Abstract
We introduce molecularevolution.org, a publicly available gateway for high-throughput, maximum-likelihood phylogenetic analysis powered by grid computing. The gateway features a garli 2.0 web service that enables a user to quickly and easily submit thousands of maximum likelihood tree searches or bootstrap searches that are executed in parallel on distributed computing resources. The garli web service allows one to easily specify partitioned substitution models using a graphical interface, and it performs sophisticated post-processing of phylogenetic results. Although the garli web service has been used by the research community for over three years, here we formally announce the availability of the service, describe its capabilities, highlight new features and recent improvements, and provide details about how the grid system efficiently delivers high-quality phylogenetic results. [garli, gateway, grid computing, maximum likelihood, molecular evolution portal, phylogenetics, web service.]
The most widely used modern statistical methods of phylogenetic inference fall into two broad classes: maximum likelihood methods and Bayesian inference methods. Depending on the number of sequences, the number of characters, and the chosen evolutionary model, both maximum likelihood and Bayesian tree inference methods can be computationally intensive, thus creating the need for strategies that speed up computation and decrease time to results. One such strategy is parallelization, which distributes a logical unit of computation over multiple processors. Maximum likelihood methods are generally more amenable to parallelization than Bayesian inference methods, since the hundreds or thousands of searches for the maximum likelihood tree and bootstrap trees that are required for a typical phylogenetic analysis may be run independently of one another. We have developed a grid computing system that features the maximum likelihood-based program garli (Genetic Algorithm for Rapid Likelihood Inference; Zwickl 2006) for high-throughput phylogenetic analysis. Here we describe this publicly available system, in particular focusing on the user-friendly garli web interface available at molecularevolution.org.
garli is an open-source phylogenetic inference program that uses the maximum likelihood criterion and a stochastic evolutionary algorithm to search for optimal solutions within the joint space of tree topologies, branch length parameter values, and model parameter values. garli was developed with the goal of increasing both the speed of maximum likelihood tree inference and the size of data sets that can be reasonably analyzed. garli 2.0 implements models for the analysis of biological sequence data (at the level of nucleotides, amino acids, or codons), as well as morphology and (not officially released) insertion–deletion characters. Version 2.0 introduced support for partitioned models, allowing simultaneous use of different data types or assignment of differing model parameters and rates to individual loci or codon positions. The program design focuses on flexibility of model choice and rigor in parameter estimation.
Searches through phylogenetic tree space may become entrapped in local optima, and therefore it is necessary to perform multiple garli searches for the tree with the highest likelihood, which we simply call the best tree. This could entail hundreds of searches depending on the difficulty of the problem. Furthermore, one typically conducts hundreds or thousands of bootstrap replicate searches to assess confidence in the bipartitions found in the best tree. Depending on the number of sequences, the number of unique alignment columns, the evolutionary models employed, various garli configuration settings, and the capability of the designated computational resource, it can take hours or even days to complete a single garli search replicate. Thus, running many search replicates in parallel on a grid computing system greatly reduces the amount of time required to complete a set of analyses.
Grid computing is a model of distributed computing that seamlessly links geographically and administratively disparate computational resources, allowing users to access them without having to consider location, operating system, or account administration (Cummings and Huskamp 2005). The Lattice Project, our grid computing system based on Globus software (Foster and Kesselman 1999), incorporates volunteer computers running boinc (Anderson 2004) as well as traditional grid computing resources such as Condor pools (Litzkow et al. 1988) and compute clusters. The architecture and functionality of the grid system is described extensively elsewhere (Bazinet 2009); fundamentally, however, The Lattice Project provides access to scientific applications (which we term grid services), as well as the means to distribute the computation required by these services over thousands of processors. In recent years, the system has been enhanced by the development of a web interface to the garli grid service (Bazinet and Cummings 2011, currently available at molecularevolution.org). The garli grid service has been used in at least 50 published phylogenetic studies, with usage having increased dramatically since the release of the garli web interface (e.g., Myers and Cummings 2003; Regier et al. 2009; Kawahara et al. 2011; Bazinet et al. 2013; Regier et al. 2013; Sohn et al. 2013, see Supplemental Material for the full list, available on Dryad http://dx.doi.org/10.5061/dryad.d7639). As of April 2, 2014, 843 distinct web service users have completed 4835 analyses comprising 2,306,159 individual garli search replicates.
