Graphical abstract
Keywords: RNA, Enzyme, Thermal stability, Molecular crowding, Dielectric constant
Highlights
-
•
Molecular crowding effects on RNA were studied using various mixed solutions.
-
•
The effect of salt concentration on ribozyme catalytic rate was altered.
-
•
The effect of salt concentration on RNA duplex stability was altered.
-
•
Dielectric constant effects are important for RNA reactions.
-
•
A method to predict nucleic acid folding in non-dilute aqueous solutions is proposed.
Abstract
Nucleic acids are useful for biomedical targeting and sensing applications in which the molecular environment is different from that of a dilute aqueous solution. In this study, the influence of various types of mixed solutions of water and water-soluble organic compounds on RNA was investigated by measuring the catalytic activity of the hammerhead ribozyme and the thermodynamic stability of an oligonucleotide duplex. The compounds with a net neutral charge, such as poly(ethylene glycol), small primary alcohols, amide compounds, and aprotic solvent molecules, added at high concentrations changed the ribozyme-catalyzed RNA cleavage rate, with the magnitude of the effect dependent on the NaCl concentration. These compounds also changed the thermodynamic stability of RNA base pairs of an oligonucleotide duplex and its dependence on the NaCl concentration. Specific interactions with RNA molecules and reduced water activity could account for the inhibiting effects on the ribozyme catalysis and destabilizing effects on the duplex stability. The salt concentration dependence data correlated with the dielectric constant, but not with water activity, viscosity, and the size of organic compounds. This observation suggests the significance of the dielectric constant effects on the RNA reactions under molecular crowding conditions created by organic compounds.
1. Introduction
Cellular activities occur in the presence of large amounts of background proteins, nucleic acids, metabolites, osmolytes, and cellular organelles. There are many reports suggesting that the molecular environment created by high concentrations of background composition is an important driving force for several cellular activities, including cell growth, metabolism, tumor generation, and age-related disease susceptibility [1–3]. The thermodynamics and kinetics of the chemical reactions occurring in a living cell are influenced by coexisting molecules through direct and indirect interactions. However, the structural stability of nucleic acids in a relevant context of living cells remains elusive. For the quantitative analysis of the influence of such non-dilute aqueous conditions, the mixed solutions of water and water-soluble organic compounds are used [4,5]. Neutral compounds are often employed to mimic the intracellular conditions where macromolecules are concentrated and the solution properties are different from pure water (e.g., reduction in the water activity). The non-dilute condition is created not only in the medium of living cells but also on the surface of a biosensor [6–9]. Poly(ethylene glycol) (PEG) is one of the most commonly used cosolutes for creating a molecularly crowded condition. It is reported that the molecular crowding by PEG increases the activity of several protein enzymes, attributed to an enhanced folding stability and increased binding with substrates caused by the excluded volume effect [10]. The polar and charged nature of nucleotides makes the molecular crowding effects on nucleic acids more complicated. For example, the thermal stability of oligonucleotide duplexes is not significantly changed in solutions containing a large PEG molecule, but decreases in solutions containing a small PEG molecule due to specific interaction with single strands and reduction in the water activity [11–14].
RNA secondary structures can be predicted by using the thermodynamic equilibrium parameters based on the nearest-neighbor model [15]. Since the structural stability of nucleic acids is strongly dependent on the salt concentration, empirical expressions have been developed for evaluating the salt concentration dependence of the thermodynamic stability of oligonucleotide structures [16,17]. Intriguingly, the salt concentration dependence of the stability of DNA duplexes changes in mixed solutions with PEG [11,18–20]. A detailed study of the nature of the salt concentration dependence is important for understanding the factors responsible for the molecular crowding effects and for predicting nucleic acid structures under non-dilute conditions. In this study, we investigated the hammerhead ribozyme activity and the thermodynamic stability of an RNA oligonucleotide duplex under the non-dilute conditions using mixed solutions of water and water-soluble neutral compounds. The hammerhead ribozyme is one of the smallest RNA catalysts that utilize metal ions to hydrolyze a target RNA sequence. Since divalent ions promote a rapid cleavage, many studies have been performed using Mg2+ [21]. The Mg2+ ions contribute to tertiary folding, and the binding is site-specific in some cases [22,23]. It is a remarkable feature that the ribozyme motif exhibits the catalytic activity when the reaction buffer contains monovalent metal ions at high concentrations instead of Mg2+ [24,25]. Monovalent ions do not supposedly play an essential role in the catalytic step, but predominantly assist in the folding of an inactive conformation with a disordered catalytic core into the catalytically active conformation [26]. A molecular dynamics simulation study has demonstrated that the binding patterns of Na+ ions necessary for the formation of a catalytically active conformation are not as rigid as those of Mg2+ [27]. The relatively nonspecific electrostatic interactions with Na+ ions are useful for assessing the influence of non-dilute conditions on RNA folding and its salt concentration dependence. The present study measures the rates of the hammerhead ribozyme-catalyzed RNA cleavage mediated by NaCl and the thermodynamic stability of a short RNA duplex, using aqueous solutions containing a high concentration of neutral additives (referred to as cosolutes), such as PEG, small primary alcohols, amide compounds, and aprotic solvent molecules. This study illustrates correlations of the dielectric constant with the magnitude of the salt concentration dependence of the cleavage rate and the base-pair stability. The results are useful when constructing a prediction model for nucleic acid structures under non-dilute conditions and also when modulating the structural stability by changing the solution composition.
2. Results
2.1. The hammerhead ribozyme reaction mediated by NaCl
RNA sequences used for the ribozyme-catalyzed RNA cleavage are the minimum sequence motif of the trans-acting hammerhead ribozyme and the substrate strand labeled with a fluorophore, as shown in Fig. 1A. The substrate RNA is site-specifically cleaved at a phosphodiester bond located in the catalytic site at the junction of three stems forming the pairwise coaxial stacking [23,26]. We confirmed that the reaction rapidly proceeded in solutions containing Mg2+ at several to several tens of millimolar, with a half-life of less than 1 min; but there was no reaction without adding the divalent metal ion to the solution. The cleavage reaction also proceeded in solutions containing NaCl at high concentrations instead of MgCl2. Although the reaction rate with NaCl was much slower than that with MgCl2, the site-specific substrate cleavage was observed (Fig. 1B). It was previously proposed that the ribozyme structure in the absence of divalent metal ions is extended with a disordered core and no pairwise coaxial stacking [26]. The binding of Na+ ions converts to the more condensed, catalytic active form in equilibrium with inactive species.
