Highlights
-
•
Discovery of a novel XIAP-inhibitory natural compound from Morus root bark (Sanggenon G).
-
•
Sanggenon G binds specific to the BIR3 domain of XIAP in a low μM range.
-
•
Sanggenon G interferes with XIAP-BIR3-substrate binding in living cells.
-
•
Sanggenon G acts as chemosensitizer in tumor cell lines with high XIAP expression.
Abbreviations: (FP-) assay, fluorescence polarization assay; ARPF-FAM, ARPF-K(5-Fam)-NH2-peptide; BIR-3, baculovirus-IAP-repeat-3; CC, column chromatography; Kd, dissociation constant; Ki, binding affinity; MAC, methanol crude extract of mulberry root bark; PCA, protein fragment complementation analysis; RLU, relative luminescence units; SG1, sanggenon G; XIAP, X-linked inhibitor of apoptosis protein
Keywords: XIAP inhibitor, Small-molecular weight, Cell permeable, Sanggenon G, Natural
Abstract
Defects in the regulation of apoptosis are one main cause of cancer development and may result from overexpression of anti-apoptotic proteins such as the X-linked inhibitor of apoptosis protein (XIAP). XIAP is frequently overexpressed in human leukemia and prostate and breast tumors. Inhibition of apoptosis by XIAP is mainly coordinated through direct binding to the initiator caspase-9 via its baculovirus-IAP-repeat-3 (BIR3) domain. XIAP inhibits caspases directly making it to an attractive target for anti-cancer therapy. In the search for novel, non-peptidic XIAP inhibitors in this study we focused on the chemical constituents of sāng bái pí (mulberry root bark). Most promising candidates of this plant were tested biochemically in vitro by a fluorescence polarization (FP) assay and in vivo via protein fragment complementation analysis (PCA). We identified the Diels Alder adduct Sanggenon G (SG1) as a novel, small-molecular weight inhibitor of XIAP. As shown by FP and PCA analyses, SG1 binds specifically to the BIR3 domain of XIAP with a binding affinity of 34.26 μM. Treatment of the transgenic leukemia cell line Molt3/XIAP with SG1 enhances caspase-8, -3 and -9 cleavage, displaces caspase-9 from XIAP as determined by immunoprecipitation experiments and sensitizes these cells to etoposide-induced apoptosis. SG1 not only sensitizes the XIAP-overexpressing leukemia cell line Molt3/XIAP to etoposide treatment but also different neuroblastoma cell lines endogenously expressing high XIAP levels. Taken together, Sanggenon G (SG1) is a novel, natural, non-peptidic, small-molecular inhibitor of XIAP that can serve as a starting point to develop a new class of improved XIAP inhibitors.
1. Introduction
Apoptosis or programmed cell death is a cellular process necessary for successful embryonic development, the maintenance of normal tissue homeostasis, the deletion of defective cells in the body and suppression of oncogenesis.
In mammals apoptosis can mainly be initiated by two pathways: the extrinsic pathway which is triggered by the ligation of death receptors on the cell surface and the intrinsic pathway which is activated by intracellular damages and cellular stress [1] and is regulated on the level of mitochondria by proteins of the BCL2 family [2,3]. Both pathways converge to a common execution phase of apoptosis that requires proteolytic activation of caspases-3 and/or -7 from their inactive zymogens [4,5]. Apoptosis execution is regulated by several major groups of molecules, inter alia by the inhibitor of apoptosis proteins (IAPs) which act as the key apoptosis regulators [6]. Therefore they are attractive molecular targets for designing entirely new classes of anticancer drugs aiming to overcome apoptosis resistance of cancer cells [7].
IAPs bind caspases and thereby interfere with apoptotic cell death signaling via death receptors or intrinsic cell death pathways. They were originally discovered in baculoviruses as suppressors of host cell apoptosis [8]. All IAP proteins share one to three common structures of baculovirus-IAP-repeat (BIR) -domains that allow them to bind and to inactivate caspases.
XIAP is the most potent inhibitor of apoptosis among the IAPs [9]. Inhibition of apoptosis by XIAP is mainly coordinated through direct binding to initiator caspase-9 via its BIR3-domain and by binding the effector caspases-3 and -7 [10]. Negative regulators of XIAP are SMAC/DIABLO and Omi, which are released from mitochondria in apoptotic cells, when the mitochondrial membrane begins to collapse. SMAC/DIABLO is the most effective XIAP inhibitor.
In several human malignancies an elevated expression of IAPs has been reported [11–14]. Tamm et al. investigated the expression of IAPs in 60 human tumor cell lines at mRNA and protein levels and found higher expression of XIAP in most cancer cell lines analyzed [15]. Increased XIAP levels have been reported for esophageal carcinoma, ovarian carcinoma, clear cell renal cancer and lymphoma [16–20]. In human prostate, non-small cell lung cancer cells and hepatocarcinoma apoptosis resistance correlates with the expression level of XIAP [21–24]. Several approaches to neutralize XIAP and to re-sensitize tumor cells to chemotherapy have been explored. In a first approach antisense oligonucleotides [25] and siRNAs [26–28], that are designed to decrease the mRNA and protein levels of XIAP, were used. Some of them are able to induce spontaneous apoptosis and to enhance chemotherapeutics-induced apoptosis in cancer cells [25,29]. The second and even more promising approach is to sensitize cancer cells to chemotherapeutic drugs by blocking XIAP’s anti-apoptotic activity by small peptidic compounds that bind into the BIR3 domain, so called SMAC-mimetics. These are usually small compounds derived from the oligopeptide sequence of the SMAC N-terminus that binds into XIAP. Most mimetics have a high affinity but due to their peptidic character they are also relatively instable and, as other peptide-based inhibitors, do not efficiently enter cells [30–32]. An alternative method is to identify small non-peptidic molecules e.g. from natural resources that mimic the SMAC interaction and can be used as effective and affordable drugs in anticancer therapy.
By using a fluorescence polarization (FP) -assay and based on empirical knowledge we focused on the herbal remedy sāng bái pí (mulberry root bark form Morus alba L.). This plant material is well known for its traditional use in Chinese medicine to treat hypertension, upper respiratory diseases and edema and to promote urination [33]. Mulberry flavonoids have been described to possess anticancer activity [34]. Until now an anticancer activity has only been reported for multi-component mixtures, e.g. aqueous Morus root bark extract induced apoptosis through inhibition of microtubule assembly [35]. Recently, Choi et al. [36] showed that inhibition of the YB-1 dependent MDR1 gene expression by the root extract decreased the viability of multidrug-resistant MCF-7/Dox cells. However there is scarce information available on chemically well-defined constituents from Morus root bark and their anticancer molecular mechanism. In this study the molecular and physiological modes of action of most promising constituents of this traditionally used herbal remedy have been assessed in silico, in vitro and in vivo revealing sanggenon G (SG1) as a non-peptidic, cell-permeable small inhibitor of XIAP.
2. Materials and methods
2.1. Plant material
The dried root bark of Morus sp. L., Moraceae, was purchased from Plantasia (Import und Vertrieb asiatischer Heilkräuter, Oberndorf, Austria). Its quality was macro- and microscopically checked according to the monograph Sang-Bái-Pi of the Chinese Pharmacopoeia. Voucher specimens (JR-20090112-A1) are deposited in the Herbarium of the Department of Pharmacognosy, Institute of Pharmacy, University of Innsbruck, Austria.
2.2. Extraction and isolation
The dried root bark (1950 g) was crushed to coarse powder and macerated exhaustively with methanol (7 l at room temperature, three times for 7 days). Upon removal of the solvent under vacuum, the methanol crude extract yielded 168.6 g (termed MAC). MAC was further defatted with petrol ether by means of liquid/liquid partitions with methanol–water (1:1) and petrol ether, and separated by silica gel column chromatography (CC) as reported previously [37,38]. Fraction 4 (MAC-4, 15.3 g) enriched in Diels–Alder adducts as monitored by thin layer chromatography (TLC) was further separated by using different chromatographic methods. The isolation procedure was performed as described previously [37,38] and afforded pure compounds SG1–SG7. Their structures have been determined by 1D and 2D NMR spectroscopy, mass spectrometry and optical rotation and compared to previously obtained data [37–39]. Purity of the isolates was determined by HPLC and 1H NMR and found to be >95%.
2.3. Cell lines, culture conditions and reagents
The acute lymphoblastic T-cell leukemia (T-ALL) lines Molt3 and Molt4, the rhabdomyosarcoma cell line OVCAR-4, the neuroblastoma cell lines SH-EP, IMR-32 and NxS2 as well as PhoenixTM packaging cells for helper-free production of amphotropic retroviruses [40] were cultured in RPMI1640 (Lonza, Basel, Switzerland) containing 10% fetal calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine (Gibco BRL, Paisley, UK) at 5% CO2 and 37 °C in saturated humidity. HTB-77 cells were cultured in McCoy’s 5a Medium Modified, supplemented with 10% fetal calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin (Gibco BRL, Paisley, UK). HEK293T cells for protein fragment complementation assays were cultured in DMEM medium (Gibco BRL, Paisley, UK). All cultures were tested routinely for mycoplasma contamination using the VenorR GeM-mycoplasma detection kit (Minerva Biolabs, Germany). For each experiment, mid-log-phase cultures were seeded in fresh medium. All cell lines used were authenticated by fingerprinting at the department of legal medicine of the Medical University of Innsbruck.
2.4. Expression vectors
The vector pHED-XIAP-iresPuro was used for stable ectopic gene expression and the plasmid pEF-XIAP-FLAG was used for transient transfections. Both vectors have been described elsewhere [41] and are a kind gift of John Silke from the Walter and Eliza Hall Institute of Medical Research, Australia.