In this article, we compare The Lattice Project to other scientific gateways and describe the features of the garli web service. In addition, we provide details about how the grid system efficiently processes computationally intensive phylogenetic analyses.
Features
The Lattice Project Compared to other Scientific Gateways
There are a number of other scientific gateways that provide bioinformatics tools and services, including those for phylogenetic analysis. These include the Cyberinfrastructure for Phylogenetic Research (cipres) Gateway (Miller et al. 2010), the University of Oslo Bioportal (Kumar et al. 2009, which has recently closed), the Cornell Computational Biology Service Unit (cbsuapps.tc.cornell.edu), Phylemon (Sánchez et al. 2011), and Mobyle (Néron et al. 2009). Although each of these other systems has proved to be of use in phylogenetic research, our grid system has some distinguishing characteristics.
GARLI version 2.0—Of the gateways supporting phylogenetic analysis, only The Lattice Project and the cipres gateways offer a garli 2.0 (Zwickl 2011) service.
Unlimited computation—The garli service on molecularevolution.org currently allows an unlimited number of submissions, up to 100 best tree or 2000 bootstrap search replicates per submission, and no resource or runtime limitations. We are able to offer this level of service due to our implementation of stringent error checking, advanced scheduling mechanisms, and inclusion of novel resources such as our public computing pool of boinc clients.
Facile user interface and resource abstraction—Fully embracing the grid computing model, the computing resources backing the garli service are abstracted from the user, facilitated by an elegant user interface. In contrast, the cipres gateway requires the user to become familiar with their computing resources and to specify their analysis in such a way that it will complete on the allocated resource (usually only a small number of processors) within an allotted period of time.
Sophisticated and relevant post-processing—The use of stochastic algorithms, multiple search replicates, and bootstrap analyses generates a large number of individual results that must be compiled and processed for evaluation and subsequent use. We perform much of this post-processing automatically, including computation of the best tree found or bootstrap majority rule consensus tree, and the calculation of various summary statistics and graphical representations (see Post-processing routines).
Large-scale public participation—The Lattice Project is the only phylogenetic analysis system that provides an easy and meaningful opportunity for public participation in research, which is achieved by using our boinc project (boinc.umiacs.umd.edu). Volunteers simply download a lightweight client to their personal computer, thus enabling it to process garli workunits for The Lattice Project. As of April 2, 2014, more than 16,956 people from 146 countries have participated.
Minimal energy usage—Emergy, the energy embodied in computing components (which includes manufacture and transportation), accounts for the majority of power consumed in computing (Raghavan and Ma 2011). Put another way, the “greenest” computer is one that is never built. Apart from a few servers for web, database, and middleware services, no hardware is purchased specifically for our grid system. The institutional resources we use are comprised largely of desktop systems and clusters purchased for other purposes (e.g., teaching labs and research, respectively), and we use these resources only when they are not being used for their primary purpose. In addition, more than 38,481 computers from the general public have been volunteered at various stages of the project. For all of these resources, the emergy investment has already been made, and our use of these resources amortizes this investment over a greater usage basis. In contrast, phylogenetic analyses through other gateways compete for limited resources on high-capacity clusters, where the jobs often do not take advantage of the high-bandwidth, low-latency interconnects and other special hardware features offered. Furthermore, the widely distributed, low-density computing model of our grid system results in almost no additional energy use for cooling compared with the substantial energy costs of cooling computer data centers.
No other openly accessible phylogenetic computing system collectively shares these attributes. Although dedicated high-performance computing resources have their place in scientific research, a substantial share of phylogenetic analyses can be performed very effectively, and more energy efficiently, by means of grid and public computing.
GARLI Web Service: User Interface and Functionality
We have recently upgraded the user interface to our grid system from a Unix command-line interface to a web-based one. This greatly reduces the entry barrier for potential non-technical users. Researchers were previously required to use command-line tools to upload data, submit analyses to a particular grid service (e.g., garli), and download subsequent results. Basic utilities were also available to query the status of jobs or cancel them.
Although the command-line interface is still available, we anticipate that the web-based interfaces to our services will generate considerably more interest; the garli web service is the first of these to be developed. The following sections describe the modes of use and the basic functionality of the garli web service on molecularevolution.org.
