Highlights
-
•
We generated a floxed allele by using paired Cas9n (nickase), gRNAs and single DNA template in mouse.
-
•
We confirmed that the floxed allele was germline transmitted and functional in F1 offspring.
-
•
A floxed allele of the isoprenoid synthase containing domain (Ispd) gene in C57BL/6N background mice was created.
-
•
This method can be used to generate knockout mice for genes that are potentially embryonic lethal.
Abbreviations: Cas, CRISPR-associated protein; Cas9n, Cas9 nickase; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeat; DSBs, double stand breaks; ES, embryonic stem cells; HDR, homology directed repair; indel, insertion and deletion mutations; lspd, isoprenoid synthase containing domain; sgRNA, single guide RNA; TALENs, transcription activator-like effector nucleases; ZFNs, zinc finger nucleases
Keywords: Cas9 nickase, Conditional allele, Mouse, Gene targeting, Ispd
Abstract
CRISPR/Cas9 technology is a highly promising genome editing tool in the mouse, potentially overcoming the costs and time required for more traditional gene targeting methods in embryonic stem (ES) cells. Recently, compared to the wildtype nuclease, paired Cas9 nickase (Cas9n) combined with single guide RNA (sgRNA) molecules has been found to enhance the specificity of genome editing while reducing off-target effects. Paired Cas9n has been shown to be as efficient as Cas9 for generating insertion and deletion (indel) mutations by non-homologous end joining and targeted deletion in the genome. However, an efficient and reliable approach to the insertion of loxP sites flanking critical exon(s) to create a conditional allele of a target gene remains an elusive goal. In this study, we microinjected Cas9n RNA with sgRNAs together with a single DNA template encoding two loxP sites flanking (floxing) exon 2 of the isoprenoid synthase containing domain (Ispd) into the pronucleus and cytoplasm of C57BL/6NCr one-cell stage zygotes. After surgical transfer, one F0 mouse expressing a conditional allele was produced (at a frequency of ∼8% of live pups born). The floxed allele was transmitted through the germline to F1 progeny, and could be successfully recombined using Cre recombinase. This study indicates that conditional targeting can be accomplished effectively using paired Cas9n and a single DNA template.
1. Introduction
Gene knockout mice have traditionally been derived by gene targeting in mouse embryonic stem (ES) cells [1,2]. Although highly effective, multiple interdependent methods and procedures are required, including design and construction of a molecular targeting vector, electroporation, culture, and selection of ES cells, screening identification, and expansion of homologously recombined ES cell clones, blastocyst microinjection of gene targeted ES cells, and production and breeding of chimeras to confirm germline transmission. All these steps are commonly used for routine generation of genome-modified “knockout” mice. This process is also time-consuming (∼7–8 months or longer) and expensive, with no guarantee of success.
Gene targeting in ES cells relies on homologous recombination, during which DNA sequence homologous to the target sequence encoded by an engineered molecular vector construct is used to align and hybridize with the endogenous sequence of the target gene. Sequence exchange of genetic material is facilitated by double stand breaks (DSBs) and rejoining of the two broken ends, thereby incorporating the targeting construct [3,4]. Unfortunately, homologous recombination occurs at a relatively low frequency and thus requires the use of drug selection when targeting in ES cells. In addition, successful germline transmission from chimeras generated after blastocyst injection of ES cells is one of the major hurdles to successfully producing knockout mice using this technology.
A new approach to try to overcome these hurdles uses DNA nucleases, such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs).In a site-specific manner, these nucleases generate DSBs which can be repaired by homology directed repair (HDR) in the presence of a single-stranded oligonucleotide or double-stranded DNA template. This approach allows for genome modification through site-specific DNA nuclease action followed by HDR. More recently, DNA nuclease technology using Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CRISPR associated protein (Cas) has been shown to have significant advantages over either ZFNs or TALENs for gene targeting in mice. CRISPR/Cas is a self-defense system found in bacteria against foreign genetic material [5,6]. Cas9 is a type II Cas protein that, in association with single strand guide RNA (sgRNA), can recognize and generate DSBs at a target DNA sequence [7]. In fact, Cas9 has been shown to bean effective genome editing tool in mammalian cells [8–10] and embryos of a variety of species, including drosophila [11,12], zebra fish [13,14], mouse and rat embryos [15–17], and non-human primates [18].