Fig. 1.
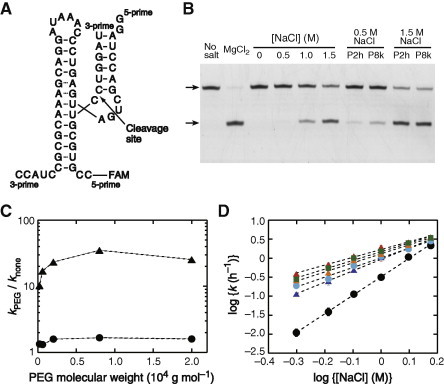
(A) RNA sequences of the hammerhead ribozyme and the FAM-labeled substrate RNA used in this study. (B) The polyacrylamide gel electrophoresis for the ribozyme reaction for 30 min with 10 mM MgCl2 or 0, 0.5, 1.0, or 1.5 M NaCl. The arrowheads indicate the uncleaved (upper) and cleaved (lower) fragments of the substrate RNA. The reactions with NaCl in the presence of PEG200 (P2h) or PEG8000 (P8k) at 20 wt% are also presented. (C) Changes in the reaction rate constant by the addition of PEG of different molecular weights, expressed as the ratio of the rate constants with (kPEG) and without PEG (knone), in the solutions containing 1.5 M (circles) or 0.5 M NaCl (triangles). (D) The NaCl concentration dependence of k in the absence (black) and presence of PEG200 (blue), PEG600 (cyan), PEG2000 (orange), PEG8000 (red), or PEG20000 (green) at 20 wt%. The correlation coefficients of a linear fit are greater than 0.995. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The observed reaction rate constant, k, was greatly changed depending on the concentration of NaCl, e.g., 2.1 h−1 with 1.5 M NaCl, 0.31 h−1 with 1 M NaCl, and 0.011 h−1 with 0.5 M NaCl. A linear correlation was obtained between the common logarithm of the rate constant (log k) and that of NaCl concentration (log [NaCl]) (Fig. S1A, in the Supporting information), suggesting that the rate-limiting step remains unchanged within the concentration range.
2.2. RNA cleavage rates in the presence of PEG
The reaction with NaCl was investigated using the solutions containing PEG at 20 wt%, comparable to the amount of biomolecules present in cells [28,29]. The effects of PEG with different average molecular weights (g mol−1) ranging from 2 × 102 to 2 × 104 (PEG200, PEG600, PEG2000, PEG8000, and PEG20000) were compared. As observed in a previous study on the reaction with Mg2+ [30], the RNA cleavage rate was increased by the addition of PEGs of different molecular weights (Fig. 1B). The rate acceleration was greater with a larger PEG, but saturated at a molecular weight exceeding 2 × 103 (Fig. 1C). Remarkably, the PEG-induced rate acceleration was more pronounced with moderate rather than high concentrations of NaCl, e.g., 35-fold acceleration of the rate constant with 0.5 M NaCl compared to a maximum acceleration of 1.7-fold with 1.5 M NaCl.
Linear dependences between log k and log [NaCl] with a correlation coefficient greater than 0.995 were obtained, as shown in Fig. 1D. The slope of the plot was changed in the PEG-containing solutions: the slope was 4.9 without cosolutes, and the slope became less steep as the size of the PEG increased, e.g., 2.1 with PEG8000. The results indicate that the PEG-induced rate accelerations, which were more significant at low NaCl concentrations, resulted from the reduced dependence on the salt concentration, and this effect is more significant with large PEG molecules.
2.3. Ribozyme reactions in other mixed solutions
We also employed the following compounds that are inert or non-inert to RNA: ethylene glycol (EG, the monomer unit of PEG); glycerol (Glyc), 1,3-propanediol (PDO), 2-methoxyethanol (MME), and 1,2-dimethoxyethane (DME) (structurally related to EG); methanol (MeOH), ethanol (EtOH), and 1-propanol (PrOH) (small primary alcohols); urea and formamide (FA) (amide compounds, known as nucleic acid denaturants); N-methylformamide (NMF), dimethylformamide (DMF), and acetamide (AcAm) (structurally related to FA); acetonitrile (AcCN), dimethyl sulfoxide (DMSO), tetrahydrofuran (THF), and 1,4-dioxane (DOX) (aprotic solvent molecules); and polysaccharide dextran (Dex) with an average molecular weight 1 × 104. Many of these compounds changed the cleavage rate (Fig. S1B), giving the rate constant range of 0.0029–3.7 h−1 with 1.5 M NaCl, as compared in Fig. 2. In particular, urea and FA significantly decreased the rate constant by 100-fold, consistent with their role as nucleic acid denaturants. The FA-like compounds of NMF, DMF, and AcAm also decreased the value by 3.9- to 6.5-fold, whereas many of the other compounds increased or had slight effects on the rate constant. Some of the cosolute effects are consistent with prior studies measuring the hammerhead ribozyme activity with Mg2+ in the mixed solutions with Glyc or MeOH [31]. Dex as a polymer molecule had a lower influence on the cleavage rate, but the small primary alcohols that do not cause a large steric effect increased the rate constant as much as large PEG molecules did. The same effects of these compounds on the ribozyme catalysis were observed when the data at low NaCl concentrations were compared. The observations suggest that factors besides the excluded volume effect have major effects. It is a remarkable finding that these cosolute molecules changed the slope of the linear plot of log k versus log [NaCl] (Fig. S1C), as summarized in Table 1. The slope values had a weak correlation with the k values at a certain NaCl concentration, e.g., 1.5 M NaCl (Fig. S1D).
Fig. 2.
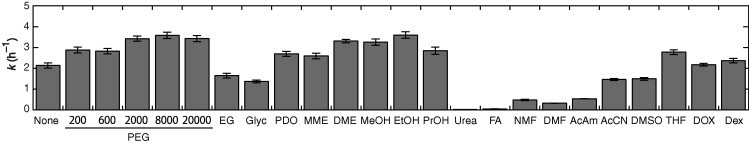
Rate constants for the ribozyme reaction in the 1.5 M NaCl solutions without and with cosolutes at 20 wt%.
Table 1.
