Highlights
-
•
Overexpression of the ZBR fragment of Emi2, but not of Emi1, induces abnormal cell division.
-
•
The Emi2 ZBR fragment impairs the association of the coactivator Cdc20 with APC/C.
-
•
The Emi2 ZBR fragment inhibits ubiquitylation by the cullin-RING of APC/C and E2C.
-
•
The Emi2 ZBR-specific residues for APC/C inhibitory activity have been identified.
Abbreviations: APC/C, anaphase-promoting complex/cyclosome; Emi2, endogenous meiotic inhibitor 2; ZBR, zinc-binding region; Cdc20, cell-division cycle protein 20; E2C, ubiquitin-conjugating enzyme E2 C or UBE2C
Keywords: APC/C, Emi2, ZBR domain, Cdc20, Ubiquitin ligase activity, UBE2C
Abstract
Anaphase-promoting complex or cyclosome (APC/C) is a multisubunit ubiquitin ligase E3 that targets cell-cycle regulators. Cdc20 is required for full activation of APC/C in M phase, and mediates substrate recognition. In vertebrates, Emi2/Erp1/FBXO43 inhibits APC/C-Cdc20, and functions as a cytostatic factor that causes long-term M phase arrest of mature oocytes. In this study, we found that a fragment corresponding to the zinc-binding region (ZBR) domain of Emi2 inhibits cell-cycle progression, and impairs the association of Cdc20 with the APC/C core complex in HEK293T cells. Furthermore, we revealed that the ZBR fragment of Emi2 inhibits in vitro ubiquitin chain elongation catalyzed by the APC/C cullin-RING ligase module, the ANAPC2–ANAPC11 subcomplex, in combination with the ubiquitin chain-initiating E2, E2C/UBE2C/UbcH10. Structural analyses revealed that the Emi2 ZBR domain uses different faces for the two mechanisms. Thus, the double-faced ZBR domain of Emi2 antagonizes the APC/C function by inhibiting both the binding with the coactivator Cdc20 and ubiquitylation mediated by the cullin-RING ligase module and E2C. In addition, the tail region between the ZBR domain and the C-terminal RL residues [the post-ZBR (PZ) region] interacts with the cullin subunit, ANAPC2. In the case of the ZBR fragment of the somatic paralogue of Emi2, Emi1/FBXO5, these inhibitory activities against cell division and ubiquitylation were not observed. Finally, we identified two sets of key residues in the Emi2 ZBR domain that selectively exert each of the dual Emi2-specific modes of APC/C inhibition, by their mutation in the Emi2 ZBR domain and their transplantation into the Emi1 ZBR domain.
1. Introduction
The post-translational modification of proteins by the covalent attachment of ubiquitin (ubiquitylation) regulates various cellular processes in eukaryotes [1,2]. The ubiquitylation reaction is catalyzed by a set of three enzymes: ubiquitin-activating enzyme, E1; ubiquitin-conjugating enzyme, E2; and ubiquitin ligase, E3. The E3 ligase functions as an adaptor to facilitate the approach of the ubiquitin-charged E2 to the target protein, and thus enables ubiquitin ligation [3]. The E3 ligase and the ubiquitin-charged E2 then perform the elongation step; i.e., the generation of the polyubiquitin chain.
Anaphase-promoting complex or cyclosome (APC/C) is a multisubunit E3 that catalyzes the polyubiquitylation of cell-cycle regulatory proteins (CCRP), in order to trigger their degradation by the 26S proteasome at specific points in the cell cycle [4,5]. The active APC/Cs comprise at least 16 individual subunits, including the coactivator proteins Cdc20 and Cdh1 [6–8]. The APC/C coactivators (Cdc20 and Cdh1) contain the C-box, KILR, and IR motifs, which bind to the core APC/C [9–14], and the WD40 repeat/WD40 domain interacts with the destruction box (D-box/DB) and the KEN box in the target proteins [15–19]. On the other hand, the APC/C subcomplex, formed between subunits 2 (ANAPC2) and 11 (ANAPC11), is the cullin-RING ligase (CRL)-type E3 module. This subcomplex (ANAPC2–11) is sufficient for the elongation step, as it promotes in vitro polyubiquitin chain formation in the presence of E1 and E2, in the absence of the target protein [20,21].
Emi1/Fbx5 (early mitotic inhibitor 1; gene symbol, FBXO5) and Emi2/Erp1/Fbx43 (endogenous meiotic inhibitor 2 or Emi1-related protein 1; gene symbol, FBXO43) constitute the Emi/Erp protein family of APC/C inhibitors against the APC/C function, which controls cell division progress [22,23]. Emi1 is expressed in many somatic tissues, and regulates the G1-to-S phase transition in proliferating cells. In contrast, the expression of Emi2 is restricted to early embryos, spermatocytes, and maturing oocytes [24–32]. Mature oocytes (unfertilized eggs) are arrested at the second metaphase of meiosis (mII) by a cytoplasmic activity, termed cytostatic factor (CSF) [33,34]. CSF prevents the APC/C-Cdc20-mediated degradation of cyclin B [35,36]. Emi2 is the pivotal component of CSF, and is required to maintain mII arrest [26,37–40]. Moreover, Emi2 regulates the timely destruction of APC/C substrates in Xenopus early embryonic divisions [31]. However, the molecular mechanism of APC/C inhibition by Emi2 is still not fully understood.
Emi1 and Emi2 share the F-box and DB motifs, the zinc-binding region (ZBR) with the in-between-RING-fingers (IBR) domain topology and the C6HC-type zinc-binding motif [22,41], and the C-terminal region with a conserved 14-residue sequence ending in the RL residues, termed the RL tail [42]. The phosphorylation status of the C-terminal region controls the interaction between Emi2 and APC/C [43]. The F-box motif (named after cyclin F) interacts with Skp1, and the F-box proteins have been characterized as components of the Skp1-cullin-F-box (SCF) ubiquitin–ligase complexes [44]. Emi1 competes, in a DB-dependent manner, with the target substrate for binding to APC/C-Cdh1 by the “pseudosubstrate inhibitory mechanism” [45,46]. In addition, the DB of Emi2 contributes to its APC/C-Cdc20 binding and morphological abnormalities [40,42].
Emi1 and Emi2 also exhibit DB-independent inhibitory activities against the polyubiquitylation of the APC/C target proteins [41,47,48]. The inhibitory activity of Emi1 requires its C-terminal region, which corresponds to the C-terminal tail of Emi2. The ZBR–RL fragment of Emi1 has inhibitory activity against polyubiquitin chain formation on the target protein by APC/C-Cdh1, in combination with E2C/UBE2C/UBCH10 (ubiquitin chain-initiating E2) and/or E2S/UBE2S (ubiquitin chain-extending E2) [41,48]. The C-terminal tails of Emi1 and Emi2 bind to APC/C, and the RL residues compete with the tail of E2S for APC/C binding [41,42,48,49].
On the other hand, the Emi1 fragments lacking the C-terminal tail do not inhibit the E2C-mediated ubiquitin chain elongation. Nevertheless, point mutations within the ZBR domain in Emi1 decrease the inhibitory activity against E2C-mediated ubiquitin chain elongation [41,48]. Moreover, mutations of the putative zinc-coordinating amino acid residues within the ZBR domain of Emi2 disrupt its CSF activity [40,47,50,51]. These results suggested that the ZBR domains of Emi1/Emi2 contribute to their inhibitory activities, although the functional mechanisms of the ZBR domain have not been clarified [52].
Emi1 and Emi2 co-immunoprecipitated with a recombinant coactivator (Cdc20 or Cdh1) in vitro [26,53]. The DB–ZBR–RL fragment of Emi1 co-immunoprecipitated with the N-terminal fragment of Cdc20 (Cdc20-NT), while the deletion of the ZBR domain impaired the Cdc20-NT binding ability [45,53] The Cdc20-NT contains the C-box motif, which is shared with Cdh1 and involved in binding to the core APC/C [9,13]. Nevertheless, Emi1 does not impair the substrate binding of APC/C [48] and the location of the ZBR domain of Emi1 is distant from that of Cdh1 on APC/C [41]. Therefore, the functional purpose of the binding of the ZBR domain of Emi1/Emi2 to Cdc20/Cdh1 remained elusive.
In the present study, we provide structural and functional information about the Emi2 ZBR domain, toward revealing the potentially novel mechanism for the inhibition of APC/C-Cdc20 activity.
2. Materials and methods
2.1. GenBank accession numbers for multiple amino acid sequence alignment
The following sequences were obtained: MmEmi2/FBXO43, NP_001074722.1; HsEmi2/FBXO43, NP_001025031; GgFBXO43, XP_418357; XlErp1/FBXO43, NP_001093338; DrFBXO43, NP_956725; MmEmi1/FBXO5, NP_080271; HsEmi1/FBXO5, NP_036309; GgFBXO5, XP_419681.3; XlEmi1/Fbxo5-a, NP_001082122; DrFBXO5, NP_001003869; HsRNF14/ARA54, NP_001188294; HsRN216/TRIAD3, NP_996994; HsRN19A/Dorfin, NP_056250; HsRNF31/ZIBRA/HOIP, NP_060469; MmPRKN2/Parkin, NP_057903; HsPRKN2/Parkin, NP_004553; HsARI1/HHARI, NP_005735; HsR144B/p53RFP, NP877434; HsCUL9/PARC, NP_055904. Abbreviations for species names are Mm, Mus musculus (mouse); Hs, Homo sapiens (human); Gg, Gallus gallus (chicken); Xl, Xenopus laevis (African clawed frog); Dr, Danio rerio (zebrafish). Multiple sequence alignments were optimized using the ClustalW program.