2.5. Production of retroviruses for infection of leukemia cells
Retrovirus supernatants were produced as described before [42]. Briefly, 6x105 PhoenixTM packaging cells were transfected with 2 μg of retroviral vectors and 1 μg of a plasmid coding for VSV-G protein using Lipofectamine2000 (Invitrogen, USA). After 48 h the virus-containing supernatants were filtered through 0.22 μm syringe filters (Sartorius, Germany) and incubated with the target cells for at least 6 h. Molt3 cells were infected with either the empty pHED-iresPuro or the pHED-XIAP-iresPuro retrovirus to generate Molt3/Ctr or Molt3/XIAP cells, respectively. The cells were bulk-selected and the expression of XIAP was verified by immunoblotting.
2.6. Immunoblotting and immunoprecipitation
For immunoblot analyses total protein was prepared from 6x106 cells resuspended in lysis buffer (50 mM HEPES-NaOH, 1% Triton X-100, 150 mM NaCl, 2 mM EDTA, 10% glycerol) with protease and phosphatase inhibitors as described before [43,44]. Protein concentration was measured using Bradford-Reagent (BioRad Laboratories, Germany). Equal amounts of total protein (50 μg/lane) were separated by SDS–PAGE.
For immunoprecipitation, 40 μl of anti-FLAG M2 magnetic beads were added to 1.5 mg of total protein lysate containing a protease inhibitor cocktail. Then SG1 or solvent was added to the lysate and incubated for 3 h at room temperature. The tubes were placed in a magnetic separator to isolate FLAG-tagged protein-bead complexes. The beads were then washed three times in 1 ml wash buffer, resuspended in SDS sample buffer and finally loaded on SDS–PAGE. Equal amounts of total protein were present in the supernatants (as demonstrated by the housekeeping control GAPDH) and equal amounts of XIAP were precipitated as proved by FLAG-antibody staining. After gel separation proteins were transferred to nitrocellulose membranes (Schleicher & Schuell, Germany) by a semi-dry blotting device (Hoefer TE70, Amersham Biosciences). Membrane blocking was performed with PBS blocking buffer containing 0.1% Tween20 and 5% nonfat dry milk, incubated with primary antibodies specific for human XIAP and caspase-8 (BD Biosciences, Heidelberg, Germany), caspase-3 (Cell signaling, Danvers, MA, USA), and caspase-9 (R&D Systems, Germany), FLAG-M2 (Sigma–Aldrich, St. Louis, MO, USA), GAPDH (Acris Antibodies, Herford, Germany) and α-tubulin (Oncogene Research Products, La Jolla, CA), washed and incubated with anti-mouse or anti-rabbit horseradish-peroxidase-conjugated secondary antibodies (GE Healthcare, Vienna, Austria). The blots were developed by enhanced chemiluminescence (GE Healthcare, Vienna, Austria) according to the manufacturer’s instructions and analyzed with an AutoChemi detection system (UVP, Cambridge, UK).
2.7. Apoptosis detection/flow cytometry
Apoptosis was measured by staining the cells with propidium-iodide (PI) and forward/sideward scatter analysis using a Cytomics FC-500 (Beckman Coulter, Vienna, Austria). 2 × 105 cells were harvested and incubated in 500 μl hypotonic PI solution containing 0.1% Triton X-100 for 4–6 h at 4 °C. Stained nuclei in the sub-G1 marker window were considered to represent apoptotic cells [45]. For measurement of AnnexinV positive cells 5 × 105 cells were harvested and stained with FITC-labeled AnnexinV (Alexis Biochemicals, San Diego, CA, USA) in AnnexinV Binding Buffer [46,47].
2.8. Production and purification of recombinant BIR3 protein
DNA encoding BIR3 of human XIAP protein was amplified by PCR and cloned into the pET30a(+) vector (Merck Millipore, Germany) via the NdeI and SalI restriction sites. Protein expression was performed in Escherichia coli BL21(DE3)pLysS (Merck Millipore, Germany). E. coli carrying the pET30a(+)-XIAP-BIR3 plasmid were cultured overnight in 25 ml of LB-medium with 50 μg/ml kanamycin at 37 °C. After 12–15 h the pre-culture was added to 500 ml of warm LB-medium and was expanded until the optical density reached OD600 0.6–0.8. Protein production was induced by 1 M IPTG. For protein isolation the bacterial pellet was resuspended in lysis buffer containing 50 mM NaH2PO4, 300 mM NaCl and 2 mM imidazole supplemented with 0.1 mg/ml lysozyme, 1 mM PMSF and 1 tablet completeMini (Roche, Mannheim, Germany) prior to use. After sonication the suspension was filtered through a 0.22 μm filter and was used for protein purification on an Äkta FPLC system (GE Healthcare, Vienna, Austria). Purification was done in two steps. Step 1: Affinity chromatography with a HisTrap HP column (GE Healthcare). After equilibration with wash buffer (20 mM NaH2PO4, 500 mM NaCl and 2 mM imidazole) protein lysate was injected onto the column at a flow rate of 0.5 ml per minute. After reaching equilibrium the protein was eluted by applying a gradient of elution buffer (20 mM NaH2PO4, 500 mM NaCl and 500 mM imidazole) from 0–100% over 10 column volumes. The elution process was monitored at UV 280 nm. The protein was further purified in step 2 by size-exclusion chromatography on a Superdex 75 10/300 GL column (GE Healthcare) in buffer containing 20 mM Tris–HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 2 mM DTT, and 10% (w/v) glycerol [48]. Protein concentration was determined at 280 nm wavelength with a NanoDrop ND-1000 UV–Vis spectrophotometer (PeqLabBiotechnologie, Erlangen, Germany).
2.9. Fluorescence polarization (FP) -assay
To determine the binding of a substance to the BIR3-domain of XIAP in vitro we established a FP-assay. Measurements were carried out in black 96-well plates with flat bottom (HVD Life Sciences, Vienna, Austria) in a chameleon plate reader (Hidex, Turku, Finland). 2 μl of substance were added to 103 μl/well XIAP/ARPF Mix containing 20 nM ARPF-K(5-Fam)-NH2-peptide (termed ARPF-FAM) and 720 nM BIR3 protein in assay buffer (100 mM KH2PO4, pH 7.5; 100 μg/ml bovine serum albumin, 0.02% sodium azide). Positive (only assay buffer and FAM-peptide) and negative (BIR3 protein and FAM-peptide) controls were analyzed on each plate. Millipolarization values (mP) were measured at an excitation wavelength of 485 nm and an emission wavelength of 530 nm. Specificity of compound binding was verified using a second FP-assay that measures the interaction between recombinant 14–3–3 sigma protein and R18 peptide [49,50].
2.10. Determination of the equilibrium dissociation constant (Kd), IC50 and binding affinity (Ki)-values
The equilibrium dissociation constant was determined by using the FP-assay as described above. Increasing concentrations of BIR3 protein from 0.1 nM to 5 μM were used. The equilibrium binding isotherm was then calculated by plotting the FP read outs in mP values as a function of the BIR3 protein concentration. The inhibition constant was calculated by nonlinear curve fitting as the concentration of BIR3 protein at which 50% of the ligand is bound. Data analysis was done with Prism 4.0 software (Graphpad Software, San Diego, CA, USA).
The IC50 value of the substance SG1 was also determined by FP-analysis. Serial dilutions of the substance were added to the master mix and polarization values were measured after 3 h of incubation. FP readings were plotted as a function of the substance concentration and the IC50 values were determined by nonlinear least-square analysis. The Ki values of the substance SG1 was calculated using the online tool “The Ki calculator” (http://sw16.im.med.umich.edu/software/calc_ki/) based on the equation of Nikolovska-Coleska [51].
2.11. Protein fragment complementation assay (PCA)
The biochemical activity of the substances in vivo was determined by PCA that measures cell-permeability and binding kinetics of the substances in living cells. 6.5 × 105 HEK293T cells were transfected with 2 μg plasmid-DNA using jetPrime® (Polyplus-transfection SA, Illkirch, France) in the following combinations: pcDNA-BIR3-F1 and pcDNA-SMAC-F2 (1 μg each), pcDNA-Reg-F1 and pcDNA-Reg-F2 (1 μg each) and pRL-Renilla (2 μg) [52]. Cells were treated 24 h after transfection with the substances in sub toxic concentrations for 3 h. Samples were split in triplicates into a white 96-well flat bottom plate (HVD Life sciences, Vienna, Austria). Coelenteracine-h (Applichem, Darmstadt, Germany) was added in a final concentration of 0.25 ng/μl, and light emission was measured by the chameleon plate reader (Hidex, Turku, Finland).
2.12. Docking studies and structure–activity relationship analysis (SAR)
Protein and ligand preparations were carried out using the software tool MOE [53] and CORINA [54]. Docking studies were performed with GOLD [55] version 5.2 using default parameters (GOLDScore, 100% search efficiency). PDB entries 2OPY [56] was selected as protein templates for human XIAP. The active site was determined by selecting all residues within 8 Å from the outside atoms of the co-crystallized smac mimicking peptide. Docking was performed without any constraints and all compounds were minimized using LigandScout’s general purpose MMFF94 implementation after docking. In order to get the most probable binding poses, a 3D pharmacophore was developed with LigandScout, [57–59] using the docking poses of the most active ligands synthesized in this work. These 3D pharmacophores were then utilized to select the docking poses that formed the basis for the described analysis. LigandScout was also used for visualization, analysis and illustrations.
2.13. Statistical analysis
For statistical analysis of experimental data the students t-test was used in all cases except for the analysis of dose dependency and the sensitization of neuroblastoma cells to chemotherapy-induced apoptosis by SG1. In these cases an ANOVA with post hoc Bonferroni-analysis was used. A 2-tailed P value ⩽0.05 was considered as significant.