Modes of use
A garli web service user may register an account or choose to remain anonymous. Anonymous users are only required to provide an email address (used to notify them of job status updates) and to fill out a captcha (Completely Automated Public Turing test to tell Computers and Humans Apart) for each job submission to prevent spam. Anonymous use of the web service is a convenient way to try out the service with minimal effort. However, registration on molecularevolution.org confers several advantages: (1) one does not have to fill out a captcha for each job submission; (2) one gains access to a file repository that can be used to store and reuse input files (Supplementary Fig. 1, available at http://dx.doi.org/10.5061/dryad.d7639; and (3) one gains the ability to view a list of their jobs and manage them.
Create job page
Submitting a garli analysis via the create job page (Supplementary Fig. 2, available at http://dx.doi.org/10.5061/dryad.d7639) consists of the following general steps: (1) specification of a job name, analysis type (best tree or bootstrap search), and number of replicates (up to 2000); (2) upload or specification of necessary input files (sequence data, starting tree, and/or constraint file); and (3) specification of model parameters and other program settings. Upon job submission, the system uses a special validation mode of the garli program to ensure that there are no problems with the user-supplied data file and the parameters specified; for example, very large data sets may require more ram than the system currently allows (i.e., 24,000 mb). garli search replicates are then scheduled to run in parallel on one or more grid system resources that meet the job requirements (e.g., that have enough ram). The user is notified by email if their job was submitted successfully or if it failed for some reason.
Job status page
The job status page (Supplementary Fig. 3, available at http://dx.doi.org/10.5061/dryad.d7639) allows a registered user to view and manage a list of their jobs. For each job listed, the following attributes are displayed: job id, job name, number of replicates complete, job status, and time the job was created. The dropdown at the top of the page allows one to filter jobs by a particular job status (“idle”, “running”, “retrieved”, “failed”, or “removed”). Finally, using the button at the bottom of the page, one may remove jobs that are no longer of interest. If the jobs to be removed are in the process of running, they will be canceled.
Job details page
When a registered user selects a particular job from the job status page, or an anonymous user enters a valid e-mail address/job id combination on the same page, the job details page is shown (Supplementary Fig. 4, available at http://dx.doi.org/10.5061/dryad.d7639). This page contains a section for job input files (both user-provided and system-generated) and a section for job output files. The job output files section always includes a zip file that contains all of the currently available output associated with the analysis. If all of the replicates for a particular analysis are complete, then the job output files section will also include the results of post-processing (see Post-processing routines).
Partitioned Analysis Specification
Support for partitioned substitution models is the most significant new feature of garli 2.0. However, partitioned analysis specification can be a relatively complicated and error-prone process. We have made the specification of modestly-sized partitioned analyses easier by introducing a guided mode that allows the user to specify the details of the partitioned analysis with graphical form elements (Supplementary Fig. 5, available at http://dx.doi.org/10.5061/dryad.d7639), rather than by manually composing a nexus sets block and garli model blocks. Guided mode is enabled once the user has selected a valid nexus data file, which the system processes with the Nexus Class Library (Lewis 2003). The user then creates one or more character sets (charsets), each consisting of a name, a start position, and an end position; charsets may also be specified by codon position using a checkbox. Once the user specifies one or more valid charsets they will be made available to be added to data subsets. Each data subset must contain at least one charset, but may contain more than one. The service currently allows the definition of up to ten data subsets in guided mode. For each data subset, a particular substitution model (or particular model parameters) may be specified. When the partitioned analysis is submitted, the service will automatically transform the charset and subset data into a nexus sets block and include it in the data file, and will likewise produce the appropriate model blocks and add them to the garli configuration file. For users who prefer to provide their own nexus sets block and garli model blocks, we provide an expert mode that allows the user to input them directly.
Post-processing Routines
Due to the difficulty of inferring large phylogenetic trees, multiple searches for the best tree are typically performed with garli. This increases the thoroughness of the search for the best tree, but the resulting large number of files and analysis results can be overwhelming. To ease the burden on the end user, our web-based system performs some post-processing routines, which include graphical and quantitative characterizations of the set of trees inferred from multiple search replicates.