2.2. Full-length cDNA clones
The coding sequences (CDSs) corresponding to Emi2, Emi1, and Cdc20 (GenBank: NP_075712) were obtained from mouse mII oocytes, as described previously [26]. The CDS of E2C was derived from the RIKEN full-length enriched mouse cDNA library (Clone ID: 1110015A16), and the CDSs for APC/C subunits ANAPC2 (GenBank: NP_780509.2), ANAPC11 (GenBank: NP_001033319), and ANAPC10 (GenBank: NP_081180) were amplified by PCR from the Mouse 17-day Embryo Marathon-Ready cDNA library (Clontech, Takara). The target DNA fragments were subcloned into the pCR2.1 TOPO vector (Invitrogen, Life Technologies) or appropriate expression vectors.
2.3. Cell culture and phenotypic analysis
HEK293T cells (RIKEN Cell Bank code: RCB2202) were cultured at 37 °C with a 5% CO2 atmosphere in Dulbecco’s modified Eagle’s medium, containing 10% (v/v) heat-inactivated bovine serum and supplemented with penicillin (10 U/ml) and streptomycin (10 μg/ml).
The target Emi2 or Emi1 cDNA fragments were inserted into the pAcGFP1-Hyg-C1 vector (Clontech, Takara) to generate the AcGFP-fusion proteins, and the plasmids were transfected into HEK293T cells using Lipofectamine LTX (Invitrogen, Life Technologies), according to the manufacturer’s instructions. At 24 h post-transfection, the medium was replaced with fresh normal culture medium, and the cells were incubated for an additional 30–48 h (2–3 days post-transfection).
To label the nuclear and chromosomal DNA for aneuploidy, live cells were stained with Hoechst 33342 (Dojindo). Mitotic morphology was observed by fluorescent microscopy (Olympus) with an Aqua Cosmos system (Hamamatsu Photonics).
For immunofluorescence (IF), HEK293T cells (2.5 × 105 cells per well) were plated onto collagen-coated square coverslips in 6-well dishes, and incubated for 18–20 h before transfection. At 2 days post-transfection, the cells were washed with PBS and fixed for 30 min at room temperature (RT) in 4% (w/v) PFA-PBS. After washing twice with PBS, the cells were permeabilized for 2 min with 50 μg/ml digitonin-PBS, washed twice again, and then quenched in 50 mM ammonium chloride-PBS for 5 min. After blocking with 0.1% gelatin-PBS for 5 min, the cells were incubated for 1–1.5 h at RT with primary antibodies diluted in 0.1% gelatin-PBS. The cells were washed three times with PBS, and subsequently incubated for 30 min at RT with Alexa Fluor-conjugated secondary antibodies (Molecular Probes, Invitrogen/Life Technologies) diluted in 0.1% gelatin-PBS. For the analysis of the subcellular localization of AcGFP-fusion constructs in mitotic metaphase cells, we performed IF using Alexa Fluor 647-labeled anti-α-tubulin antibodies (Santa Cruz Biotechnology) against the mitotic spindle and Hoechst 33422-labeled chromosomes, and visualized the cells with an LSM 510 confocal laser scanning microscope (Carl Zeiss). For the colocalization analysis of AcGFP-Emi2 ZBR–RL and the core APC/C, we used Alexa Fluor 647-labeled anti-ANAPC2 antibodies (Santa Cruz Biotechnology), and subsequently collected sequential z-stack images by using the LSM 510 system in the multi-track mode. The 3D reconstruction was created by sequentially capturing images within a series of z-planes.
To characterize the cell cycle phase by FACS (fluorescence activated cell sorting), the transfectants were fixed with 1% paraformaldehyde (PFA)-PBS for 30 min at 4 °C, washed twice with PBS, and centrifuged. The cell pellets were subsequently resuspended in cold 70% ethanol and incubated for 2 h at 4 °C. These fixed cells were stained with propidium iodide (PI), and analyzed by FACSCalibur and the CELLQuest software (BD Biosciences).
2.4. Western blotting (WB) of cell extracts
APC/C inhibition in cells expressing the ZBR–RL fragment of Emi2 was examined by comparing the CCRP levels between the AcGFP-Blank vector and the AcGFP-Emi2 fragment fusion transfectants. At 2 days post-transfection, the cells were collected, washed once with PBS, resuspended in lysis buffer [20 mM Tris–HCl buffer (pH 8.0), containing 1% Triton X-100, 150 mM NaCl, and protease inhibitor mixture (Roche Applied Science)], and incubated on ice for 30 min. The resulting cell lysates were centrifuged at 15,000 rpm for 15 min at 4 °C, fractionated by SDS–PAGE, and electroblotted onto a PVDF membrane. The membranes were subsequently blocked with skim milk prepared with TBS-Tween, and immunodetection was performed using the Immobilon Chemiluminescent Substrate Kit (Merck Millipore). To assess APC/C inhibition in cells expressing the AcGFP-Emi2 ZBR–RL, the following primary antibodies were employed for WB: anti-CycB1 antibody (V152; MBL) and anti-securin antibody (DCS-280; MBL) to measure endogenous CycB1 and securin levels, respectively; anti-GFP antibody (JL-8; Clontech, Takara) to monitor AcGFP fusion protein expression; and anti-α-tubulin antibody (DM1A; Sigma–Aldrich) as a loading control.
2.5. Immunoprecipitation (IP) assays
The in vitro binding of the Emi2 fragment to the substrate recognition catalytic module subunits of APC/C was analyzed by co-IP (Supplementary Fig. S1A). Mouse cDNAs were subcloned into the T7 expression vector, pcDNA3.1/myc-His (Invitrogen, Life Technologies), to generate Myc-tagged prey proteins: ANAPC2, ANAPC2-CT (C-terminus; residues 337–837 of mouse ANAPC2, corresponding to the cullin-homology region), ANAPC11, ANAPC10, and Cdc20. To generate the double HA-tagged bait protein, the cDNA corresponding to the F-box–DB–ZBR–RL fragment of mouse Emi2 (residues 251–641) was subcloned into the tag sequence-modified pcDNA3.1/myc-His vector. Recombinant proteins were produced by T7-driven in vitro synthesis, using the TNT Quick Coupled Transcription/Translation System (Promega), and were partially purified using the MagZ Protein Purification System (Promega), according to the manufacturer’s recommendations. Subsequently, 100 μl portions of the MagZ-purified bait proteins, in 1 ml of binding buffer [20 mM Tris–HCl buffer (pH 8.0), containing 150 mM NaCl, and 0.01% NP-40], were incubated with anti-HA affinity resin (Roche Applied Science) for 2 h. The unbound species were removed by washing, and the bait proteins were subsequently mixed and incubated with the prey proteins for 18–20 h at 4 °C. Immunocomplexes were collected by centrifugation (16,000g at 4 °C for 5 min), washed, and separated by 10% SDS–PAGE, and the labeled proteins were detected by WB with anti-HA (16B12; Covance) or anti-Myc (9B11; Cell Signaling Technology) antibodies.
To test the binding of the AcGFP-Emi2 fragment fusion proteins to APC/C and/or Cdc20, a binding assay was performed using cell extracts from HEK293T transfectants (Fig. 2C and D). HEK293T cells were transfected in 6-well plates. At 2 days post-transfection, the cells were lysed in 500 μl of IP buffer [50 mM Tris–HCl buffer (pH 8.0), 150 mM NaCl, 0.1% NP-40, and Complete EDTA-free Protease Inhibitor Cocktail (Roche Applied Science)] per plate. To pre-clear the lysates, Protein G Mag Sepharose (GE Healthcare) magnetic beads were added, and the lysates were incubated at 4 °C for 45 min on an orbital shaker. Subsequently, the pre-cleared lysates were subjected to co-IP. Following an incubation with 0.5 μl of anti-Cdc20 (mouse monoclonal antibody AR12; MBL) or anti-GFP (Living Colors Full-Length A.v. Polyclonal Antibody; Clontech, Takara) for 45 min at 4 °C with rotation, Protein G magnetic beads were added and incubated for 45 min at 4 °C with rotation. For the detection of co-immunoprecipitates, the magnetic beads were isolated, washed four times with IP buffer, and subjected to WB using antibodies against Cdc20 (rabbit polyclonal antibody; Santa Cruz Biotechnology), GFP (mouse monoclonal antibody JIL-8; Clontech, Takara), and ANAPC3/Cdc27 (mouse monoclonal antibody C-4; Santa Cruz Biotechnology).