Compared with traditional gene targeting in ES cells, Cas9 is potentially faster, easier to design, and a less expensive method for genome modification. A potential drawback as a result of off-target genetic mutations induced by promiscuous cutting by Cas9 nuclease can be spurious and unintended phenotypic consequences [19,20]. To address this issue, a mutation altering one amino acid at position D10A introduced into wildtype Cas9 replaced its double-strand nuclease activity with single-strand cutting (“nickase”) activity at the DNA target site [21,22]. Another impact of this change in enzymatic activity is that Cas9 nickase (Cas9n) co-associated with two complementary, or paired sgRNAs is reported to have a lower frequency of off-target effects compared to wildtype Cas9 [23–26].
Cas9 and Cas9n have been used to generate insertions, deletions, and exon replacements in genetically modified mice after injection into one-cell stage zygotes [27,28]. However, there are few reports using site-specific Cas9n to generate more complex alleles, such as conditional-ready (floxed) alleles, in the mouse genome. Floxed alleles, which retain wildtype (normal) expression until after being acted upon by a site-specific recombinase in a time- or tissue/cell type-specific manner [29] have significant advantages over general gene knockout approaches, such as when a whole body gene knockout causes embryonic lethality [30]. The flexibility and versatility of conditional alleles for studying gene function is reflected in the NIH’s KnockOut Mouse Project (KOMP),in which most of the genes are conditionally targeted in ES cells using the Cre/lox system [31].
Efforts to date include two separate Cas9 targeting events guided by their associated sgRNAs and two loxP-containing DNA oligos to insert loxP sites in F0 mice after zygote injection [28].However, it has not been reported whether a functional conditional allele generated in this fashion transmits through the germline. The efficiency of this approach is the sum of the efficiencies of each of 2 separate Cas9and homologous recombination events. In contrast, a different approach using a paired Cas9n and a single DNA template incorporating loxP sites flanking a critical exon should be more efficient. Further, using paired Cas9n instead of 2 separate Cas9sshould reduce the incidence of off-target effects and further enhance the process, especially in a high-throughput production pipeline. If successful, this approach could be of significant benefit to advancing genetic studies and biomedical research.
To test this hypothesis, in the present experiments we co-injected paired Cas9n with sgRNAs and a single DNA template into C57BL/6NCrl zygotes to generate F0 mouse expressing a conditional allele of the isoprenoid synthase domain containing (Ispd) gene. We observed a genotypically-confirmed floxed allele and sequence-verified targeting efficiency in 1 F0 mouse (∼8% of live pups born). We confirmed germline transmission and functional recombination of the floxed allele. This approach demonstrates the relative ease and reliability of generating conditional alleles using Cas9nand a single DNA template.
2. Results
2.1. Gene targeting strategy
The Ispd gene contains 10 exons spanning 307.9 kb on chromosome 12. To maximize cleavage at the target site [26], complementary sgRNAs (sgRNA 1 and sgRNA 2) separated by a 20 bp spacer were designed (Fig. 1A). The targeting vector encoded a single DNA template consisting of 1.9 kb arms of homologous genomic sequence immediately upstream (5′) and downstream (3′) of exon 2 flanked by two loxP sites, for a total size of 4.4 kb. This design strategy was chosen because following Cre recombination, excision of exon 2 will lead to a frameshift mutation followed by nonsense-mediated mRNA decay, resulting in premature translation of ISPD protein, and a knockout of the Ispd gene.
Fig. 1.