Post-processing generates a textual summary for all analyses (Supplementary Fig. 6, available at http://dx.doi.org/10.5061/dryad.d7639). This file contains the following general information: (1) the data file used; (2) the number of replicates performed; (3) the cumulative garli runtime; and (4) suggestions for citing the garli web service (omitted from Supplementary Fig. 6, available at http://dx.doi.org/10.5061/dryad.d7639). The analysis summary for a best tree search also contains summary statistics that characterize the distribution of log-likelihood scores and symmetric tree distances (Robinson and Foulds 1981, absolute and normalized), as well as estimates of the number of search replicates required to recover the best tree topology at three probability levels (see Calculating the required number of GARLI search replicates).
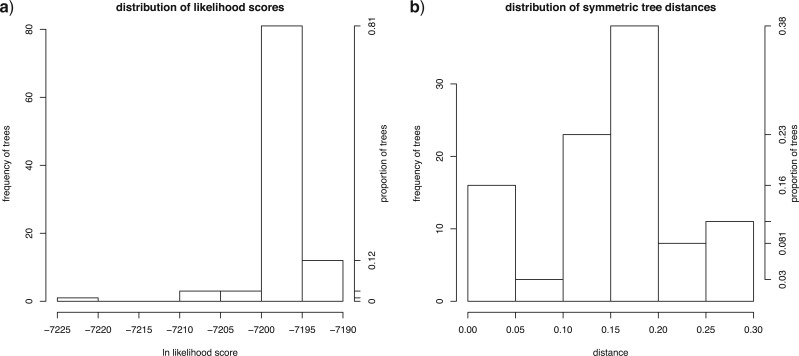
In the case of a best tree search, post-processing generates the following files in addition to the analysis summary: (1) a nexus file containing the single tree with the highest likelihood score; (2) a file containing all of the trees found across search replicates, as well as a file containing only the unique trees found (both files in nexus format); (3) a file containing a sorted list of the likelihood scores of the trees found by the analysis and a file containing a sorted list of the likelihood scores of the unique trees found; (3) a pdf file showing the distribution of likelihood scores among trees (Fig. 1a); and (4) a pdf file showing the distribution of symmetric tree distances (Fig. 1b).
Figure 1.
Properties of trees from multiple search replicates for a representative analysis using garli. a) The distribution of likelihood scores. b) The distribution of symmetric tree distances (as a fraction of the maximum possible value for the data set). Both measures are given as frequency and proportion.
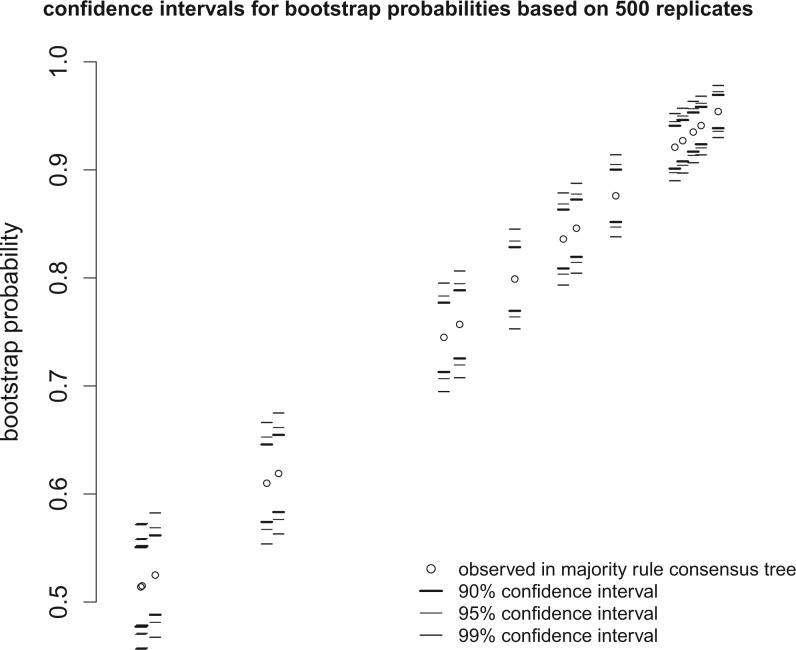
In the case of a bootstrap analysis, post-processing uses DendroPy (Sukumaran and Holder 2010) to generate the following files in addition to the analysis summary: (1) a nexus file containing all of the bootstrap trees from the analysis; (2) a nexus file containing the majority rule bootstrap consensus tree with bootstrap probability values embedded; (3) a pdf file showing the 0.90, 0.95, and 0.99 confidence intervals for the bootstrap probabilities observed in the majority rule bootstrap consensus tree, calculated using the formulas given in Hedges (1992) (Fig. 2); and (4) a table giving the 0.90, 0.95, and 0.99 confidence intervals for the bootstrap probabilities observed in the majority rule bootstrap consensus tree.
