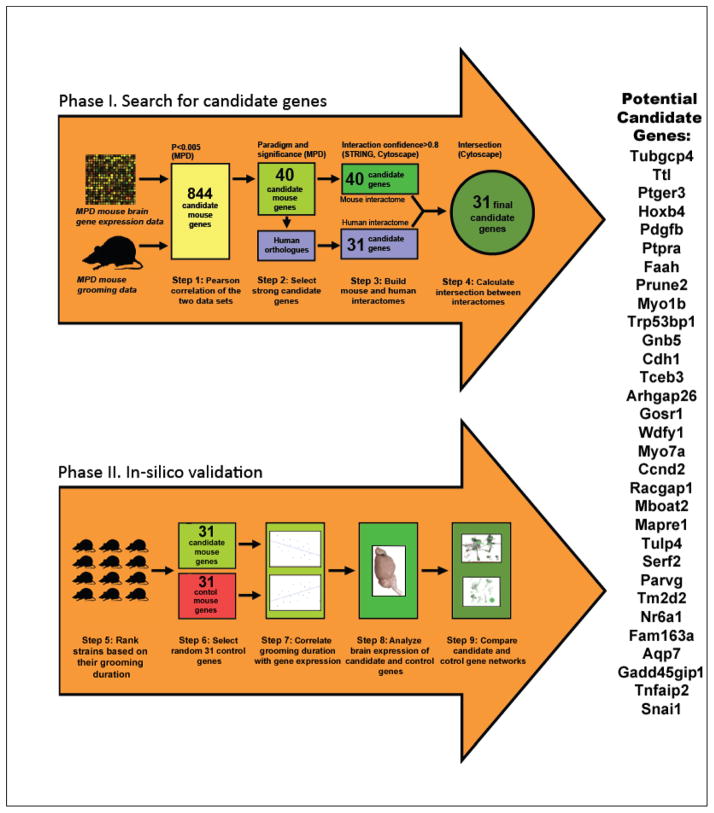
Figure 1. Flowchart summarizing the methodology of selecting and analyzing candidate grooming genes.
During Phase I, the Mouse Phenome Database (MPD) brain expression microarray data and behavioral data were selected from several murine anxiety paradigms and a wide spectrum of mouse strains. Step 1 used the MPD toolbox to generate Pearson correlation coefficients between these two data sets yielding 844 mouse genes that correlated with self-grooming (P<0.005). In step 2, this list of genes was prioritized by 1) how many sets of grooming data from different behavioral paradigms a gene was correlated with, and 2) the strength of their Pearson correlations, yielding a total of 40 candidate genes, 31 of which shared human orthologs (step 3) and were selected for interactome analyses (step 4). During Phase II, we ranked the strains based on their grooming duration data (step 5). We then selected 31 random control genes (step 6) and correlated the grooming data with gene expression of the 31 candidate and randomly selected control genes (step 7). See text for details on the selection of control genes for this study. Because the expression data from this study were from the whole brain, the Allen Brain Atlas (ABA) provided further data for the regional expression patterns of both control and candidate genes (step 8). The STRING database of protein-protein interactions was used to create an interaction network (interaction confidence ≥0.8) of the protein products of these genes in mice, visualized using Cytoscape. A similar network was generated using orthologous human proteins and Cytoscape, to calculate the intersection between the mouse and human interactomes, resulting in a shared interactome containing only the nodes and edges conserved for both mice and humans (step 9).
Figure 1 outlines the overall methodological approach used in this study, summarizing its phases and steps. The rationale of Phase I is using correlational analyses from two MPD projects for a step-by-step dissection and identification of potential candidate genes and creating an integrated ‘translational’ molecular network for these genes. Phase II of this projects aims to provide an in-silico validation for Phase I, assessing known functions of the selected candidate genes, their expression in relation to regional distribution in the brain, and correlation with grooming behavior. Collectively, this approach allowed us to identify an interconnected network of candidate genes (which have expression levels that correlate with grooming behavior, display altered patterns of expression in key brain areas related to grooming and underlie important functions in the brain) which are therefore likely to represent potential neural targets for mouse grooming behavior.
2.2. Phase I. The search for candidate genes
General approach and generation of candidate genes (Step 1)
The Brown laboratory’s 2004 study (Brown1 project in MPD) contains grooming duration data for male and female mice of the 10 weeks-old 129S1/SvImJ, A/J, AKR/J, BALB/cByJ, BALB/cJ, C3H/HeJ, C57BL/6J, CAST/EiJ, DBA/2J, FVB/NJ, MOLF/EiJ and SJL/J strains (Brown, Gunn, 2004). Only male mouse data were used in the present experiment, to eliminate potential confounds associated with using mixed-sex cohorts (Table 1). There were marked strain differences in grooming duration in the four behavioral tests, with C57BL/6J mice showing the longest, and MOLF/EiJ, BALB/cJ, BALBc/ByJ and FVB/NJ strains showing lower grooming. Since strain differences in grooming behavior were not the main focus of this analysis, this aspect will not be discussed here. Note, however, that the MPD enables a fast comparison of all grooming scores in the Brown et al. 2004 study (Brown, Gunn, 2004), and is publicly available for further evaluation. The reliability of these grooming data was first assessed using the MPD toolbox, to determine whether the strain means for duration of grooming in the open field, elevated plus-maze, elevated zero-maze and light-dark box tests of anxiety were significantly correlated. Briefly, from grooming phenotypical data (Brown1 MPD project) for each of the four tests we chose “Other tools/toolbox”, and used “Correlations and relationships between phenotypes” option to “Search all MPD for correlated phenotypes”, selecting the Brown1 project from the drop-down menu. Tables 1 and 2 show the correlations for male mouse grooming duration in 12 inbred strains in the four different behavioral tests. Completed globally for all strains and separately within each strain and using strain means, these results generally show positive correlations between grooming duration data in different novelty-based anxiety tests.
Table 1.
Mouse Phenome Database-derived correlational analyses (Pearson R) of Brown et al. (2004) grooming duration data in male mice of multiple inbred strains tested in the open field test (OFT), light-dark box (LDB), elevated plus-maze (EPM) and elevated zero-maze (EZM) tests.
| Tests | Strains | LDB | EPM | EZM |
|---|---|---|---|---|
|
| ||||
| OFT | 129S1/SvImJ | 0.51# N = 12 | − 0.08 N = 12 | 0.34 N = 10 |
| A/J | 0.54* N = 20 | −0.11 N = 19 | −0.08 N = 6 | |
| AKR/J | 0.59** N = 19 | 0.43# N = 19 | @ | |
| BALB/cByJ | 0.24 N = 12 | −0.06 N = 12 | −0.03 N = 12 | |
| BALB/cJ | −0.22 N = 12 | −0.23 N = 14 | −0.18 N = 10 | |
| C3H/HeJ | 0.35 N = 16 | −0.13 N = 16 | $ | |
| C57BL/6J | 0.49# N = 13 | −0.03 N = 13 | 0.49 N = 10 | |
| CAST/EiJ | −0.04 N = 10 | 0.10 N = 10 | @ | |
| DBA/2J | 0.11 N = 13 | 0.07 N = 13 | 0.88*** N = 10 | |
| FVB/NJ | −0.18 N = 12 | 0.14 N = 12 | 0.22 N = 12 | |
| MOLF/EiJ | −0.10 N = 11 | 0.94*** N = 11 | @ | |
| SJL/J | −0.38 N = 12 | 0.35 N = 12 | @ | |
|
| ||||
| LDB | 129S1/SvImJ | −0.18 N = 12 | 0.74* N = 10 | |
| A/J | - | −0.28 N = 20 | −0.21 N = 6 | |
| AKR/J | 0.45 # N = 19 | @ | ||
| BALB/cByJ | −0.09 N = 12 | 0.31 N = 12 | ||
| BALB/cJ | 0.70* N = 12 | 0.20 N = 10 | ||
| C3H/HeJ | −0.21 N = 18 | $ | ||
| C57BL/6J | 0.38 N = 13 | 0.018 N = 10 | ||
| CAST/EiJ | 0.25 N = 12 | @ | ||
| DBA/2J | 0.22 N = 13 | −0.22 N = 10 | ||
| FVB/NJ | −0.23 N = 12 | −0.23 N = 12 | ||
| MOLF/EiJ | −0.18 N = 11 | @ | ||
| SJL/J | −0.54# N = 12 | @ | ||
|
| ||||
| EPM | 129S1/SvImJ | −0.08 N = 10 | ||
| A/J | - | −0.25 N = 6 | ||
| AKR/J | @ | |||
| BALB/cByJ | −0.23 N = 12 | |||
| BALB/cJ | 0.05 N = 10 | |||
| C3H/HeJ | $ | |||
| C57BL/6J | −0.34 N = 10 | |||
| CAST/EiJ | @ | |||
| DBA/2J | 0.06 N = 10 | |||
| FVB/NJ | 0.58* N = 12 | |||
| MOLF/EiJ | @ | |||
| SJL/J | @ | |||
Correlations were not performed for some strains due to an insufficient sample size (n<3; $) or very low grooming activity (resulting in a lack of data variability; @).
P<0.05;
P<0.01;
P<0.001;
P<0.0001,
P = 0.05–0.1 (trend);
NS – not significant (p>0.05).
Table 2.
Correlation (Pearson R) of mean grooming duration across 12 strains of mice, listed in Table 1, between the elevated plus-maze (EPM) and zero-maze (EZM), light-dark box (LDB) and open field (OFT) tests.
| EPM | LDB | EZM | |
|---|---|---|---|
| OFT | 0.40 | 0.37 | −0.01 |
| EPM | 0.61* | 0.28 | |
| LDB | 0.40 |
Correlations with the EZM used 11 strains as C3H/HeJ were not tested on the EZM in the Brown et al. project;
P<0.05.
Gene expression data from microarray studies from the Tabakoff laboratory, also available on the MPD (Tabakoff1 project), used an Affymetrix GeneChip Mouse Genome 430 2.0 containing 39,985 probesets to analyze whole-brain mRNA expression in multiple inbred strains of male mice (Tabakoff, Bhave, 2007) of the same age (10 weeks) as in Brown et al. (2004) study. For each strain, 4–6 replications were performed in order to minimize random variation between subjects, and the Robust Microchip Average expression measure was used to normalize the values for each gene for a given mouse strain (see (Irizarry et al., 2003) for details). To parallel grooming behavior with gene expression, the strain means for grooming data (Brown, Gunn, 2004) on each behavioral test were correlated with the mean whole-brain mRNA expression data (Tabakoff, Bhave, 2007) for male mice of the same strains using the MPD correlational toolbox. Briefly, from grooming phenotypical data (Brown 1 project) we chose “Other tools/toolbox”, and used its “Correlations and relationships between phenotypes and genotype or gene expression” option to “Find correlated gene expression probesets” in the “brain_Tabakoff1” project (selected from the drop-down menu). Note, however, that the MPD user interface undergoes regular modifications, and its future online versions and menu options may differ from those used in this 2012 study. A stringent level of significance (P<0.005) was used for this procedure, yielding significant correlations between 1028 mRNA probesets and grooming duration, of which 881 were located in known gene areas, in total accounting for 844 different mouse genes.