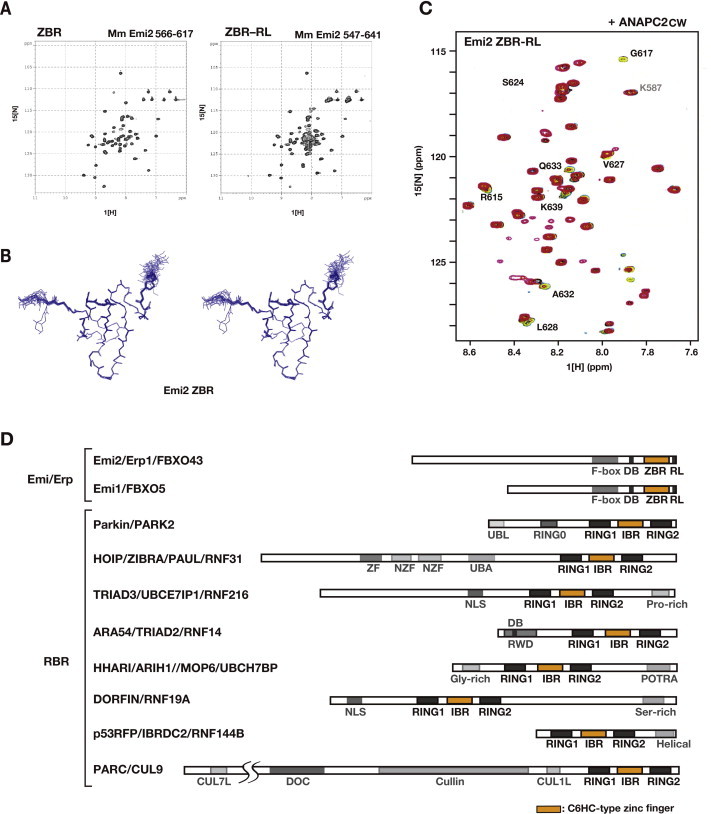
Fig. 2.
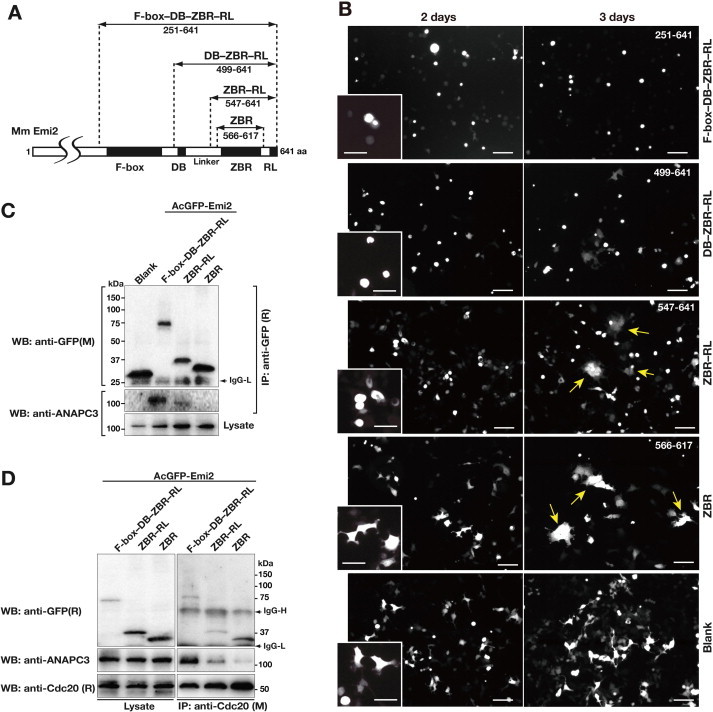
Cellular assays using a series of AcGFP-Emi2 fragment fusion constructs. (A) Diagram of the Emi2 C-terminal constructs. Numbers correspond to amino acid positions of mouse (Mm) Emi2. (B) Morphological phenotypes of HEK293T cells transfected with AcGFP-Emi2 fragment fusion constructs (see also panel A) at 2- and 3-days post-transfection. Scale bar, 100 μm. The boxes in the bottom left corners show close-up views of the cells at 2 days post-transfection. Scale bar, 50 μm. Yellow arrows indicate multinucleated cells and giant cells. (C) and (D) Interactions between endogenous APC/C and/or Cdc20 with a series of AcGFP-Emi2 fragment fusion constructs in the cell. Cell extracts from the indicated HEK293T transfectants were used for co-IPs with an anti-GFP antibody (C) or an anti-Cdc20 antibody (D). WBs of the immunoprecipitates (IP) and cell lysates were probed with the indicated antibodies. M, mouse monoclonal antibodies; R, rabbit polyclonal antibodies. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.6. Ubiquitylation assay
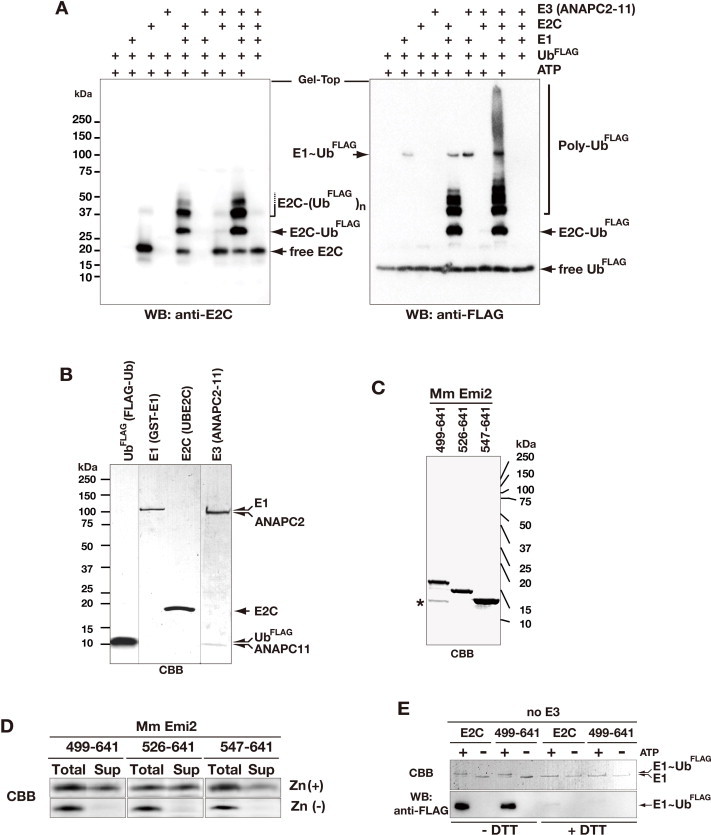
In vitro ubiquitylation assays were performed in a volume of 25 μl. For the experiments shown in Fig. 5, reaction mixtures containing 10 ng/μl GST-E1 (Calbiochem, Merk Millipore), 20 ng/μl E2C (UBE2C), 20 ng/μl HT-ANAPC2–11 (prepared using a baculovirus expression system [21], and 100 ng/μl FLAG-tagged ubiquitin (Sigma–Aldrich) were incubated with 40 ng/μl of Emi2 C-terminal fragments at 30 °C for 0, 5, 15, or 45 min, in 20 mM HEPES-NaOH (pH 7.4), 5 mM MgCl2, and 5 mM ATP. For subsequent experiments using the cell-free-synthesized ANAPC2CW–11 complex as the minimum CRL E3 module of APC/C (mCRLA) shown in Fig. 6, reaction mixtures containing 2 ng/μl GST-E1, 5 ng/μl E2C, 40 ng/μl mCRLA, and 50 ng/μl FLAG-tagged ubiquitin (Sigma–Aldrich) were incubated with 30 ng/μl of Emi2 fragments at 30 °C for 0, 5, or 15 min, in 20 mM HEPES-NaOH (pH 8.0), containing 1 mM MgCl2 and 2 mM ATP. The reactions were stopped by adding 25 μl of 3 × SDS–PAGE sample loading buffer [5% (w/v) SDS, 250 mM DTT, 15% (v/v) glycerol, 140 mM Tris–HCl (pH 6.8), and 0.01% (w/v) bromophenol blue] and incubated at 95 °C for 3 min. The ubiquitylation reactions were resolved by SDS–PAGE on a 10–20% gradient gel, followed by WB using anti-FLAG antibodies (M2; Sigma–Aldrich) and detection with an Immobilon Chemiluminescence Detection Kit (Merck Millipore). Luminescent signals corresponding to polyubiquitin chains were acquired with the Luminescent Image Analyzer LAS-3000 (Fujifilm, GE Healthcare), and were quantified using the Multi Gauge image analysis program (Fujifilm, GE Healthcare).
Fig. 5.
