Highlights
-
•
It is important to understand how alkaliphilic prokaryotes thrive at high pH.
-
•
An interesting issue is their ability to cope with bioenergetics at high pH.
-
•
We show that four genetically similar strains adopt different biochemical behaviors.
-
•
Two of the strains show a functional redundancy of the terminal part of the respiratory chain.
-
•
Biochemical data correlate with the expression of cytochrome c oxidase and quinol oxidase genes (heme-copper types).
Keywords: Bacillus clausii, Alkaliphilic bacilli, Bioenergetics, Terminal oxidases, Cyanide sensitivity, Respiratory chain
Abstract
A comparative analysis of terminal respiratory enzymes has been performed on four strains of Bacillus clausii used for preparation of a European probiotic. These four strains originated most probably from a common ancestor through early selection of stable clones for industrial propagation. They exhibit a low level of intra-specific diversity and a high degree of genomic conservation, making them an attractive model to study the different bioenergetics behaviors of alkaliphilic bacilli. The analysis of the different bioenergetics responses has been carried out revealing striking differences among the strains. Two out of the four strains have shown a functional redundancy of the terminal part of the respiratory chain. The biochemical data correlate with the expression level of the mRNA of cytochrome c oxidase and quinol oxidase genes (heme-copper type). The consequences of these different bioenergetics behaviors are also discussed.
1. Introduction
Alkaliphilic prokaryotes offer a wealth of opportunities to understand the mechanisms by which organisms thrive at high pH, as well as for the isolation of natural products [14]. Bacillus species are among the most frequently found aerobic eubacterial alkaliphiles. The mechanisms of adaptation of alkaliphilic microorganisms to alkaline environments have been intensively studied [9,14]. Among several mechanisms of alkaline adaptation, solute transport systems that reduce intracellular pH, cell wall structure, buffering capacity and alkali stability of cytoplasmic components have been entertained. One of the most interesting issue is the capability of alkaliphilic prokaryotes to cope with the bioenergetics function [9].
Bacillus clausii is a gram–positive, aerobic, endospore–forming, facultative alkaliphilic rod bacterium. Its relevant characteristics are oxidase and catalase enzymes expression, starch hydrolysis, gelatine hydrolysis, nitrate reduction and growth from 30 to 50 °C in NaCl up to 10%. The G + C content is around 43% [21].
The respiratory chain components of various alkaliphilic Bacillus species have been characterized. NADH dehydrogenases and succinate dehydrogenases have been purified and characterized and the presence of bc-type enzymes is well recognized [9,7,14,17]. The presence of diverse types of terminal oxidases such as the pH-regulated caa3 oxidase of Bacillus pseudofirmus OF4 [24,11], the similar enzyme purified from Bacillus polygonii YN-1 [10], or the aco3 oxidase of Bacillus cohnii YN2000 [31], has been described. The presence of terminal oxidases which belong to the bd-type family has also been shown in alkaliphilic bacilli [8].
The cyanide sensitivity of oxygen uptake in some alkaliphilic bacilli has been reported. In the obligate alkaliphilic B. polygonii YN-1 the cyanide sensitive component attributed to the caa3-type terminal oxidase represents only 10% of the total oxygen consumption [10]. These findings have led to the proposal that cyanide insensitive terminal respiratory component(s) together with catalase are of major importance in the respiratory mechanism of these bacteria.
In this work a comparative analysis of terminal respiratory enzymes has been performed on four closely related strains of B. clausii used for preparation of a European probiotic. The fact that these four strains belong to a unique genospecies, displaying a low level of intra-specific diversity compared with that observed for the reference strains of B. clausii, and the high degree of genomic conservation through the time [25], make these strains a unique model to study the differences in bioenergetics behaviors employed among closely related strains of an alkaliphilic bacterium. These strains originated most probably from a common ancestor through early selection of stable clones for industrial propagation. The principal focus of this work is the analysis of the different bioenergetics responses adopted by very similar strains of a single species.
2. Materials and methods
2.1. Strains and culture conditions
B. clausii strains O/C, N/R, SIN and Tetra have been obtained from Sanofi-Aventis. These strains are characterized by the resistance to the following antibiotics: novobiocin + rifampin (strain N/R), chloramphenicol (strain O/C), streptomycin + neomycin (strain SIN) and tetracycline (strain Tetra). Moreover, they were all resistant to erythromycin, cephalosproins and cycloserine, kanamycin, tobramycin, and amikacin [20]. The strains have been maintained on LB-agar plates. For the biochemical studies bacteria were inoculated in LB supplemented with 100 mM tricine, pH 7.9, at an OD600 of 0.01 from a stationary pre-culture. Growth was performed in 2 l flasks containing 500 ml of broth at 37 °C in an orbital shaker with radius of 5 cm at 150 rpm.
2.2. Oxygen consumption measurements
Cells were harvested by centrifugation at 3000 g for 5 min at 25 °C and re-suspended in LB–tricine to a final OD600 of 10. The endogenous respiratory activity was determined in an oxygraphic chamber containing 1 ml of LB–tricine at 25 °C, using 50 μl of the bacterial suspension. The cyanide sensitivity of the endogenous respiratory activity was measured in the range 0–10 mM [30]. Protein concentration was determined by the biuret method.