Analysis of candidate genes (Steps 2–5)
After identifying genes whose expression strongly correlated with grooming duration (P<0.005), we ranked these genes based on the number of behavioral paradigms in which they significantly correlated with grooming. All genes significantly correlating with grooming in more than one behavioral test (e.g., Tubgcp4, Ttl, Ptger3, Hoxb4, Pdgfb, Ptpra, Faah) were first included in our analysis as independently reconfirmed in several different behavioral models. Next, we ranked the remaining genes as potential candidate genes based on the absolute size of the Pearson correlation coefficient between grooming duration and mRNA expression (R nearest 1 or −1 in one of the four tests). In order to obtain a manageable number of genes for network analysis, we limited our search to the first 40 genes, allowing us to ensure adequate statistical power and avoid false positives. Due to the translational nature of our study, we further focused on the 31 candidate genes which were present in both mice and humans (Fig. 1).
GoMiner software (Zeeberg et al., 2003) was used to analyze the function of the candidate genes and determine any known role of these genes in brain function and neurobehavioral disorders. To complement these analyses, the Mouse Genome Informatics (MGI) database (Shaw, 2009) provided aberrant phenotypes of genetically modified mice (relevant for each of the candidate genes), allowing additional insight into gene-behavior interactions for the group of candidate genes identified in our study (Table 3). Protein-specific BLAST searches enabled further comparison of the homology between candidate mouse gene products and their human orthologs (Table 3), and the Ontological Discovery Environment (ODE) (Baker et al., 2009) and Drug Related Gene Database (DRG) (Gardner et al., 2008) were used to further search published gene expression studies linking candidate genes to mouse neural phenotypes. As shown in Table 3, bioinformatics-based analysis of the 31 genes present in both mice and humans revealed several interesting patterns, including 4 genes that encode tubulin-associated proteins (Tubgcp4, Ttl, Racgap and Mapre1), and 5 genes related to either actin or myosin (Pdgfb, Myo1b, Cdh1, Myo7a and Parvg). Finally, the Protein database (Pruitt et al., 2007) and sequence analysis using Hum-mPLoc (Shen and Chou, 2007) were also used in this study to characterize cellular location of protein products of the selected candidate gene (Fig. 3E).
Table 3.
A list of potential candidate genes selected for further analysis based on high correlation of grooming duration in the open field test (OFT), light-dark box (LDB), elevated plus-maze (EPM) and elevated zero-maze (EZM) tests, with brain expression microarray data.
| Tests and Pearson correlation, P | Gene name | Selected gene ontology | Selected mammalian phenotypes from genetically modified mice(Shaw, 2009) | Human orthologs (and % homology) | Selected phenotypes from the Drug Related Gene Database and the Ontological Discovery Environment |
|---|---|---|---|---|---|
| Genes selected based on high correlation with grooming activity observed independently in two different tests | |||||
| EZM R=0.89 P<0.0005 OFT R=−0.83 P<0.001 |
Tubgcp4 (tubulin, γ-complex associated protein 4) | Cytoplasm, microtubule, cytoskeleton organization | NA | TUBGCP 4 (99 %) | Ethanol preference in mice (Rodriguez et al., 1994); expression in cerebellum linked with activity in open field test (Philip et al., 2010) and varies across early development in mice (Kagami and Furuichi, 2001) (may have potential implications for motor phenotypes, reward pathways and neurodevelopmental disorders) |
| EZM R =0.87 P<0.0005 OFT R=−0.75 P<0.005 |
Ttl (Tubulin tyrosine ligase) | ATP binding, ligase activity, microtubule cytoskeleton organization, regulation of axon extension | Abnormal telencephalon development, abnormal neuron differentiation | TTL (96%) | Increased expression in nucleus accumbens following chronic cocaine in rats (Renthal et al., 2009); expression in striatum correlates with distance traveled in rats (Philip, Duvvuru, 2010); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications for motor phenotypes, reward pathways and neurodevelopmental disorders) |
| EZM R=−0.83 P<0.005 OFT R=−0.77 P<0.005 |
Ptger3 (prostaglandin E receptor 3 subtype EP3) | Activation of phospholipase C via G-protein-coupled receptor signaling | Abnormal body temperature regulation and pain threshold | PTGER3 (86%) | Expression in cerebellum linked with activity in the open field test after cocaine in mice (Philip, Duvvuru, 2010); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications for motor phenotypes, reward pathways and neurodevelopmental disorders) |
| EPM R=−0.82 P<0.005 EZM R=−0.75 P<0.005 |
Hoxb4 (homeobox B4) | Stem cell division, transcription and regulation, DNA-dependent, sequence specific DNA binding | Decreased body size and lower survivor rate | HOXB4 (90%) | Associated with ethanol consumption in mice (Mulligan et al., 2006); cerebellum expression associated with increased vocalization threshold (Philip, Duvvuru, 2010) and expression across early development in mice (Kagami and Furuichi, 2001) (may have potential implications for motor phenotypes, reward pathways and neurodevelopmental disorders) |
| EZM R=0.82 P<0.005 OFT R=0.75 P<0.005 |
Pdgfb (platelet derived growth factor, B polypeptide) | Cytoskeleton organization, cell projection assembly, activation of fibroblast growth factor receptor signaling pathway | Capillary aneurisms, edema endothelial hyperplasia | PDGFB (90%) | Associated with ethanol consumption in mice (Mulligan, Ponomarev, 2006); expression in neocortex associated with hyperactivity after ethanol consumption in mice (Philip, Duvvuru, 2010); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications for motor phenotypes, reward pathways and neurodevelopmental disorders) |
| LDB R=0.76 P<0.005 OFT R=0.76 P<0.005 |
Ptpra (protein tyrosine phosphatase receptor type A) | Positive regulation of oligodendrocyte differentiation, integral to membrane, phosphatase activity | Decreased anxiety-related response, hypoactivity, abnormal spatial learning | PTPRA (96%) | Associated with ethanol consumption in mice (Mulligan, Ponomarev, 2006); decreased expression in striatum following acute morphine and increased expression following chronic morphine in mice (Korostynski et al., 2007); decreased expression in dorsolateral prefrontal cortex following chronic crack cocaine in humans (Lehrmann et al., 2003); expression in hippocampus correlates with base-line activity during fear conditioning in mice (Philip, Duvvuru, 2010); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications for motor phenotypes, reward pathways and neurodevelopmental disorders) |
| LDB R=−0.76 P<0.005 OFT R=0.75 P<0.005 |
Faah (fatty acid amide hydrolase) | Degradation of bioactive fatty acid amides | Hypoactivity, increased alcohol consumption, analgesia | FAAH (84%) | Decreased expression in nucleus accumbens following chronic cocaine in mice (Renthal, Kumar, 2009) (may have potential implications for reward pathways) |
| Genes selected based on significant very high correlation with grooming activity observed in a specific single behavioral model | |||||
| EZM R=0.96 P<0.000005 |
Prune2 (prune homologue 2-Drosophila) | Induction of apoptosis, phosphatase activity | NA | PRUNE2 (68%) | NA |
| EZM R=0.96 P<0.000005 |
Myo1b (myosin 1B) | Actin binding, motor activity, myosin complex, ATP binding | NA | MYO1B (96%) | Decreased expression in nucleus accumbens following chronic cocaine in mice (Renthal, Kumar, 2009); brain expression correlated with activity in open field test in mice (Philip, Duvvuru, 2010); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001)(may have potential implications for motor phenotypes, reward pathways and neurodevelopmental disorders) |
| EZM R=0.95 P<0.000005 |
Trp53bp1 (transformation related protein 53 binding protein 1 | Response to DNA damage stimulus, regulation of transcription | Decreased body weight, postnatal growth retardation | TP53BP1 (81%) | NA |
| EZM R=0.95 P<0.00001 |
Gnb5 (guanine nucleotide binding protein beta 5) | G-protein coupled receptor protein signaling pathway, signal transducer activity | Seizures, small body size, ataxia, impaired motor coordination | GNB5 (99%) | Associated with activation of mesolimbic dopamine reward pathway after acute ethanol in mice (Kerns et al., 2005); increased expression in nucleus accumbens following chronic cocaine in mice (Renthal, Kumar, 2009); decreased expression in striatum following acute cocaine in rats (Paletzki et al., 2008); expression correlates with novel environment exploration in mice (Philip, Duvvuru, 2010)(may have potential implications for motor phenotypes and reward pathways) |
| OFT R=−0.95 P<0.000005 |
Cdh1 (cadherin 1) | Calcium ion binding, cell-cell junction, regulation of caspase, actin cytoskeleton | Decreased hair follicle number, abnormal skin | CDH1 (82%) | Decreased expression following chronic cocaine (Lehrmann, Oyler, 2003) and increased expression following chronic crack cocaine in dorsolateral prefrontal cortex in humans (Lehrmann, Oyler, 2003); expression modulated by nicotine in several brain regions in mice (Wang et al., 2008); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications for reward pathways and neurodevelopmental disorders) |
| EZM R=0.94 P<0.00005 |
Tceb3 (transcription elongation factor B (SIII), polypeptide 3) | Regulation of transcription from RNA polymerase II promoter | Delayed brain development | TCEB3 (81%) | Associated with ethanol consumption in mice (Mulligan, Ponomarev, 2006); differentially expressed in nucleus accumbens 24 h following ethanol consumption in mice (Mulligan, Ponomarev, 2006); expression modulated by nicotine in several brain regions in mice (Wang, Gutala, 2008); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications in reward pathways and neurodevelopmental disorders) |