Dependence of the rate constant of the ribozyme-catalyzed RNA cleavage on the NaCl concentration.
| Cosolute | Slope | Cosolute | Slope | Cosolute | Slope |
|---|---|---|---|---|---|
| None | 4.9 | PDO | 2.7 | NMF | 4.7 |
| PEG200 | 3.0 | MME | 2.6 | DMF | 3.4 |
| PEG600 | 2.5 | DME | 2.1 | AcAm | 4.7 |
| PEG2000 | 2.4 | MeOH | 2.3 | AcCN | 3.1 |
| PEG8000 | 2.1 | EtOH | 2.2 | DMSO | 3.3 |
| PEG20000 | 2.3 | PrOH | 2.5 | THF | 2.8 |
| EG | 4.5 | Urea | 6.1 | DOX | 2.9 |
| Glyc | 4.5 | FA | 5.6 | Dex | 3.6 |
Values represent the slope of the plot of log k versus log [NaCl] obtained using the mixed solutions with a cosolute at 20 wt%. Each error value is within 0.2.
2.4. Stability of RNA base pairs in the mixed solutions
We also investigated the thermodynamic stability of RNA base pairs in the mixed aqueous solutions. In general, the base-pair formation is accompanied by the association of metal ions, and the ions preferentially interact with the phosphate groups through the Coulomb interaction, without obvious site specificity [32]. We prepared an 11-mer RNA duplex 5′-AUCCAGCGCGC-3′/5′-GUGCGCUGGAU-3′, derived from the base-pair sequence formed by the hammerhead ribozyme and the substrate RNA. Circular dichroism (CD) spectra of the duplex displayed no obvious change in the signal in the mixed solutions (Fig. S2A), suggesting that the double-helical structure is not significantly affected by the cosolutes although the spectra are not particularly sensitive to subtle or local conformational changes.
The values of the free energy change for the duplex formation determined by UV melting curve analysis showed that no cosolutes, excluding those with PEG8000 (phase separation at high temperatures), PEG20000 (phase separation at high temperatures), and THF (evaporated at high temperatures) for which measurements could not be obtained, increased the duplex stability at 1 M NaCl (Fig. S2B). As found in the ribozyme study, the amide compounds had the strongest effect, e.g., FA decreased the Tm by 9 °C and −ΔG° by 2.8 kcal mol−1 (Fig. 3A), consistent with previous reports that the stability of DNA duplexes was decreased in the presence of FA or urea due to interactions with unpaired bases through the formation of multiple hydrogen bonds [33–35]. EG, EG-like compounds (Glyc, PDO, MME, and DME), primary alcohols (MeOH, EtOH, and PrOH), and aprotic compounds (AcCN, DMSO, THF, and DOX), which increased or had slight effects on the RNA cleavage rate, significantly decreased the value of −ΔG°. Small PEG molecules decreased the duplex stability (1.6 kcal mol−1 by PEG200 and 0.9 kcal mol−1 by PEG600), which can be explained by specific interactions with single-stranded bases [13] and reduction in the water activity [14]. Small cosolutes may act as osmolytes that reduce the activity of water and increase the osmotic pressure [36,37]. The solutions containing PEG or the amide compounds have moderate water activities (0.94−0.96), and those containing EG, the EG-like compounds, the small primary alcohols, and the aprotic compounds have relatively low values (0.93 or less). We found a weak correlation of the −ΔG°, excluding those of the amide compounds, with the logarithm of the water activity, while no correlation was observed with the viscosity (Fig. S3A and B). In addition, the k values of the ribozyme reaction showed no clear correlation with these solution property parameters (Fig. S3C and D), and there was no strong correlation between the values of k and −ΔG° (Fig. S3E).
Fig. 3.
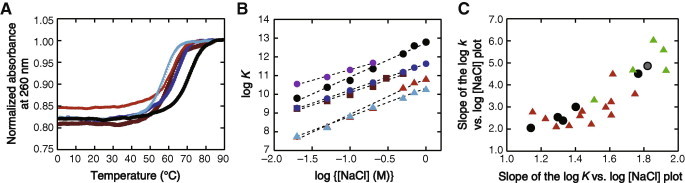
(A) Melting curves of the 11-mer RNA duplex in the 1 M NaCl solutions without (black) and with PEG200 (blue), EtOH (brown), FA (red), or DMF (cyan) at 20 wt%. (B) Plots of log K versus log [NaCl]. The data for PEG8000 (purple) at high NaCl concentrations were not available due to phase separation. The correlation coefficients of a linear fit are greater than 0.990. (C) Plots between the NaCl concentration dependence of log k and that of log K. The data using PEG and EG are indicated by black symbols, and those using the amide compounds and other compounds are indicated in green and red, respectively. The data using the solution without cosolutes are indicated in gray. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Some organic compounds may cause microphase separation or decrease the solubility of NaCl, which could increase the effective concentration of NaCl. The excluded volume effect of large PEG molecules also increases the effective concentration of NaCl, and the effect becomes more significant when a large PEG is used. These effects on the salt concentration are expected to enhance the duplex stability; however the studied mixed solutions did not increase the stability of the RNA duplex, excepting those containing large PEG molecules at low concentrations of NaCl. Fig. 3B shows the comparison of the effect of the duplex stability on the NaCl concentration. Linear dependences between log K (= −ΔG°/2.303RT, where R is the gas constant and T is the absolute temperature) and log [NaCl] with a correlation coefficient greater than 0.990 were obtained, where the data for the small primary alcohols at high salt concentrations and those for large PEG molecules causing phase separation at high salt concentrations and high temperatures were omitted from the linear regression analysis. The slope value 1.8 in the absence of cosolutes was within those previously obtained for DNA and RNA structures, e.g., ranging from 0.6 to 4 which varied depending on the length and structure [19,20,38]. The slope changed when using the mixed solutions: for example, the slope value decreased to 1.1 in the PEG8000-containing solution. The reduction in the dependence on the salt concentration was also reported in previous studies using DNA duplexes with PEG [11,18–20] and RNA tertiary structures with trimethylamine oxide (TMAO) [38]. Our results show that many of the mixed solutions changed the magnitude of the salt concentration dependence of the duplex stability. In contrast, the slope value did not decrease in the FA- or urea-containing solutions. This property is important to keep their role as nucleic acid denaturants at low salt concentrations. Taken together with the ribozyme data, there is found a correlation between the NaCl concentration dependence of log k and that of log K (Fig. 3C) even though the ribozyme activity depends not only on the stability of secondary structures but also on tertiary interactions.