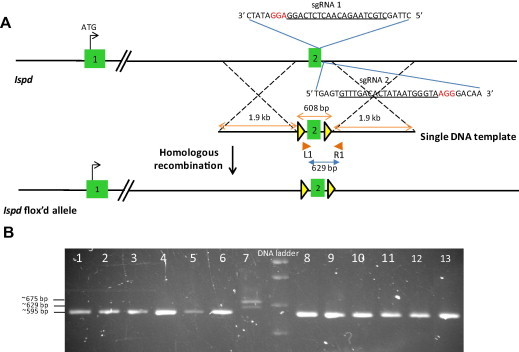
Generation of an Ispd conditional allele by Cas9n. (A) Schematic diagram of first 2 exons of the wildtype Ispd allele with relative position of single DNA template used for homologous recombination by Cas9n/sgRNA; sgRNA target sites in exon2 are underlined (sgRNA 1 and 2) with protospacer-adjacent motif (PAM) sequence shown in red font; single DNA template consisting of two loxP sites (yellow triangle) surroundingexon2 of Ispd (608 bp) flanked by arms of genomic homology(∼1.9 kb); PCR primers L1 and R1 (orange triangles) used for genotype screening of F0 offspring; a conditional, floxed exon 2 allele is generated after homologous recombination of the single DNA template at the target locus. (B) 1% Agarose gel showing amplicons (wildtype band 595 bp; mutant bands 629 and >629 bp) generated by PCR screening to discriminate wildtype pups (shown in lanes 1–6 and 8–13) from those in which the single DNA template had inserted into the genome (shown in lane 7); DNA from pup 7 shows two PCR amplicons, one of which (629 bp) is the expected size for a targeted insertion of one loxP site, and the other (∼675 bp) indicates the presence of an indel mutation (1 kb DNA ladder). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.2. Generation of genome-modified mice with conditional allele
The pronuclei and cytoplasm of 108 C57Bl/6N zygotes were injected with paired Cas9nwith associated sgRNAs, and the single DNA template under continuous pneumatic pressure. Following injection, zygotes were developed to 2-cell stage embryos overnight in a 5.5% CO2 incubator before surgical transfer into the oviducts of female CD1 pseudopregnant recipient mice. Thirteen F0 pups (18% birthrate) were born (Table 1). We designed a simple short range PCR to quickly screen offspring to discriminate wildtype pups from those in which the single DNA template had inserted into the genome (Fig. 1A, primers L1 & R1). Of these, 1 pup (founder 7) exhibited bi-allelic mutation of the intended target site (Fig. 1B), as indicated by the absence of a wildtype PCR amplicon (595 bp) and the presence of a mutant PCR amplicon (629 bp) corresponding to the expected size of the homologous region in the single DNA template and one loxP site. A second larger mutant PCR amplicon (>629 bp) was also identified represented an indel mutation.
Table 1.
Microinjection data for Cas9n generation of a conditional allele of Ispd by homologous recombination in mice.
| Number of injected zygotes | Zygotes surviving injection (% of injected) | Number of embryos transferred (% of surviving zygotes) | Number of F0 pups born (% of transferred embryos) | Number of F0 pups with flox’d allele (% of pups born) |
|---|---|---|---|---|
| 108 | 85 (79%) | 73 (86%) | 13 (18%) | 1 (8%) |
2.3. Confirmation of Ispd floxed allele
The genomic positioning of primers for rapid PCR screening of offspring could not be used to distinguish between random insertions and homologous recombination of the single DNA template at the targeted locus. Therefore, to definitively confirm conditional targeting of the Ispd allele and to discern between the 2 mutant amplicons identified in founder pup 7, we designed longer PCR amplified regions using primer pairs homologous to genomic sequence external to the targeting construct (Fig. 2). In one PCR reaction, primer L2 located upstream of the 5′ homology arm sequence and primer R1 positioned at the 3′ end of exon 2 (Fig. 2A) produced a PCR product (∼2.8 kb) which was subcloned by T/A cloning for DNA sequencing (Fig. 2B). Out of 18 clones, 12 were correctly targeted with loxP flanked exon 2 of one allele (Fig. 2C). The other 6 clones identified an allele containing only one loxP site (downstream of exon2) and a 95 bp insertion at the nick site targeted by sgRNA1 (Supplementary Fig. S1). In a second PCR reaction, primer L3 located downstream of exon 2 and primer R2 located downstream of the 3′ homology arm sequence (Fig. 2A) produced a PCR product (∼2.2 kb) that was subcloned by T/A cloning for DNA sequencing (Fig. 2B). As predicted, all seven clones showed a single loxP site downstream of exon 2. The combined DNA sequencing data of ∼2.8 kb and ∼2.2 kb amplicons from the 2 PCR reactions described above confirmed thatexon2 in one allele of Ispd was successfully floxed (flanked by loxP sites) while the complementary allele incorporated only one loxP site.