| EZM R=0.94 P<0.00005 |
Arhgap26 (rho GTPase activating protein 26 | NA | NA | ARHGAP26 (97%) | Associated with nicotine dependence in humans (Drgon et al., 2009); expression in neocortex correlates with distance traveled in mice (Philip, Duvvuru, 2010) (may have potential implications in motor phenotypes and reward pathways) |
| EZM R=0.94 P<0.00005 |
Gosr1 (golgi SNAP receptor complex member 1) | Golgi membrane, protein transport, SNAP receptor activity | NA | GOSR1 (98%) | Associated with ethanol consumption in mice (Mulligan, Ponomarev, 2006); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications in reward pathways and neurodevelopmental disorders) |
| EZM R=−0.89 P<0.0005 |
Wdfy1 (WD repeat and FYVE domain containing 1) | Metal ion binding, early endosome | NA | WDFY1 (97%) | Increased expression in nucleus accumbens following chronic cocaine in rats (Renthal, Kumar, 2009); ethanol preference in mice (Rodriguez, Plomin, 1994); variable expression in nucleus accumbens following ethanol consumption in rats (Bell et al., 2009) (may have potential implications in reward pathways) |
| EZM R=0.93 P<0.00005 |
Myo7a (myosin VIIA) | Melanosome, synapse, binding, cytoskeleton, nucleotide binding, auditory receptor cell differentiation | Abnormal hair cell morphology, small body size, circling, hyperactivity, altered anxiety | MYO7A (96%) | Decreased expression in the nucleus accumbens with chronic cocaine in rats (Renthal, Kumar, 2009); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications in reward pathways and neurodevelopmental disorders) |
| EZM R=0.93 P<0.00005 |
Ccnd2 (cyclin D2) | Cell cycle and division, G1 S transition of mitotic cell cycle, up-regulation of cell proliferation | Abnormal cerebellar morphology, absent Purkinje cells, motor incoordination, increased ethanol intake | CCND2 (90%) | Associated with ethanol consumption in mice (Mulligan, Ponomarev, 2006); ethanol preference in mice (Rodriguez, Plomin, 1994); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications in reward pathways and neurodevelopmental disorders) |
| OFT R=−0.93 P<0.00005 |
Racgap1 (Rac GTPase-activating protein 1) | α,β,γ-tubulin binding, cytokinesis, actomyosin contractile ring assembly | Premature death | RACGAP1 (84%) | Hippocampus expression correlates with Dowel test time in mice (Philip, Duvvuru, 2010); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications in motor phenotypes and neurodevelopmental disorders) |
| EZM R=0.93 P<0.00005 |
Mboat2 (membrane O-acetyltransferase domain containing 2) | Endoplasmic reticulum, phospholipid biosynthesis | NA | MBOAT2 (87%) | Associated with ethanol consumption in mice (Mulligan, Ponomarev, 2006) (may have potential implications in reward pathways) |
| OFT R=−0.92 P<0.00005 |
Mapre1 (microtubule-associated protein, RP/EB family member 1) | Cell projection, microtubule plus end binding, mitosis, centrosome | NA | MAPRE1 (97%) | Associated with ethanol consumption in mice (Mulligan, Ponomarev, 2006); expression altered in hippocampus after experimental brain injury (Matzilevich et al., 2002) and correlates with hippocampal neurogenesis in mice (Philip, Duvvuru, 2010); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications in reward pathways and neurodevelopmental disorders) |
| EZM R=0.92 P<0.00005 |
Tulp4 (tubby like protein 4) | Intracellular signal transduction | NA | TULP4 (95%) | Increased expression in nucleus accumbens after chronic cocaine in mice (Renthal, Kumar, 2009); altered brain expression after ethanol treatment in rats (Kerns, Ravindranathan, 2005); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications in reward pathways and neurodevelopmental disorders) |
| OFT R=−0.91 P<0.00005 |
Serf2 (small EDRK-rich factor 2) | NA | NA | SERF2 (100%) | Expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications in reward pathways) |
| EZM R=−0.91 P<0.0005 |
Parvg (parvin, gamma) | Actin binding, cell adhesion, cell junction, cytoskeleton | Abnormal retinal and astrocyte morphology, gliosis, aberrant blood brain barrier | PARVG (80%) | Increased expression in nucleus accumbens during chronic cocaine in mice (Renthal, Kumar, 2009) (may have potential implications in reward pathways) |
| EZM R=0.90 P<0.0005 |
Tm2d2 (TM2 domain containing 2) | Integral to membrane | NA | TM2D2 (90%) | Cerebellar expression correlates with activity in the open field in mice (Philip, Duvvuru, 2010); associated with consumption of ethanol in mice (Mulligan, Ponomarev, 2006); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications in motor phenotypes, reward pathways and neurodevelopmental disorders) |
| EZM R=−0.89 P<0.0005 |
Nr6a1 (nuclear receptor subfamily 6, group A member 1) | Down-regulated transcription from RNA polymerase II promoter, protein homodimerization, sequence-specific DNA binding | Abnormal brain development and cranial nerve morphology, lower testosterone | NR6A1 (96%) | Cerebellar expression correlates with activity in the light-dark box and time spent in zero maze quadrants in mice (Philip, Duvvuru, 2010) (may have potential implications in motor phenotypes) |
| EZM R=−0.90 P<0.0005 |
Fam163a (family with sequence similarity 163, member A) | Integral to membrane | NA | FAM163A (85%) | NA |
| OFT R=−0.89 P<0.0001 |
Aqp7 (aquaporin 7) | Porin activity, glycerol transport, water transport | Abnormal kidney physiology, hypoglycemia | AQP7 (77%) | Associated with ethanol consumption in mice (Mulligan, Ponomarev, 2006); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications in reward pathways and neurodevelopmental disorders) |
| EZM R=−0.89 P<0.0005 |
Gadd45gip1 (growth arrest DNA-damage-inducible, gamma interacting protein 1) | Negative regulator of cell cycle progression | Embryonic growth retardation, lethality | GADD45GIP1 (78%) | Increased expression in nucleus accumbens during chronic cocaine in mice (Renthal, Kumar, 2009), differential expression in striatum in several inbred mouse strains (Korostynski et al., 2006) (may have potential implications for motor phenotype and reward pathways) |
| EZM R=0.89 P<0.0005 |
Tnfaip2 (tumor necrosis factor, alpha-induced protein 2) | Dentritic cell marker, modulation of inflammation and angiogenesis | Aberrant angiogenesis | TNFAIP2 (71%) | NA |
| OFT R=−0.89 P<0.0005 |
Snai1 (snail homolog 1 Drosophila) | Hair follicle morphogenesis, down-regulation of cell differentiation | Open neural tube, abnormal cell migration | SNAI1 (88%) | Increased expression in D1 mutant mice in the caudoputamen (Zhang et al., 2005); expression varies in cerebellum across early development in mice (Kagami and Furuichi, 2001) (may have potential implications in reward pathways) |
Over 800 genes had significant correlations with grooming duration using P<0.005. Potential candidate genes were then ranked based on the number of behavioral tests in which gene expression correlated significantly with grooming behavior. The remaining genes were then ranked based on the strength of their Pearson correlation between grooming duration and expression in one test. Gene ontology information was gathered from GoMiner, mouse phenotypes were selected from Mouse Genome Informatics (MGI) database (Shaw, 2009), and protein-specific BLAST provided the degree of homology between candidate mouse proteins and their human orthologs. The final column includes relevant neurological information from the Drug Related Gene Database (DRG) (Gardner, Akil, 2008) and the Ontological Discovery Environment (ODE) (Baker, Jay, 2009). Genes which highly correlated with grooming activity in the EPM, OFT and or EZM, but do not have human orthologs, include A230056J06Rik, A930015D, Cml3, A630012P03Rik, 4930402C16Rik, Gm11738, 1110005A03Rik, AI853106 and 1700023L04Rik (|R|=0.76–0.95, P<0.0005–0.00001), and were not assessed here. NA – information is not available.
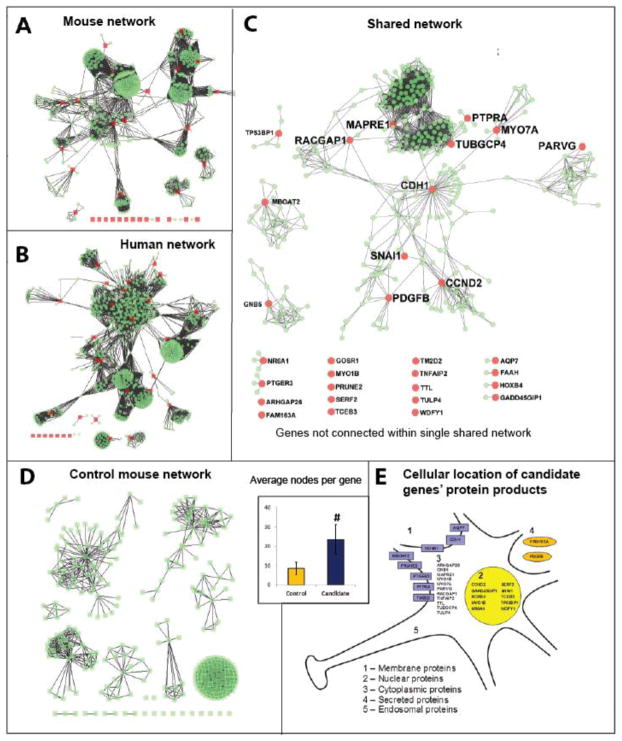
Figure 3. Network-based analysis of mouse candidate genes, their human orthologs and shared networks (see Fig. 1 for general rationale).
A: Generating mouse grooming interactome using Cytoscape with data from grooming duration (Brown, Gunn, 2004) and whole brain expression microarray (Tabakoff, Bhave, 2007). Mouse grooming duration data from 12 strains in four behavioral tests were correlated using the MPD Pearson R toolkit with whole brain expression microarray data, generating 844 genes (step 1 in Fig. 1). The 40 most promising genes were selected for further analysis (step 2). Using the 31 genes present both in mice and humans, the STRING database generated a list of all proteins known to interact (Interaction Confidence ≥0.8). Cytoscape software then allowed these proteins to be displayed graphically in a user-friendly network (step 3). The 31 candidate gene products are displayed in red, while interacting proteins are displayed in green and the edges are in black. Twenty-two genes had interaction data available at an Interaction Confidence ≥0.8 (while 9 did not), and 15 of the candidate genes maintained connectivity within a single network.
B: Generating human interactome based on human orthologs of mouse candidate genes identified in panel A. Using the 31 genes present in both mice and humans, the STRING database generated a list of all proteins known to interact (Interaction Confidence ≥0.8). Cytoscape software then allowed these proteins to be displayed graphically in a user-friendly network (Step 3). Interaction data were unavailable for 7 genes at an Interaction Confidence ≥ 0.8; the remaining 20 human orthologs remained connected within a single network (step 3).