2.3. Membrane preparation
Cells were harvested by centrifugation at 5000 g for 10 min at 25 °C. The pellet was washed twice in a buffer containing 50 mM Tris–Cl, pH 7.4, 50 mM NaCl, 1 mM PMSF and 0.1 mM benzamidine. The pellet was stored at −80 °C until use. After thawing, the pellet from 500 ml of culture was suspended in 10 ml of washing buffer supplemented with 5 mg ml−1 of lysozyme. After 90 min at 37 °C with shacking, cells were broken by an osmotic shock obtained by adding to the sample 10 volumes of cold distilled water supplemented with 0.2 mM PMSF, 0.1 mM benzamidine and few grains of DNAse I. Shocked samples were incubated at 37 °C for 15 min and homogenized on ice. Samples were centrifuged at 40,000 g for 30 min at 4 °C. The pellet was suspended in a buffer containing 20 mM Tris–Cl pH 8.0, 1 mM EDTA, 0.2 mM PMSF and 0.01 mM benzamidine. The membrane extract was solubilized by adding 4 mg (mg of proteins)−1 of Triton X100. The solubilization was performed first at room temperature for 30 min and then overnight at 4 °C. The solubilized membranes were obtained by centrifugation at 100,000 g for 1 h at 4 °C.
2.4. Spectral analysis
For the determination of the different type of heme content, solubilized membranes were mixed 1 ÷ 1 (vol/vol) with an aqueous solution of pyridine (40% vol/vol) supplemented with 200 mM NaOH and 100 mM K3Fe(CN)6. Samples were reduced by an excess of sodium dithionite. The extinction coefficients were as in Ref. [2].
Membrane spectra were obtained by mixing the solubilized membrane extract with a buffer containing 20 mM Tris–Cl pH 8.0 and 0.01% (w/vol) Triton X100. Absolute spectra were recorded at room temperature (25 °C) against the buffer absorbance. The reduced membranes were obtained by addition of few grains of sodium dithionite; for the CO-reduced conditions, CO was carefully bubbled in the cuvette containing the reduced sample for 5 min. The CO-reduced spectra were stable for at least 1 h. Three different replicate experiments were performed for each strain and time-point. These experimentally collected spectra were then averaged and normalized to 1 (mg of solubilized proteins)−1 for the successive data analysis.
On the experimentally obtained spectra eigenvector decomposition was performed to in order to resolve the dataset’s spectral components. This has been done essentially as described in Refs. [23] and [4]. Briefly, average spectra were arranged in a matrix M whose columns represent the different strains at each considered time-point (12 columns) and each row represent a wavelength (from 400 to 650). After centroid subtraction, the correlation matrix C was obtained and eigenvalues and eigenvectors of C are calculated by solving the equation
with the usual convention λ1 ⩾ λ2 ⩾ λ3 ⩾ ··· ⩾ λ12. Eigenspectra (i.e. the fundamental component spectra) are obtained by calculating the matrix
where T means transposition. The cleaned dataset matrix was obtained using only the significant columns of Y and V (i.e. only the components representing the major sample variance) and the zero padding strategy.
2.5. DNA and RNA procedures
To obtain genomic DNA, 3 ml of culture was centrifuged at 13,000 g for 3 min. The pellets were resuspended in 1 ml of 10 mM Tris HCl, 10 mM EDTA, 100 mM NaCl, 2% (v/v) SDS, and 400 mg ml–1 proteinase K; the mixture was incubated at 56 °C for 30 min. After phenol and chloroform steps, DNA was precipitated and finally resuspended in sterile water.
PCR reactions were performed with PCR Master Mix (Promega, Southampton, UK) by using 50 ng of genomic DNA as a template. The PCR reactions with primers shown in Table 1 were performed under the following conditions for 30 cycles: 45 s at 95 °C, 45 s at 55 °C and 1 min at 72 °C. The amplified products were separated on 1% agarose gel, stained by ethidium bromide and purified from the gel using the PCR purification kit (Qiagen, West Sussex, UK). The nucleotide sequences of both strands of purified PCR fragments were determined by cycle sequencing on an automated ABI 310 sequencer using the Big Dye Terminator kit according to the manufacturer’s instructions (PE Applied Biosystems).
Table 1.
Oligonucleotide primers used in PCR and RT real-time PCR experiments.