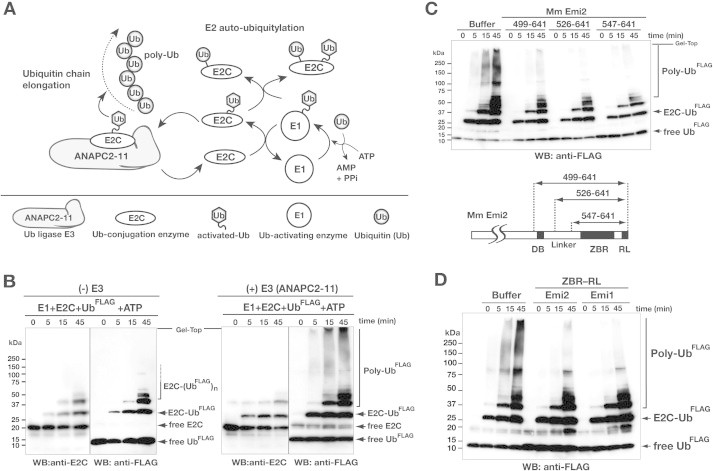
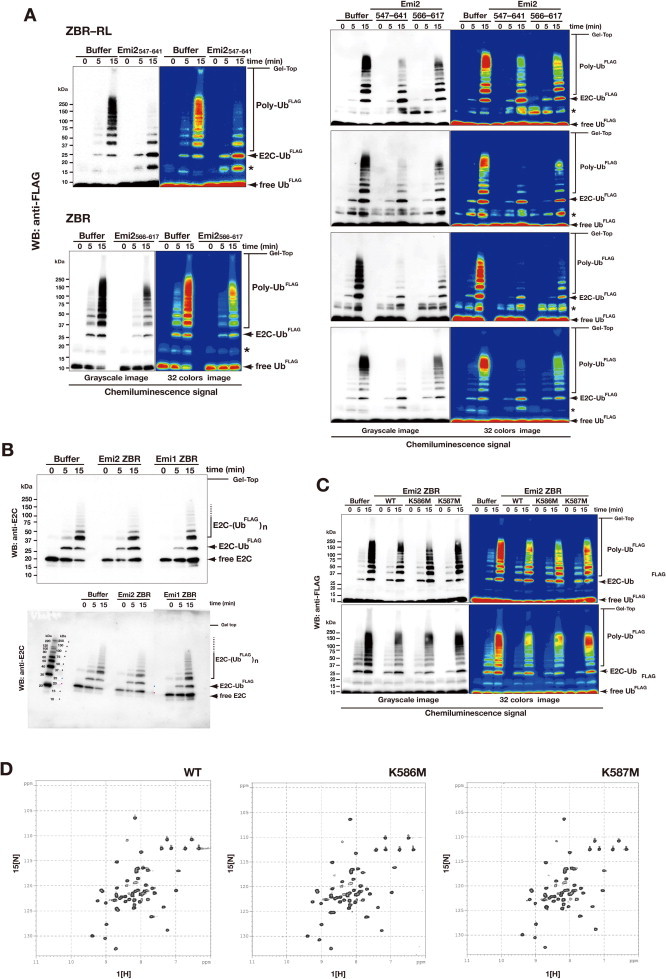
The ZBR–RL fragment of Emi2 inhibits ubiquitin chain elongation by ANAPC2–11 in combination with E2C. (A) Schematic diagram showing the core enzymatic reaction of APC/C for polyubiquitin chain (poly-Ub) formation. E1 and the combination of E2C with E3 ligase (ANAPC2–11) generates activated ubiquitin (Ub) for ubiquitin chain elongation. Details of the ubiquitylation factors are described in the Section 3. (B) In vitro ubiquitylation reactions using N-terminal FLAG-tagged ubiquitin (UbFLAG) and the E1-E2C-E3 system shown in panel A. UbFLAG adducts and UbFLAG chains were detected by anti-FLAG Western blotting (WB). The WB with the anti-E2C antibody shows the auto-ubiquitylation activity of E2C. (C) Inhibition of ubiquitin chain elongation by the indicated Emi2 C-terminal fragments. Numbers correspond to amino acid positions of mouse (Mm, Mus musculus) Emi2. (D) Inhibition of ubiquitin chain elongation by the ZBR–RL fragments from Mm Emi2 (residues 547–641) and Mm Emi1 (residues 326–421).
Fig. 6.
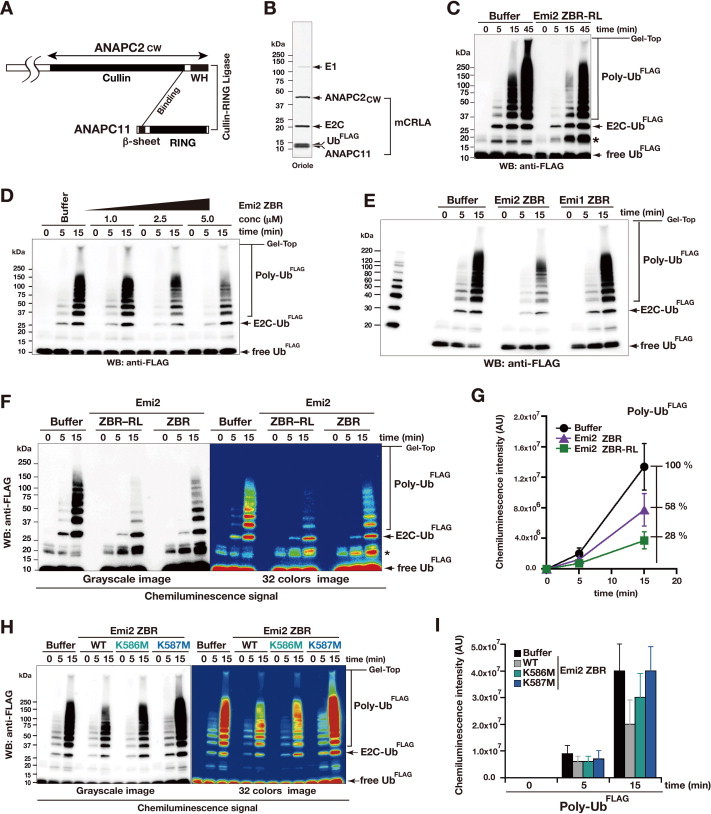
Inhibition of ubiquitin chain elongation by the Emi2 ZBR-domain fragment. (A) Diagram representing the minimized CRL-type E3 module of APC/C, mCRLA. (B) Purified protein components of the in vitro ubiquitylation system using mCRLA. The SDS–PAGE gel was stained with Oriole fluorescent gel stain. (C) In vitro ubiquitylation inhibition assay of the Emi2 ZBR–RL fragment, using the mCRLA system. Bands marked with asterisks contain ubiquitin dimers and partially ubiquitylated Emi2 fragments. (D) WB showing the concentration-dependent inhibitory effect of the ZBR fragment of Emi2 against the mCRLA-mediated ubiquitin chain elongation. (E) In vitro assay for the inhibitory activities of the ZBR fragments from mouse Emi2 (residues 566–617) and Emi1 (residues 345–396) against ubiquitin elongation by the mCRLA system. (F) and (G) Comparison of the inhibitory activities of the Emi2 ZBR–RL and ZBR fragments against ubiquitin chain elongation. (F) WB bands, visualized using a luminescent image analyzer system. (G) Graphs representing the production of poly-Ub, determined by measuring the chemiluminescent-signal intensities (arbitrary units, AU) of WBs, using the Image Gauge software. Baseline levels were adjusted by subtracting the time-zero value from all other time-point values. Each graph represents the mean ± SEM (n = 6; the other 5 WBs are shown in Supplementary Fig. S5A). (H) WB showing the effects of the K576M and K587M mutations on the inhibition of ubiquitin chain elongation by the Emi2 ZBR fragment. (I) Production levels of poly-Ub, representing the chemiluminescent intensities (AU) of the bands shown in panel H. Results represent the mean ± SEM of 3 independent experiments (the other two WBs are shown in Supplementary Fig. S5C).
To examine the effects of the Emi2 C-terminal fragments on E1-catalyzed ubiquitin activation, reactions were performed in the absence of E2 and E3 and analyzed by WB. For the detection of non-covalent binding, E1∼Ub (Supplementary Fig. S4E), reaction aliquots were quenched in SDS–PAGE loading buffer under reducing (+DTT) or non-reducing (−DTT) conditions, fractionated by SDS–PAGE and visualized by Coomassie Brilliant Blue (CBB) staining or WB.
To analyze the ubiquitylation sensitivity of AcGFP-Emi2 ZBR mutants in cell, HEK293T transfectants were lysed in IP buffer at 3 days post-transfection. Following IP using mouse monoclonal antibodies, FK2 (BIOMOL, Enzo Life Sciences) or anti-GFP (M) mixed with Protein G magnetic beads, the IP samples were subjected to WB using a rabbit polyclonal anti-GFP antibody (R).
For the preparation of ubiquitin adducts and ubiquitin chain linkages for mass spectrometry analyses, the reaction products were resolved by SDS–PAGE and visualized using a SilverQuest Silver Staining Kit (Invitrogen, Life Technologies), according to the manufacturer’s protocol, or by standard CBB staining. Gel regions containing proteins were excised and digested with trypsin (Sigma–Aldrich) for 20 h at 37 °C. The resulting peptides were analyzed by liquid chromatography–electrospray ionization tandem mass spectrometry (LC–ESI-MS/MS).