2.5. Correlation with the dielectric constant
We then considered the relative dielectric constant, ɛr. An inclusion of an organic compound having a low dielectric constant lowers the dielectric constant of a solution [39,40]. Most of the studied compounds decrease the dielectric constant, but some amide compounds have a unique dielectric property: the addition of FA or NMF increases the value of ɛr while DMF decreases the value. The studied solutions have values ranging from 55 to 100 (Fig. S4). Although the creation of a uniform dielectric medium is assumed and the dielectric constant decreases with the salt concentration and the temperature [41], there is some correlation between the values of ɛr−1 and k of the ribozyme reaction (Fig. 4A). More strikingly, the inverse of ɛr correlated well with the NaCl concentration-dependent data given in Table 1: the slope value decreases as ɛr−1 increases, except in the case of NMF (ɛr−1 = 0.010), as shown in Fig. 4B. The deviation for the NMF-containing solution having the highest dielectric constant might be due to the overestimation of ɛr caused by a nonhomogeneous distribution in the solution. Furthermore, the x-intercept of the log k versus log [NaCl] plots also showed a correlation with the ɛr−1 (Fig. 4C). It is emphasized that the data points obtained using the mixed solutions with amide compounds, but excluding NMF, fell along the same lines of the correlation plots. The data obtained using the solutions with different cosolute amounts ranging from 5 to 20 wt% and the binary mixture solutions of PEG200, PEG8000, EtOH, and FA followed the same lines of the plots, but no correlation was observed with the water activity or viscosity (Fig. S5). The same correlations with the ɛr−1 were also found for the duplex stability regardless of the cosolute property of interacting with RNA, but no correlation was observed between the values of −ΔG° and ɛr−1 (Fig. 4D–F).
Fig. 4.
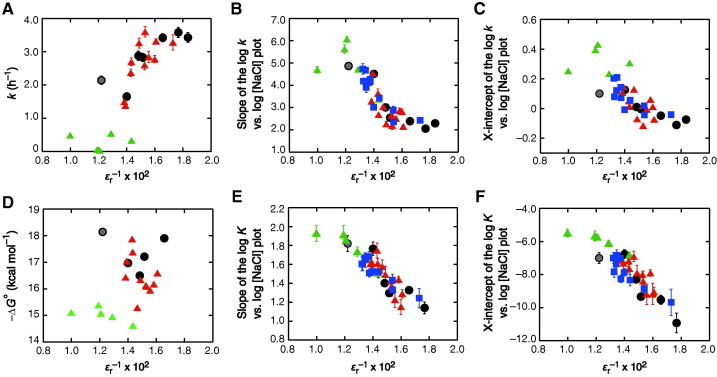
(A) Plots of k obtained with 1.5 M NaCl versus ɛr−1 of the solutions. Symbols are the same as in Fig. 3C. (B and C) Plots of the slope or x-intercept of the log k versus log [NaCl] plot against ɛr−1. Symbols are the same as in Fig. 3C, and the data using the solutions with different cosolute amounts (5 or 10 wt%, given in Fig. S5) and the binary mixture solutions of PEG200 and PEG8000, PEG200 and EtOH, PEG200 and FA, or EtOH and FA, each at 10 wt%, are indicated in blue. The correlation coefficients of a linear fit, excluding the data using NMF (ɛr−1 = 0.010), are 0.861 in panel B and 0.772 in panel C. (D) Plots between −ΔG° of the RNA duplex at 1 M NaCl and ɛr−1. The data using PEG8000, PEG20000, and THF are excluded because of the phase separation or evaporation at high temperature during thermal melting. Symbols are the same as in Fig. 3C. (E and F) Plots of the slope or x-intercept of the log K versus log [NaCl] plot against ɛr−1. The correlation coefficients of a linear fit, excluding the data using NMF, are 0.906 in panel E and 0.888 in panel F. Symbols are the same as in panels B and C. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Discussion
3.1. Influence of the mixed solutions on the ribozyme catalytic activity
This study investigated the RNA cleavage rate of the hammerhead ribozyme and the thermodynamic stability of RNA base pairs in the solutions containing NaCl. The experiments were performed with Na+ rather than Mg2+ because of several benefits. First, monovalent ions are less likely to associate with cosolute molecules. Second, the association of Na+ ions is mostly nonspecific through electrostatic interactions. The ion binding enhances the activity of the hammerhead ribozyme by increasing the population of the catalytically active conformation relative to inactive conformations. Third, Na+ ions weakly but cooperatively associate with RNA, which can be analyzed by a linear regression method presented in Figs. 1D and 3B. The plot of the NaCl concentration dependence provides a steeper slope than the case of MgCl2 because a greater number of monovalent ions are required for the charge neutralization than when using divalent ions [42]. This facilitates the comparison of the salt concentration-dependent data obtained using different solutions.
The studies on the effect of PEG on the hammerhead ribozyme showed increased rates of RNA cleavage in the solutions containing large PEG molecules. The results are consistent with the excluded volume effect that may increase the effective concentration and restrict the degree of freedom of RNA molecules [10]. However, the k value did not continuously increase with the molecular weight of PEG. The result suggests that the simple excluded volume model cannot fully explain the PEG effects. The k values also have no clear correlation with the water activity, viscosity, or RNA base-pair stability of the 11-mer duplex, but have some correlation with the ɛr−1 (Fig. 4A). A simple interpretation of the correlation between the rate constant and the dielectric constant is that the transition state is electrostatically stabilized in less-polar media. However, since the rate of RNA cleavage with monovalent ions is controlled by the folding process rather than by the catalytic step, it is more likely that the low dielectric environment enhances the folding into the catalytically active conformation mediated by sodium ion binding (discussed later). It has been reported that the RNA cleavage rates of the hairpin ribozyme and the lead-dependent ribozyme (leadzyme) are accelerated in the mixed solutions with PEG (MW = 4 × 102): the acceleration of the hairpin ribozyme reaction with Mg2+ was attributed to the stabilization of the catalytically active conformation under the PEG-mediated osmotic pressure, and not to the influence of the dielectric property [43,44]. The acceleration of the leadzyme reaction with Pb2+ was also attributed to the osmotic pressure effects by PEG [45]. On the other hand, the study for the group I intron ribozyme motif showed that the PEG (MW = 1 × 103)-containing solution favored a more compact structure of the large RNA molecule and enabled the folding with lower Mg2+ concentrations, attributed to the dominant influence of the excluded volume effects [46]. It is possible that the origin of the PEG effect varies among the different ribozyme motifs, due to the differences in the structure and stability of RNA, molecular sizes of RNA and PEG, rate-limiting step, and contribution of ions and waters to the reactions.