| Primer name | Gene | Sequence (5′-3′) |
|---|---|---|
| PCR experiment | ||
| CydAF1 | cydA | TGCTGTTTGGCACGTCAATG |
| CydAR1 | cydA | GTGCGATTAAGAGCCATTT |
| FQUI | qoxD | TATGGGTAATGTCGCAAACC |
| RQUI | qoxD | GCAATGATAATGGCAAACCA |
| FCOX | coxC | CGGATGACCATTTGAGTAAGC |
| RCOX | coxC | ATGATAAACACCGTTGCCCA |
| RPOF1 | rpoB | CCGCATTTGGAGAAAAATGT |
| RPOR6 | rpoB | AAAAACGGAATACATGACGTCG |
| S12F3 | rpsL | AACCAGTTAATCCGCAAAGG |
| S12R4 | rpsL | GGTTTTGAACACCAGCGTG |
| Real-time PCR experiments | ||
| Cyd1A-F | cydA | TGCTGTTTGGCACGTCAATG |
| Cyd1A-R | cydA | CCAAGAAGGATGGCAAGCC |
| Cox-F | coxC | GCGGAGAGACCATCATGTTTG |
| Cox-R | coxC | ACGAGCGGCAGTGTGAAC |
| Qox-F | qoxD | TAGTAGCGTGAGGACAATTGAGAG |
| Qox-R | qoxC | GGCGTATATTTGCTAGGAATGGAG |
| RpoB-F | rpoB | ATGGTGGAGACGGAATCGTTC |
| RpoB-R | rpoB | AACGGCATATCTTCTTCTGGC |
| RpoD-F | rpoD | AGAGAAAATGGCTGCGTTTGAAC |
| RpoD-R | rpoD | TCTTCGTCTTTGGCTACTTGTGC |
For RT real-time PCR experiments, total RNA was isolated from exponential, early stationary and stationary phases of B. clausii growing cultures, using the RNeasy midi-kit (Qiagen). Residual genomic DNA was removed by DNase I digestion (Roche, Mannheim, Germany). Total RNA (1 μg) was reverse transcribed by using random hexamers (250 ng) with AMV reverse transcriptase-RNase H minus (New England Biolabs). Semi-quantitative analysis of the qoxD–qoxC, coxC and cydA transcripts, normalized to rpoB encoding RNA polymerase beta subunit and rpoD encoding RNA polymerase sigma factor, was performed by RT real-time PCR with the iQ SYBRgreen Supermix (BioRad) on a Biorad iCycler iQ instrument (Table 1 for primer sequences). About 10% of each RT reaction was used to run real-time PCR reactions with: primers Qox-F/Qox-R, specific for a region between qoxD and qoxC genes; Cox-F/Cox-R specific for the coxC gene; Cyd1A-F/Cyd1A-R specific for cydA gene; RpoB-F/RpoB-R specific for rpoB gene; RpoD-F/RpoD-R specific for rpoD gene. The real-time PCR conditions were: 20′′ at 94 °C, 30′′ at 59 °C, 45′′ at 72 °C for 45 cycles, followed by a melt curve cycle.
3. Results
3.1. Oxygen uptake measurements
For each strain endogenous respiration was measured at three different growth phases. The times analyzed were 4.5, 16.5 and 29 h, when the cultures were, respectively, in the early exponential, exponential and pre-steady state phases. In the culture conditions used the optical density (not shown), as well as the total protein yield (Fig. 1A) increased during the growth times explored.
Fig. 1.
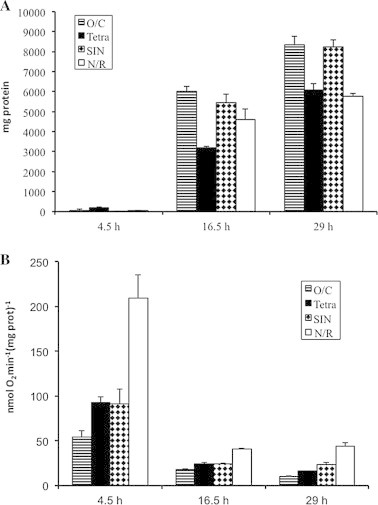
B. clausii strains Tetra, N/R SIN and O/C protein yield and endogenous respiratory activity. Panel A: B. clausii strains O/C, Tetra SIN and N/R protein yield. Bacteria were grown in batch as described under Section 2. Reported values are the averages from at least three independent experiments; error bars represent 1 SD. Panel B: B. clausii strains O/C, Tetra SIN and N/R endogenous respiration. The oxygen consumption activity has been determined as reported in Section 2. Oxygen consumption measurements have been performed on at least three independent cultures for each strain and for each time point; error bars represent 1 SD.
The endogenous respiratory activity measured as nmol O2 (mg of protein)−1 was maximal during the early exponential phase of growth and declined markedly during the subsequent phases (Fig. 1B). Beside this general trend, the four B. clausii strains presented different behaviors. In particular, the strain N/R exhibited a specific respiratory activity significantly higher with respect to the other three strains at all the time intervals explored. On the other hand, the strain O/C, showing the best performance with respect to growth yield, exhibited the lowest specific respiratory activity.
The cyanide sensitivity titration curves of the respiratory activity showed biphasic slopes in all the four strains, reaching almost complete inhibition at the highest inhibitor concentrations (see Figs. 1S–12S of Supplementary data). At 29 h of growth the residual activities of the endogenous respiration in the presence of 10 mM of KCN were around 14% for the O/C strain, 6% for the Tetra strain, 4% for the SIN strain and only 2% for the N/R strain.
Closer inspection of the titration curves of cyanide inhibition in the μM range for the N/R and O/C strains revealed opposite changes in the cyanide sensitivity in relation with the growth phases (Fig. 2). While the cyanide sensitivity for the N/R strain increased going from the initial to the late phase of the exponential growth, the O/C sensitivity to cyanide inhibition decreased markedly going from the initial to the late growth phase (see also Figs. 1S–12S). This behavior cannot be ascribed to the presence of bd-type oxidases. Such observed threshold effects in the μM KCN range can be ascribed to a functional redundancy of the terminal heme-copper oxidases, which has been previously reported in Bacillus subtilis [30].
Fig. 2.