C: Generating shared network of candidate genes (Step 4). Using mouse (A) and human (B) interactomes generated previously in Step 3, Cytoscape calculated the intersection between them to identify the candidate genes and interactors that were conserved between the two species (Step 4). This network displays only the nodes and edges that are present in both networks. Of the 31 genes conserved in both species, 10 remained connected within a single network. The genes in this network are highly correlated with grooming behavior, interact within a small network, and are prominent in both mice and humans. Further analysis for these genes can be found in Table 3 (also see steps 5–8 in Fig. 1).
D: Generating the control gene network. In order to assess the candidate gene network, the control genes network was generated using the same approach (step 9), showing little connectivity between the genes, as can be assessed visually by comparing ‘candidate’ mouse interactome (A) with ‘control’ interactome (this panel). Further analyses of the average number of interactors per network showed that the mouse interactome of candidate genes tended to have more nodes per gene, compared to the mouse interactome of candidate genes (#P<0.08, trend, U-test), suggesting a generally higher functional interconnectedness compared to the randomly selected control genes.
E: Diagram showing cellular location of protein products of 31 candidate genes (A). Cellular location of these proteins was established based on protein sequence from Protein database (Pruitt, Tatusova, 2007) and sequence analysis using Hum-mPLoc (Shen and Chou, 2007).
2.3. Phase II. In-silico validation
Selection of control genes (Step 6)
To examine the validity of the procedure used to select candidate genes in Phase I, we used a random approach to select a group of control genes (see similar methods of selecting control genes to link gene activity to behavioral phenotype used in published literature (Mignogna and Viggiano, 2010)). For the present study, using the list of probes contained on the Affymetrix GeneChip Mouse Genome 430 2.0 microarray, we selected every 500th probe (e.g., 500, 1000, 1500), which resulted in 31 control probes that targeted a gene-coding area, which were present in both mice and humans, and were not part of the 844 ‘putative’ candidate genes correlated with grooming duration in the previous step (Fig. 1). The control genes selected for this study included Cdkn2d, Trpm7, Sult2b1, Tnfaip1, N6amt2, Bmp7, Tbc1d1, Tspan8, Chrna4, Rps6kb2, Lipe, Csnk1g2, Rhbdd1, Slc27a4, Lpxn, Map2k7, Srek1, Fmn1, Txndc1, Nfam1, Syt11, Alkbh4, Ppp1r14c, Wwox, Sf3a3, Ppm1l, Cotl1, Gpr183, Erbb2ip, Lpp and Zfp879 (based on ABA data, all these genes are expressed in the mouse brain, and therefore were appropriate to use as control for this study).
Correlation of strain rankings of grooming duration and gene expression (Steps 5 and 7)
In order to validate the selection criteria used to generate candidate genes, the grooming duration measurements for each strain were compared with the gene expression of each strain. The Brown et al. data provided grooming duration for 12 strains which we ranked from 1 to 12, based on their results in 4 separate behavioral paradigms (the C3H/HeJ mice were not tested in the elevated zero maze in the Brown et al. study, and their strain rank for grooming was calculated based on 3 other behavioral tests). The four ranks for each strain/test were then averaged across all tests, enabling us to organize the 12 mouse strains according to their overall grooming duration rank ranging from 1 to 12 (Fig. 2A).
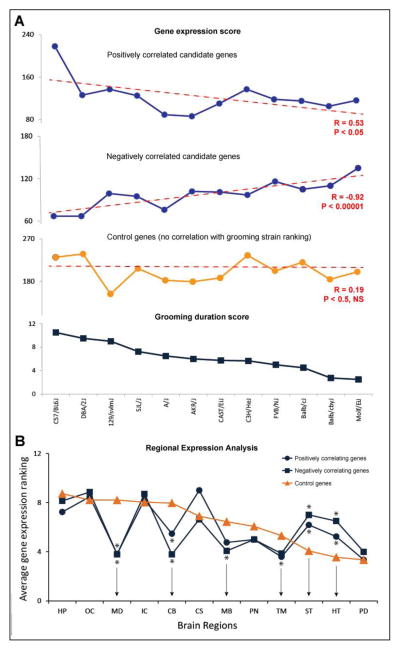
Figure 2. Analysis of strain ranking and regional gene expression.
A: Comparison of strain grooming duration with strain gene expression. Raw behavioral data were used to rank the 12 mouse strains based on their grooming duration in 4 behavioral tests. C57BL/6J mice groomed the most while the MOLF/EiJ strain groomed the least. Genes were divided into three groups: positively correlating with grooming (17 genes; top panel), negatively correlating with grooming (14 genes; middle panel), and randomly selected control genes (31 genes; bottom panel). Microarray data allowed the expression of the genes in these three groups to be summed for each strain, to be used for correlation analyses (using Spearman correlation) with the strain ranking of grooming duration.
B: Regional expression analysis of data obtained using the Allen Brain Atlas (ABA) for C57BL/6J mouse strain. The following brain areas (selected based on ABA pre-defined brain sectioning) were included in this analysis: HP = hippocampal formation; OC = olfactory cortex; MD = medulla; IC = isocortex; CB = cerebellum; CS = cortical subplate; MB = midbrain; PN = pons; TM = thalamus; ST = striatum; HT = hypothalamus; PD = pallidus. To investigate the expression patterns of candidate genes in the C57BL/6J mouse strain (showing robust grooming responses in Brown et al. 2004 behavioral study), we used the ABA data to establish the raw expression scores for 31 candidate and 31 control genes across 12 regions of the mouse brain (see Methods for details of selecting control genes). Based on their Pearson correlations with grooming duration, the candidate genes were again divided into two groups, positively and negatively correlating with grooming, in order to investigate whether different brain regions differentially affect expression data. The expression values for each gene were numbered 1–12 (with 12 indicating the region of highest expression for a given gene and 1 the lowest expression) and averaged for each brain region. Control genes expression ranking score differed significantly from both the positively correlated and negatively correlated genes in 6 brain regions (P<0.05, U-test vs. the respective ranking scores of control genes) – medulla, cerebellum, midbrain, thalamus, striatum and hypothalamus, all strongly implicated in the regulation of mouse grooming behavior.
The Tabakoff laboratory’s microarray data provided expression values for our candidate and control genes in each of the 12 mouse strains (genes with multiple probes targeting the same gene were averaged to obtain a single value per gene). The expression values for each gene were ranked from 1 (lowest) to 12 (highest), to match with the number of mouse strains used in this study. At this point, based on their initial ‘strain’ Pearson correlation coefficients, as explained in Phase I, the candidate genes were divided into positively correlating with mouse grooming (17 genes) or negatively correlating with mouse grooming (14 genes), in addition to 31 control genes. For each mouse strain, we then calculated the total rank of expression of genes from each group separately, i.e., positively correlating (range: 17–204), negatively correlating (range: 14–168) and control genes (range = 31–372). Once the strain ranking for grooming and gene expression were calculated, we applied Spearman correlation coefficient to further analyze these data. As the strains were ranked from highest to lowest grooming duration, we graphed the gene expression data for each strain (Fig. 2A). The genes positively correlating with grooming trended downward (i.e., the lower the grooming duration of a given strain, the weaker the gene expression; Spearman R = 0.53; P<0.05). The genes which negatively correlated with grooming showed the opposite pattern (Fig. 2A; Spearman R = −0.92; P<0.00001). In contrast, as shown in Fig. 2A, the control genes showed no significant correlation between strain gene expression and strain grooming duration (Spearman R = 0.19; P<0.5, NS).
Notably, the C57BL/6J mice had the highest grooming duration in the behavioral tests, consistent with earlier observation of robust grooming behavior in this common inbred mouse strain (Kalueff and Tuohimaa, 2004). In this strain, the genes which positively correlated with grooming were highly expressed in the brain while the genes with negative correlation were expressed at low levels, thereby supporting the genes’ selection criteria described above, and the suitability of this strain for further analyses and validation.
Regional expression analysis using the Allen Brain Atlas (Step 8)
Since the microarray data used here (Tabakoff, Bhave, 2007) provided only whole-brain expression data, the regional expression of candidate and control genes was assessed using the ABA expression data for the C57BL/6J strain (Lein, Hawrylycz, 2007). Notably, of the 12 strains investigated in this analysis, the C57BL/6J mice displayed the longest grooming duration, supporting the use of this strain in dissecting the expression patterns of our candidate genes. The ABA contains RNA expression values from 12 different regions and multiple genes across the entire genome (Lein, Hawrylycz, 2007). If the ABA gene expression data contained multiple experiments for the same gene, data were averaged across experiments to obtain a single value per gene for each brain region. Expression data were unavailable for 3 candidate genes (Gosr1, Tm2d2, Racgap1) and 2 control genes (Srek1, Tnfaip1). Because some genes are expressed at high levels across the brain while other genes have uniformly lower expression, we converted each raw expression score into a rank from 1 to 12, giving each gene equal weighting, regardless of their raw expression levels. This strategy was first applied to a cohort of randomly selected control genes, reflecting the expression patterns of the entire genome. Next, the candidate genes were divided into two groups (as described previously), including genes positively or negatively correlating with grooming duration. The brain structures where candidate and control genes were expressed differently provided us with potential regions of importance for mouse grooming. Analyzing the average expression for each brain area, the highest deviation in expression between control and candidate genes occurred in the medulla, but not in the areas usually not implicated in the grooming phenotypes of mice, such as the olfactory cortex and pons (Fig. 2B). Overall, the candidate genes that positively correlated with grooming differed significantly (by U-test) from the control cohort in 6 regions (medulla, cerebellum, midbrain, thalamus, striatum and hypothalamus), while the negatively correlated candidate genes differed from the control genes in these same 6 regions (Fig. 2B). Interestingly, the positively and negatively correlated candidate genes showed similar trends in expression across different brain regions, implicating these genes in grooming in a striking contrast to randomly selected control genes (Fig. 2B).