3. Results
3.1. In vitro screening for the identification of natural substances that specifically bind to the BIR3 domain of XIAP
For an automated medium throughput screening we established a FP-assay platform to monitor the binding of the fluorophore-labeled peptide ARPF-FAM to the protein BIR3. The ARPF peptide is derived from the AVPI peptide which is the main binding sequence of the natural XIAP antagonist SMAC/DIABLO to BIR3. The ARPF peptide shows, compared to AVPI a higher affinity to the XIAP-BIR3 domain which is achieved by replacement of the two amino acids valine and isoleucine to arginine and phenylalanine [60]. The binding of ARPF-FAM to BIR3 results in the decrease of peptide rotation and hence in a measurable increase in polarization upon excitation with polarized light. By binding of a compound with a higher affinity to BIR3 the amount of unbound highly rotating ARPF-FAM peptide increases resulting in a changed polarization plane of the emitted light and in a reduction of mP signal.
In the search for small molecular natural compounds able to interact with the BIR3 domain of XIAP, we screened in house available herbal extracts for the selection of promising starting material for phytochemical investigation. The most pronounced reduction of the mP value in the FP-screening was obtained for the methanol crude extract of mulberry root bark (MAC). This result together with data about the anticancer potential of this herbal remedy [33–35] prompted us to further investigate its constituents. Fraction MAC-4 obtained from column chromatography and enriched in Diels–Alder adducts pointed to those ingredients in the multi-component mixture most likely to be responsible for the selective BIR3 binding affinity (Fig. 1A). Compounds from MAC-4 have been isolated and identified as sanggenon G (SG1), sanggenon D (SG2), kuwanon L (SG3), sanggenon C (SG4), an equivalent mixture of the isobars moracin O and P (SG5), sanggenon B (SG6) and sanggenol A (SG7). Apart from SG5 and SG7, all of these isolates bear the characteristic feature of Diels–Alder adducts composed of a chalcon and a flavan moiety linked via a cyclohexene ring (Fig. 1B). The in vitro screening of SG1–7 identified sanggenon G (SG1) and kuwanon L (SG3) as those ingredients with the most pronounced reduction of the mP value in the FP-screening (Fig. 1A and C). Compounds SG1 and SG3 bind specifically to the BIR3 domain of XIAP which is demonstrated by the reduction of the mP signal to 40% of the control. Further SG1 did not affect protein–protein interaction in a second FP-assay system that measures the binding of an R18 peptide to recombinant 14–3–3 sigma protein. In addition also the unspecific competitor R18 does not quench the mP signal in the XIAP-BIR3 assay setup (Fig. 1C). To investigate the differences in in vitro binding to XIAP-BIR3 in greater depth we analyzed the protein-inhibitor interactions at the level of BIR3 domain by docking studies.
Fig. 1.
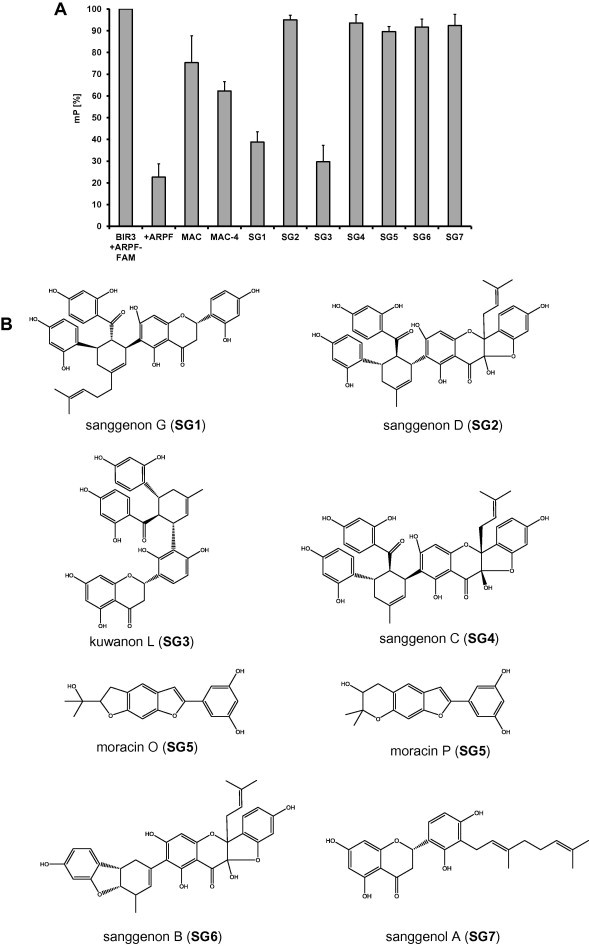
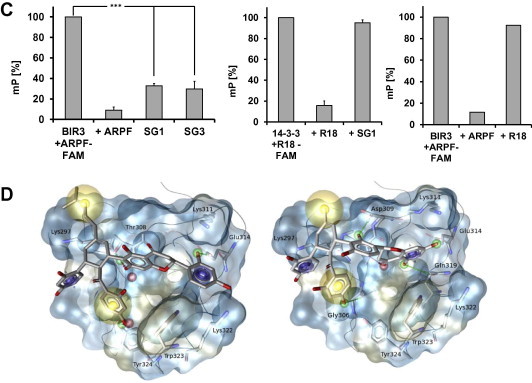
SG1 binds to the BIR3 domain of XIAP in vitro. (A) Screening of mulberry root bark extracts (MAC, MAC-4, SG1–SG7; 0.2 μg/μl) for binding to the ARPF domain of SMAC/DIABLO in vitro by FP-assay. MAC (methanol crude extract); MAC-4 (enriched extract). (B) Chemical structures of isolated constituents from mulberry root bark. (C) Proof of specificity of the BIR3-ARPF interaction by FP-assay. Specific binding of the natural compound SG1 and SG3 (200 μM) to the BIR3 domain of XIAP was analyzed by FP-assay (left panel). An unspecific competitor R18 (2 μM) does not displace the probe AbuRPF-K(5-Fam)-NH2 (FAM-ARPF, 20 nM) from BIR3 protein (720 nM). The specific competitor ARPF (2 μM) displaces the probe almost completely. Shown are means ± s.e.m. of three independent experiments. Statistical analysis was done using Student’s unpaired t-test, ∗∗∗P < 0.01 compared to corresponding control. (D) Proposed binding mode for compounds SG1 (A) and SG3 (B) in XIAP (PDB code 2OPY). Yellow spheres indicate lipophilic areas, red/green arrows hydrogen bonds, and cyan discs cation–π interactions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. In silico validation of natural substances that specifically bind to the BIR3 domain of XIAP
The crystal structure of XIAP reveals a BIR3 binding domain at the surface of the enzyme [56]. PDB [61] entry 2OPY [56] was selected as template and the protein structure was processed with CCDC’s software GOLD [55] for docking experiments. After docking, each docked conformation was geometrically optimized in the binding site and prioritized using a 3D pharmacophore model explaining the most plausible docking poses.
As a result, a comparable binding mode was identified for compounds SG1 and SG3 (Fig. 1D). The cyclohexene ring of both compounds was found in a central position, in the neighborhood of Thr308. From there, the ligand can orient its peripheral moieties in four directions towards different regions of the BIR3 binding domain. The dihydroxybenzoyl groups of both compounds SG1 and SG3 point towards Tyr324 (Fig. 1D, pointing downwards). In this region, the aromatic ring of each ligand is stabilized by a hydrophobic contact with the Trp323, and a hydrogen bond can be observed between the p-hydroxyl group of each ligand and either the head of Tyr324 or the backbone carbonyl of Gly306. The dihydroxyphenyl moiety of each ligand orients its hydroxyl group towards the solvent, while the aromatic ring is stabilized by a π–cation interaction with the positively charged head of Lys297 (Fig. 1D, pointing to the left side). The remaining lipophilic substitution of the central cyclohexene moiety forms hydrophobic contact with the side chain of Thr308 (Pointing upward in Fig. 1D). The forth moiety of ligands SG1 and SG3 (oriented to the right in Fig. 1D) combine a 5,7-dihydroxychroman-4-one covalently linked to a dihydroxyphenyl group. Although these moieties occur in reverse order in SG1 and SG3, similar interactions are found with the enzyme. In the first ring, an H-bond donor in the ligand interacts with the hydroxyl group of the Tyr 308 and/or the Asp309. In the second ring, an H-bond with the side chains of Glu 314 and can equally stabilize the elucidated binding mode. Additionally, a π–cation interaction could be identified between the second aromatic ring and Lys311 and Lys322. The reported interactions that were shared in both binding modes were compiled in a 3D-pharmacophore using LigandScout [57–59]. Interestingly, no docking poses generated for all other compounds investigated in this study could be matched to the above-described 3D model. This result is in agreement with the low inhibitory potency against XIAP experimentally determined for these molecules.
3.3. Compound SG1 but not SG3 interferes with binding of the SMAC/DIABLO-derived AVPI peptide to the XIAP-BIR3 domain in living cells
To measure binding dynamics of the SMAC/DIABLO-derived peptide AVPI to the XIAP-BIR3 domain and possible effects of BIR3-targeting compounds on this complex formation we set up an in vivo analysis based on the protein fragment complementation principle (Fig. 2A). For this assay the BIR3 domain of XIAP and the AVPI peptide of SMAC/DIABLO were fused each to a structural fragment of the Renilla-luciferase. If the fragments of the luciferase get in close proximity a functional luciferase is formed and light is emitted. This complex formation is reversible, which means that if a natural compound is able to compete off the AVPI-peptide the light emission is reduced. SG1 significantly reduced XIAP-BIR3/AVPI-SMAC complex formation at 2.9 μM (P < 0.05) to about 56%, thereby proofing that this substance is membrane permeable and in vivo active, SG3 failed to reduce the bioluminescence signal specifically and significantly (Fig. 2B). SG1 and SG3 neither interact with the regulatory subunits of the protein kinase A [52] nor with the Renilla protein demonstrating that the SG1-mediated reduction of the XIAP-BIR3/AVPI-SMAC complex formation is specific (Fig. 2B). The XIAP inhibitor embelin [7] was used as a control. Embelin diminishes the relative luminescence units (RLU) to 50% (Fig. 2C). Differences in the chemical structure between the two compounds SG1 and SG3 point to the different linkage between the flavan- and chalcon moiety (SG1, position 6 of the flavonoids moiety; SG3, position 3’), but also to the additional isoprenyl group in compound SG1 (at the cyclohexene ring). The latter difference distinctly confers to an increased hydrophobicity (Fig. 1B) which may increase the ability of this compound (in contrast to SG3) to transduce cytoplasm membranes.