2.7. Protein synthesis and purification
The proteins composing the full-length ANAPC2–11 complex, His6-tagged ANAPC2 and untagged ANAPC11, were coexpressed in Sf9 cells using the Bac-to-Bac Baculovirus Expression System (Invitrogen, Life Technologies) and copurified according to the method of Huang et al. [21], with the exception that TALON resin (Clontech, Takara) was used for the first capture step, instead of Ni-Agarose. The Emi2 fragments, E2C and mCRLA, were synthesized using a cell-free protein expression system [54] and subsequently purified using an AKTA 10S system (GE Healthcare), as follows. The protein fragments were purified by first adsorbing each reaction mixture to a HisTrap HP column (GE Healthcare), which was washed with concentration gradient buffer [50 mM Tris–HCl buffer (pH 8.0) containing 1 M NaCl and 10 mM imidazole]. The protein was eluted with a concentration gradient of imidazole (from 10 to 500 mM) in elution buffer [50 mM Tris–HCl buffer (pH 8.0) containing 0.5 M NaCl], and the imidazole was removed by overnight dialysis at 4 °C in wash buffer. The His-tag was removed by an incubation at 4 °C for 20 h with TEV protease. The resulting untagged proteins were purified by ion exchange chromatography using a HiTrap column (GE Healthcare) and gel filtration using a HiLoad Superdex 75 pg 16/60 column (GE Healthcare), with the final buffer [20 mM Tris–HCl buffer (pH 8.0), containing 150 mM NaCl, and 2 mM DTT]. In the cases, where the synthesized protein region that contained a zinc-finger domain (mCRLA and ZBRs), ZnCl2 was added to the purification buffers at a final concentration of 20 μM.
The concentrations of the purified proteins were determined by measuring the absorbance at 280 nm with a NanoDrop spectrophotometer (Thermo Scientific).
2.8. Nuclear magnetic resonance (NMR) spectroscopy and structure calculations
For the determination of the Emi2 ZBR domain structure by NMR, the protein samples were prepared as [13C, 15N]-labeled proteins using a cell-free protein synthesis system, and were subsequently purified and concentrated in NMR analysis buffer [20 mM Tris–HCl (pH 6.5), 100 mM NaCl, 1 mM DTT, 1 mM iminodiacetic acid (IDA), 50 μM ZnCl2, 0.02% (w/v) NaN3, and 10% (v/v) 2H2O]. We solved the solution structure of the Emi2 ZBR domain by standard NMR methods [55,56].
NMR experiments were performed at 298 K, using Bruker AVANCE spectrometers equipped with triple-axis gradients operating at 600 and 800 MHz, with the 13C- and 15N-doubly labeled samples in Shigemi susceptibility-matched tubes. The 1H, 15N, and 13C chemical shifts were referenced to the frequency of the 2H lock resonance of water. A series of 2D and 3D standard triple resonance NMR experiments were recorded [56], and 2D [1H, 15N]-HSQC and 3D HBHA(CO)NH, HNCA, HN(CO)CA, HNCACB, and CBCA(CO)NH spectra were used for sequence-specific backbone assignments. Side-chain assignments were obtained using 2D [1H, 15N]-HSQC and 3D HBHA(CO)NH, H(CCCO)NH, (H)CC(CO)NH, HCCH-COSY, HCH-TOCSY, and (H)CCH-TOCSY spectra. For consistency, 3D 15N- and 13C-edited NOESY-HSQC spectra were also obtained, to check the chemical shift assignments. All NMR data were processed with the NMRPipe program [57]. Linear prediction and zero filling were used in the indirect dimensions, to obtain higher resolution. The KUJIRA [58] and NMRView [59] programs were used for NMR spectral analysis. NOE data from 15N- and 13C-edited 3D NOESY spectra were used for the structure calculations. Automated NOE cross-peak assignments [60] and structure calculations with torsion angle dynamics were performed using the CYANA1.0.7 software package. Dihedral angle restraints were derived using the TALOS program [61], and structures were validated using PROCHECK-NMR [62]. Graphical analysis was performed with the MOLMOL or PyMol programs.
More than 20 NOE distance restraints per residue—including 364 long-range distance restraints and the restraints for coordinating the zinc ion—were used for the final structure calculations with the CYANA 2.1 program [63]. The final structures were energy-refined with the AMBER12 program [64], using the generalized Born model and the Amber ff99SB force field, as described previously [65]. The final 20 energy-minimized conformers that represent the solution structure of the Emi2 ZBR domain are well defined and show excellent agreement with the experimental data (Supplementary Table S1; Supplementary Fig. S2B). The precision of the structure is indicated by the RMSD values of the mean coordinates of 0.31 Å for the backbone and 1.11 Å for all heavy atoms of residues A569–R615. The quality of the structure is also reflected by the fact that 80.8% of the (ϕ, ψ) backbone torsion angle pairs were found in the most favored region, and 19.3% were within the additionally allowed regions of the Ramachandran plot, as determined by the PROCHECK-NMR program [62].
2.9. NMR chemical shift perturbation analysis
For the NMR chemical shift perturbation experiment (Fig. 3C; Supplementary Fig. S2C), the ANAPC2 fragment corresponding to the cullin domain and winged-helix (ANAPC2CW) was prepared as a un-labeled protein using a cell-free system, and was subsequently purified and concentrated in NMR analysis buffer [20 mM Tris–HCl buffer (pH 7.0), containing 100 mM NaCl, 1 mM DTT, 1 mM IDA, 50 μM ZnCl2, 0.02% (w/v) NaN3, and 10% (v/v) 2H2O]. 2D [1H, 15N]-HSQC spectra of the Emi2 ZBR–RL fragment were recorded while increasing the peptide concentration relative to that of the [15N, 13C]-labeled Emi2 fragments (35 μM), to final molar ratios of 1:0.1 to 1:1.0. The 2D-HSQC spectra collected at each titration point were normalized, and the chemical shift changes were calculated.
Fig. 3.
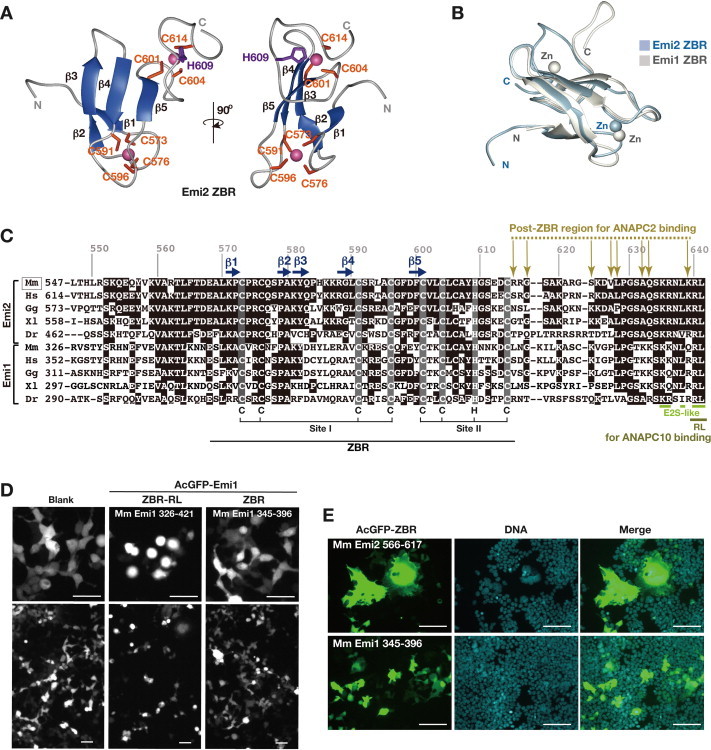
Structural and functional comparisons of the Emi1 and Emi2 ZBR domains. (A) and (B) Solution structure of the Emi2 ZBR domain, determined by NMR spectroscopy. (A) Ribbon-diagram representation of the lowest energy structures, showing two views from different angles. Secondary structure elements (β-strands) are colored ultramarine blue. Magenta-colored balls represent zinc ions. Numbered positions (orange and violet) indicate the residues coordinating the zinc ions. (B) Superimposition of the solution structure of the Emi1 ZBR domain (PDB ID, 2M6 N; Frye et al. [41]) and that of the Emi2 ZBR domain (PDB ID, 2RT9; the present study). (C) Multiple sequence alignment of the ZBR–RL regions from Emi1 and Emi2 orthologs. Mm, Mus musculus (mouse); Hs, Homo sapiens (human); Gg, Gallus gallus (chicken); Xl, Xenopus laevis (African clawed frog); Dr, Danio rerio (zebrafish). Identical residues are shown in black. Horizontal ultramarine blue arrows represent β-strands, and the residues in shaded boxes indicate zinc-coordinating residues, related to panel A. Ocher arrows indicate ANAPC2CW-interacting sites in the ZBR–RL region from mouse Emi2, based on NMR chemical shift perturbation data, related to Supplementary Fig. S2C. The lime line indicates the E2S-like sequence. The olive dashed line indicates the RL residues involved in binding to ANAPC10 [49]. (D) Morphological phenotypes of HEK293T cells transfected with AcGFP-fusion constructs of the Mm Emi1 ZBR–RL and ZBR fragments at 3 days post-transfection. Scale bar, 50 μm. (E) Cells expressing AcGFP-Emi2 ZBR and AcGFP-Emi1 ZBR at 3 days post-transfection, stained with Hoechst 33342 for nuclear DNA. Scale bar, 50 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.10. Structural data deposition
The Protein Data Bank (PDB) ID for the solution structure of the Emi2 ZBR domain reported in this paper is 2RT9. The chemical shift assignments have been deposited in the BioMagResBank (BMRB) database (accession code, 11529).