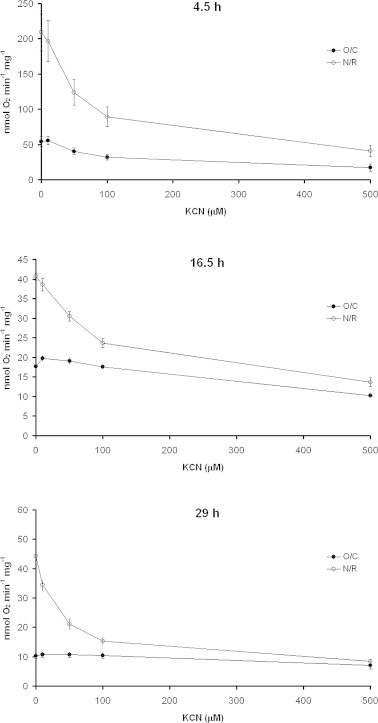
Cyanide sensitivity titration curves. The cyanide sensitivity titration curves for the strains N/R (open circles) and O/C (filled circles) are reported. Titrations were performed as described in Section 2 after 4.5 (upper panel), 16.5 (middle panel) and 29 (lower panel) hours of growth. Reported values are the averages of at least three independent experiments; error bars represent 1 SD. For the full set of the cyanide sensitivity titration curves see supplementary data.
3.2. Identification and transcription of terminal oxidases genes
The identification of the genes coding for subunits of the heme-Cu cytochrome c oxidase, the heme-Cu quinol oxidase and bd quinol oxidases was performed by PCR followed by sequencing of the amplified products. The B. clausii KSM-K16 (Accession number: AP006627) most conserved regions of the genes coxD and coxB (coding for cytochrome c oxidase subunits II and IV, respectively), qoxD and qoxA (coding for heme-Cu quinol oxidase subunits II and IV respectively) and cydA (coding for bd-type quinol oxidase subunit I) were taken as a reference to generate primers pairs (Table 1). Sequence analysis of the PCR products confirmed the presence of the same terminal oxidases genes annotated in B. clausii KSM-K16 genome in all the four B. clausii strains analyzed in this study (data not shown).
To estimate the transcript levels of both the identified heme-Cu terminal oxidases genes during the growth, RT real-time-PCR experiments were carried out. In the experiment shown in Fig. 3 transcript levels of the heme Cu-oxidases genes were evaluated after 4.5, 16.5 and 29 h of growth by using specific primers. Data were normalized with respect to the transcript levels of rpoB and rpoD.
Fig. 3.
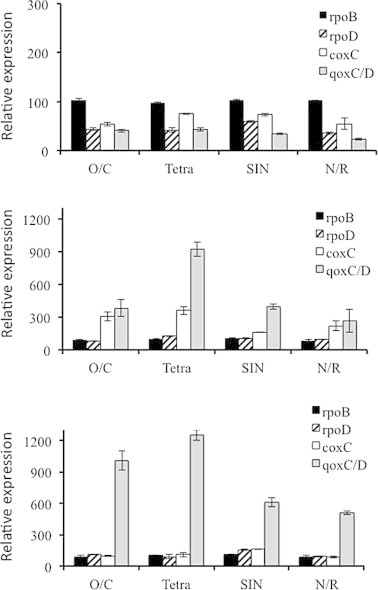
Relative expression analysis of the terminal respiratory enzymes. Relative expression analysis of qoxC/D and of coxC genes in each of the four B. clausii strains after 4.5 (upper panel), 16.5 (middle panel), and 29 (lower panel) hours of growth. The expression level of the transcripts was analyzed by real-time PCR. Normalization was performed with the rpoB transcript which was set as 100 in the first experiment. The efficiency of the rpoB as a reference gene was validated using rpoD as a second reference gene. The average values and standard deviations in three real-time PCR analyses are shown.
The general transcription trend during the three growth phases did not present macroscopic differences among the four strains, with heme Cu-oxidase genes mRNA levels peaking, respectively, during the exponential and pre-steady state phases. However, several differences were observed between the strains. In particular, coxC and qoxC/D mRNA levels were higher in O/C and Tetra with respect to SIN and N/R strains in pre-steady state and stationary phases respectively. qoxC/D mRNA levels were also higher in Tetra than in the other strains during pre-steady state. The higher expression levels of the quinol oxidase genes observed in O/C and Tetra during the stationary phase were consistent with the functional redundancy described above. Based on the mRNA levels, the heme-Cu quinol oxidase appears to be the main oxidase at 29 h of growth. Transcript levels of bd-type quinol oxidase were practically negligible (at least 50–100 times less with respect the rpoB transcript level) and were substantially unchanged during the three growth phases considered (not shown).
3.3. Difference spectroscopy
In order to quantify the amount of different types of hemes, difference pyridine-hemochrome spectra (dithionite reduced minus oxidized) were performed. These hemochrome extracts and their spectral analysis were necessary since the exact extinction coefficients of the various B. clausii cytochromes are currently unknown. The results of these determinations are reported in the Table 2. The total heme content (a + b + c) correlated well with the endogenous respiratory activity. The strain N/R, which displayed the highest respiratory activity in all tested growth phases, showed the highest total heme content, referred to total proteins.
Table 2.