Fig. 2.
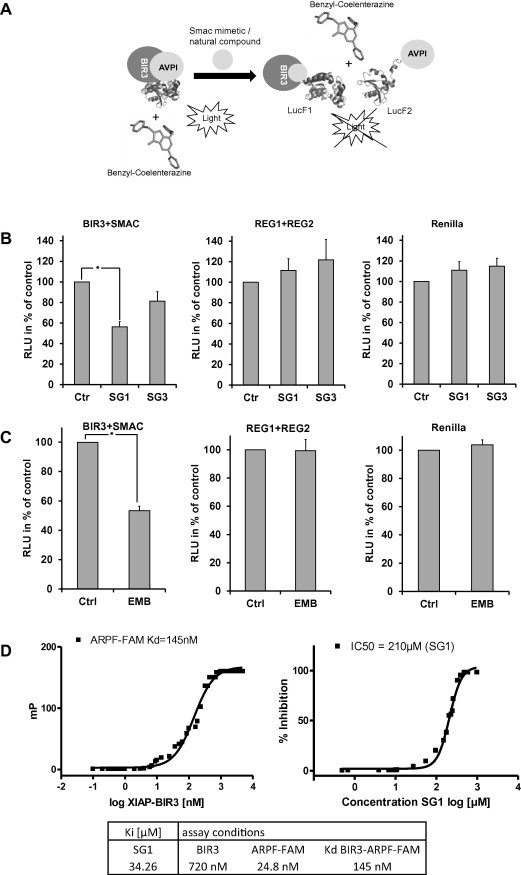
SG1 binds to the BIR3 domain of XIAP in vivo. (A) Schematic representation of the biosensor used in the protein fragment complementation assay. The BIR3 domain of XIAP and the AVPI peptide of SMAC/DIABLO are fused to structural fragments of the Renilla-luciferase. If the fragments of the luciferase get in close proximity a functional luciferase is formed and light is emitted under addition of the substrate. If a natural compound is able to compete off the AVPI peptide the light emission is reduced. (B) Biological activity of SG1in vivo is shown by PCA assay. SG1 displaces the AVPI-peptide from the BIR3 domain of XIAP in HEK293T cells thereby reducing luciferase activity. The dissociation of the BIR3-SMAC complex by addition of SG1 (2.9 μM) results in a reduction of the Renilla-luminescence of 44%.SG1 does neither reduce Renilla-luminescence of the control for unspecific protein interactions (REG1 + REG2) nor does it quench the Renilla-luminescence itself. Shown are means ± s.e.m. of three independent experiments. Statistical analysis was done with the Student’s unpaired t-test, ∗P < 0.05 compared to corresponding control. RLU (relative luminescence units). (C) Biological activity of the control substance embelin in vivo is shown by PCA. Embelin (3.4 μM) has a similar effect on XIAP-BIR3-LucF1/AVPI-LucF2-luciferase activity as SG1. Neither the Renilla-luminescence (right panel) nor the controls for unspecific protein interaction (middle panel) were affected by embelin. Shown are means ± s.e.m. of three independent experiments. Statistical analysis was done with the Student’s unpaired t-test, ∗P < 0.05 compared to corresponding control. (D) Biochemical characterization of SG1: Binding isotherm of AbuRPF-K(5-Fam)-NH2 to the XIAP-BIR3 domain. The probe (24.8 nM) was incubated with increasing concentrations of the recombinant XIAP-BIR3 domain (0–15 μM). The Ki-value of SG1 was calculated using the equation of Nikolovska-Coleska (51), based upon the measured IC50-value, the Kd-value of the probe and the XIAP-BIR3 complex, and the concentration of protein and probe in the competition assay (720 nM BIR3, 24.8 nM probe).
3.4. Biochemical characterization of SG1
As SG1 was able to inhibit the binding of the AVPI-peptide to XIAP-BIR3 in living cells, we next analyzed the binding properties of SG1 to XIAP-BIR3. To determine the IC50 for SG1 we first measured the dissociation constant (Kd) for the protein/ligand pair XIAP-BIR3/ARPF-FAM using a constant concentration of the probe and titrating with the XIAP-BIR3 domain protein at increasing concentrations significantly above the expected Kd of the protein-probe pair. Fig. 2D (left graph) shows nonlinear least-square fits to a single-site binding model for a saturation experiment in which the XIAP-BIR3 concentration varied from 0 to 15 μM at constant ARPF-FAM concentration. The obtained Kd-value of binding between the ARPF-FAM and the XIAP-BIR3 domain was determined to be 145 nM respective to our assay conditions. Of note Nikolovska-Coleska et al. published for the same ligand–protein pair using different assay conditions (differences in assay temperature and recombinant BIR3 protein) a Kd-value of 38 nM.
For the determination of the IC50-value of substance SG1 a competitive binding experiment with constant concentrations of BIR3 protein and FAM-labeled probe was used. SG1 was then titrated with increasing concentrations and the IC50 value was determined from the graph to be 210 μM using nonlinear least-square analysis (Fig. 2D, right graph).
The binding affinity (Ki) -value of SG1 was calculated using the equation of Nikolovska-Coleska [51] based upon the measured IC50-value, the Kd-value of the probe and the XIAP-BIR3 complex and the concentration of the protein and probe in the competition assay. The Ki-value for SG1 at our assay conditions was 34.26 μM (Fig. 2D). The Ki of embelin, at our conditions 22.3 μM, was reported to be 0.4 μM using a slightly different BIR3 protein and different measurement conditions [7].
3.5. SG1 treatment of Molt3/XIAP cells enhances caspase activation
In a next step we generated Molt3/XIAP leukemia cells that ectopically express human XIAP to generate a model system for XIAP-imposed drug resistance. For that purpose Molt3 cells were infected with a retrovirus for constitutive expression of XIAP. Overexpression of XIAP was verified by immunoblot (Fig. 3A). Molt3/XIAP cells are less sensitive to etoposide-induced apoptosis (100 ng/ml) compared to mock-infected Molt3/Ctr control cells as measured by propidium iodide (PI) FACS analyses (Fig. 3A). For caspase activation studies Molt3/Ctr and Molt3/XIAP cells were treated with etoposide (100 ng/ml) and then analyzed for caspase activation by immunoblot. Addition of SG1 (14.4 μM) in combination with etoposide enhances caspase-9 activation compared to the Molt3/Ctr and Molt3/XIAP cells treated with etoposide alone. Addition of the substance alone does not enhance caspase-9 activation (Fig. 3B). Treatment of Molt3/XIAP cells with SG1 and etoposide further leads to enhanced caspase-8 cleavage compared to Molt3/Ctr and Molt3/XIAP cells treated with etoposide alone and reaches nearly the same extend than in Molt3/Ctr cells treated with both substances (Fig. 3C). Enhanced caspase-3 cleavage can also be monitored in Molt3/XIAP cells that have been treated with SG1 and etoposide compared to the Molt3/XIAP cells treated only with etoposide. Nevertheless caspase-3 cleavage does not reach the same amount as in Molt3/Ctr cells (Fig. 3D).
Fig. 3.
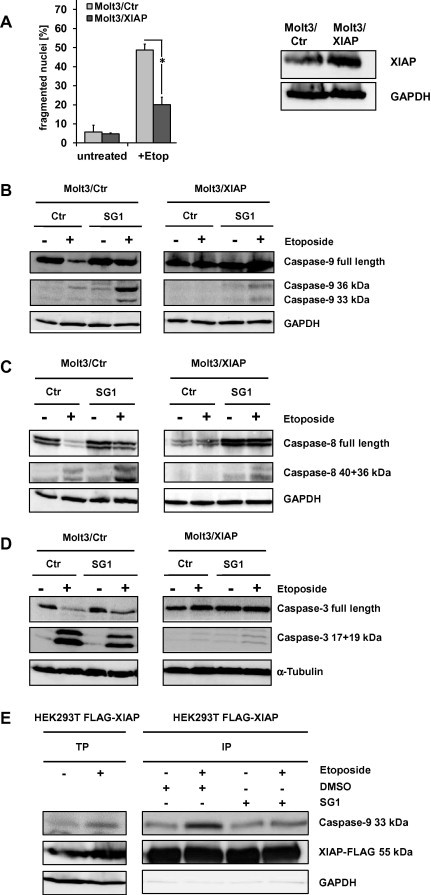
Treatment of Molt3/XIAP cells with SG1 enhances caspase activation. (A) Molt3/XIAP cells are more resistant to etoposide-induced apoptosis than Molt3/Ctr cells. Molt3/Ctr and Molt3/XIAP cells were treated with etoposide (100 ng/ml) for 72 h and then subjected to FACS analyses of PI-stained nuclei. Ectopic XIAP expression in Molt3/XIAP cells is shown by immunoblot. GAPDH is used as loading control. (B) Treatment of XIAP overexpressing cells with substance SG1 leads to enhanced caspase-9 activity. Molt3/Ctr and Molt3/XIAP cells were treated with SG1 (14.4 μM) alone and in combination with etoposide (130 nM) for 24 h. Caspase-9 activation was determined by immunoblot. GAPDH served as loading control. The immunoblot was performed three times (independent experiments). Shown is one exemplary figure. (C) Molt3/Ctr and Molt3/XIAP cells were incubated with 130 nM etoposide and 14.4 μM SG1 either alone or in combination for 24 h. Caspase-8 cleavage was then analyzed by immunoblot. GAPDH was used as loading control. The immunoblot was performed three times (independent experiments). Shown is one exemplary figure. (D) Caspase-3 cleavage was analyzed by immunoblot in Molt3/Ctr and Molt3/XIAP cells which were treated with SG1 (14.4 μM) alone and in combination with etoposide (130 nM) for 24 h. α-Tubulin was used as loading control. The immunoblot was performed three times (independent experiments). Shown is one exemplary figure. (E) HEK293T cells were transfected with FLAG-tagged XIAP and treated with etoposide (130 nM), 14.4 μM SG1 or DMSO for 18 h and subjected to co-immunopurification using anti-flag beads. Immunoprecipitates were analyzed for presence of caspase-9 by immunoblot. TP (total protein input), IP (immunoprecipitate), SN (supernatant). Areas containing the expected bands were cropped and assembled to the final figure. This applies for all immunoblots shown here.