Fig. 2.
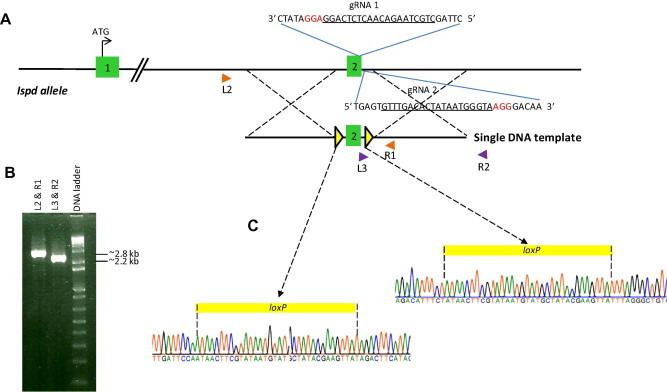
Long range PCR strategy to confirm homologous recombination of the single DNA template at the Ispd locus. (A) Relative positions of primer pairs L2 & R1 (orange triangles) and L3 & R2 (purple triangles) used to PCR amplify genomic DNA from pup 7; note that primers L2 and R2 are “outside” (upstream and downstream) of the homology region of the single DNA template used as the targeting vector (other details already described in Fig. 1). (B) PCR products amplified from the two primer sets (∼2.8 kb, L2 & R1; ∼2.2 kb, L3 & R2) were cloned for DNA sequencing. (C) DNA sequence of the 2 PCR amplicons spanning the targeted locus confirmed the presence of two intact loxP sites flanking (upstream and downstream) exon2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.4. Confirmation of germline transmission and functionality of the floxed allele
The first litter (F1) from breeding founder pup 7 (female) and C57BL/6N (male) produced one pup. Allele-specific PCR of DNA extracted from tail tissue confirmed germline transmission of the floxed Ispd allele (Fig. 3A). To confirm functionality of the conditional allele, genomic DNA from the F1 offspring was treated with recombinant Cre protein in vitro. Compared to wildtype genomic DNA, allele-specific PCR confirmed recombination of the floxed allele and the presence of the expected circular deletion product (Fig. 3B). These results further support the use of paired Cas9n and a single DNA template to achieve germline transmission of a functional floxed allele.
Fig. 3.
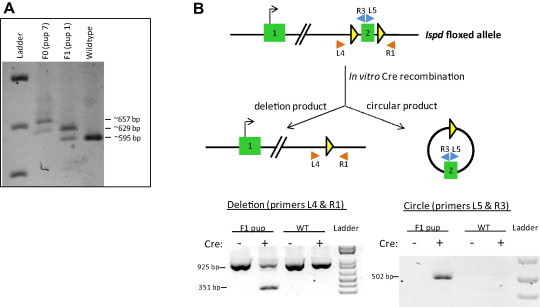
Germline transmission and in vitro Cre recombination of the Ispd conditional allele. (A) 1% Agarose gel showing PCR amplification of genomic DNA from F0 pup 7 (∼629 bp and ∼675 bp), F1 pup 1 (∼595 bp and ∼629 bp) and a wildtype pup (∼595 bp) using PCR primers L1 & R1 (same as in Fig. 1A). (B) After treatment with Cre recombinase in vitro, genomic DNA from F1 pup 1 used as template for PCR amplification with primers L4 and R1 flanking the floxed exon 2 produced a deletion product with two amplicons, a Cre-recombined band (∼351 bp) and a wildtype band (∼925 bp); PCR primers R3 and L5 amplified the circular DNA product (∼502 bp) containing the expected single loxp site and exon 2 resulting after Cre recombinationtreatment.