Total heme content of the B. clausii strains. The total heme content (heme a + b + c) of the four B. clausii strains at 4.5 and 29 h of growth has been determined as described in Section 2. Reported values are in [(nmol of heme) (mg of total protein)−1]. Reported values are means of three experiments.
| Strain | Hours | Total heme types a + b + c | Heme type a | Heme type b | Heme type c |
|---|---|---|---|---|---|
| O/C | 4.5 | 22 | 6.0 | 10.4 | 5.6 |
| 29 | 15 | 4.4 | 6.9 | 3.7 | |
| Tetra | 4.5 | 26 | 7.6 | 11.7 | 6.7 |
| 29 | 9 | 2.1 | 3.3 | 3.6 | |
| SIN | 4.5 | 33 | 7.7 | 15.2 | 10.1 |
| 29 | 6 | 2.2 | 3.5 | 0.4 | |
| N/R | 4.5 | 61 | 13.9 | 29.8 | 17.3 |
| 29 | 21 | 6.2 | 11.1 | 3.7 |
The heme quantification assay showed differences in heme ratios during growth progression. A large difference between SIN and Tetra strains, which displayed an (heme b)/(heme c) ratio after 29 h of growth of 9.6 and 0.9 respectively was observed. In all strains, spectral evidence for d-type cytochromes (630 nm) was missing.
In order to gain further insight into the membrane cytochrome content, solubilized membrane spectra have been analyzed. The reduced minus oxidized spectra of membranes reveal the strain peculiar cytochrome pattern dynamics. Even if the overall spectral pattern dynamics could appear complex at a first sight, it should be noted that the entire dataset, i.e. the spectra of each strain at every time-point, can be obtained by linear combination of only three eigenspectra, which are reported in Fig. 4. These eigenspectra account for 98.9% of the overall variance of the experimental dataset. This means that the residual spectral components are absolutely unimportant from a quantitative point of view. As can be seen in Fig. 4, the first component, which contributes to roughly the 50% of the overall spectral variance, is composed by heme a-, b- and c-type containing cytochromes. The second component appears strikingly similar to the spectra of bc1-type cytochromes, plus trace amount of a-containing cytochromes, and the third component contain c- and a-type cytochromes plus a small deep in the b-type cytochromes. This last feature means that when the third component increases, we observe in the final spectra an increase in the c- and a- containing cytochromes together with a decrease of b-type cytochromes. It is important to stress out that no spectral evidence of d-type cytochromes can be observed (characterized by a peak and a deep centered respectively at 625 and 650 nm). This is true also for the not shown minor spectral components.
Fig. 4.
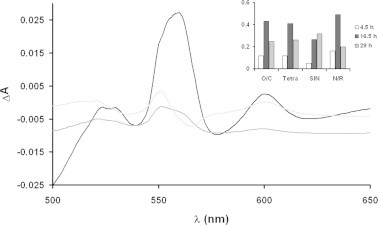
Spectral components in the reduced minus oxidized conditions. The α-band region of the eigenspectra for the reduced minus oxidized spectral dataset are reported. Reported spectra are the first (black), the second (gray) and third (light gray) components. The inset reports the first eigenvector entries (i.e. relative to the first spectral component, black line in the main plot) ordered by strain and culture time.
Spectra of the cleaned dataset are used to obtain, for each strain, time-difference spectra, which are reported in Fig. 5. These time-difference spectra permit to appreciate the dynamical changes of the cytochrome pattern. One important finding is that two strains behave rather identically from the point of view of cytochrome dynamics, namely O/C and Tetra. These two strains show an increase in heme c-, b- and a-containing cytochromes going from 4.5 to 16.5 h of growth, followed by a marked decrease in the heme c- and a-containing cytochromes in the 29 minus 16.5 h of growth difference spectra. N/R strain shows an increase in b- and a-containing cytochromes in the 16.5 minus 4.5 h difference spectra, followed by a marked decrease in c-, b- and a- containing cytochromes in the 29 minus 16.5 h spectra (this last pattern implies that already in the 4.5 h time point N/R membranes contain a large amount of cytochromes, consistent with the high respiratory activity of this strain). SIN strain instead shows an increase of all type of cytochromes in the time-difference spectra, and this suggests to consider this strain as a very peculiar strain with respect to all the other ones. This cytochrome pattern dynamics relate also with the first eigenvector entries (see inset in Fig. 4) which suggest a pattern (from the first to the second time-point and from the second to the third one) of increase–decrease of the first eigenspectrum component for all strains but SIN, for which an increase–increase pattern is observed.
Fig. 5.
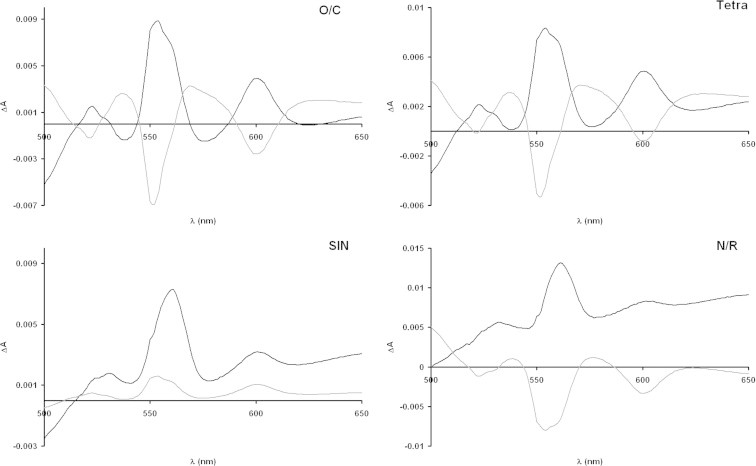
Time-difference spectra. The difference spectra at time 16.5 minus 4.5 h (black line) and 29 minus 16.5 h (gray line) for each strain are reported. Difference spectra are obtained using the reconstructed dataset as detailed in Section 2.