3.2. Prediction method in the mixed solutions
It is convenient for practical use to predict nucleic acid structures and their stability under non-dilute conditions. The thermodynamic parameters based on the nearest-neighbor interaction model can be used for the predictions of the hybridization energy and secondary structures of nucleic acids [15], and we have previously found the applicability of the nearest-neighbor model for DNA structure formations in several mixed solutions [14]. It was reported that EG, MeOH, EtOH, PrOH, urea, FA, DMF, DMSO, and DOX destabilized a short RNA duplex, and there was found no good correlation between the duplex stability and the viscosity, dipole moment, surface tension, or solubility of the nucleotide adenine [47]. We demonstrated in this study that there was no correlation between the −ΔG° of the 11-mer RNA duplex and the dielectric constant (Fig. 4D). These results are probably due to significant energetic contributions from specific interactions with several cosolutes (e.g., amide compounds) and from base pairing in a relatively low water activity environment (e.g., EG, Glyc, and small primary alcohols).
The dependence of the duplex stability on the NaCl concentration was characterized by a linear correlation between log K and log [NaCl], as shown in Fig. 3B. Although increasing the ionic strength may replace cosolute molecules that possibly accumulate at the nucleic acid surface, the linear correlations suggest that the replacements could have only minor effects on the data analysis. To evaluate the salt concentration dependence, empirical expressions that take into account the number of phosphate charges contributing to base pairing and the Coulombic end effects in ion accumulation have been developed [16,17]. Our results demonstrate that the salt concentration dependence changes as a function of the dielectric constant (Fig. 4E), which could have some relevance to the phosphate charge effect and the Coulombic end effect. Based on the finding, the value of −ΔG° of an RNA duplex under non-dilute conditions may be predicted by considering a possibility of specific interactions with coexisting compounds, water activity effects, and dielectric constant effects, for example, by using the following calculation: −ΔG° at a certain salt concentration in a non-dilute solution = (−ΔG° at a standard condition, e.g., 1 M NaCl in a dilute solution) + (energy penalty due to specific interactions and the reduced water activity) + (a term involving the dielectric constant, possibly combined with the phosphate charge effect and the end effect) × log[salt]. More comprehensive studies using other sequences and concerning the energy penalty and sequence- and length-dependent dielectric constant effects will contribute to nucleic acid folding predictions under non-dilute conditions.
3.3. Dielectric constant effects on nucleic acids
Based on the polyelectrolyte model of nucleic acids, the magnitude of the dependence of duplex stability on the salt concentration is related to the thermodynamic degree of ion accumulation near nucleic acids [48–50]. Our finding that mixed solutions changed the NaCl concentration dependence (the slope of the linear plots in Fig. 3B) suggests altered binding properties of salt ions to RNA molecules in the solution conditions created by organic compounds. It can therefore be assumed that the linear correlations between the salt concentration dependence data and the ɛr−1 indicate the significance of electrostatic ion binding to RNA, mediated by Coulomb interaction that is inversely proportional to the dielectric constant. In addition to the slope of the linear plots, their x-intercept values have a correlation with the ɛr−1 (Fig. 4C and F). The environment with a low dielectric constant would enhance the efficiency of ion binding to RNA molecules, and is favorable both for the ribozyme catalysis and for the duplex formation with a lower amount of NaCl, and one of such compounds is PEG having a molecular weight of more than several thousands. In contrast, urea and FA providing a medium of high dielectric constant effectively destabilize the RNA structures in both high and low salt concentrations. Their role as nucleic acid denaturants could be attributed not only to the formation of hydrogen bonding with single-stranded bases, but also to the property of not creating a low dielectric constant condition.
It is noted that the values of the dielectric constant may not accurately reflect the local environment near the RNA surface. However, it is reported that ions distant from an RNA surface interact significantly with an RNA, and long-range interactions are suggested to be important in ion-induced stabilization for RNA structures [51]. Our observations imply that ions at some distance from the RNA surface, influenced by the bulk dielectric constant, play an important role in the stabilization of the RNA structures: sodium ions are distributed around the phosphate backbone and strong electronegative positions in the active site for both the reactant and activated precursor states of the hammerhead ribozyme [27], and a number of sodium ions bind primarily to the phosphate groups of a base-paired duplex [32]. Importance of the dielectric constant on nucleic acid reactions involving metal ion binding has also been proposed for the multivalent ion-mediated DNA condensation and aggregation [52,53], thermal stability of a DNA G-quadruplex structure formed by coordination of K+ ions [54], structural changes in a functionally important bulge region of the group II intron ribozyme and its Mg2+ binding properties [55], and dimerization of RNA or DNA hairpins induced by metal ion binding [56]. A relatively low dielectric constant of about 50 or even less, which is significantly lower than the value for pure water of approximately 80, is suggested for the inside of intact cells [57,58]. A computer simulation study predicts a reduced dielectric constant values in the range of 30–50 under the molecular crowding condition [59]. In addition to the intracellular environment, the mobility and arrangements of water near the surface of nucleic acid biosensors are highly restricted, and thus the dielectric constant of the medium of a nucleic acid probe is significantly lower than that of a dilute solution. There would be important contributions of the low dielectric constant environment inside of cells and near the surface of a biosensor to the ribozyme activity and the stability of nucleic acid structures. The molecular crowding effects on nucleic acids are well explained by the excluded volume effect, the osmotic pressure effect, or specific interactions, and it would also be important to consider the effects of the change in the dielectric constant in the molecular crowding studies using mixed solutions with organic compounds.