Network analysis using STRING database (Step 9)
Examining the functionality of the genes in our study, we used the STRING database (Szklarczyk, Franceschini, 2011) containing known and predicted protein-protein interactions to analyze the protein products of the 31 candidate genes present in both humans and mice. Several other studies have already utilized protein-protein interaction networks to make predictions about the role of a gene and its potential phenotypes (Lage et al., 2007, Wang and Marcotte, 2010). The STRING database calculated all direct interactions between these 31 candidate protein products and the rest of the proteome, generating a network of protein-protein interactions. The confidence that a given protein-protein interaction represents a functional relationship is reported by the STRING database as an Interaction Confidence ranging from 0 to 1. To increase the predictive power of this network, we generated a protein-protein interactome for the 31 candidate protein products using a stringent Interaction Confidence of at least 0.8, a threshold that is high enough to manage false positives and is commonly used in the literature (Kim et al., 2010, Rybarczyk-Filho et al., 2011). Cytoscape software (Cline et al., 2007) was used to visualize these interactions in a web of nodes and edges, organized for visualization using a layout algorithm (Fig. 3A). Of the 31 candidate mouse gene-products, 9 did not have known interactors exceeding the interaction confidence threshold, while 15 remained connected within a single network. For comparative purposes, a similar network was also generated for the control genes (Fig. 3B).
The same procedure was next applied to the respective human orthologs of these proteins, using an interaction confidence of ≥ 0.8 (Fig. 3A). Cytoscape generated and visualized the interactome of 31 human gene candidates, where 7 did not have known interactors at the chosen interaction confidence, and 20 remained connected in a single network. We then used Cytoscape to assess the overlap between the mouse and human interactomes, yielding a final network of interacting proteins present in both species (Fig. 3A). Ten of the 31 gene candidates remained interconnected within a single network, representing promising translational targets to study grooming based on correlation between behavioral phenotypes, brain gene expression and integration within the larger cross-species protein interactome (Fig. 3A, Table 3). Finally, in order to assess the differences in connectivity between candidate and control genes, we constructed a mouse interactome for our control genes and performed an unpaired Wilcoxon-Mann-Whitney U-test comparing the number of connectors per node between the networks of mouse candidate and control genes. Overall, the networks differed both qualitatively (with control genes appearing less interconnected in the graphic form) and quantitatively, as the candidate gene networked showed a trend (P<0.08) to more connectors per node, compared to the respective control gene network (Fig. 3B).
3. Discussion
The present study is the first comprehensive in-silico analysis combining behavioral and genomic data to examine mouse grooming behavior. An increased understanding of this important phenotype is likely to lead to insights into complex neurobehavioral disorders, such as autism and obsessive-compulsive disorder, OCD) (Berridge et al., 2005, Bienvenu et al., 2009, Crawley, 2007, Feusner et al., 2009, Rapoport, 1991, Shmelkov et al., 2010, Silverman et al., 2010, Swedo et al., 1989, Welch et al., 2007, Yang and Lu, 2011). In addition, this ‘proof-of-concept’ approach can easily be adapted to other complex traits in mice, as well as can be applied to grooming and other complex behaviors in various model organisms and humans.
As already mentioned, brain expression microarray results initially provided 844 genes with the expression significantly correlating with mouse grooming behavior. Since these genes have been selected with a high stringency (P<0.005), we first chose genes with high significance demonstrated independently in several behavioral tests, then selecting the remaining candidates based on the strength of the correlation in a single test. By selecting the top 40 genes, we were able to generate a highly integrated web of candidate genes and their interactors, revealing easily visualized, potentially novel interactions for mouse grooming behavior (Fig. 3A). Selecting genes with highly homologous and similarly interconnected human orthologs further supported the translational potential of the candidate genes identified in this study (Fig. 3A). To ensure that our candidate genes yielded a robust and meaningful network, the protein-protein interactions in the mouse and human interactomes were generated with a stringent Interaction Confidence (Fig. 3A). Since human and mouse genomes share a high degree of homology (Boguski, 2002), many of the pathways and interactions in humans are expected to be present in mice.
Representing a prominent phenotype sensitive to various genetic, behavioral and pharmacological manipulations (Angrini et al., 1998, Greer and Capecchi, 2002, Kalueff et al., 2005, Kalueff and Tuohimaa, 2005c), rodent grooming is a complex, highly organized behavior that can be further dissected for an in-depth analysis of centrally-controlled neurophenotypes (Kyzar, Gaikwad, 2011). The current study has generated a list of putative genes for the further study of mouse self-grooming behavior, representing a promising step in understanding of the genetic control of multifaceted behavioral domains. This information may help elucidate the relatively unknown neural and molecular mechanisms of self-grooming and other patterned motor responses, including pathological stereotypic behavior in OCD, attention deficit/hyperactivity disorder, schizophrenia and autism spectrum disorder (Chao et al., 2010, Mahone et al., 2004, Nayate et al., 2011).
In general, as genetic contributions to baseline anxiety/activity levels, motor coordination and other factors may modulate mouse grooming behavior through multiple mechanisms, each of the candidate genes may influence baseline grooming activity, or self-grooming in response to novelty stress. For example, the mammalian phenotype for prostaglandin E receptor 3 (Ptger3) mutation includes an abnormal body temperature and impaired pain threshold (Ushikubi et al., 1998), which may produce variation in thermoregulation and subsequently affect baseline grooming behavior. While myosin VIIA (Myo7a) mutation primarily leads to vestibular dysfunction, numerous reports reveal comorbidity between balance disorders and anxiety in both rodents (Kalueff et al., 2008, Shefer et al., 2010) and humans (Alvord, 1991, Balaban and Thayer, 2001), consistent with altered anxiety phenotypes in Myo7a mice (Shefer, Gordon, 2010).
Importantly, a number of cytoskeletal genes were associated with grooming behavior in this study. While cytoskeletal proteins are well-known for their role in cellular organization (Kellogg et al., 1994, Misteli, 2001), recent evidence has implicated actin- and myosin-related proteins in more complex phenomena, such as receptor trafficking, dendritic plasticity and sensorimotor gating (Bosch and Hayashi, 2011, Fradley et al., 2005, Yuen and Yan, 2009). Certain cytoskeletal genes are likely to be differentially regulated in various brain areas, leading to increased divergence and specialized functions in neurons. Therefore, variation in synaptic receptor expression, driven by cytoskeletal mechanisms, may contribute to the observed strain differences in grooming activity. This mechanism is only beginning to be recognized by the field, as very few studies have focused on the multifaceted role of cytoskeletal genes in complex behavioral and physiological domains. The importance of actin in chromatin remodeling has been well documented (Ferrai et al., 2009, Obrdlik et al., 2007, Percipalle and Visa, 2006), possibly explaining why the actin-associated proteins Pdgfb, Myo1b, Cdh1, Myo7a and Parvg were implicated by this study (Table 3). The presence and interconnectedness of the cytoskeletal proteins Tubgcp4, Pdgfb, Cdh1, Racgap1, Myo7a, Mapre1 and Parvg in the shared interactome (Fig. 3A) further suggest their role in various processes related to grooming behavior.
Our analysis also produced some unexpected results, as several notable genes implicated in compulsive grooming and OCD-like behavior (Slitrk5 and Sapap3) in rodents (Shmelkov, Hormigo, 2010, Ting and Feng, 2011, Welch, Lu, 2007, Yang and Lu, 2011) and humans (Bienvenu, Wang, 2009, Boardman et al., 2011, Zuchner et al., 2009) were not identified here. This may be due to the detrimental effects arising from the genes’ mutation or knockout, leading to the disruption of striatal neuronal differentiation and neurotransmission (Shmelkov, Hormigo, 2010, Welch, Lu, 2007, Yang and Lu, 2011), whereas we focused on the gene expression of wild-type inbred mice whose brain function has not been disrupted through genetic modification. While genes identified in genetically modified animals may not be involved in the normal self-grooming behavior, their disruption affects corticostriatal circuitry, which can indirectly evoke aberrant grooming. Specifically, the selective over-activation of the orbitofrontal cortex, abnormalities in striatal anatomy/cell morphology, and alterations in glutamate receptor composition that accompany a mutation (Shmelkov, Hormigo, 2010), may disrupt key neural pathways involved in normal grooming behavior.
Likewise, while our analysis revealed the role of the transcription factor homeobox B4 (Hoxb4) in grooming, we did not observe a correlation of the more widely reported Hoxb8 gene, linked to compulsive grooming and other OCD-like behaviors (Chen, Tvrdik, 2010, Greer and Capecchi, 2002, Yang and Lu, 2011). The Hox genes are arranged in the genome in a collinear fashion, with the Hoxb genes clustered together on chromosome 17 (Chambeyron and Bickmore, 2004). Hoxb4 and Hoxb8 are both DNA sequence-specific transcription factors responsible for various developmental processes, including hematopoesis. Interestingly, the aberrant grooming in Hoxb8 mutant mice has recently been linked to defective hemopoietic-derived microglia (Chen, Tvrdik, 2010). Thus, the possible role of a hematopoietic gene Hoxb4 in mouse grooming in this study is congruent with the hematopoitic hypothesis of aberrant grooming in mice (Chen, Tvrdik, 2010). Several of our candidate genes have previously been linked to psychiatric disorders, some of which are closely related to pathological grooming. For example, Ptpra knockout mice display defects in neuronal migration, sensorimotor gating and habituation to startle response, thereby linking Ptpra to schizophrenia (Takahashi et al., 2011). They also show altered anxiety phenotypes (Skelton et al., 2003), whereas its human ortholog PTPRA resides in the 20p13 region which has repeatedly been linked to psychosis (Fanous et al., 2008, Teltsh et al., 2008). As already suggested in the literature (Audet, Goulet, 2006, Isingrini et al., 2011, Papaleo et al., 2011), grooming responses in mice may represent traits highly relevant to schizophrenia, anxiety and depression, and our results are in line with this notion.
Previous work has implicated abnormal brain development in several complex neuropsychiatric disorders. For example, aberrant neuronal migration and callosal hypoplasticity are commonly reported in schizophrenia (Connor et al., 2011, Knochel et al., 2012), whereas autistic patients show underdevelopment of the cerebellum and migration defects (Verhoeven et al., 2010). Analysis of the 31 shared genes generated by this study in mice and humans implicates these genes in brain development (Table 3), including abnormal cerebellar morphology (Ccnd2), abnormal neuronal migration (Snai1), abnormal brain development (Tceb3, Nr6a1) and impaired neuronal differentiation (Ttl). Moreover, genes selected for the analyses have different numbers of known interactors (Fig. 3A), many of which form key nodes in our shared interactome (e.g., Tubgcp4, Pdgfb, Cdh1, Gnb5, Racgap1, Myo7a, Ccnd2, Mapre1, Parvg). The genes Tubgcp4, Racgap1 and Mapre1 were mentioned previously because of their involvement with tubulin, while Cdh1, Myo7a and Parvg interact with actin and may play a role in chromatin remodeling. Examining public databases for Ccnd2 revealed its importance in the development of the cerebellum (Table 3), where Gnb5 is also important, since Gnb5 knockout mice have abnormal cerebellar development and motor incoordination (Zhang et al., 2011).