Beside the overall cytochrome pattern, it is interesting to analyze the CO-binding cytochromes, which are related to the oxygen-reactive cytochromes. This kind of analysis could be useful for the evaluation of the heme contained in the terminal aerobic oxidases. Membrane spectra in the form CO-reduced minus reduced have been determined for the entire dataset, as detailed above. Eigenspectra analysis of this dataset shows that, also in this case, the entire dataset can be described as a linear combination of only three components, which account for the 96.7% of the overall variance in the experimental data (Fig. 6). As it can be seen, there are no evidences for the presence of bd-type enzymes. These enzymes are characterized, in the CO-reduced minus reduced spectra, by a bathochromic shift of the heme d band with a maximum at 644 nm and a minimum at 624 nm and a W-shaped appearance in the Soret region with minima centered at 429 and 445 nm [3]. No evidence of this type of cytochrome can be observed in the eigenspectra set, including also the not shown small components. As reported in Fig. 6, the overall experimental dataset can be described by a (major) component with minima at 443 and 434 nm, and two separated components with minima at 444 nm (the second component) and at 430 nm (the third one). These findings rule out, from a spectral point of view, the possibility that significant amounts of bd-type oxidase are present in the four strains in the explored culture conditions. Our conclusion is that the heme-copper terminal oxidases of the B. clausii strains analyzed in this work are aa3 (minimum centered at 443–444 nm) or bo3 (minimum centered at 430–434 nm), even if the presence of ao3 containing enzymes (particularly in the O/C strain) cannot be excluded (not shown). This type of enzyme has been described in alkaliphilic bacilli [26,31,6]. The temporal evolution of these spectra shows that all strains, but SIN which shows only the presence of o-type cytochromes, start at 4.5 h with both type of enzymes (aa3 and bo3). At the last time-point two strains, namely O/C and Tetra, show only the presence of aa3 terminal oxidase, while SIN and N/R exhibit both o3- and a3-containing enzymes.
Fig. 6.
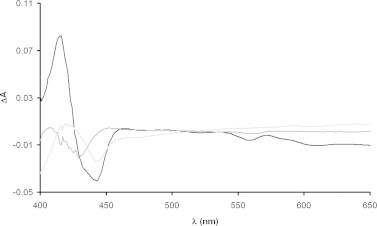
Spectral components in the CO-reduced minus reduced conditions. The visible region eigenspectra for the CO-reduced minus reduced spectral dataset are reported. Reported spectra are the first (black), the second (gray) and third (light gray) components.
4. Discussion
The respiratory function of four strains of the alkaliphilic B. clausii (N/R, SIN, Tetra, O/C) was characterized in detail, particularly with respect to the terminal oxidases of the respiratory chain. As expected, all the four strains showed a decrease in the specific respiratory activity during the later phases of growth due to the fact that in the later phase ATP and reducing power requirement for the biosynthetic pathways are lower. It can be noted that the strain specific oxygen uptake data correlated with the total heme (i.e. a + b + c) content. All strains started with a higher specific respiratory activity, and ended with a lower one; also the strain specific global heme content (see Table 2) behaved rather similarly (higher at 4.5 h and lower at the last time point). N/R strain exhibited the highest specific respiratory activity (see Fig. 1B), although it was not the strain with the highest protein yield (Fig. 1A). This suggests that in this strain the respiratory chain presents a low thermodynamic efficiency and/or more energy is dissipated, both in the oxidative phosphorylation system as well as in other metabolic pathways, with respect to the cognate strains. The opposite trend was observed with O/C exhibiting the highest protein yields but the lowest specific respiratory activity.
The four strains of B. clausii are characterized by the presence in their genome of genes coding for a heme-Cu cytochrome c oxidase, a heme-Cu quinol oxidase as well as a bd quinol oxidase. According to the real-time PCR data, both the heme-Cu cytochrome c oxidase as well as the quinol oxidase genes were both expressed during the early phase of growth. In the later growth phase (16.5 h) the expression of the quinol oxidase became predominant over that of the cytochrome c oxidase gene whose expression declined (Fig. 3). The heme-Cu quinol oxidase was also prevalent at 29 h of growth suggesting that the limited energy demand in the stationary phase results in a respiratory chain with lower thermodynamic efficiency. The bd quinol oxidase genes were expressed at negligible level in all of the four strains at least in the culture conditions used in this work. This finding is also substantiated by spectral analysis in the reduced minus oxidized conditions (Fig. 4) as well as by the study of CO-binding cytochromes (Fig. 6).
A detailed analysis of the respiratory chain shows that the O/C and Tetra strains have adopted very similar bioenergetics behaviors, which, on the other hand, are significantly different from those in SIN and N/R strains. This finding was supported by the heme content analysis (Table 2 and inset in Fig. 4) and the time-difference spectra (Fig. 5). This last kind of data showed an astonishing similar time-dependent cytochrome pattern in the strain O/C and Tetra. The presence of threshold effects clearly indicates that some strains, in particular phases of growth, have a functional redundancy of the terminal part of the respiratory chain. Furthermore, it might be considered that transcription of the heme-Cu oxidases genes in O/C and Tetra strains was consistently higher with respect to the other two strains, i.e. SIN and N/R. It is conceivable that higher expression level of the terminal oxidases in O/C and Tetra strains can be responsible for the threshold effects observed.