4. Conclusions
We have found that many types of mixed solutions change the rate of the ribozyme-catalyzed RNA cleavage and its dependence on the concentration of NaCl. The solutions also affect the stability of RNA base pairs and its dependence on the salt concentration. Specific interactions with RNA molecules (e.g., urea and FA) and the reduced water activity (e.g., EG, Glyc, and small primary alcohols) can explain their inhibiting effects on the ribozyme catalysis and decreasing effects on the duplex stability, but they do not explain the changes in their NaCl concentration dependence. We found good correlations of the dielectric constant with the salt concentration dependence of the ribozyme reaction rate and the RNA base-pair stability. The observation provides useful insights when constructing a prediction model for RNA structures under non-dilute conditions and also when modulating the stability by changing the solution composition.
5. Materials and methods
5.1. Preparation of RNA oligonucleotides and buffer solutions
The hammerhead ribozyme sequence derived from Schistosoma mansoni was enzymatically prepared as previously described [30]. The substrate RNA labeled with 6-carboxyfluorescein (FAM) at the 5′-end and the 11-mer RNA oligonucleotides, purified by high-performance liquid chromatography (HPLC), were purchased from Hokkaido System Science.
All reagents used to prepare buffer solutions were purchased from Wako with the following exceptions: 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and the disodium salt of ethylenediamine-N,N,N′,N′-tetraacetic acid (Na2EDTA) from Dojindo, PEG8000 and dextran with an average molecular weight 1 × 104 from Sigma, and 2-methoxyethanol and 1,2-dimethoxyethane from TCI. All reagents were used without further purification. Cosolute molecules were added to buffer solutions at 20 wt%, unless otherwise mentioned. No obvious precipitation and phase separation were observed, excluding the experiments using large PEG molecules at high salt concentrations and high temperatures.
5.2. Ribozyme-catalyzed RNA cleavage
The RNA cleavage rate of the hammerhead ribozyme was measured under the single-turnover conditions, in which the ribozyme (2 μM) was present in excess relative to the substrate (100 nM). The reaction was performed at 37 °C using a buffer solution consisting of 50 mM HEPES and 1 mM Na2EDTA at pH 7.0. No significant contamination of divalent metal ions was confirmed from the experiments using higher concentrations of Na2EDTA.
The RNA solution was annealed in the buffer solution at 60 °C and then incubated at 37 °C for 10 min, followed by the addition of salts to start the reaction, during which the addition of NaCl changed the solution pH by no more than 0.13. The quenched reaction mixture was loaded onto a 20% polyacrylamide gel (acrylamide:bisacrylamide = 19:1) containing 7 M urea. After the gel electrophoresis, the fluorescence emission from the FAM-labeled RNA strands was visualized and quantified by a fluorescent scanner (FLA-5100, Fuji) using a 473-nm excitation laser and a 520-nm emission filter.
The observed rate constant, k, for the ribozyme-catalyzed cleavage of the substrate RNA was determined from a plot of the RNA fraction cleaved at the correct site, f, versus time, t, fitted to the single-exponential equation of f = f0 + (fmax − f0) [1 − exp(−kt)], where f0 is the fraction at time zero and fmax is the fraction at the endpoint of the reaction. The fitting was performed using KaleidaGraph 4.1 (Synergy). A linear approximation of the equation was used to calculate the rate constant of very slow reactions.
5.3. CD spectra of an RNA duplex
CD spectra were obtained using a spectropolarimeter (J-820, JASCO). All spectra were measured at 5 °C using an RNA strand concentration of 40 μM in a buffer consisting of 1 M NaCl, 10 mM Na2HPO4, and 1 mM Na2EDTA at pH 7.0. The RNA solution was heated to 80 °C and cooled at the rate of 2 °C min−1 prior to use.
5.4. Thermodynamic stability of an RNA duplex
The thermal melting curve of the RNA duplex was obtained by monitoring the absorption at 260 nm using a spectrophotometer (UV-1800, Shimadzu). The melting curve was measured using an RNA strand concentration of 2 μM in the phosphate buffer. The heating rate was 1 °C min−1, and the cuvette was sealed with an adhesive sheet to prevent evaporation. The melting temperature, Tm, at which half of the duplex structure was denatured, the association equilibrium constant, K, and the free energy change, ΔG°, at 37 °C were determined from the melting curve [60].
5.5. Properties of the mixed aqueous solutions
The water activity was determined by the osmotic pressure method using vapor phase osmometry (5520XR pressure osmometer, Wescor) or freezing point depression osmometry (Typ Dig.L osmometer, Halbmikro). The relative dielectric constant was calculated according to the equation reported by Oster or experimentally determined using a fluorescent probe, 1-anilino-8-naphthalene sulfonate, as previously described [56]. The solution viscosity was measured with a viscometer (SV-10 vibro viscometer, A&D). These solution property parameters were measured using the buffer solutions containing 1 M NaCl and 20 wt% cosolute at 25 °C, unless otherwise mentioned.
Acknowledgements
We thank Akiko Matsuyama and Junpei Ueno for technical assistance. We also thank Prof. Philip C. Bevilacqua (Department of Chemistry, Pennsylvania State University, USA) for critical reading and English editing. This work was supported in part by Grants-in-Aid for Scientific Research from JSPS (Japan Society for the Promotion of Science) (No. 24550200) and MEXT (Ministry of Education, Culture, Sports, Science and Technology in Japan)-Supported Program for the Strategic Research Foundation at Private Universities, 2009-2014.
Appendix A. Supplementary data
Supporting Information.