3. Results and discussion
3.1. Overexpression of the Emi2 ZBR–RL fragment results in morphological abnormalities and increased G2/M content
In the case of the mitotic cell-cycle regulator Emi1, the minimal region required for in vitro APC/C inhibition is reportedly its ZBR–RL region, encompassing the ZBR domain and the C-terminal RL residues [48]. For a comparison between Emi1 and Emi2, we examined the effects of the exogenous expression of the corresponding ZBR–RL fragment of Emi2 on mitotic events. Under normal proliferation conditions, the human embryonic kidney cell-line 293T (HEK293T) undergoes a dramatic cell shape change during the mitotic phase/M phase, which is a relatively short period in the cell cycle period (less than 1 h). Therefore, we transfected HEK293T cells with the plasmid vector expressing the Emi2 ZBR–RL fragment as a fusion protein with the Aequorea coerulescens green fluorescent protein (AcGFP), under the control of the cytomegalovirus-immediate early (CMV-IE) promoter, a strong and constitutively active promoter.
The cells expressing AcGFP-Emi2 ZBR–RL exhibited sphere- or ball-like shapes at two days post-transfection (Fig. 1A). A fluorescence-activated cell sorting (FACS) analysis revealed that the cells expressing AcGFP-Emi2 ZBR–RL exhibited a significantly increased population in the G2/M phase and an increased DNA content (Fig. 1B). Subcellular analyses with simultaneous visualization of the mitotic spindle and chromosomes confirmed that the sphere-shaped cells were in M-phase (Fig. 1C). In addition, the characteristic accumulation of AcGFP-Emi2 ZBR–RL at the pericentrosomal region of the mitotic spindle was observed. Western blotting (WB) indicated that the levels of cyclin B1 (CycB1) and securin (M-phase substrates of APC/C) in the cell extracts from the AcGFP-Emi2 ZBR–RL transfectants were higher than those of the blank control (Fig. 1D). APC/C is localized to the mitotic spindle in M phase and at the nucleus in interphase [66,67] and AcGFP-Emi2 ZBR–RL exhibited similar subcellular distributions. ANAPC2 exists in the cell as a stable complex with ANAPC11, for the cullin-RING ligase (CRL)-type E3 catalytic module of APC/C. Notably, 3D-reconstruction images of the sphere-shaped M-phase cells revealed that the AcGFP-Emi2 ZBR–RL immunofluorescence (IF) signals overlap with those of ANAPC2 around the condensed chromosome in the mitotic spindle (Fig. 1E). After culturing for 24 h further, multinucleated giant cells were frequently seen in the AcGFP-Emi2 ZBR–RL transfectants (Fig. 1F). Collectively, these results showed that the overexpression of the Emi2 ZBR–RL fragment in HEK293T cells leads to mitotic abnormalities (a schematic representation is shown in Fig. 1G).
Fig. 1.
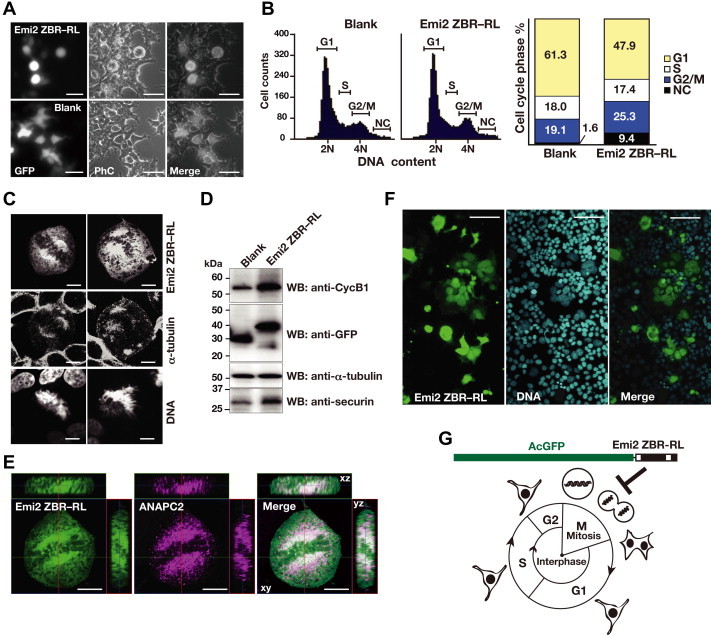
Exogenous expression of the Emi2 ZBR–RL fragment in proliferating cells causes abnormal cell division. (A) Morphological properties of unfixed HEK293T cells transfected with AcGFP-Emi2 ZBR–RL and AcGFP-Blank control vector. At 2 days post-transfection, cells were observed by exposure to blue light (GFP) and phase contrast imaging (PhC). The panels on the right show merged images. Scale bar, 50 μm. (B) Fluorescence-activated cell sorting (FACS) analysis of AcGFP-positive cells at 2 days post-transfection, using propidium iodide DNA staining to determine cell cycle phase: G1, S, or G2/M. NC, not categorized. A stacked bar graph showing the percentage of cells in each phase of the cell cycle is displayed on the right. (C) Subcellular analysis of sphere-shaped cells expressing AcGFP-Emi2 ZBR–RL. Mitotic spindles were visualized with Alexa Fluor 633-labeled anti-α-tubulin antibodies (middle) and Hoechst 33422-stained chromosomal DNA (bottom). Scale bar, 10 μm. (D) Western blots (WBs) showing endogenous CycB1 and securin levels in cell extracts from AcGFP transfectants at 2 days post-transfection. Alpha-tubulin served as a loading control. (E) 3D confocal microscopy images of cells expressing AcGFP-Emi2 ZBR–RL (green) at M-phase. Alexa Fluor 647-labeled anti-ANAPC2 served as an APC/C marker (magenta). The dark shaded area in the middle of the cell corresponds to the region of condensed chromosomes in the mitotic spindle. The images represent the xy section with 2 perpendicular lines: the horizontal line (top) in the xz plane and the vertical line (side) in the yz plane. The central box corresponds to the position of the currently displayed xy plane. Scale bar, 10 μm. (F) Cells expressing AcGFP-Emi2 ZBR–RL at 3 days post-transfection, stained with Hoechst 33342 for nuclear DNA. Scale bar, 100 μm. (G) Diagram of major morphological changes observed during the cell cycle progression of HEK293T cells and the inhibition of mitosis by the AcGFP-Emi2 ZBR–RL fusion construct used in the transfection assays. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. The ZBR fragment of Emi2 induces abnormal cell division and inhibits Cdc20 binding to the core APC/C
We compared the ZBR–RL fragment with the F-box–DB–ZBR–RL, DB–ZBR–RL, and ZBR fragments (Fig. 2A), with respect to the effects of their expression on mitotic events. First, we confirmed, by a co-immunoprecipitation (co-IP) assay using in vitro-translated proteins (Supplementary Fig. S1), that the F-box–DB–ZBR–RL fragment of Emi2 binds to the full-length ANAPC2, as reported for Emi1 [48], and its C-terminal cullin-like sequence fragment. Most of the transfectants expressing the AcGFP-fused F-box–DB–ZBR–RL or DB–ZBR–RL fragment exhibited sphere- or ball-like shapes at 2 days post-transfection, and stopped cell proliferation (Fig. 2B). The cells expressing the ZBR fragment at 3 days post-transfection formed giant cells, due to the failure of nuclear division, as also detected with the ZBR–RL fragment at 2–3 days post-transfection (Fig. 2B, right). Therefore, the cell-division inhibiting activities are in the order ZBR < ZBR–RL < DB–ZBR–RL ⩽ F-box–DB–ZBR–RL. It should be emphasized here that the ZBR fragment also exerted the cell-division inhibiting activity, although it was weaker than those of the longer fragments.
The F-box–DB–ZBR–RL and DB–ZBR–RL fragments exhibited stronger phenotypes than that of the ZBR–RL fragment (Fig. 2B). Correspondingly, we found that the AcGFP-fused Emi2 F-box–DB–ZBR–RL fragment binds to APC/C more tightly than the ZBR–RL fragment in co-IP analyses of cell extracts of the transfectants with an anti-GFP antibody (Fig. 2C). In contrast, the binding of the ZBR fragment to APC/C was not detectable by this analysis (Fig. 2C). These results are consistent with the previous report [42] that the C-terminal tail containing the RL residues of Emi2 is sufficient for its APC/C binding.