3. Discussion
To our knowledge, this is the first study to demonstrate the successful generation of a conditional knockout (floxed) allele in mice using paired Cas9n with a single DNA template expressing a loxP-flanked critical exon for targeting by homologous recombination. DNA cleavage induced by paired Cas9n enabled the single DNA template to undergo homologous recombination at the target site. Because this was a relatively small study, only 1 pup (8% of live born offspring) expressed the mutant allele. Further, although low, the overall efficiency (∼1% of injected zygotes) was within the range for other HDR and homologous recombination targeting experiments using Cas9 [28]. Using paired Cas9n and a single DNA template is simpler and easier than previously described methods using two separate cutting events using Cas9 and two donor oligonucleotides [28]. Further, an earlier study used Cas9 and a single DNA template to generate conditional alleles in rats. [32]. In addition, compared to Cas9, fewer off-target effects makes the use of Cas9n and a single DNA template as in our experiment a compelling approach to the production of conditional alleles in mice. In many previous publications, Cas9 and Cas9n have been used to generate mutant mice on genetically hybrid and mixed backgrounds [25,27,28]. Our study focused on Cas9n targeting in an inbred (C57Bl/6NCrl) strain that is particularly desirable strain for high-throughput phenotyping studies of mouse models for human disease. Our experiments in C57BL/6N show that paired Cas9n can be used to generate conditional alleles in inbred strains. Further, our study confirms not only functionality of the floxed allele but also germline transmission of the targeted allele.
We chose to conduct this pilot study using the Ispd gene because a conditional allele for this gene has not yet been described in the literature, nor has it been targeted by the large International Mouse Phenotyping Consortium (IMPC) production and phenotyping projects currently underway (http://www.mousephenotype.org/). Mutations in the ISPD gene in humans have been associated with Walker–Warburg Syndrome, a neurodevelopmental disorder [33,34]. A mutant Ispd mouse containing a premature stop codon in exon 1 leading to a truncated protein was generated by chemical-induced random mutagenesis using N-Ethyl-N-Nitrosourea (ENU) [35]. Homozygous mutants died at birth due to respiratory failure, and E18 embryos exhibited defects in the formation of the dorsal funiculus [33]. Conditional mutagenesis is particularly useful to study genes in which homozygous deletion mutation results in embryonic lethality. Our study exemplifies the application of a simpler and easier approach using paired Cas9n for creating a germline competent and functional conditional allele for potentially embryonic lethal genes in mice.
4. Materials and methods
4.1. Preparation of sgRNA transcripts and targeting vector
DNA oligos were obtained from Life Technologies (Grand Island, NY). Oligos were heated to 95 °C and then cooled to room temperature. The self-annealing oligo duplex was cloned into linearized T7 gRNA vector (System Biosciences, Mountain view, CA USA), and sequence verified by DNA sequencing. The sgRNA template for in vitro transcription (IVT) was prepared by PCR amplification of Phusion high fidelity DNA polymerase (NEB Biolabs, Ipwich, MA) followed by PCR cleanup reaction (Qiagen, Hilden, Germany). The sgRNA transcripts were generation by IVT synthesis kit (System Biosciences) and the quality of sgRNA transcripts were analyzed by NanoDrop (Thermo Fisher Scientific (Waltham, MA) and Bioanalyzer instrument (Agilent Technologies, Inc., Santa Clara, CA). The gene targeting vector encoding a single DNA template containing a floxed Ispd exon 2 was generated by GeneArt® Gene Synthesis (GeneArt, Regensburg) and sequence verified. The single DNA template was released from the gene targeting vector by restriction enzyme XhoI (NEB Biolabs) and was gel purified from agarose gel (Lonza, Rockland, ME) before microinjection.
4.2. Microinjection
C57BL/6NCr female mice (3–4 weeks olds) were super ovulated and mated overnight with C57Bl/6NCr male mice (>7 weeks old). Zygotes were harvested from the ampullae of super ovulated females and were placed in KSOM medium (Millipore, Billerica, MA) before microinjection. Microinjection was performed in M2 medium (Sigma, St Louis, MO) using a micromanipulator (Narishige) and microscope (Nikon). The Cas9n (System Biosciences), sgRNA and a single DNA template were co-injected into the pronucleus and cytoplasm of 108 zygotes. The final injection concentration in the mixture was 10 ng/μl Cas9n, 5 ng/μl of each sgRNA and 10 ng/μl of the single DNA template. After injection and incubation in 5.5% CO2 at 37 °C overnight for 24 h, surviving 2-cell stage embryos were transferred to CD1 recipients via oviduct transfer.