3.6. Treatment of Molt3/XIAP cells with SG1 reactivates caspase-9 by releasing it from XIAP
In XIAP overexpressing cell lines apoptosis execution is often blocked by the XIAP-mediated inhibition of caspase-9. To test whether the apoptosis inducing potential of SG1 can be ascribed to the competition of caspase-9 for the caspase-9 binding site of XIAP, we performed a co-immunoprecipitation experiment. Therefore we prepared cell lysates from HEK293T cells transfected with an expression vector for FLAG-tagged XIAP, which were cultured in presence/absence of etoposide for 16 h to initialize a mitochondrial death signal. The lysates were split and incubated with either SG1 or solvent and to directly assess whether SG1 can displace caspase-9 from XIAP. As shown in Fig. 3E, an increased amount of caspase-9 can be co-precipitated in etoposide treated cells compared to controls. SG1 (14.4 μM) significantly reduces the amount of co-precipitated caspase-9. This indicates that SG1 is able to release caspase-9 from its antagonist XIAP and thereby SG1 sensitizes to apoptosis triggered via caspase-9.
3.7. SG1-treatment of XIAP-overexpressing Molt3 cells inhibits cell growth and enhances etoposide-induced apoptosis
To further investigate the physiological effects of SG1 we treated Molt3/XIAP cells with etoposide and 14.4 μM SG1 alone and in combination and analyzed cell growth over 96 h. As shown in Fig. 4A the combination treatment leads to a significant reduction of cell growth in Molt3/XIAP cells compared to the treatment with etoposide alone. In Molt3/Ctr cells no further growth reduction could be achieved due to their lower XIAP levels resulting in a decreased resistance to etoposide-induced apoptosis. SG1 alone leads to a lower reduction of cell growth in both cell lines. To determine the impact of SG1 on apoptosis induction we treated Molt3/Ctr and Molt3/XIAP cells with SG1 (14.4 μM) alone and in combination with etoposide (100 ng/ml) and subjected them to flow cytometric measurements of apoptotic cells either by AnnexinV staining or by analysis of propidium-iodide stained nuclei (Fig. 4B). Overexpression of XIAP significantly reduced cell death by the chemotherapeutic agent etoposide (P < 0.05). The compound SG1 per se only slightly increased the number of apoptotic cells, whereas SG1 significantly increased apoptosis in etoposide-treated, XIAP-overexpressing cells (P < 0.05). In contrast, no sensitizing effect can be observed for SG3 (Fig. S1). The sensitizing effect of SG1 to etoposide-induced apoptosis in Molt3/XIAP cells is dose-dependent, as the number of AnnexinV positive cells rises with increasing concentrations of SG1 (Fig. 4C).
Fig. 4.
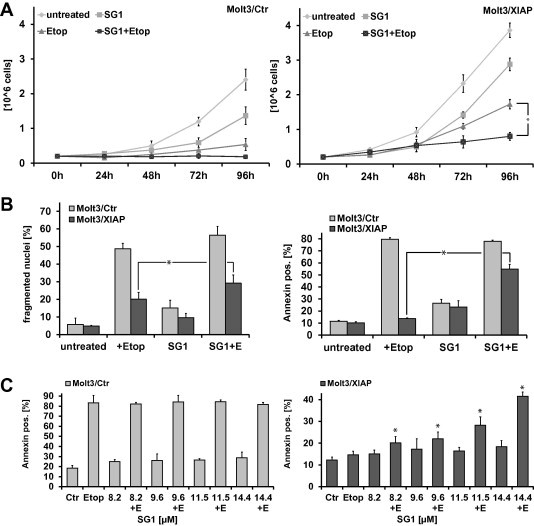
Treatment of Molt3/XIAP cells with SG1 inhibits cell growth and enhances etoposide-induced apoptosis in a dose-dependent manner. (A) The compound SG1 inhibits growth of the XIAP-overexpressing cell line Molt3/XIAP in combination with etoposide. Molt3/Ctr and Molt3/XIAP cells were incubated with SG1 (14.4 μM) alone and in combination with etoposide (130 nM) for the indicated time points. Cells were counted with a Casy cell counter (Roche Diagnostics, Mannheim, Germany). Shown are means ± s.e.m. of three independent experiments. Statistical analysis was done with the Student’s unpaired t-test, ∗P < 0.05 compared to corresponding controls. (B) The compound SG1 sensitizes Molt3/XIAP cells to etoposide-induced apoptosis. Molt3/Ctr and Molt3/XIAP cells were treated with SG1 (14.4 μM) alone and in combination with etoposide (130 nM) for 72 h and were then subjected to FACS analyses of PI-stained nuclei. For analysis of apoptotic cells by AnnexinV staining, cells were incubated with 130 nM etoposide and 14.4 μM SG1, either alone or in combination, for 24 h and then analyzed for AnnexinV positive cells. Shown are means ± s.e.m. of three independent experiments. Statistical analysis was done with the Student’s unpaired t-test, ∗P < 0.05 compared to corresponding control. (C) For dose–response curves Molt3/Ctr and Molt3/XIAP cells were treated with increasing concentrations of SG1 (8.2–14.4 μM) alone and in combination with 130 nM etoposide for 24 h. Apoptotic cells were analyzed by AnnexinV staining. Shown are means ± s.e.m. of three independent experiments.
3.8. Compound SG1 does not induce cIAP1 degradation
To test whether SG1 interferes also with the function of other IAPs such as cIAP1, we treated the cIAP1-inhibitor sensitive cell lines OVCAR-4 and HTB-77 with SG1 and used as a positive control embelin. The cell lines OVCAR-4 and HTB-77 undergo spontaneous apoptosis when these cells are treated with an inhibitor specific for cIAP1 [62]. Inhibitor treatment should also cause rapid depletion of cIAP1 protein levels [62]. As shown in Fig. 5A treatment of HTB-77 cells with 6.8 μM SG1 and OVCAR-4 cells with 17 μM SG1 does not elevate apoptosis. In a next step we compared the effect of SG1 on cIAP1 and cIAP2 protein expression with the smac mimetics LCL-161 (Novartis) and TL32711 (Birinapant, Tetralogic) (Fig. 5B). The same results are achieved with the XIAP-inhibitor embelin (Fig. S2). This suggests that SG1, like embelin, neither inhibits cIAP1 / cIAP2 function nor induces the degradation of these IAPs, which supports the notion that SG1 predominantly targets XIAP and neutralizes its function in malignant cells.
Fig. 5.
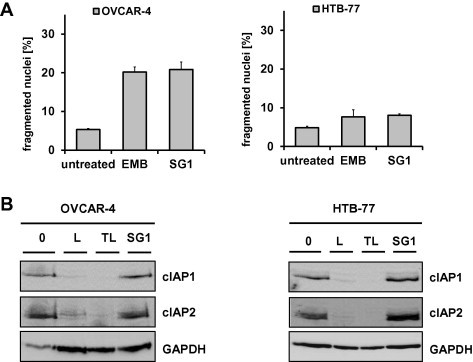
Proof of specificity of SG1 for XIAP in the cIAP1-inhibitor sensitive cell lines OVCAR-4 and HTB-77. (A) The cIAP1-inhibitor sensitive OVCAR-4 and HTB-77 cells were incubated with SG1 (8.5 μM) for 48 h and then subjected to FACS analysis of PI-stained nuclei. Shown are means ± s.e.m. of three independent experiments. Embelin, a known specific XIAP inhibitor, served as control. (B) OVCAR-4 and HTB-77 cells were treated with SG1 (8.5 μM) for 4 h and then subjected to immunoblot analysis of cIAP1 protein levels. The smac mimetics LCL-161 (L) and TL32711 (TL) which both neutralize cIAP1/2 were used as a controls for cIAP1 degradation. GAPDH served as loading control.
3.9. Sensitization of neuroblastoma cells to chemotherapy-induced apoptosis by SG1
To determine whether SG1 is able to sensitize different neuroblastoma cell lines with naturally high XIAP levels (Fig. S3) to etoposide-induced apoptosis we treated SH-EP, IMR-32 and NxS2 cells with 10 μM SG1 and with different concentrations of etoposide (250 μM, 750 μM, 1270 μM). In all three cell lines treatment with SG1 induces apoptosis as measured by PI-FACS-analyses (Fig. 6A–C). IMR-32 cells are only partially sensitized by SG1, whereas SH-EP and NxS2 cell lines were sensitized to cell death by SG1 in combination with all doses of etoposide. The murine neuroblastoma cell line NxS2 was most sensitive to SG1-treatment (P < 0.01). The chemosensitizing effect of SG1 in these cell lines can be explained by their elevated XIAP-expression compared to the leukemia cell line Molt4 which has significant lower XIAP levels (Fig. S3) and does not show increased apoptosis levels after treatment with SG1 (data not shown).
Fig. 6.