Interestingly, we have not observed high levels of cyanide insensitivity, in contrast to previous reports on other alkaliphilic strains [10,14]. The cyanide–insensitive oxygen uptake was higher in the O/C strain, but never above the 15% of the total. The other strains exhibited a still lower level of cyanide insensitivity (2–6% of the total). This residual oxygen consumption could be ascribed to various mechanisms, like ROS production. But this can also relate to different contents or mobilities of the quinones. In fact, in the non-alkaliphilic B. subtilis, stress-induced cyanide insensitivity has been related to profound changes in the lipid membrane composition [18]. However, in B. clausii, the presence of particular processes postulated for others alkaliphilic strains [10] can be excluded.
It is possible that these different bioenergetics behaviors may be the consequences of mutations in key genes, which mark the different strains and are involved in antibiotic resistance. Indeed chromosomal mutations conferring resistance to rifampin and streptomycin have been associated with functional alterations in RNA polymerase and ribosomal function respectively [22,12,16,13,5]. There is evidence that these mutations may lead to activation of (p)ppGpp-regulated bacterial responses and have significant effects on carbon, energy and secondary metabolism, and bacterial fitness [29,27,5,1].
Bacteria are characterized by extensive intra-species variation, which can be related to sequence differences, to the presence or absence of whole genes or clusters of genes [15], as well as to different expression behaviors in the assembly pathways [28,19]. The branched bacterial respiratory chains, modularly assembled by components with various thermodynamic efficiencies, determine a significant adaptive flexibility. Although a higher cytochrome c content, as observed for example for the Tetra strain, generally implies a greater proton per electron ratio, this could not be directly related to the bioenergetics efficiency of the strains. This because many factors contribute to determine the overall bioenergetic efficiency of the strains. Factors such as ATP synthase content and more generally membrane coupling efficiency should also be taken into account. These four strains represent an ideal object for studying the different bioenergetics behaviors adopted by almost identical genomes.
Acknowledgments
This work was supported as part of the Project MIUR-FAR DM23154 “Laboratorio pubblico-privato per lo sviluppo di processi e prodotti innovativi nel settore dei farmaci antinfettivi (Laboratorio interdisciplinare Farmaci Antinfettivi: LIFA)”.
A. Gaballo, L.L. Palese and S. Papa conceived and designed the project; A. Abbrescia, P.L. Martino, D. Panelli, A. Gaballo and L.L. Palese acquired the data; A. Gaballo, L.L.Palese, P. Alifano and A.M. Sardanelli analyzed and interpreted the data; L.L. Palese and A. Gaballo wrote the paper.
Contributor Information
L.L. Palese, Email: luigileonardo.palese@uniba.it.
A. Gaballo, Email: antonio.gaballo@nano.cnr.it.
Appendix A. Supplementary data
This supplementary material contains Figs. 1S–12S.
References
- 1.Alifano P. Fitness cost of antibiotic resistance. In: Gualerzi C.O., Brandi L., Fabbretti A., Pon C.L., editors. Antibiotics: Targets, Mechanisms and Resistance. Wiley-VCH Verlag GmbH & Co. KGaA; 2014. pp. 109–132. [Google Scholar]
- 2.Berry E.A., Trumpower B.L. Simultaneous determination of hemes a, b and c from pyridine hemochrome spectra. Anal. Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 3.Borisov V.B. Interaction of bd-type quinol oxidase from Escherichia coli and carbon monoxide: heme d binds CO with high affinity. Biochemistry (Moscow) 2008;73:14–22. doi: 10.1134/s0006297908010021. [DOI] [PubMed] [Google Scholar]
- 4.Bossis F., Palese L.L. Amyloid beta(1–42) in aqueous environments: effects of ionic strength and E22Q (Dutch) mutation. Biochim. Biophys. Acta. 2013;1834:2486–2493. doi: 10.1016/j.bbapap.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Carata E., Peano C., Tredici S.M., Ferrari F., Talà A. Phenotypes and gene expression profiles of Saccharopolyspora erythraea rifampicin-resistant (rif) mutants affected in erythromycin production. Microb. Cell Fact. 2009;8:e18. doi: 10.1186/1475-2859-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denda K., Oshima A., Fukumori H. Structural analyses of the deduced amino acid sequences of a novel type heme-copper terminal oxidase, cytochrome aco3, from alkalophilic Bacillus YN-2000. Can. J. Microbiol. 2001;47:1075–1081. [PubMed] [Google Scholar]
- 7.Gilmour R., Krulwich T.A. Purification and characterization of the succinate dehydrogenase complex and CO – reactive b – type cytochromes from the facultative alkaliphile Bacillus firmus OF4. Biochim. Biophys. Acta. 1996;1276:57–63. doi: 10.1016/0005-2728(96)00028-x. [DOI] [PubMed] [Google Scholar]
- 8.Gilmour R., Krulwich T.A. Construction and characterization of a mutant of alkaliphilic Bacillus firmus OF4 with a disrupted cta operon and purification of a novel cytochrome bd. J. Bacteriol. 1997;179:863–870. doi: 10.1128/jb.179.3.863-870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hicks D.B., Krulwich T.A. The respiratory chain of alkaliphilic bacilli. Biochim. Biophys. Acta. 1995;1229:303–314. doi: 10.1016/0005-2728(95)00024-d. [DOI] [PubMed] [Google Scholar]