The Cdc20-bound APC/C (APC/C-Cdc20) controls the G2/M transition in cell division. The F-box–DB–ZBR–RL fragment of Emi2 is sufficient for mII arrest [50], and actually binds to the APC/C coactivator Cdc20 in vitro (Supplementary Fig. S1). Therefore, we examined whether the Emi2 ZBR fragment interacts with Cdc20 in the cell, by a co-IP analysis with an anti-Cdc20 antibody that precipitates both Cdc20 and Cdc20-bound APC/C. In fact, all of the F-box–DB–ZBR–RL, ZBR–RL, and ZBR fragments co-precipitated with Cdc20 (Fig. 2D). On the other hand, the binding of the ZBR fragment to APC/C was not detectable by the co-IP analysis of cell extracts of the AcGFP-Emi2 fragment transfectants with an anti-GFP antibody (Fig. 2C). These results indicated that the ZBR domain is responsible for Cdc20 binding. Intriguingly, the Cdc20 bound with the ZBR fragment hardly co-precipitated with APC/C (Fig. 2D). We speculate that the mechanism underlying the phenotype at 3 days post-transfection of the Emi2 ZBR fragment may be that the ZBR fragment binds to Cdc20, and dissociates it from APC/C.
On the other hand, the ZBR–RL and F-box–DB–ZBR–RL fragments exhibited weaker Cdc20 separation activities, probably because the C-terminal tail connects the ZBR·Cdc20 complex to ACP/C, and the F-box–DB region reinforces the interaction (Fig. 2C). In general, the F-box and DB motifs mediate the recognition of targets, such as CCRP, by the E3 ubiquitin ligases, including APC/C, for degradation by the ubiquitin–proteasome system (UPS) in cells. The F-box and DB motifs may trigger the initial binding of Emi2 to the Cdc20-bound APC/C to enhance the inhibitory mechanism further, rather than exerting inhibition by the classical pseudosubstrate mechanism.
3.3. Structural and functional analyses of the Emi2 ZBR–RL region
The [1H-15N] HSQC spectra of the Emi2 ZBR–RL fragment showed well-dispersed resonances, with sharp resonances clustered around 8.3 ppm (1H chemical-shift value). After the main-chain resonance assignment, the region spanning residues 566–617 of mouse Emi2 (the ZBR fragment) produced well-dispersed resonances with uniform signal intensities (Supplementary Fig. S2A, left). In contrast, the sharp resonances clustered around 8.3 ppm originated from the C-terminal residues 618–641, suggesting that this region is flexible and disordered (Supplementary Fig. S2A, right).
The solution structure of the Emi2 ZBR fragment was determined by multidimensional NMR spectroscopy (Fig. 3A; Supplementary Fig. S2B; Supplementary Table S1). The N-terminus of the Emi2 ZBR domain comprises two successive β-hairpins: the first is composed of β1 (K571–P572) and β2 (P579–A580), and the second is composed of β3 (K581–Q583) and β4 (R588–L590). The first zinc ion is coordinated with four ligand residues (C573 and C576 at the tip of the first β-hairpin, and C591 and C596 at the tip of the second β-hairpin). The two β-hairpins are tightly bound to each other, and form a scissor-like structure. The β5 strand (D599–C601) associates with the β4 strand in an anti-parallel manner, to form the C-terminal GAG knuckle-like zinc-binding site, including the C601, C604, H609, and C614 residues (Fig. 3A and C). The Emi2 ZBR domain adopts the same topology as the Emi1 ZBR domain (Fig. 3B).
The co-IP and IF analyses both indicated that the Emi2 ZBR–RL fragment interacts with ANAPC2 (Fig. 1E; Supplementary Fig. S1A). NMR titration experiments, using 13C- and 15N-labeled Emi2 ZBR–RL, revealed that the R615, G617, S624, V627, L628, A632, Q633, and K639 residues in the Emi2 ZBR–RL region interact with the cullin–winged-helix subdomain of ANAPC2 (Fig. 3C; Supplementary Fig. S2C). On the other hand, no chemical shift changes were observed for the NMR peaks corresponding to the RL residues at the C-terminus, which is required for ANAPC10 binding [42,49]. Therefore, the residues 615–627, corresponding to the region between the ZBR domain and the C-terminal RL residues [the “post-ZBR” (PZ) region], shown with a dashed line in Fig. 3C, are involved in the binding of Emi2 to ANAPC2. According to the cryo-EM analyses of APC/C, which indicated that ANAPC10 connects to ANAPC2 (ANAPC2–11 subcomplex) within intact APC/C [18,68,69], we consider that the RL residues at the C-terminus of Emi2 bind to the ANAPC10, whereas the PZ region interacts with the cullin-winged helix subdomain of ANAPC2.
This architecture of the ZBR domain and the C-terminal tail consisting of the PZ region and the RL residues is evolutionally conserved in the Emi/Erp family (Fig. 3C; Supplementary Fig. S2D). We examined the effects of the over-expression of the ZBR–RL and ZBR fragments of Emi1 on cell division. The cells expressing the AcGFP-fused Emi1 ZBR–RL fragment exhibited sphere- or ball-like shapes at three days post-transfection (Fig. 3D). However, in contrast to the phenotypes of cells expressing the ZBR fragment of Emi2, almost all of the cells expressing the ZBR fragment of Emi1 lacked the abnormal mitotic phenotypes, including giant cell formation (Fig. 3E). These results suggested that the roles of the ZBR domains are different between Emi2 and Emi1.
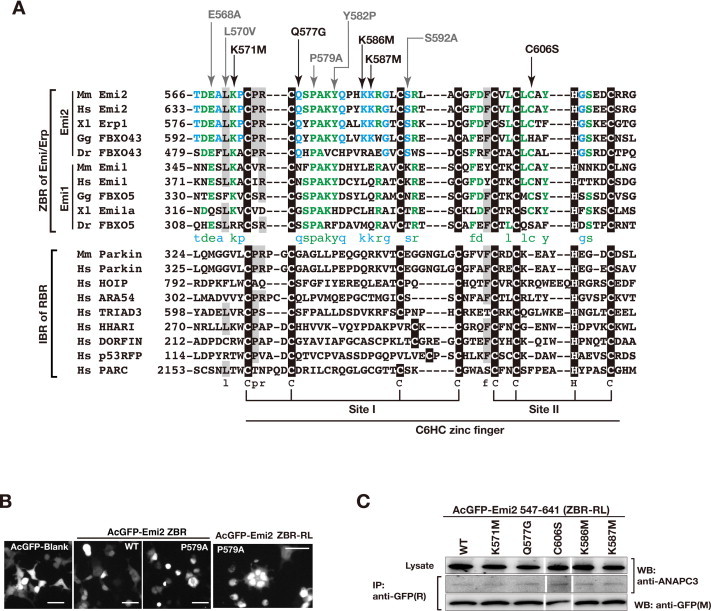
3.4. Amino acid residues important for the cell-division inhibiting activity of the Emi2 ZBR domain
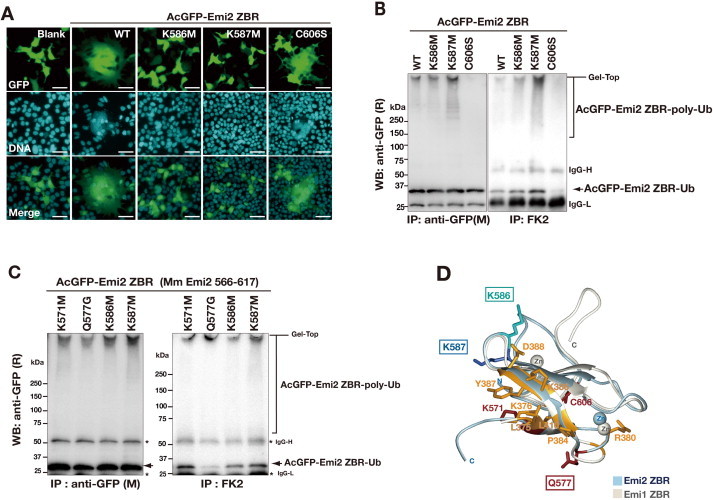
To identify the residues in the ZBR domain of Emi2 that are important for the cell-division inhibiting activity, we introduced single amino acid substitutions within the ZBR domain in the ZBR–RL and ZBR fragments. The well-conserved and exposed amino acid residues among the Emi/Erp ZBR domains were selected on a structural basis, and the zinc-coordination sites were excluded (Supplementary Fig. S3A). The effects of these mutations on the abnormal mitotic phenotype of HEK293T cells transfected with AcGFP-Emi2 ZBR–RL, the appearance of multinucleated cells at three days post-transfection (Figs. 1F, 2B), were analyzed as shown in Fig. 4A. The expression levels of the mutants were similar to that of the wild type (WT), and the following residues (in the order of effectiveness) reduced the abnormal mitotic phenotype: K586 M > K587 M ⩾ K571 M > Q577G ≈ C606S. These amino-acid substitutions within the ZBR domain did not impair the ZBR–RL·APC/C binding (Supplementary Fig. S3C).