Figure 2.
Confidence intervals associated with the bootstrap probabilities observed in the majority rule consensus tree computed from 500 garli bootstrap replicates. Confidence intervals are given for three probabilities (0.90, 0.95, and 0.99).
Calculating the required number of GARLI search replicates
Our post-processing routines for a best tree search include the calculation of the number of search replicates necessary to guarantee a particular probability (e.g., 0.95) of recovering the tree topology with the highest observed likelihood score (Regier et al. 2009). This statistic, based on properties of the binomial distribution, is calculated using the number of replicates that find the same best topology (x), where “same topology” is defined as having symmetric distance from the best topology equal to zero.
For example, if the topology of the best tree out of 100 is unique among all topologies found (x = 1), then 298 replicates are required in order to recover the best topology with a probability of at least 0.95 (Fig. 3). Of course, it is entirely possible that upon running 298 replicates, the statistical estimate would be revised upwards; e.g., if the topology of the best tree were still unique among the set of topologies, then yet more replicates would be required.
Figure 3.
Relationship between the number of search replicates (out of 100) returning the same topology as that of the best tree found and the estimated number of search replicates necessary to guarantee a particular probability of recovering that topology. Estimates are given at three probabilities (0.90, 0.95, 0.99).
This statistical estimate of the number of search replicates required to guarantee a particular probability of obtaining the best tree found is intended to inform users about the joint behavior of their data and the garli search algorithm, and consequently how many search replicates they should perform. This introduces an objective decision process into the analysis design that eliminates guesswork and the need to evaluate intermediate output, thus saving investigator time and improving analytical results. It also reduces waste of grid resources and energy by suggesting that the user run only the number of replicates needed.
Eventually, we intend to have the system automatically and adaptively run the appropriate number of search replicates on behalf of the user. It may also be possible to do something similar for bootstrap replicates, perhaps based on a desired level of precision (Fig. 2) or other criteria (Pattengale et al. 2010).
System Performance
The performance of any distributed computing system depends on how efficiently its resources are utilized. We have implemented a number of scheduling optimizations that enable efficient use of our grid computing resources (Bazinet 2009). These include a round-robin scheduling algorithm to distribute load evenly among resources; a scheme for benchmarking resources and prioritizing job assignments so that faster resources receive jobs before slower resources; use of predicted job runtime to ensure that long-running jobs are placed on resources where they are unlikely to be interrupted; and a mechanism for combining many short-running jobs into a single job with an “optimal” aggregate runtime to maximize system throughput. These last two features depend on a framework we developed for garli runtime prediction using random forests (Bazinet and Cummings 2011), a machine learning method. We have improved this framework so that the runtime prediction model is continuously updated as new jobs are run. In supplemental material (http://dx.doi.org/10.5061/dryad.d7639) we describe two system performance improvements in some detail: use of optimal-length jobs for grid computing, and automatic measurement of resource throughput.