4.3. Use of animals
All mice were housed in a specific pathogen free vivarium with a 14 h on/10 h off light cycle (7 am on and 9 pm off). Female pseudopregnant mice undergoing surgical transfer of embryos into the uterus were anesthetized using Avertin (1.2%, 2,2,2, tribromoethanol, Sigma, St Louis, MO)at a dose of 0.017 ml/g of body weight administrated intraperitoneally. For post-operative pain relief, 0.1 ml of 0.03 mg/ml buprenorphine hydrochloride (Buprenex, Reckitt Becnkiser Healthcare, Hull, England) was administered intraperitoneally. Euthanasia was performed by CO2 asphyxiation and cervical dislocation according to the 2013 AVMA Guidelines on Euthanasia. The care, use, and disposition of all mice used in this study were reviewed and approved by the Institutional Animal Care and Use Committee of the University of California, Davis.
4.4. Genotyping of founders
At 10 days of age, toe snips of surviving F0 mice were harvested and genomic DNA was extracted using the DNeasy blood and tissue kit (Qiagen). The target region was PCR amplified by DNA polymerase (Agilent Technologies, Inc) followed by DNA sequencing for screening F0 offspring. In pups showing positive PCR reactions for the single DNA template, genomic DNA was further PCR amplified to produce amplicons that were subsequently cloned by TOPO T/A subcloning (Invitrogen, Carlsbad, CA) followed by DNA sequencing.
4.5. In vitro Cre recombination
1 μg genomic DNA was mixed with 10 units of recombinant Cre recombinase (NEB Biolabs) in 1X Cre recombinase reaction buffer (supplementary with Cre recombinase) with final volume of 40 μl. The mixture was incubated in 37 °C for 1 h and then the reaction was heat inactivated at 70 °C for 10 min. For all targets, 2 μl of Cre reaction mix was used as a PCR template for PCR amplification with gene-specific primers. Primers (L4 and R1) were used for detection of deletion products, while primers (R3 and L5) were used for detection of the circular DNA byproduct remaining after Cre-mediated recombination.
Acknowledgments
The authors acknowledge funding for this project in part from NIH grant 5U42OD011175 KOMP Phase II Mouse Production and Cryopreservation and from the Mouse Biology Program, University of California, Davis.
Appendix A. Supplementary data
Supplementary figure.
References
- 1.Thomas K.R., Capecchi M.R. Site-directed mutagenesis by gene targeting in mouse embryo- derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 2.Mansour SL, Thomas KR, Deng CX, Capecchi MR (1990). Introduction of a lacZ reporter gene into the mouse int-2 locus by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 87, 7688–7692. [DOI] [PMC free article] [PubMed]
- 3.Johnson RD, Jasin M (2001). Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc.Trans. 29, 196–201. [DOI] [PubMed]
- 4.Dudas A., Chocanec M. DNA double-strand break repair by homologous recombination. Mutat. Res. 2004;566:131–167. doi: 10.1016/j.mrrev.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Horvath P, Barrangou R (2010). CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170. [DOI] [PubMed]
- 6.Bhaya D., Davison M., Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 7.Barrangou R. RNA-mediated programmable DNA cleavage. Nat. Biotech. 2012;30:836–838. doi: 10.1038/nbt.2357. [DOI] [PubMed] [Google Scholar]
- 8.Cho S.W., Kim S., Kim J.M., Kim J. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotech. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 9.Mali P., Yang L., Esvelt K.M., Arch J, Guell M., DiCarlo J.E., Norville J.E., Church G.M. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. [DOI] [PMC free article] [PubMed]
- 10.Wang R., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2013;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gratz S.J., Cummings A.M., Nguyen J.N., Hamm D.C., Donohue L.K., Harrison M.M., Wildonger J., O’Connor-Giles K.M. (2013). Genome engineering of Drosophila with the CRISPR RNA-guided CAS9 nuclease. Genetics 194, 1029–1035. [DOI] [PMC free article] [PubMed]
- 12.Ren X., Sun J., Housden B.E., Hu Y., Roesel C., Lin S., Liu L.P., Yang Z., Mao D., Sun L. et al. (2013). Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc. Natl. Acad. Sci. USA 110, 19012–19017. [DOI] [PMC free article] [PubMed]