- 10.Higashibata A., Fujiwara T., Fukumori Y. Studies on the respiratory system in alkaliphilic Bacillus: a proposed new respiratory mechanism. Extremophiles. 1998;2:83–92. doi: 10.1007/s007920050046. [DOI] [PubMed] [Google Scholar]
- 11.Janto B., Ahmed A., Ito M., Liu J., Hicks D.B. Genome of alkaliphilic Bacillus pseudofirmus OF4 reveals adaptations that support the ability to grow in an external pH range from 7.5 to 11.4. Environ. Microbiol. 2011;13:3289–3309. doi: 10.1111/j.1462-2920.2011.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin D.J., Gross C.A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 13.Korzheva N., Mustaev A., Kozlov M., Malhotra A., Nikiforov V. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 14.Krulwich T.A. Alkaliphilic prokaryotes. In: Dworkin M., Falkow S., Rosemberg E., Schleifer K.H., Stackebrandt E., editors. vol. 2. Springer; 2006. pp. 283–308. (The Prokaryotes). Third ed. [Google Scholar]
- 15.Lan R., Reeves P.R. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 2000;8:396–401. doi: 10.1016/s0966-842x(00)01791-1. [DOI] [PubMed] [Google Scholar]
- 16.Landick R., Stewart J., Lee D.N. Amino acid changes in conserved regions of the beta-subunit of Escherichia coli RNA polymerase alter transcription pausing and termination. Genes Dev. 1990;4:1623–1636. doi: 10.1101/gad.4.9.1623. [DOI] [PubMed] [Google Scholar]
- 17.Liu J., Krulwich T.A., Hicks D.B. Purification of two putative type II NADH dehydrogenases with different substrate specificities from alkaliphilic Bacillus pseudofirmus OF4. Biochim. Biophys. Acta. 2008;1777:453–461. doi: 10.1016/j.bbabio.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lobasso S., Palese L.L., Angelini R., Corcelli A. Relationship between cardiolipin metabolism and oxygen availability in Bacillus subtilis. FEBS Open Bio. 2013;3:151–155. doi: 10.1016/j.fob.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mavridou D.A., Ferguson S.J., Stevens J.M. Cytochrome c assembly. IUBMB Life. 2013;65:209–216. doi: 10.1002/iub.1123. [DOI] [PubMed] [Google Scholar]
- 20.Mazza P., Zani F., Martelli P. Studies on the antibiotic resistance of Bacillus subtilis strains used in oral bacteriotherapy. Boll. Chim. Farm. 1992;131:401–408. [PubMed] [Google Scholar]
- 21.Nielsen P., Fritze D., Priest F.G. Phenetic diversity of alkaliphilic Bacillus strains: proposal for nine new species. Microbiology. 1995;141:1745–1761. [Google Scholar]
- 22.Ovchinnikov Y.A., Monastyrskaya G.S., Guriev S.O., Kalinina N.F., Sverdlov E.D. RNA polymerase rifampicin resistance mutations in Escherichia coli: sequence changes and dominance. Mol. Gen. Genet. 1983;190:344–348. doi: 10.1007/BF00330662. [DOI] [PubMed] [Google Scholar]
- 23.Palese L.L. Protein dynamics: complex by itself. Complexity. 2013;18:48–56. [Google Scholar]
- 24.Quirk P.G., Hicks D.B., Krulwich T.A. Cloning of cta operon from alkaliphilic Bacillus firmus OF4 and characterization of the pH-regulated cytochrome caa3 oxidase it encodes. J. Biol. Chem. 1993;268:678–685. [PubMed] [Google Scholar]
- 25.Senesi S., Celandroni F., Tavanti A., Ghelardi E. Molecular characterization and identification of Bacillus clausii strains marketed for use in oral bacteriotherapy. Appl. Env. Microbiol. 2001;67:834–839. doi: 10.1128/AEM.67.2.834-839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sone N., Fujiwara Y. Effects of aeration during growth of Bacillus stearothermophilus on proton pumping activity and change of terminal oxidases. J. Biochem. 1991;110:1016–1021. doi: 10.1093/oxfordjournals.jbchem.a123671. [DOI] [PubMed] [Google Scholar]
- 27.Talà A., Wang G., Zemanova M., Okamoto S., Ochi K. Activation of dormant bacterial genes by Nonomuraea sp. strain ATCC 39727 mutant-type RNA polymerase. J. Bacteriol. 2009;191:805–814. doi: 10.1128/JB.01311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thony-Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microbiol. Mol. Biol. Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vigliotta G., Tredici S.M., Damiano F., Montinaro M.R., Pulimeno R. Natural merodiploidy involving duplicated rpoB alleles affects secondary metabolism in a producer actinomycete. Mol. Microbiol. 2005;55:396–412. doi: 10.1111/j.1365-2958.2004.04406.x. [DOI] [PubMed] [Google Scholar]
- 30.Villani G., Capitanio N., Bizzoca A., Palese L.L., Carlino V. Effects of site-directed mutagenesis of protolytic residues in subunit I of Bacillus subtilis aa3-600 quinol oxidase. Role of lysine 304 in proton translocation. Biochemistry. 1999;38:2287–2294. doi: 10.1021/bi980774g. [DOI] [PubMed] [Google Scholar]
- 31.Yumoto I., Takahashi S., Kitagawa T., Fukumori Y., Yamanaka T. The molecular features and catalytic activity of CuA – containing aco3 – type cytochrome c oxidase from a facultative alkalophilic Bacillus. J. Biochem. 1993;114:88–95. doi: 10.1093/oxfordjournals.jbchem.a124145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This supplementary material contains Figs. 1S–12S.