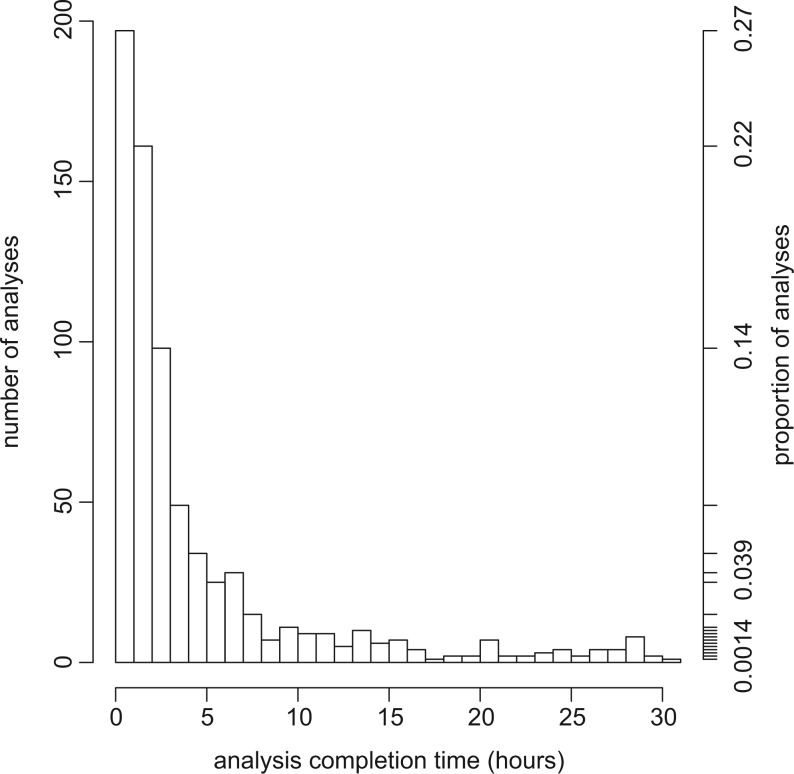
It is important to keep in mind that our grid system is designed for high-throughput computing rather than high-performance computing. As a result, while any one analysis might run more quickly on a dedicated high-performance platform, the system described here allows many such analyses to run concurrently and still complete in a relatively modest amount of time (Fig. 4). In addition, use of a high-performance system may not necessarily yield decreased time to results once allocation processes, system availability, queue waiting times, scheduling policies, and other considerations commonly associated with the use of high-performance resources are factored in. The high-throughput computing gateway at molecularevolution.org is well matched to the requirements of many typical phylogenetic analyses, and it has already proven useful to many researchers conducting maximum likelihood phylogenetic analyses using garli 2.0.
Figure 4.
Completion times of 719 analyses submitted to the garli web service for a recent six month period (2013-07-23 to 2014-01-23). Despite great variation in analysis parameters (e.g., data matrix size, substitution model used, number of replicates requested), ≈97% of analyses were completed in less than 24 hours.
Supplementary material
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.d7639.
Funding
This work was supported by the National Science Foundation (DBI-0755048).
Acknowledgments
We thank Barry Dutton, Yevgeny Deviatov, and Derrick Hinkle for their efforts developing various aspects of the grid system and the garli web service; Charles Mitter for developing the statistical determination of the number of required garli search replicates; and Mike Landavere, Christopher Camacho, Ahmed El-Haggan, Wasay Taha Mohammed Abdul, Patrick Beach, Matthew Kweskin, Kevin Hildebrand, and Fritz McCall for connecting and administering grid system resources. We also thank the associate editor and one anonymous reviewer for their helpful suggestions.
References
- Anderson D.P. Proceedings of the 5th IEEE/ACM International Workshop on Grid Computing GRID '04 IEEE Computer Society. Washington, DC, USA: 2004. BOINC: A system for public-resource computing and storage; pp. 4–10. [Google Scholar]
- Bazinet A.L. The Lattice Project: A Multi-model Grid Computing System. College Park: University of Maryland; 2009. Master's thesis. [Google Scholar]
- Bazinet A.L., Cummings M P. 2011 IEEE International Symposium on Parallel and Distributed Processing Workshops and PhD Forum. 2011. Computing the tree of life: Leveraging the power of desktop and service grids; pp. 1896–1902. [Google Scholar]
- Bazinet A.L., Cummings M.P., Mitter K.T., Mitter C.W. Can RNA-Seq resolve the rapid radiation of advanced moths and butterflies (Hexapoda: Lepidoptera: Apoditrysia)? An exploratory study. PLoS ONE. 2013;8:e82615. doi: 10.1371/journal.pone.0082615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings M., Huskamp J. Grid computing. EDUCAUSE Review. 2005;40:116–117. [Google Scholar]
- Foster I., Kesselman C. Globus: a toolkit-based grid architecture. In: Foster I., Kesselman C., editors. The grid: blueprint for a new computing infrastructure Morgan-Kaufmann. 1999. pp. 259–278. [Google Scholar]
- Hedges S.B. The number of replications needed for accurate estimation of the bootstrap P value in phylogenetic studies. Mol. Biol. Evol. 1992;9:366–9. doi: 10.1093/oxfordjournals.molbev.a040725. [DOI] [PubMed] [Google Scholar]