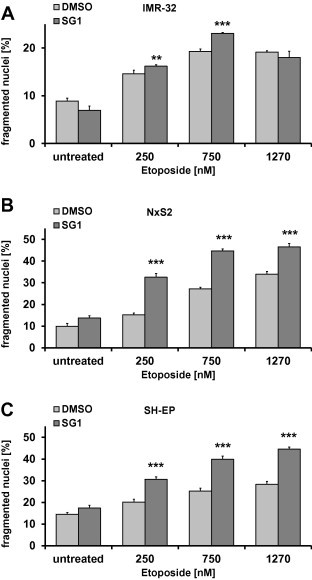
Induction of apoptosis by SG1 in neuroblastoma cell lines. (A–C) IMR-32, NxS2 and SH-EP cells were treated with 10 μM SG1 and 250, 750 and 1270 nM etoposide. Apoptosis was determined by flow cytometry of PI stained nuclei after 48 h. Shown are means ± s.e.m. of three independent experiments. For statistical analysis ANOVA with post hoc Bonferroni-analysis was used. A 2-tailed P value ⩽0.05 was considered as significant.
4. Discussion
Much effort has been invested into the development of XIAP-inhibitors due to the fact that XIAP is a central inhibitor of inducer- and executor-caspases and is overexpressed in many different types of tumors [15–20]. This has led to the idea that XIAP is a promising therapeutic target for personalized cancer medicine. Most inhibitors, some of them are already in preclinical phase I and phase II trials (TL32711, Tetralogic Pharmaceuticals; LCL161, Novartis; AEG35156, Aegra Therapeutics) [29], are peptidic structures derived from the processed N-terminus of the cellular XIAP-inhibitor SMAC/DIABLO that is released from mitochondria upon cell death induction.
In this study we identified the natural substances sanggenon G (SG1) and kuwanon L (SG3) as novel, non-peptidic, small-molecular weight inhibitors of XIAP. The compound SG1 and the structurally-related compound SG3 were identified as components of an extract from Morus alba root bark (MAC) in a FP-based screening platform for extracts and their chemical constituents that bind the proto-oncogene XIAP (Fig. 1A). As shown by FP-analyses and docking studies SG1 binds specifically to the BIR3 domain of XIAP in vitro (Fig. 1C). The binding affinity of this natural compound to XIAP was determined to be 34.26 μM at our assay conditions. This value is in the upper range of active substances but has to be seen in the context of the XIAP-inhibitor embelin that was used as a reference compound. For embelin the Ki-value was reported to be 0.4 μM [7]. In our experimental setting we determined the Ki-value of embelin to be 22.25 μM. These differences to other assay systems are most likely due to different experimental conditions, mainly influenced by temperature, the concentration of the FAM-labeled ARPF-peptide and recombinant XIAP-BIR3 protein as well as differences in the sequence and position of the His-tag on the BIR3-protein which is located C-terminal in our case. Whereas Nikolovska-Coleska et al. expressed a recombinant, N-terminal His-tagged XIAP-BIR3 consisting of the sequence Gly241-His356 [51], the protein used in our experiments was slightly larger (Phe238-Glu359), which may also affect binding properties. These differences are also reflected by the Kd-value between BIR3 protein and the ARPF-FAM-peptide that we measured. In our setting the Kd-value was 145 nM. In the literature this value varies between 20 nM, [60] 38 nM, [51] and 77.6 nM [51]. To classify the XIAP-inhibiting potency of SG1, we referenced the measured Ki-value to the Ki-value of the control substance embelin, which suggests that SG1 binds with a similar affinity to XIAP-BIR3 as embelin. Importantly, PCA-analyses (Fig. 2B) that directly assess protein–protein interaction in living cells revealed a significant reduction of the bioluminescence signal to 56% by as low as 2.9 μM SG1. This significantly increased in vivo effect might reflect the lower affinity of the AVPI-peptide to XIAP-BIR3 and is possibly due to effective import into cells or accumulation in the cytoplasm. It clearly demonstrates that the in vivo efficacy of SG1 to dissociate XIAP-BIR3/AVPI complexes is much higher than the in vitro measured binding efficiency. The effective concentration of SG1 in this assay is the same as for embelin (3.4 μM) suggesting that SG1 has a similar biological activity.
By use of the PCA assay we further showed that SG1 binds to XIAP-BIR3 protein in vivo and it competes for the same binding site as SMAC/DIABLO in living cells. This in vivo binding is a crucial feature for a potential use of SG1 as a drug candidate because it already demonstrates that the compound efficiently enters the cytoplasm of the cell and directly interferes with XIAP function. This is a main advantage of SG1 compared to other compounds especially the peptide-based ones that are limited in cell permeability and therefore have to be coupled to carrier peptides [63].
In contrast to SG1, the compound SG3 did not show a significant effect in the PCA-sensor (Fig. 2B) and also did not act as a chemosensitizer in XIAP-overexpressing cells (Fig. S1), suggesting that despite the structural similarity and in vitro activity, compound SG3 does not enter cells efficiently. This is most probably due to the missing isoprenyl group in contrast to SG1, thus losing the mandatory hydrophobicity. Intriguingly, further Diels–Alder adducts isolated from this herbal remedy, namely compounds SG2, SG4, and SG6 did not show an activity in our in vitro assay. Non Diels–Alder adducts, i.e. moracin O and P (SG5) belonging to the chemical class of benzofurans and sanggenol A (SG7), a simple flavanone with a geranyl moiety in position 3’, neither contribute to the bioactivity of MAC. These results are in line with the in silico docking studies revealing that no docking poses exist that match the 3D-pharmacophore model. Although the chemical diversity within the investigated Diels–Alder adducts and their sample number do not allow an in depth structure activity relationship, it is obvious that the inactive members of this compound class share a common chemical feature, namely an additional dihydrofuran by cyclization in position 2, 3, 2’ of the flavonoid scaffold forming a rigid condensed four ring system (Fig. 1B). Accordingly, we hypothesize that the structural features of the flavano-chalcon moieties which are linked via a cyclohexene-ring system by Diels–Alder addition need a maintained flexibility of the phenyl group (ring B) in the flavonoid scaffold to act as ligands of the XIAP-BIR3 domain. This is only given in compounds SG1 and SG3. Further, a sufficient hydrophobicity as given by the additional isoprenyl group in compound SG1 seems to be mandatory for cell permeability.
A second evidence for the high potency of SG1 is that treatment of the XIAP-overexpressing cell line Molt3/XIAP with 14.4 μM SG1 synergizes with etoposide to inhibit cell growth and to induce apoptosis as demonstrated by PI-FACS and AnnexinV staining (Fig. 4A-B). This dose dependent effect in a range from 8.2 to 14.4 μM (Fig. 4C) suggests a similar biological activity as the broadly used XIAP inhibitor LBW242 (10 μM) [11,64] which is already in clinical trial phase I [29] and was used in a number of different experimental studies such as to enhance vincristine-induced apoptosis in NxS2 murine neuroblastoma cells [11].
This sensitizing effect was observed not only in the XIAP-overexpressing leukemia cell line Molt3/XIAP but also in different neuroblastoma cell lines endogenously expressing high XIAP levels (Fig. S3). Treatment of the neuroblastoma cell lines SH-EP, IMR-32 and NxS2 with SG1 results in a significant induction of apoptosis as measured by PI-FACS-analyses, which is very promising for future therapeutical use of SG1 (Fig. 6A–C). The combined data suggest that SG1 overcomes the death-protective effect of XIAP and restores sensitivity of cells to etoposide-induced apoptosis.
To further investigate the effects of SG1 in detail we analyzed activation of caspase-3, -8 and -9 (Fig. 3B–D). The inhibitory effect of XIAP overexpression on the activation of caspase-3 and caspase-8 in etoposide treated cells was clearly demonstrated by a significantly reduced presence of cleavage products in the immunoblot (Fig. 3B–D). This suggests that caspase-8 and caspase-3 cleavage occurs downstream of outer mitochondrial membrane permeabilization and caspase-9 activation in these cells and is also inhibited by XIAP-overexpression. Caspase-3 also activates caspase-9 in an amplifying feedback loop [65]. As XIAP binds both, caspase-9 and caspase-3 via its BIR3 and BIR2 domains, respectively, high XIAP expression interferes also with this processing of caspase-9 by caspase-3 as it was observed in the immunoblot analysis. XIAP associates with the active, proteolytically processed caspase-9/APAF1 holoenzyme [10,65] and prevents it from activating downstream caspases like caspase-3 and -7. In Molt3/Ctr cells etoposide treatment mainly triggers the mitochondrial death pathway resulting in caspase-9 activation and a subsequent increase of partly-processed caspase-9 that in turn is sequestered by elevated XIAP-levels in Molt3/XIAP cells. Therefore, increased amounts of processed caspase-9 in complex with XIAP can be detected during etoposide treatment as it is also demonstrated in Fig 3E. The observation that SG1 did not reduce basal caspase-9/XIAP-interaction might be explained by the fact that we adjusted SG1 concentrations to a level that per se only partially reduces long term survival (Fig4 A) and only slightly increases apoptotic cell death (Fig 4B).
Neutralization of XIAP might also facilitate cell death induced by other death triggers such as death ligands as previously demonstrated for TRAIL-induced cell death in leukemia cells [30]. Apoptosis sensitization by SG1 partially compensated XIAP-mediated inhibition, which was also reflected by increased caspase activation: treatment with SG1 leads to enhanced caspase activation of caspases-9, -8 and -3 in Molt3/XIAP cells. Additionally, SG1 treatment releases caspase-9 from inhibition by XIAP by competing with caspase-9 for the same binding site on XIAP. As shown by co-immunoprecipitation, treatment with SG1 displaces caspase-9 from XIAP. Taken together these results indicate that SG1 acts as a chemosensitizer in drug-resistant XIAP-overexpressing cancer cells.
To further elucidate the specificity of SG1 we tested whether it would induce apoptosis spontaneously in the cell lines HTB-77 and OVCAR-4. These cells have been shown to undergo apoptosis after addition of a cIAP1 inhibitor [62]. However, treatment with SG1 does not lead to spontaneous apoptosis induction and no spontaneous degradation of cIAP1 was detected by immunoblotting contrasting to the smac mimetics LCL-161 and TL32711 (Fig. 5A + B). We conclude from these results that SG1 specifically targets XIAP.