References
- 1.Johansson G., Walter H. Partitioning and concentrating biomaterials in aqueous phase systems. Int. Rev. Cytol. 2000;192:33–60. doi: 10.1016/s0074-7696(08)60521-5. [DOI] [PubMed] [Google Scholar]
- 2.Al-Habori M. Macromolecular crowding and its role as intracellular signalling of cell volume regulation. Int. J. Biochem. Cell Biol. 2001;33:844–864. doi: 10.1016/s1357-2725(01)00058-9. [DOI] [PubMed] [Google Scholar]
- 3.Vazquez A., Oltvai Z.N. Molecular crowding defines a common origin for the Warburg effect in proliferating cells and the lactate threshold in muscle physiology. PLoS One. 2011;6:e19538. doi: 10.1371/journal.pone.0019538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou H.X., Rivas G., Minton A.P. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakano S., Miyoshi D., Sugimoto N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem. Rev. 2014;114:2733–2758. doi: 10.1021/cr400113m. [DOI] [PubMed] [Google Scholar]
- 6.Peterson A.W., Wolf L.K., Georgiadis R.M. Hybridization of mismatched or partially matched DNA at surfaces. J. Am. Chem. Soc. 2002;124:14601–14607. doi: 10.1021/ja0279996. [DOI] [PubMed] [Google Scholar]
- 7.Ricci F., Lai R.Y., Heeger A.J., Plaxco K.W., Sumner J.J. Effect of molecular crowding on the response of an electrochemical DNA sensor. Langmuir. 2007;23:6827–6834. doi: 10.1021/la700328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jayaraman A., Hall C.K., Genzer J. Computer simulation study of probe-target hybridization in model DNA microarrays: effect of probe surface density and target concentration. J. Chem. Phys. 2007;127:144912. doi: 10.1063/1.2787618. [DOI] [PubMed] [Google Scholar]
- 9.Nakano S., Kanzaki T., Nakano M., Miyoshi D., Sugimoto N. Measurements of the binding of a large protein using a substrate density-controlled DNA chip. Anal. Chem. 2011;83:6368–6372. doi: 10.1021/ac201312d. [DOI] [PubMed] [Google Scholar]
- 10.Minton A.P. Molecular crowding: analysis of effects of high concentrations of inert cosolutes on biochemical equilibria and rates in terms of volume exclusion. Methods Enzymol. 1998;295:127–149. doi: 10.1016/s0076-6879(98)95038-8. [DOI] [PubMed] [Google Scholar]
- 11.Goobes R., Kahana N., Cohen O., Minsky A. Metabolic buffering exerted by macromolecular crowding on DNA–DNA interactions: origin and physiological significance. Biochemistry. 2003;42:2431–2440. doi: 10.1021/bi026775x. [DOI] [PubMed] [Google Scholar]
- 12.Nakano S., Karimata H., Ohmichi T., Kawakami J., Sugimoto N. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J. Am. Chem. Soc. 2004;126:14330–14331. doi: 10.1021/ja0463029. [DOI] [PubMed] [Google Scholar]
- 13.Knowles D.B., LaCroix A.S., Deines N.F., Shkel I., Record M.T., Jr. Separation of preferential interaction and excluded volume effects on DNA duplex and hairpin stability. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12699–12704. doi: 10.1073/pnas.1103382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano S., Yamaguchi D., Tateishi-Karimata H., Miyoshi D., Sugimoto N. Hydration changes upon DNA folding studied by osmotic stress experiments. Biophys. J. 2012;102:2808–2817. doi: 10.1016/j.bpj.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markham N.R., Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.SantaLucia J., Jr. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shkel I.A., Record M.T., Jr. Effect of the number of nucleic acid oligomer charges on the salt dependence of stability (ΔG°37) and melting temperature (Tm): NLPB analysis of experimental data. Biochemistry. 2004;43:7090–7101. doi: 10.1021/bi036225e. [DOI] [PubMed] [Google Scholar]
- 18.Spink C.H., Chaires J.B. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry. 1999;38:496–508. doi: 10.1021/bi9820154. [DOI] [PubMed] [Google Scholar]
- 19.Karimata H., Nakano S., Sugimoto N. Effects of polyethylene glycol on DNA duplex stability at different NaCl concentrations. Bull. Chem. Soc. Jpn. 2007;80:1987–1994. [Google Scholar]
- 20.Nakano S., Wu L., Oka H., Karimata H.T., Kirihata T., Sato Y., Fujii S., Sakai H., Kuwahara M., Sawai H., Sugimoto N. Conformation and the sodium ion condensation on DNA and RNA structures in the presence of a neutral cosolute as a mimic of the intracellular media. Mol. BioSyst. 2008;4:579–588. doi: 10.1039/b718806d. [DOI] [PubMed] [Google Scholar]
- 21.Stage-Zimmermann T.K., Uhlenbeck O.C. Hammerhead ribozyme kinetics. RNA. 1998;4:875–889. doi: 10.1017/s1355838298980876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blount K.F., Uhlenbeck O.C. The structure–function dilemma of the hammerhead ribozyme. Annu. Rev. Biophys. Biomol. Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- 23.Martick M., Scott W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray J.B., Seyhan A.A., Walter N.G., Burke J.M., Scott W.G. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 25.O’Rear J.L., Wang S., Feig A.L., Beigelman L., Uhlenbeck O.C., Herschlag D. Comparison of the hammerhead cleavage reactions stimulated by monovalent and divalent cations. RNA. 2001;7:537–545. doi: 10.1017/s1355838201002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bassi G.S., Murchie A.I., Walter F., Clegg R.M., Lilley D.M. Ion-induced folding of the hammerhead ribozyme: a fluorescence resonance energy transfer study. EMBO J. 1997;16:7481–7489. doi: 10.1093/emboj/16.24.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee T.S., Giambasu G.M., Sosa C.P., Martick M., Scott W.G., York D.M. Threshold occupancy and specific cation binding modes in the hammerhead ribozyme active site are required for active conformation. J. Mol. Biol. 2009;388:195–206. doi: 10.1016/j.jmb.2009.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int. Rev. Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- 29.Ellis R.J. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 30.Nakano S., Karimata H.T., Kitagawa Y., Sugimoto N. Facilitation of RNA enzyme activity in the molecular crowding media of cosolutes. J. Am. Chem. Soc. 2009;131:16881–16888. doi: 10.1021/ja9066628. [DOI] [PubMed] [Google Scholar]