Furthermore, there were several limitations in our study. First, while we used correlational in-silico analyses, more specific studies are needed to investigate the functionality of identified genes in mouse grooming behaviors. Also, since our study utilized whole-brain microarray data, this limitation may be further resolved using region-specific gene expression analyses, empowered by sophisticated databases, such as the ABA (Lein, Hawrylycz, 2007). Far from providing an expansive and complete list of genes associated with mouse grooming, the approach described in this study offers a rapid, cost-effective and promising way to find new targets for important neurobiological functions (see (Stewart, Gaikwad, 2011) for review). Furthermore, while our study focused on quantity (duration) data, mouse grooming is a complex behavior with an important sequential (patterning) microstructure (Kalueff et al., 2007). Therefore, future studies may elucidate the correlation between gene expression and sequencing of mouse grooming. Again, because correlations in biological systems do not necessarily represent functional interrelationships between different phenomena or processes, future integrative research (currently underway in our laboratory) will have to assess in-depth the exact causal pathways of aberrant grooming examined here. Finally, epigenetic factors play an important role in the regulation of activity of various genes (Fish et al., 2004, Meaney and Szyf, 2005, Rothbart and Posner, 2005, Sheese et al., 2007, Voelker et al., 2009, Weaver et al., 2007). Therefore, further characterization of the genes generated by our method, as well as analysis of their epigenetic regulation and gene x environment interactions, may provide important clues in understanding the neurobiology of grooming behavior and identifying targets for modulating complex patterned behavior across a number of model species. While the link between genes and behavior remains a major challenge in modern biological psychiatry, our study may offer one of potential large-scale, data-mining approaches to address these aspects.
Highlights.
Mouse grooming is an important, evolutionarily conserved behavioral phenotype
This in-silico study uses open gene expression and behavioral databases
We identify an interconnected network of candidate genes implicated in grooming
Our results show the utility of large-scale data-mining and in-silico modeling
Acknowledgments
The study was supported by Tulane University Pilot funds, Tulane Synergy and Provost’s Scholarly Enrichment grants to AVK, as well as by ZENEREI Institute. The authors thank R. Gunn (Dalhousie University) for her help with this project. This study used several open-access databases, including the Allen Brain Atlas of gene activity in the C57BL/6J mouse brain (ABA, http://mouse.brain-map.org/; http://uri.neuinfo.org/nif/registry/nif-0000-00508); GoMiner (to classify genes in microarray experiments, http://discover.nci.nih.gov/gominer/; http://uri.neuinfo.org/nif/registry/nif-0000-21181), the Gene Ontology (GO, used by GoMiner to provide detailed gene information, http://www.geneontology.org/; http://uri.neuinfo.org/nif/registry/nif-0000-02915); the Mouse Genome Informatics (MGI) database of mutant and transgenic mouse models (http://www.informatics.jax.org; http://uri.neuinfo.org/nif/registry/nif-0000-00096); the Mouse Phenome Database (MPD, a repository of genotypic and phenotypic data, which allows for genotype-phenotype association, http://www.jax.org/phenome; http://uri.neuinfo.org/nif/registry/nif-0000-03160); the Phenogen Informatics (a microarray data repository and analysis tool allowing users to research candidate genes, http://phenogen.ucdenver.edu/PhenoGen/); the Drug Related Gene Database (DRG, describing differences in gene expression as a function of drug exposure, https://confluence.crbs.ucsd.edu/display/NIF/DRG; http://uri.neuinfo.org/nif/registry/nif-0000-37443) and the Search Tool for the Retrieval of Interacting Genes (STRING, database containing all known and predicted interactions between proteins in over 1000 different species, http://string.embl.de/; http://uri.neuinfo.org/nif/registry/nif-0000-03503). We also used Cytoscape (http://cytoscape.org; http://uri.neuinfo.org/nif/registry/nif-0000-30404) to visualize protein-protein interaction networks, and the Ontological Discovery Environment (ODE, http://ontologicaldiscovery.org/; http://uri.neuinfo.org/nif/registry/nif-0000-00517) as a freely accessible phenotype-centered database with integrated analysis and visualization tools. In addition to homepage URL, we provided the Uniform Resource Identifier (URI) for the databases used in this study, directing the user to the original source regardless of its current URLs. This information was provided here to recognize the value and importance of open-source, publicly accessible scientific information (Cheung et al., 2009). The Protein database (Pruitt, Tatusova, 2007) and sequence analysis using Hum-mPLoc (Shen and Chou, 2007) were also used in this study to characterize cellular location of protein products of the selected candidate gene (Fig. 3E). Grooming data was collected with the help of Nicola Hoffman, Lisa Currie, Vicki Savoie and Martin Williamson. Behavioral data collection was funded by a grant from NSERC of Canada to REB. Generous funds from the AstraZeneca R&D Boston were used to defray cost of mice through the Mouse Phenome Project (The Jackson Laboratory, Bar Harbor, ME). Behavioral equipment was purchased with funds from an NSERC equipment grant to REB.
Abbreviations
- ABA
Allen Brain Atlas
- DRG
Drug Related Gene Database
- EPM
The elevated plus-maze test
- EZM
The elevated zero-maze test
- GO
Gene Ontology
- LDB
The light-dark box test
- MPD
Mouse Phenome Database
- MGI
Mouse Genome Informatics
- OCD
Obsessive-compulsive disorder
- ODE
Ontological Discovery Environment
- OFT
The open field test
- STRING
Search Tool for the Retrieval of Interacting Genes/Proteins
- URI
Uniform Resource Identifier
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akil H, Martone ME, Van Essen DC. Challenges and opportunities in mining neuroscience data. Science. 2011;331:708–12. doi: 10.1126/science.1199305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvord LS. Psychological status of patients undergoing electronystagmography. J Am Acad Audiol. 1991;2:261–5. [PubMed] [Google Scholar]
- Angrini M, Leslie JC, Shephard RA. Effects of propranolol, buspirone, pCPA, reserpine, and chlordiazepoxide on open-field behavior. Pharmacol Biochem Behav. 1998;59:387–97. doi: 10.1016/s0091-3057(97)00457-7. [DOI] [PubMed] [Google Scholar]
- Audet MC, Goulet S, Dore FY. Repeated subchronic exposure to phencyclidine elicits excessive atypical grooming in rats. Behav Brain Res. 2006;167:103–10. doi: 10.1016/j.bbr.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Baker EJ, Jay JJ, Philip VM, Zhang Y, Li Z, Kirova R, et al. Ontological Discovery Environment: a system for integrating gene-phenotype associations. Genomics. 2009;94:377–87. doi: 10.1016/j.ygeno.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban CD, Thayer JF. Neurological bases for balance-anxiety links. J Anxiety Disord. 2001;15:53–79. doi: 10.1016/s0887-6185(00)00042-6. [DOI] [PubMed] [Google Scholar]
- Bell RL, Kimpel MW, McClintick JN, Strother WN, Carr LG, Liang T, et al. Gene expression changes in the nucleus accumbens of alcohol-preferring rats following chronic ethanol consumption. Pharmacol Biochem Behav. 2009;94:131–47. doi: 10.1016/j.pbb.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B, Saba LM, Hornbaker CK, Kechris KJ, Hoffman P, Tabakoff B. Genetical genomic analysis of complex phenotypes using the PhenoGen website. Behav Genet. 2011;41:625–8. doi: 10.1007/s10519-010-9427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergner CL, Smolinsky AN, Hart PC, Dufour BD, Egan RJ, Laporte JL, et al. Mouse models for studying depression-like states and antidepressant drugs. Methods Mol Biol. 2010;602:267–82. doi: 10.1007/978-1-60761-058-8_16. [DOI] [PubMed] [Google Scholar]
- Berridge KC, Aldridge JW, Houchard KR, Zhuang X. Sequential super-stereotypy of an instinctive fixed action pattern in hyper-dopaminergic mutant mice: a model of obsessive compulsive disorder and Tourette’s. BMC Biol. 2005;3:4. doi: 10.1186/1741-7007-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhave SV, Hornbaker C, Phang TL, Saba L, Lapadat R, Kechris K, et al. The PhenoGen informatics website: tools for analyses of complex traits. BMC Genet. 2007;8:59. doi: 10.1186/1471-2156-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienvenu OJ, Wang Y, Shugart YY, Welch JM, Grados MA, Fyer AJ, et al. Sapap3 and pathological grooming in humans: Results from the OCD collaborative genetics study. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:710–20. doi: 10.1002/ajmg.b.30897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman L, van der Merwe L, Lochner C, Kinnear CJ, Seedat S, Stein DJ, et al. Investigating SAPAP3 variants in the etiology of obsessive-compulsive disorder and trichotillomania in the South African white population. Compr Psychiatry. 2011;52:181–7. doi: 10.1016/j.comppsych.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Boguski MS. Comparative genomics: the mouse that roared. Nature. 2002;420:515–6. doi: 10.1038/420515a. [DOI] [PubMed] [Google Scholar]