The combined data indicate that sanggenon G (SG1) is a promising new XIAP inhibitor of natural origin that acts as a chemosensitizer in XIAP-expressing cancer cells by neutralizing the pro-survival function of XIAP via binding to its BIR3 domain. The chemical structure of SG1 might serve as a starting point to develop a new class of improved, non-peptidic XIAP inhibitors that may be used in combination therapy to reduce doses of chemotherapeutic drugs and therapy side effects.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
The authors thank Gerhard Gaedicke for support and valuable discussion, John Silke for donating plasmids and Soeren Heiderstadt for technical assistance in re-isolation of natural compounds. This work was supported by grants by the COMET Center ONCOTYROL, which is funded by the Austrian Federal Ministries BMVIT/BMWFJ (via FFG) and the TirolerZukunftsstiftung/Standortagentur Tirol, by the ”Kinderkrebshilfe Tirol und Vorarlberg”, the “Kinderkrebshilfe Südtirol-Regenbogen”, the “Krebshilfe Südtirol”, the “SVP-Frauen”, the “Provita Kinderleukämie Stiftung” and the “MFF-Tirol”. The Tyrolean Cancer Research Institute and this study are supported by the “Tiroler Landeskrankenanstalten Ges.m.b.H. (TILAK)” and the Tyrolean Cancer Society.
Contributor Information
Judith M. Rollinger, Email: Judith.Rollinger@uibk.ac.at.
Petra Obexer, Email: Petra.Obexer@i-med.ac.at.
Michael J. Ausserlechner, Email: Michael.J.Ausserlechner@i-med.ac.at.
Appendix A. Supplementary data
Supplementary materials.
Supplementary materials.
References
- 1.Fulda S., Debatin K.M. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006;25(34):4798–4811. doi: 10.1038/sj.onc.1209608. [DOI] [PubMed] [Google Scholar]
- 2.Andersen J.L., Kornbluth S. The tangled circuitry of metabolism and apoptosis. Mol. Cell. 2013;49(3):399–410. doi: 10.1016/j.molcel.2012.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartman M.L., Czyz M. Anti-apoptotic proteins on guard of melanoma cell survival. Cancer Lett. 2013;331(1):24–34. doi: 10.1016/j.canlet.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Bratton S.B., Salvesen G.S. Regulation of the Apaf-1-caspase-9 apoptosome. J. Cell Sci. 2010;123(Pt 19):3209–3214. doi: 10.1242/jcs.073643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghavami S., Hashemi M., Ande S.R., Yeganeh B., Xiao W., Eshraghi M. Apoptosis and cancer: mutations within caspase genes. J. Med. Genet. 2009;46(8):497–510. doi: 10.1136/jmg.2009.066944. [DOI] [PubMed] [Google Scholar]
- 6.Strasser A., O’Connor L., Dixit V.M. Apoptosis signaling. Annu. Rev. Biochem. 2000;69:217–245. doi: 10.1146/annurev.biochem.69.1.217. [DOI] [PubMed] [Google Scholar]
- 7.Nikolovska-Coleska Z., Xu L., Hu Z., Tomita Y., Li P., Roller P.P. Discovery of embelin as a cell-permeable, small-molecular weight inhibitor of XIAP through structure-based computational screening of a traditional herbal medicine three-dimensional structure database. J. Med. Chem. 2004;47(10):2430–2440. doi: 10.1021/jm030420+. [DOI] [PubMed] [Google Scholar]
- 8.Roulston A., Marcellus R.C., Branton P.E. Viruses and apoptosis. Annu. Rev. Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- 9.Salvesen G.S., Duckett C.S. IAP proteins: blocking the road to death’s door. Nat. Rev. Mol. Cell Biol. 2002;3(6):401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 10.Eckelman B.P., Salvesen G.S., Scott F.L. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7(10):988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eschenburg G., Eggert A., Schramm A., Lode H.N., Hundsdoerfer P. Smac mimetic LBW242 sensitizes XIAP-overexpressing neuroblastoma cells for TNF-alpha-independent apoptosis. Cancer Res. 2012;72(10):2645–2656. doi: 10.1158/0008-5472.CAN-11-4072. [DOI] [PubMed] [Google Scholar]
- 12.Chen X., Wang T., Yang D., Wang J., Li X., He Z. Expression of the IAP protein family acts cooperatively to predict prognosis in human bladder cancer patients. Oncol. Lett. 2013;5(4):1278–1284. doi: 10.3892/ol.2013.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saleem M., Qadir M.I., Perveen N., Ahmad B., Saleem U., Irshad T., et al. (2013). Inhibitors of apoptotic proteins: new targets for anti-cancer therapy. Chem. Biol. Drug Des. [DOI] [PubMed]
- 14.Li F., Yin X., Luo X., Li H.Y., Su X., Wang X.Y. Livin promotes progression of breast cancer through induction of epithelial–mesenchymal transition and activation of AKT signaling. Cell. Signal. 2013;25(6):1413–1422. doi: 10.1016/j.cellsig.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Tamm I., Kornblau S.M., Segall H., Krajewski S., Welsh K., Kitada S. Expression and prognostic significance of IAP-family genes in human cancers and myeloid leukemias. Clin. Cancer Res. 2000;6(5):1796–1803. [PubMed] [Google Scholar]
- 16.Zhang S., Ding F., Luo A., Chen A., Yu Z., Ren S. XIAP is highly expressed in esophageal cancer and its downregulation by RNAi sensitizes esophageal carcinoma cell lines to chemotherapeutics. Cancer Biol. Ther. 2007;6(6):973–980. doi: 10.4161/cbt.6.6.4195. [DOI] [PubMed] [Google Scholar]
- 17.Ramp U., Krieg T., Caliskan E., Mahotka C., Ebert T., Willers R. XIAP expression is an independent prognostic marker in clear-cell renal carcinomas. Hum. Pathol. 2004;35(8):1022–1028. doi: 10.1016/j.humpath.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Akyurek N., Ren Y., Rassidakis G.Z., Schlette E.J., Medeiros L.J. Expression of inhibitor of apoptosis proteins in B-cell non-Hodgkin and Hodgkin lymphomas. Cancer. 2006;107(8):1844–1851. doi: 10.1002/cncr.22219. [DOI] [PubMed] [Google Scholar]
- 19.Mansouri A., Zhang Q., Ridgway L.D., Tian L., Claret F.X. Cisplatin resistance in an ovarian carcinoma is associated with a defect in programmed cell death control through XIAP regulation. Oncol. Res. 2003;13(6–10):399–404. doi: 10.3727/096504003108748410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizutani Y., Nakanishi H., Li Y.N., Matsubara H., Yamamoto K., Sato N. Overexpression of XIAP expression in renal cell carcinoma predicts a worse prognosis. Int. J. Oncol. 2007;30(4):919–925. [PubMed] [Google Scholar]
- 21.Berezovskaya O., Schimmer A.D., Glinskii A.B., Pinilla C., Hoffman R.M., Reed J.C. Increased expression of apoptosis inhibitor protein XIAP contributes to anoikis resistance of circulating human prostate cancer metastasis precursor cells. Cancer Res. 2005;65(6):2378–2386. doi: 10.1158/0008-5472.CAN-04-2649. [DOI] [PubMed] [Google Scholar]
- 22.Nomura T., Yamasaki M., Nomura Y., Mimata H. Expression of the inhibitors of apoptosis proteins in cisplatin-resistant prostate cancer cells. Oncol. Rep. 2005;14(4):993–997. [PubMed] [Google Scholar]
- 23.Shi Y.H., Ding W.X., Zhou J., He J.Y., Xu Y., Gambotto A.A. Expression of X-linked inhibitor-of-apoptosis protein in hepatocellular carcinoma promotes metastasis and tumor recurrence. Hepatology. 2008;48(2):497–507. doi: 10.1002/hep.22393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golovine K., Makhov P., Uzzo R.G., Kutikov A., Kaplan D.J., Fox E. Cadmium down-regulates expression of XIAP at the post-transcriptional level in prostate cancer cells through an NF-kappaB-independent, proteasome-mediated mechanism. Mol. Cancer. 2010;9:183. doi: 10.1186/1476-4598-9-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y., Cherton-Horvat G., Dragowska V., Baird S., Korneluk R.G., Durkin J.P. Antisense oligonucleotides targeting XIAP induce apoptosis and enhance chemotherapeutic activity against human lung cancer cells in vitro and in vivo. Clin. Cancer Res. 2003;9(7):2826–2836. [PubMed] [Google Scholar]
- 26.Hehlgans S., Petraki C., Reichert S., Cordes N., Rodel C., Rodel F. (2013). Double targeting of survivin and XIAP radiosensitizes 3D grown human colorectal tumor cells and decreases migration. Radiother. Oncol. [DOI] [PubMed]
- 27.Kunze D., Kraemer K., Erdmann K., Froehner M., Wirth M.P., Fuessel S. Simultaneous siRNA-mediated knockdown of antiapoptotic BCL2, Bcl-xL, XIAP and survivin in bladder cancer cells. Int. J. Oncol. 2012;41(4):1271–1277. doi: 10.3892/ijo.2012.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao L.P., Song J.L., Yi X.P., Li Y.X. Double inhibition of NF-kappaB and XIAP via RNAi enhances the sensitivity of pancreatic cancer cells to gemcitabine. Oncol. Rep. 2013;29(4):1659–1665. doi: 10.3892/or.2013.2246. [DOI] [PubMed] [Google Scholar]
- 29.Fulda S., Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat. Rev. Drug Discov. 2012;11(2):109–124. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- 30.Fulda S., Wick W., Weller M., Debatin K.M. Smac agonists sensitize for Apo2L/TRAIL- or anticancer drug-induced apoptosis and induce regression of malignant glioma in vivo. Nat. Med. 2002;8(8):808–815. doi: 10.1038/nm735. [DOI] [PubMed] [Google Scholar]
- 31.Arnt C.R., Chiorean M.V., Heldebrant M.P., Gores G.J., Kaufmann S.H. Synthetic Smac/DIABLO peptides enhance the effects of chemotherapeutic agents by binding XIAP and cIAP1 in situ. J. Biol. Chem. 2002;277(46):44236–44243. doi: 10.1074/jbc.M207578200. [DOI] [PubMed] [Google Scholar]
- 32.Yang L., Mashima T., Sato S., Mochizuki M., Sakamoto H., Yamori T. Predominant suppression of apoptosome by inhibitor of apoptosis protein in non-small cell lung cancer H460 cells: therapeutic effect of a novel polyarginine-conjugated Smac peptide. Cancer Res. 2003;63(4):831–837. [PubMed] [Google Scholar]
- 33.Bensky D., Clavey S., Stöger E. Chinese Herbal Medicine: Materia Medica. 3rd ed. Seattle, WA: Eastland Press; (2004).