- 31.Feig A.L., Ammons G.E., Uhlenbeck O.C. Cryoenzymology of the hammerhead ribozyme. RNA. 1998;4:1251–1258. doi: 10.1017/s1355838298980943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng Y., Korolev N., Nordenskiold L. Similarities and differences in interaction of K+ and Na+ with condensed ordered DNA. A molecular dynamics computer simulation study. Nucleic Acids Res. 2006;34:686–696. doi: 10.1093/nar/gkj434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blake R.D., Delcourt S.G. Thermodynamic effects of formamide on DNA stability. Nucleic Acids Res. 1996;24:2095–2103. doi: 10.1093/nar/24.11.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong J., Capp M.W., Anderson C.F., Record M.T. Preferential interactions in aqueous solutions of urea and KCl. Biophys. Chem. 2003;105:517–532. doi: 10.1016/s0301-4622(03)00112-1. [DOI] [PubMed] [Google Scholar]
- 35.Hong J., Capp M.W., Anderson C.F., Saecker R.M., Felitsky D.J., Anderson M.W., Record M.T., Jr. Preferential interactions of glycine betaine and of urea with DNA: implications for DNA hydration and for effects of these solutes on DNA stability. Biochemistry. 2004;43:14744–14758. doi: 10.1021/bi049096q. [DOI] [PubMed] [Google Scholar]
- 36.Parsegian V.A., Rand R.P., Rau D.C. Osmotic stress, crowding, preferential hydration, and binding: a comparison of perspectives. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3987–3992. doi: 10.1073/pnas.97.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blandamer M.J., Engberts J.B., Gleeson P.T., Reis J.C. Activity of water in aqueous systems; a frequently neglected property. Chem. Soc. Rev. 2005;34:440–458. doi: 10.1039/b400473f. [DOI] [PubMed] [Google Scholar]
- 38.Lambert D., Leipply D., Draper D.E. The osmolyte TMAO stabilizes native RNA tertiary structures in the absence of Mg2+: evidence for a large barrier to folding from phosphate dehydration. J. Mol. Biol. 2010;404:138–157. doi: 10.1016/j.jmb.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akhadov Y.Y. Pergamon Press; Oxford, UK: 1980. Dielectric Properties of Binary Solutions: A Data Handbook. [Google Scholar]
- 40.Arnold K., Herrmann A., Pratsch L., Gawrisch K. The dielectric properties of aqueous solutions of poly(ethylene glycol) and their influence on membrane structure. Biochim. Biophys. Acta. 1985;815:515–518. doi: 10.1016/0005-2736(85)90381-5. [DOI] [PubMed] [Google Scholar]
- 41.Levy A., Andelman D., Orland H. Dielectric constant of ionic solutions: a field-theory approach. Phys. Rev. Lett. 2012;108:227801. doi: 10.1103/PhysRevLett.108.227801. [DOI] [PubMed] [Google Scholar]
- 42.Nakano S., Fujimoto M., Hara H., Sugimoto N. Nucleic acid duplex stability: influence of base composition on cation effects. Nucleic Acids Res. 1999;27:2957–2965. doi: 10.1093/nar/27.14.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tobe S., Heams T., Vergne J., Herve G., Maurel M.C. The catalytic mechanism of hairpin ribozyme studied by hydrostatic pressure. Nucleic Acids Res. 2005;33:2557–2564. doi: 10.1093/nar/gki552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herve G., Tobe S., Heams T., Vergne J., Maurel M.C. Hydrostatic and osmotic pressure study of the hairpin ribozyme. Biochim. Biophys. Acta. 2006;1764:573–577. doi: 10.1016/j.bbapap.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 45.Giel-Pietraszuk M., Barciszewski J. Hydrostatic and osmotic pressure study of the RNA hydration. Mol. Biol. Rep. 2012;39:6309–6318. doi: 10.1007/s11033-012-1452-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kilburn D., Roh J.H., Guo L., Briber R.M., Woodson S.A. Molecular crowding stabilizes folded RNA structure by the excluded volume effect. J. Am. Chem. Soc. 2010;132:8690–8696. doi: 10.1021/ja101500g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hickey D.R., Turner D.H. Solvent effects on the stability of A7U7p. Biochemistry. 1985;24:2086–2094. doi: 10.1021/bi00329a042. [DOI] [PubMed] [Google Scholar]
- 48.Anderson C.F., Record M.T., Jr. Salt–nucleic acid interactions. Annu. Rev. Phys. Chem. 1995;46:657–700. doi: 10.1146/annurev.pc.46.100195.003301. [DOI] [PubMed] [Google Scholar]
- 49.Record M.T., Jr., Zhang W., Anderson C.F. Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: a practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts. Adv. Protein Chem. 1998;51:281–353. doi: 10.1016/s0065-3233(08)60655-5. [DOI] [PubMed] [Google Scholar]
- 50.Leipply D., Lambert D., Draper D.E. Ion–RNA interactions: thermodynamic analysis of the effects of mono- and divalent ions on RNA conformational equilibria. Methods Enzymol. 2009;469:433–463. doi: 10.1016/S0076-6879(09)69021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soto A.M., Misra V., Draper D.E. Tertiary structure of an RNA pseudoknot is stabilized by “diffuse” Mg2+ ions. Biochemistry. 2007;46:2973–2983. doi: 10.1021/bi0616753. [DOI] [PubMed] [Google Scholar]
- 52.Rau D.C., Parsegian V.A. Direct measurement of the intermolecular forces between counterion-condensed DNA double helices. Evidence for long range attractive hydration forces. Biophys. J. 1992;61:246–259. doi: 10.1016/S0006-3495(92)81831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arscott P.G., Ma C., Wenner J.R., Bloomfield V.A. DNA condensation by cobalt hexammine (III) in alcohol–water mixtures: dielectric constant and other solvent effects. Biopolymers. 1995;36:345–364. doi: 10.1002/bip.360360309. [DOI] [PubMed] [Google Scholar]
- 54.Smirnov I.V., Shafer R.H. Electrostatics dominate quadruplex stability. Biopolymers. 2007;85:91–101. doi: 10.1002/bip.20609. [DOI] [PubMed] [Google Scholar]
- 55.Furler M., Knobloch B., Sigel R.K.O. Influence of decreased solvent permittivity on the structure and magnesium (II)-binding properties of the catalytic domain 5 of a group II intron ribozyme. Inorg. Chim. Acta. 2009;362:771–776. [Google Scholar]
- 56.Nakano S., Hirayama H., Miyoshi D., Sugimoto N. Dimerization of nucleic acid hairpins in the conditions caused by neutral cosolutes. J. Phys. Chem. B. 2012;116:7406–7415. doi: 10.1021/jp302170f. [DOI] [PubMed] [Google Scholar]
- 57.Asami K., Hanai T., Koizumi N. Dielectric properties of yeast cells. J. Membr. Biol. 1976;28:169–180. doi: 10.1007/BF01869695. [DOI] [PubMed] [Google Scholar]
- 58.Tanizaki S., Clifford J., Connelly B.D., Feig M. Conformational sampling of peptides in cellular environments. Biophys. J. 2008;94:747–759. doi: 10.1529/biophysj.107.116236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harada R., Sugita Y., Feig M. Protein crowding affects hydration structure and dynamics. J. Am. Chem. Soc. 2012;134:4842–4849. doi: 10.1021/ja211115q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mergny J.L., Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.