- 13.Chang N., Sun C., Gao L., Zhu D., Xu X., Zhu X., Xiong J, Xi J.J. (2013). Genome editing with RNA-guided Cas9 nuclease in Zebrafish embryos. Nature23, 465–472. [DOI] [PMC free article] [PubMed]
- 14.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR & Joung JK (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature 31, 227–229. [DOI] [PMC free article] [PubMed]
- 15.Mashiko D., Fujihara Y., Satouh Y., Miyata H., Isotani A., Ikawa M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 2013;3:1–6. doi: 10.1038/srep03355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen B., Zhang J., Wu H., Wang J., Ma K., Li Z., Zhang X., Zhang P., Huang X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;5:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X. et al. (2013) Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 8, 681–683. [DOI] [PubMed]
- 18.Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., Kang Y., Zhao X., Si W., Xiang A.P. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2013;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 19.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nuclease in human cells. Nat. Biotechnol. 2013;9:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR (2013). High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. l9, 839–843. [DOI] [PMC free article] [PubMed]
- 21.Gasiunas G., Barrangou R., Horvath P., Siksnys V. (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 109, E2579–E2586. [DOI] [PMC free article] [PubMed]
- 22.Ran F., Hsu P.D., Wright J., Agarwala V., Scott D., Zhang F. Genome engineering the CRISPR-Cas9 system. Nat. Protoc. 2013;11:2281–2308. doi: 10.1038/nprot.2013.143. (21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;9:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ran F.A., Hsu P.D., Lin C., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L., Wang L., Hodgkins A., Iyer V., Huang X. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 27.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:1–9. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H., Wang H., Shivalila C.S., Cheng A.W., Shi L., Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediate genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu H., Marth J.D., Orban P.C., Mossmann H., Rajewsky K. Deletion of a DNA polymerase beta gene targeting segment in T cells using cell type specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 30.Rajewsky K., Gu H., Kuhn R., Betz U.A.K., Muller W., Roes J., Schwenk F. Conditional gene targeting. J. Clin. Invest. 1996;98:600–603. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skarnes, W.C., Rosen, B., West, A.P., Koutsourakis, M., Bushell, W., Iyer, V., Mujica, A.O., Thomas, M., Harrow J., Cox T. et al. (2011). A conditional knockout resource for the genome-widestudy of mouse gene function. Nature 474, 337–342. [DOI] [PMC free article] [PubMed]
- 32.Ma Y., Zhang X., Shen B., Lu Y., Chen W., Ma J., Bai L., Huang X., Zhang L. Generating rats with conditional alleles using CRISPR/Cas9. Cell Res. 2014;24:122–125. doi: 10.1038/cr.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright K.M., Lyon K.A., Leung H., Leahy D.J., Ma L., Ginty D.D. Dystroglycan organizes axon guidance cue localization and axonal pathfinding. Neuron. 2012;76:913–944. doi: 10.1016/j.neuron.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roscioli T., Kamsteeg E.J., Buysse K., Maystadt I., Van Reeuwijk J., Van Den Elzen C., Van Beusekom E., Riemersma M., Pfundt R., Vissers L.E. Mutations in ISPD cause Walker–Warburg syndrome and defective glycosylation of α-dystroglycan. Nat. Genet. 2012;44:581–585. doi: 10.1038/ng.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiler T., Lee H., Lommel M., Yoshida-Moriguchi T., De Bernabe D.B., Venzke D., Cirak S., Schachter H., Vajsar J., Voit T. ISPD loss-of-function mutations disrupt dystroglycan O-mannosylation and cause Walker–Warburg syndrome. Nat. Genet. 2012;44:575–580. doi: 10.1038/ng.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure.