- 34.Nomura T., Fukai T., Hano Y., Yoshizawa S., Suganuma M., Fujiki H. Chemistry and anti-tumor promoting activity of Morus flavonoids. Prog. Clin. Biol. Res. 1988;280:267–281. [PubMed] [Google Scholar]
- 35.Nam S.Y., Yi H.K., Lee J.C., Kim J.C., Song C.H., Park J.W. Cortex mori extract induces cancer cell apoptosis through inhibition of microtubule assembly. Arch. Pharmacal. Res. 2002;25(2):191–196. doi: 10.1007/BF02976562. [DOI] [PubMed] [Google Scholar]
- 36.Choi Y.K., Cho S.G., Choi H.S., Woo S.M., Yun Y.J., Shin Y.C. JNK1/2 activation by an extract from the roots of Morus alba L. reduces the viability of multidrug-resistant MCF-7/Dox cells by inhibiting YB-1-dependent MDR1 expression. Evidence-based Complementary Alternative Med. 2013;2013:741985. doi: 10.1155/2013/741985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rollinger J.M., Bodensieck A., Seger C., Ellmerer E.P., Bauer R., Langer T. Discovering COX-inhibiting constituents of Morus root bark: activity-guided versus computer-aided methods. Planta Med. 2005;71(5):399–405. doi: 10.1055/s-2005-864132. [DOI] [PubMed] [Google Scholar]
- 38.Rollinger J.M., Spitaler R., Menz M., Marschall K., Zelger R., Ellmerer E.P. Venturia inaequalis-inhibiting Diels-Alder adducts from Morus root bark. J. Agric. Food Chem. 2006;54(22):8432–8436. doi: 10.1021/jf061871g. [DOI] [PubMed] [Google Scholar]
- 39.Fukai T., Pei Y.-H., Nomura T., Xu C.-Q., Wu L.-J., Chen Y.-J. Isoprenylated flavanones from Morus cathayana. Phytochemistry. 1998;47(2):273–280. [Google Scholar]
- 40.Grignani F., Kinsella T., Mencarelli A., Valtieri M., Riganelli D., Grignani F. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 1998;58(1):14–19. [PubMed] [Google Scholar]
- 41.Silke J., Hawkins C.J., Ekert P.G., Chew J., Day C.L., Pakusch M. The anti-apoptotic activity of XIAP is retained upon mutation of both the caspase 3- and caspase 9-interacting sites. J. Cell Biol. 2002;157(1):115–124. doi: 10.1083/jcb.200108085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geiger K., Hagenbuchner J., Rupp M., Fiegl H., Sergi C., Meister B. FOXO3/FKHRL1 is activated by 5-aza-2-deoxycytidine and induces silenced caspase-8 in neuroblastoma. Mol. Biol. Cell. 2012;23(11):2226–2234. doi: 10.1091/mbc.E11-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obexer P., Hagenbuchner J., Rupp M., Salvador C., Holzner M., Deutsch M. P16INK4A sensitizes human leukemia cells to FAS- and glucocorticoid-induced apoptosis via induction of BBC3/Puma and repression of MCL1 and BCL2. J. Biol. Chem. 2009;284(45):30933–30940. doi: 10.1074/jbc.M109.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagenbuchner J., Kuznetsov A., Hermann M., Hausott B., Obexer P., Ausserlechner M.J. FOXO3-induced reactive oxygen species are regulated by BCL2L11 (Bim) and SESN3. J. Cell Sci. 2012;125(Pt 5):1191–1203. doi: 10.1242/jcs.092098. [DOI] [PubMed] [Google Scholar]
- 45.Hagenbuchner J., Ausserlechner M.J., Porto V., David R., Meister B., Bodner M. The anti-apoptotic protein BCL2L1/Bcl-xL is neutralized by pro-apoptotic PMAIP1/Noxa in neuroblastoma, thereby determining bortezomib sensitivity independent of prosurvival MCL1 expression. J. Biol. Chem. 2010;285(10):6904–6912. doi: 10.1074/jbc.M109.038331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ausserlechner M.J., Salvador C., Deutschmann A., Bodner M., Viola G., Bortolozzi R., et al. (2013). Therapy-resistant acute lymphoblastic leukemia (ALL) cells inactivate FOXO3 to escape apoptosis induction by TRAIL and Noxa. Oncotarget. [DOI] [PMC free article] [PubMed]
- 47.Hagenbuchner J., Kuznetsov A.V., Obexer P., Ausserlechner M.J. BIRC5/Survivin enhances aerobic glycolysis and drug resistance by altered regulation of the mitochondrial fusion/fission machinery. Oncogene. 2013;32(40):4748–4757. doi: 10.1038/onc.2012.500. [DOI] [PubMed] [Google Scholar]
- 48.Veisova D., Rezabkova L., Stepanek M., Novotna P., Herman P., Vecer J. The C-terminal segment of yeast BMH proteins exhibits different structure compared to other 14-3-3 protein isoforms. Biochemistry. 2010;49(18):3853–3861. doi: 10.1021/bi100273k. [DOI] [PubMed] [Google Scholar]
- 49.Du Y., Khuri F.R., Fu H. A homogenous luminescent proximity assay for 14–3–3 interactions with both phosphorylated and nonphosphorylated client peptides. Curr. Chem. Genom. 2008;2:40–47. doi: 10.2174/1875397300802010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du Y., Masters S.C., Khuri F.R., Fu H. Monitoring 14–3–3 protein interactions with a homogeneous fluorescence polarization assay. J. Biomol. Screen. 2006;11(3):269–276. doi: 10.1177/1087057105284862. [DOI] [PubMed] [Google Scholar]
- 51.Nikolovska-Coleska Z., Wang R., Fang X., Pan H., Tomita Y., Li P. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal. Biochem. 2004;332(2):261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 52.Stefan E., Aquin S., Berger N., Landry C.R., Nyfeler B., Bouvier M. Quantification of dynamic protein complexes using Renilla luciferase fragment complementation applied to protein kinase A activities in vivo. Proc. Natl. Acad. Sci. U.S.A. 2007;104(43):16916–16921. doi: 10.1073/pnas.0704257104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pomerantz J.L., Baltimore D. Two pathways to NF-kappaB. Mol. Cell. 2002;10(4):693–695. doi: 10.1016/s1097-2765(02)00697-4. [DOI] [PubMed] [Google Scholar]
- 54.Sadowski J., Gasteiger J., Klebe G. Comparison of automatic three-dimensional model builders using 639 X-ray structures. J. Chem. Inf. Comput. Sci. 1994;34(4):1000–1008. [Google Scholar]
- 55.Jones G., Willett P., Glen R.C., Leach A.R., Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267(3):727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 56.Wist A.D., Gu L., Riedl S.J., Shi Y., McLendon G.L. Structure-activity based study of the Smac-binding pocket within the BIR3 domain of XIAP. Bioorg. Med. Chem. 2007;15(8):2935–2943. doi: 10.1016/j.bmc.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 57.Wolber G., Dornhofer A., Langer T. Efficient overlay of small organic molecules using 3D pharmacophores. J. Comput. Aided Mol. Des. 2006;20(12):773–788. doi: 10.1007/s10822-006-9078-7. [DOI] [PubMed] [Google Scholar]
- 58.Wolber G., Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2004;45(1):160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 59.Wolber G., Seidel T., Bendix F., Langer T. Molecule-pharmacophore superpositioning and pattern matching in computational drug design. Drug Discov. Today. 2008;13(1–2):23–29. doi: 10.1016/j.drudis.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 60.Kipp R.A., Case M.A., Wist A.D., Cresson C.M., Carrell M., Griner E. Molecular targeting of inhibitor of apoptosis proteins based on small molecule mimics of natural binding partners. Biochemistry. 2002;41(23):7344–7349. doi: 10.1021/bi0121454. [DOI] [PubMed] [Google Scholar]
- 61.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vince J.E., Wong W.W., Khan N., Feltham R., Chau D., Ahmed A.U. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131(4):682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 63.Sun H., Nikolovska-Coleska Z., Yang C.Y., Qian D., Lu J., Qiu S. Design of small-molecule peptidic and nonpeptidic Smac mimetics. Acc. Chem. Res. 2008;41(10):1264–1277. doi: 10.1021/ar8000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petrucci E., Pasquini L., Bernabei M., Saulle E., Biffoni M., Accarpio F. A small molecule SMAC mimic LBW242 potentiates TRAIL- and anticancer drug-mediated cell death of ovarian cancer cells. PLoS One. 2012;7(4):e35073. doi: 10.1371/journal.pone.0035073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Srinivasula S.M., Hegde R., Saleh A., Datta P., Shiozaki E., Chai J. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature. 2001;410(6824):112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials.
Supplementary materials.