- Bosch M, Hayashi Y. Structural plasticity of dendritic spines. Curr Opin Neurobiol. 2011;22:383–8. doi: 10.1016/j.conb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RE, Gunn RK, Schellinck HS, Wong AA, O’Leary TP. Mouse Phenome Database. The Jackson Laboratory; Bar Harbor, Maine USA: Mar, 2007. Anxiety, exploratory behavior, and motor activity in 13 strains of mice. MPD: 94. World Wide Web (URL: http://www.jax.org/phenome. [available 18 March 2008]. 2004. [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–30. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–9. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SK, Tvrdik P, Peden E, Cho S, Wu S, Spangrude G, et al. Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell. 2010;141:775–85. doi: 10.1016/j.cell.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K-H, Prud’hommeaux E, Wang Y, Stephens S. Semantic Web for Health Care and Life Sciences: a review of the state of the art. Briefings in Bioinformatics. 2009;10:111–3. doi: 10.1093/bib/bbp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–82. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor CM, Crawford BC, Akbarian S. White matter neuron alterations in schizophrenia and related disorders. Int J Dev Neurosci. 2011;29:325–34. doi: 10.1016/j.ijdevneu.2010.07.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN. Mouse behavioral assays relevant to the symptoms of autism. Brain Pathol. 2007;17:448–59. doi: 10.1111/j.1750-3639.2007.00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denmark A, Tien D, Wong K, Chung A, Cachat J, Goodspeed J, et al. The effects of chronic social defeat stress on mouse self-grooming behavior and its patterning. Behav Brain Res. 2010;208:553–9. doi: 10.1016/j.bbr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- Drgon T, Montoya I, Johnson C, Liu QR, Walther D, Hamer D, et al. Genome-wide association for nicotine dependence and smoking cessation success in NIH research volunteers. Mol Med. 2009;15:21–7. doi: 10.2119/molmed.2008.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estanislau C. Cues to the usefulness of grooming behavior in the evaluation of anxiety in the elevated plus-maze. Psych Neurosci. 2012;5:105–12. [Google Scholar]
- Fanous AH, Neale MC, Webb BT, Straub RE, O’Neill FA, Walsh D, et al. Novel linkage to chromosome 20p using latent classes of psychotic illness in 270 Irish high-density families. Biol Psychiatry. 2008;64:121–7. doi: 10.1016/j.biopsych.2007.11.023. [DOI] [PubMed] [Google Scholar]
- Fentress JC. Interrupted ongoing behaviour in two species of vole (Microtus agrestis and Clethrionomys britannicus). II. Extended analysis of motivational variables underlying fleeing and grooming behaviour. Anim Behav. 1968;16:154–67. doi: 10.1016/0003-3472(68)90125-5. [DOI] [PubMed] [Google Scholar]
- Fentress JC. The tonic hypothesis and the patterning of behavior. Ann N Y Acad Sci. 1977;290:370–95. doi: 10.1111/j.1749-6632.1977.tb39739.x. [DOI] [PubMed] [Google Scholar]
- Fentress JC. Expressive contexts, fine structure, and central mediation of rodent grooming. Ann N Y Acad Sci. 1988;525:18–26. doi: 10.1111/j.1749-6632.1988.tb38592.x. [DOI] [PubMed] [Google Scholar]
- Ferrai C, Naum-Ongania G, Longobardi E, Palazzolo M, Disanza A, Diaz VM, et al. Induction of HoxB transcription by retinoic acid requires actin polymerization. Mol Biol Cell. 2009;20:3543–51. doi: 10.1091/mbc.E09-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feusner JD, Hembacher E, Phillips KA. The mouse who couldn’t stop washing: pathologic grooming in animals and humans. CNS Spectr. 2009;14:503–13. doi: 10.1017/s1092852900023567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- File SE, Mabbutt PS, Walker JH. Comparison of adaptive responses in familiar and novel environments: modulatory factors. Ann N Y Acad Sci. 1988;525:69–79. doi: 10.1111/j.1749-6632.1988.tb38596.x. [DOI] [PubMed] [Google Scholar]
- Fish EW, Shahrokh D, Bagot R, Caldji C, Bredy T, Szyf M, et al. Epigenetic programming of stress responses through variations in maternal care. Ann N Y Acad Sci. 2004;1036:167–80. doi: 10.1196/annals.1330.011. [DOI] [PubMed] [Google Scholar]
- Fradley RL, O’Meara GF, Newman RJ, Andrieux A, Job D, Reynolds DS. STOP knockout and NMDA NR1 hypomorphic mice exhibit deficits in sensorimotor gating. Behav Brain Res. 2005;163:257–64. doi: 10.1016/j.bbr.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Gardner D, Akil H, Ascoli GA, Bowden DM, Bug W, Donohue DE, et al. The neuroscience information framework: a data and knowledge environment for neuroscience. Neuroinformatics. 2008;6:149–60. doi: 10.1007/s12021-008-9024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer JM, Capecchi MR. Hoxb8 is required for normal grooming behavior in mice. Neuron. 2002;33:23–34. doi: 10.1016/s0896-6273(01)00564-5. [DOI] [PubMed] [Google Scholar]
- Grubb SC, Maddatu TP, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res. 2009;37:D720–30. doi: 10.1093/nar/gkn778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart PC, Bergner CL, Smolinsky AN, Dufour BD, Egan RJ, Laporte JL, et al. Experimental models of anxiety for drug discovery and brain research. Methods Mol Biol. 2010;602:299–321. doi: 10.1007/978-1-60761-058-8_18. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isingrini E, Belzung C, d’Audiffret A, Camus V. Early and late-onset effect of chronic stress on vascular function in mice: a possible model of the impact of depression on vascular disease in aging. Am J Geriatr Psychiatry. 2011;19:335–46. doi: 10.1097/JGP.0b013e318202bc42. [DOI] [PubMed] [Google Scholar]
- Kagami Y, Furuichi T. Investigation of differentially expressed genes during the development of mouse cerebellum. Brain Res Gene Expr Patterns. 2001;1:39–59. doi: 10.1016/s1567-133x(01)00007-2. [DOI] [PubMed] [Google Scholar]
- Kalueff AV, Aldridge JW, LaPorte JL, Murphy DL, Tuohimaa P. Analyzing grooming microstructure in neurobehavioral experiments. Nat Protoc. 2007;2:2538–44. doi: 10.1038/nprot.2007.367. [DOI] [PubMed] [Google Scholar]
- Kalueff AV, Ishikawa K, Griffith AJ. Anxiety and otovestibular disorders: linking behavioral phenotypes in men and mice. Behav Brain Res. 2008;186:1–11. doi: 10.1016/j.bbr.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Kalueff AV, Lou YR, Laaksi I, Tuohimaa P. Increased grooming behavior in mice lacking vitamin D receptors. Physiol Behav. 2004;82:405–9. doi: 10.1016/j.physbeh.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Kalueff AV, Lou YR, Laaksi I, Tuohimaa P. Abnormal behavioral organization of grooming in mice lacking the vitamin D receptor gene. J Neurogenet. 2005;19:1–24. doi: 10.1080/01677060590949683. [DOI] [PubMed] [Google Scholar]
- Kalueff AV, Tuohimaa P. Contrasting grooming phenotypes in C57Bl/6 and 129S1/SvImJ mice. Brain Res. 2004;1028:75–82. doi: 10.1016/j.brainres.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Kalueff AV, Tuohimaa P. Contrasting grooming phenotypes in three mouse strains markedly different in anxiety and activity (129S1, BALB/c and NMRI) Behav Brain Res. 2005a;160:1–10. doi: 10.1016/j.bbr.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Kalueff AV, Tuohimaa P. The grooming analysis algorithm discriminates between different levels of anxiety in rats: potential utility for neurobehavioural stress research. J Neurosci Methods. 2005b;143:169–77. doi: 10.1016/j.jneumeth.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Kalueff AV, Tuohimaa P. Mouse grooming microstructure is a reliable anxiety marker bidirectionally sensitive to GABAergic drugs. Eur J Pharmacol. 2005c;508:147–53. doi: 10.1016/j.ejphar.2004.11.054. [DOI] [PubMed] [Google Scholar]
- Kellogg DR, Moritz M, Alberts BM. The centrosome and cellular organization. Annu Rev Biochem. 1994;63:639–74. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, et al. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–66. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Kim E, Kwon EY, Jang HS, Hur CG, Choi MS. Network analysis of hepatic genes responded to high-fat diet in C57BL/6J mice: nutrigenomics data mining from recent research findings. J Med Food. 2010;13:743–56. doi: 10.1089/jmf.2009.1350. [DOI] [PubMed] [Google Scholar]
- Knochel C, Oertel-Knochel V, Schonmeyer R, Rotarska-Jagiela A, van de Ven V, Prvulovic D, et al. Interhemispheric hypoconnectivity in schizophrenia: Fiber integrity and volume differences of the corpus callosum in patients and unaffected relatives. Neuroimage. 2012;59:926–34. doi: 10.1016/j.neuroimage.2011.07.088. [DOI] [PubMed] [Google Scholar]
- Koide T, Takahashi A. Mouse Phenome Database Web Site. The Jackson Laboratory; Bar Harbor, Maine USA: [February 2011]. 2006. Temporal recording of open field behavior in 12 inbred strains of mice, mostly wild-derived (Mishima) strains. MPD:359. World Wide Web (URL: http://www.jax.org/phenome. [Google Scholar]
- Korostynski M, Kaminska-Chowaniec D, Piechota M, Przewlocki R. Gene expression profiling in the striatum of inbred mouse strains with distinct opioid-related phenotypes. BMC Genomics. 2006;7:146. doi: 10.1186/1471-2164-7-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostynski M, Piechota M, Kaminska D, Solecki W, Przewlocki R. Morphine effects on striatal transcriptome in mice. Genome Biol. 2007;8:R128. doi: 10.1186/gb-2007-8-6-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyzar E, Gaikwad S, Roth A, Green J, Pham M, Stewart A, et al. Towards high-throughput phenotyping of complex patterned behaviors in rodents: focus on mouse self-grooming and its sequencing. Behav Brain Res. 2011;225:426–31. doi: 10.1016/j.bbr.2011.07.052. [DOI] [PubMed] [Google Scholar]
- Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–16. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- Lake J, Donahue L, Davisson MT. Mouse Phenome Database Web Site. The Jackson Laboratory; Bar Harbor, Maine USA: [October 2007]. 2005. Behavioral and neuromuscular phenotypes, C57BL/6J-Chr#A/NaJ mouse chromosome substitution panel (23 strains). MPD: 190. World Wide Web (URL: http://www.jax.org/phenome. [Google Scholar]
- Lehrmann E, Oyler J, Vawter MP, Hyde TM, Kolachana B, Kleinman JE, et al. Transcriptional profiling in the human prefrontal cortex: evidence for two activational states associated with cocaine abuse. Pharmacogenomics J. 2003;3:27–40. doi: 10.1038/sj.tpj.6500146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Mahone EM, Bridges D, Prahme C, Singer HS. Repetitive arm and hand movements (complex motor stereotypies) in children. J Pediatr. 2004;145:391–5. doi: 10.1016/j.jpeds.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Matzilevich DA, Rall JM, Moore AN, Grill RJ, Dash PK. High-density microarray analysis of hippocampal gene expression following experimental brain injury. J Neurosci Res. 2002;67:646–63. doi: 10.1002/jnr.10157. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Szyf M. Environmental programming of stress responses through DNA methylation: life at the interface between a dynamic environment and a fixed genome. Dialogues Clin Neurosci. 2005;7:103–23. doi: 10.31887/DCNS.2005.7.2/mmeaney. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignogna P, Viggiano D. Brain distribution of genes related to changes in locomotor activity. Physiology & Behavior. 2010;99:618–26. doi: 10.1016/j.physbeh.2010.01.026. [DOI] [PubMed] [Google Scholar]
- Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–5. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A. 2006;103:6368–73. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayate A, Tonge BJ, Bradshaw JL, McGinley JL, Iansek R, Rinehart NJ. Differentiation of High-Functioning Autism and Asperger’s Disorder Based on Neuromotor Behaviour. J Autism Dev Disord. 2011 doi: 10.1007/s10803-011-1299-5. [DOI] [PubMed] [Google Scholar]
- Ng L, Bernard A, Lau C, Overly CC, Dong HW, Kuan C, et al. An anatomic gene expression atlas of the adult mouse brain. Nat Neurosci. 2009;12:356–62. doi: 10.1038/nn.2281. [DOI] [PubMed] [Google Scholar]
- Obrdlik A, Kukalev A, Percipalle P. The function of actin in gene transcription. Histol Histopathol. 2007;22:1051–5. doi: 10.14670/HH-22.1051. [DOI] [PubMed] [Google Scholar]
- Paletzki RF, Myakishev MV, Polesskaya O, Orosz A, Hyman SE, Vinson C. Inhibiting activator protein-1 activity alters cocaine-induced gene expression and potentiates sensitization. Neuroscience. 2008;152:1040–53. doi: 10.1016/j.neuroscience.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaleo F, Silverman JL, Aney J, Tian Q, Barkan CL, Chadman KK, et al. Working memory deficits, increased anxiety-like traits, and seizure susceptibility in BDNF overexpressing mice. Learn Mem. 2011;18:534–44. doi: 10.1101/lm.2213711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Visa N. Molecular functions of nuclear actin in transcription. J Cell Biol. 2006;172:967–71. doi: 10.1083/jcb.200512083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, et al. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav. 2010;9:129–59. doi: 10.1111/j.1601-183X.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–5. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport JL. Recent advances in obsessive-compulsive disorder. Neuropsychopharmacology. 1991;5:1–10. [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, 3rd, Maze I, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–48. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez LA, Plomin R, Blizard DA, Jones BC, McClearn GE. Alcohol acceptance, preference, and sensitivity in mice. I. Quantitative genetic analysis using BXD recombinant inbred strains. Alcohol Clin Exp Res. 1994;18:1416–22. doi: 10.1111/j.1530-0277.1994.tb01444.x. [DOI] [PubMed] [Google Scholar]
- Rothbart MK, Posner MI. Genes and experience in the development of executive attention and effortful control. New Dir Child Adolesc Dev. 2005:101–8. doi: 10.1002/cd.142. [DOI] [PubMed] [Google Scholar]
- Rybarczyk-Filho JL, Castro MA, Dalmolin RJ, Moreira JC, Brunnet LG, de Almeida RM. Towards a genome-wide transcriptogram: the Saccharomyces cerevisiae case. Nucleic Acids Res. 2011;39:3005–16. doi: 10.1093/nar/gkq1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs BD. The development of grooming and its expression in adult animals. Ann N Y Acad Sci. 1988;525:1–17. doi: 10.1111/j.1749-6632.1988.tb38591.x. [DOI] [PubMed] [Google Scholar]
- Sears WR, McCombe PF, Sasso RC. Kinematics of Cervical and Lumbar Total Disc Replacement. 2006;18:117–29. [Google Scholar]
- Shaw DR. Searching the Mouse Genome Informatics (MGI) resources for information on mouse biology from genotype to phenotype. Curr Protoc Bioinformatics. 2009;Chapter 1(Unit 1):7. doi: 10.1002/0471250953.bi0107s05. [DOI] [PubMed] [Google Scholar]
- Sheese BE, Voelker PM, Rothbart MK, Posner MI. Parenting quality interacts with genetic variation in dopamine receptor D4 to influence temperament in early childhood. Dev Psychopathol. 2007;19:1039–46. doi: 10.1017/S0954579407000521. [DOI] [PubMed] [Google Scholar]
- Shefer S, Gordon CR, Avraham KB, Mintz M. Progressive vestibular mutation leads to elevated anxiety. Brain Res. 2010;1317:157–64. doi: 10.1016/j.brainres.2009.12.059. [DOI] [PubMed] [Google Scholar]
- Shen HB, Chou KC. Hum-mPLoc: an ensemble classifier for large-scale human protein subcellular location prediction by incorporating samples with multiple sites. Biochem Biophys Res Commun. 2007;355:1006–11. doi: 10.1016/j.bbrc.2007.02.071. [DOI] [PubMed] [Google Scholar]
- Shmelkov SV, Hormigo A, Jing D, Proenca CC, Bath KG, Milde T, et al. Slitrk5 deficiency impairs corticostriatal circuitry and leads to obsessive-compulsive-like behaviors in mice. Nat Med. 2010;16:598–602. doi: 10.1038/nm.2125. 1p following. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Tolu SS, Barkan CL, Crawley JN. Repetitive self-grooming behavior in the BTBR mouse model of autism is blocked by the mGluR5 antagonist MPEP. Neuropsychopharmacology. 2010;35:976–89. doi: 10.1038/npp.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelton MR, Ponniah S, Wang DZ, Doetschman T, Vorhees CV, Pallen CJ. Protein tyrosine phosphatase alpha (PTP alpha) knockout mice show deficits in Morris water maze learning, decreased locomotor activity, and decreases in anxiety. Brain Res. 2003;984:1–10. doi: 10.1016/s0006-8993(03)02839-7. [DOI] [PubMed] [Google Scholar]
- Spruijt BM, van Hooff JA, Gispen WH. Ethology and neurobiology of grooming behavior. Physiol Rev. 1992;72:825–52. doi: 10.1152/physrev.1992.72.3.825. [DOI] [PubMed] [Google Scholar]
- Stewart A, Gaikwad S, Hart P, Kyzar E, Roth A, Kalueff A. Experimental models of anxiolytic drug discovery in the era of omes and omics. Expert Opinion on Drug Discovery. 2011;6:755–69. doi: 10.1517/17460441.2011.586028. [DOI] [PubMed] [Google Scholar]
- Swedo SE, Rapoport JL, Leonard H, Lenane M, Cheslow D. Obsessive-compulsive disorder in children and adolescents. Clinical phenomenology of 70 consecutive cases. Arch Gen Psychiatry. 1989;46:335–41. doi: 10.1001/archpsyc.1989.01810040041007. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–8. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Bhave S, Saba L. Mouse Phenome Database Web Site. The Jackson Laboratory; Bar Harbor, Maine USA: [September 2010]. 2007. Whole brain gene expression in males of 20 inbred strains of mice. MPD: 354. World Wide Web (URL: http://www.jax.org/phenome. [Google Scholar]
- Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, et al. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7:70. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Nielsen KS, Aleksic B, Petersen S, Ikeda M, Kushima I, et al. Loss of Function Studies in Mice and Genetic Association Link Receptor Protein Tyrosine Phosphatase alpha to Schizophrenia. Biol Psychiatry. 2011;70:626–35. doi: 10.1016/j.biopsych.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teltsh O, Kanyas K, Karni O, Levi A, Korner M, Ben-Asher E, et al. Genome-wide linkage scan, fine mapping, and haplotype analysis in a large, inbred, Arab Israeli pedigree suggest a schizophrenia susceptibility locus on chromosome 20p13. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:209–15. doi: 10.1002/ajmg.b.30591. [DOI] [PubMed] [Google Scholar]
- Ting JT, Feng G. Neurobiology of obsessive-compulsive disorder: insights into neural circuitry dysfunction through mouse genetics. Curr Opin Neurobiol. 2011 doi: 10.1016/j.conb.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushikubi F, Segi E, Sugimoto Y, Murata T, Matsuoka T, Kobayashi T, et al. Impaired febrile response in mice lacking the prostaglandin E receptor subtype EP3. Nature. 1998;395:281–4. doi: 10.1038/26233. [DOI] [PubMed] [Google Scholar]
- Verhoeven JS, De Cock P, Lagae L, Sunaert S. Neuroimaging of autism. Neuroradiology. 2010;52:3–14. doi: 10.1007/s00234-009-0583-y. [DOI] [PubMed] [Google Scholar]
- Voelker P, Sheese BE, Rothbart MK, Posner MI. Variations in catechol-O-methyltransferase gene interact with parenting to influence attention in early development. Neuroscience. 2009;164:121–30. doi: 10.1016/j.neuroscience.2009.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gutala R, Hwang YY, Kim JM, Konu O, Ma JZ, et al. Strain- and region-specific gene expression profiles in mouse brain in response to chronic nicotine treatment. Genes Brain Behav. 2008;7:78–87. doi: 10.1111/j.1601-183X.2007.00328.x. [DOI] [PubMed] [Google Scholar]
- Wang PI, Marcotte EM. It’s the machine that matters: Predicting gene function and phenotype from protein networks. J Proteomics. 2010;73:2277–89. doi: 10.1016/j.jprot.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, D’Alessio AC, Brown SE, Hellstrom IC, Dymov S, Sharma S, et al. The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: altering epigenetic marks by immediate-early genes. J Neurosci. 2007;27:1756–68. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JM, Lu J, Rodriguiz RM, Trotta NC, Peca J, Ding JD, et al. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature. 2007;448:894–900. doi: 10.1038/nature06104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Dai M, Buckner J, Mirel B, Song J, Athey B, et al. Cross-domain neurobiology data integration and exploration. BMC Genomics. 2010;11 (Suppl 3):S6. doi: 10.1186/1471-2164-11-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XW, Lu XH. Molecular and cellular basis of obsessive-compulsive disorder-like behaviors: emerging view from mouse models. Curr Opin Neurol. 2011;24:114–8. doi: 10.1097/WCO.0b013e32834451fb. [DOI] [PubMed] [Google Scholar]
- Yuen EY, Yan Z. Dopamine D4 receptors regulate AMPA receptor trafficking and glutamatergic transmission in GABAergic interneurons of prefrontal cortex. J Neurosci. 2009;29:550–62. doi: 10.1523/JNEUROSCI.5050-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhang L, Tang Y, Zhang Q, Lou D, Sharp FR, et al. Repeated cocaine administration induces gene expression changes through the dopamine D1 receptors. Neuropsychopharmacology. 2005;30:1443–54. doi: 10.1038/sj.npp.1300680. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Pandey M, Seigneur EM, Panicker LM, Koo L, Schwartz OM, et al. Knockout of G protein beta5 impairs brain development and causes multiple neurologic abnormalities in mice. J Neurochem. 2011;119:544–54. doi: 10.1111/j.1471-4159.2011.07457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchner S, Wendland JR, Ashley-Koch AE, Collins AL, Tran-Viet KN, Quinn K, et al. Multiple rare SAPAP3 missense variants in trichotillomania and OCD. Mol Psychiatry. 2009;14:6–9. doi: 10.1038/mp.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]