- Kawahara A., Ohshima I., Kawakita A., Regier J., Mitter C., Cummings M., Davis D., Wagner D., De Prinis J., Lopez-Vaamonde C. Increased gene sampling provides stronger support for higher-level groups within gracillariid leaf mining moths and relatives (Lepidoptera: Gracillariidae) BMC Evol. Biol. 2011;11:182. doi: 10.1186/1471-2148-11-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Skjaeveland A., Orr R.J.S., Enger P., Ruden T., Mevik B.-H., Burki F., Botnen A., Shalchian-Tabrizi K. AIR: a batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinformatics. 2009;10:357. doi: 10.1186/1471-2105-10-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.O. NCL: a C + + class library for interpreting data files in NEXUS format. Bioinformatics. 2003;19:2330–2331. doi: 10.1093/bioinformatics/btg319. [DOI] [PubMed] [Google Scholar]
- Litzkow M., Livny M., Mutka M. Distributed Computing Systems, 1988., 8th International Conference on. 1988. Condor–a hunter of idle workstations; pp. 104–111. [Google Scholar]
- Miller M., Pfeiffer W., Schwartz T. Gateway Computing Environments Workshop (GCE), 2010. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees; pp. 1–8. [Google Scholar]
- Myers D., Cummings M. Necessity is the mother of invention: a simple grid computing system using commodity tools. J. Parallel Distr. Com. 2003;63:578–589. [Google Scholar]
- Néron B., Ménager H., Maufrais C., Joly N., Maupetit J., Letort S., Carrere S., Tuffery P., Letondal C. Mobyle: a new full web bioinformatics framework. Bioinformatics. 2009;25:3005–11. doi: 10.1093/bioinformatics/btp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattengale N.D., Alipour M., Bininda-Emonds O.R.P., Moret B.M.E., Stamatakis A. How many bootstrap replicates are necessary? J. Comput. Biol. 2010;17:337–54. doi: 10.1089/cmb.2009.0179. [DOI] [PubMed] [Google Scholar]
- Raghavan B., Ma J. HotNets. 2011. The energy and emergy of the Internet; p. 9. [Google Scholar]
- Regier J.C., Mitter C., Zwick A., Bazinet A.L., Cummings M.P., Kawahara A.Y., Sohn J.-C., Zwickl D.J., Cho S., Davis D.R., Baixeras J., Brown J., Parr C., Weller S., Lees D.C., Mitter K.T. A large-scale, higher-level, molecular phylogenetic study of the insect order Lepidoptera (moths and butterflies) PLoS ONE. 2013;8:e58568. doi: 10.1371/journal.pone.0058568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regier J.C., Zwick A., Cummings M.P., Kawahara A.Y., Cho S., Weller S., Roe A., Baixeras J., Brown J.W., Parr C., Davis D.R., Epstein M., Hallwachs W., Hausmann A., Janzen D.H., Kitching I.J., Solis M.A., Yen S.-H., Bazinet A.L., Mitter C. Toward reconstructing the evolution of advanced moths and butterflies (Lepidoptera: Ditrysia): an initial molecular study. BMC Evol. Biol. 2009;9:280. doi: 10.1186/1471-2148-9-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.R., Foulds L.R. Comparison of phylogenetic trees. Math. Biosci. 1981;53:131–147. [Google Scholar]
- Sánchez R., Serra F., Tárraga J., Medina I., Carbonell J., Pulido L., de María A., Capella-Gutíerrez S., Huerta-Cepas J., Gabaldón T., Dopazo J., Dopazo H. Phylemon 2.0: a suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Res. 2011;39:W470–W474. doi: 10.1093/nar/gkr408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J.-C., Regier J.C., Mitter C., Davis D., Landry J.-F., Zwick A., Cummings M.P. A molecular phylogeny for Yponomeutoidea (Insecta, Lepidoptera, Ditrysia) and its implications for classification, biogeography and the evolution of host plant use. PLoS ONE. 2013;8:e55066. doi: 10.1371/journal.pone.0055066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran J., Holder M.T. DendroPy: a Python library for phylogenetic computing. Bioinformatics. 2010;26:1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
- Zwickl D.J. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. The University of Texas at Austin; 2006. Ph.D. thesis. [Google Scholar]
- Zwickl D.J. 2011. GARLI 2.0 https://www.nescent.org/wg_garli/main_page.