Fig. 4.
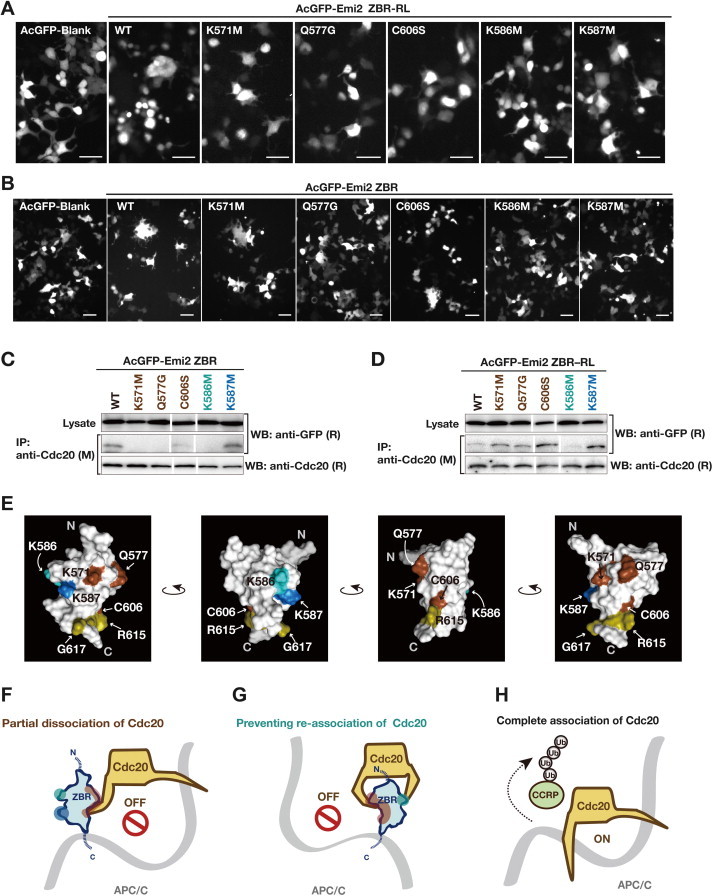
Mutations of Emi2 ZBR domain surface residues. (A) and (B) Effects of single amino acid substitutions of the ZBR domain within AcGFP-Emi2 ZBR–RL and AcGFP-Emi2 ZBR on the abnormal mitotic phenotype in HEK293T cells at 3 days post-transfection. Morphological properties of the cells expressing the indicated mutants, the ZBR–RL fragment (A), or the ZBR fragment (B). WT, wild type sequence. Scale bar, 50 μm. (C) and (D) Interactions between endogenous Cdc20 and the ZBR mutants of AcGFP-Emi2 fragments in the cell. Cell extracts from the indicated HEK293T transfectants were used for co-IP with an anti-Cdc20 antibody. White lines indicate the gaps between lanes. (E) Characterization of amino acid residues on the surface of the Emi2 ZBR domain. (F–H) Hypothetical mechanism for the Emi2 ZBR-induced abnormal cell division phenotype, by destabilizing the association of Cdc20 with the APC/C core complex. The ZBR domain of Emi2 binds to the APC/C coactivator Cdc20, partially dissociates it from the core APC/C (red-brown) (F), and furthermore prevents the re-association of APC/C·Cdc20 (cyan) (G). Complete association of Cdc20 with APC/C (active mode) for the cell cycle regulatory protein (CCRP) ubiquitylation, illustrated (H) for comparison. The ZBR domain has a Cdc20-independent APC/C inhibitory activity (cobalt blue). The C-terminal region of contributes to anchoring the ZBR domain to the cullin-RING module of APC/C (ocher), shown in Fig. 3C. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The ZBR fragment of Emi2 lacking the C-terminal tail also exerted the cell-division inhibiting activity, although it was weaker than that of the ZBR–RL fragment (Figs. 2B, 3E). Mutations of the ZBR fragment actually affected the Emi2 ZBR-induced mitotic abnormality, in the order K586 M > K587 M ⩾ Q577G > C606S > K571 M (Fig. 4B). An analysis of binding in cells revealed that Cdc20 co-precipitated with the ZBR fragment, but hardly co-precipitated with APC/C (Fig. 2D), indicating that the Cdc20·ZBR complex negligibly interacts with APC/C. We then examined whether these mutations impaired the binding of the ZBR fragment to Cdc20, by an in vivo co-IP analysis. The K571M, Q577G, K586M, and C606S mutants hardly bound to Cdc20, whereas K587M bound to Cdc20 as well as the WT (Fig. 4C). Therefore, four residues, K571, Q577, K586, and C606, but not K587, are involved in the Cdc20 binding. Thus, the effects of the K571M, Q577G, K586M, and C606S mutations on the Emi2 ZBR-induced mitotic abnormality are correlated with the effects on the Cdc20 binding.
In the case of the ZBR–RL fragment, not only K587M but also the K571M, Q577G, and C606S mutants co-immunoprecipitated with Cdc20 (Fig. 4D), indicating that the K571M, Q577G, and C606S mutants are anchored to APC/C through the C-terminal tail, consisting of the PZ region and the RL residues, and Cdc20 binds normally to the core APC/C. The K586M mutation impairs the Cdc20·APC/C interaction in both the ZBR and ZBR–RL fragments (Fig. 4C and D), while this mutation most strongly reduces the Emi2 ZBR-induced mitotic abnormality. Interestingly, K571, Q577, and C606, but not K586, are located on one side of the domain surface (Fig. 4E). Therefore K586 seems to contribute robustly to prevent Cdc20 re-association with the core APC/C, but minimally to the putative interaction with Cdc20 in the APC/C-Cdc20.
In contrast to the results reported by Wu et al. (2007) that Emi2 can bind to the APC/C core complex in the absence of Cdc20 [70], Sako et al. (2014) recently indicated that Cdc20 is required for the APC/C-Emi2 interaction before cytostatic arrest release in unfertilized Xenopus eggs [49]. In our present study, the Cdc20 bound with the ZBR fragment hardly co-precipitated with APC/C (Fig. 3B) and the PZ region of Emi2 is likely to connect the ZBR·Cdc20 complex to ACP/C (Fig. 3C). Nevertheless, overexpression of the ZBR fragment of Emi2 induces cell division abnormalities (Figs. 2B, 3E). Therefore, we propose a model for the inhibition of normal M-phase progression. First, the Emi2 ZBR domain binds to Cdc20, and at least partly disrupts the association of Cdc20 with APC/C (Fig. 4F). Subsequently, the Emi2 ZBR domain separates Cdc20 from the APC/C core complex (Fig. 4G), thus preventing their complete association for the full activation of APC/C-Cdc20 (Fig. 4H).
Finally, the effect of the K587M mutation on the Emi2 ZBR-induced mitotic abnormality (Fig. 4A and B) is distinct from those of the other mutations, as the K587M mutant retains the ability to bind Cdc20 (Fig. 4C and D). The side chain of K587 is oriented oppositely, as compared with that of K586 (Fig. 4E, the second panel). Therefore, the Emi2 ZBR-induced mitotic abnormality is the consequence of two different mechanisms. In this context, it was recently reported that the ZBR–RL region of Emi1 inhibits the E2C-mediated ubiquitin chain elongation with the intact APC/C polyubiquitylation system [41,48].
3.5. The ZBR–RL fragment of Emi2 inhibits the ubiquitin chain elongation by the ANAPC2–11 subcomplex in combination with E2C
To examine the Cdc20-independent APC/C inhibition by the ZBR–RL fragment of Emi2, we performed the in vitro ubiquitylation assay using the purified recombinant APC/C core enzymatic components. The fundamental ubiquitylation reaction was performed by the set of three enzymes, E1, E2C, and E3, without a specific substrate and its recognition factors (Fig. 5A and B; Supplementary Fig. S4). The E3 catalytic subcomplex of APC/C, ANAPC2–11, was prepared using a baculovirus expression system, according to the method reported by Huang et al. [21]. The ubiquitin chain-initiating E2, E2C (also known as UBE2C or UbcH10), is the genetically validated E2 partner for APC/C [71,72], and E2C interacts with ANAPC2 [20,73]. In this reaction, we could observe E2C-mediated ubiquitin chain elongation, and distinguish it from the auto-ubiquitylation of E2C (Fig. 5B; Supplementary S4A). A mass spectrometry analysis of the poly-ubiquitin linkage type in our system indicated that K11 and K48 are the major linkages, with the former more abundant than the latter. Both ubiquitin linkage types are produced in CycB1 polyubiquitylation by the combined activities of the intact APC/C and E2C in vitro [74].
This in vitro polyubiquitylation assay unambiguously revealed that the Emi2 fragment corresponding to the ZBR–RL region inhibits the ubiquitin chain elongation in the absence of the natural target protein, the coactivator Cdc20, ANAPC10, and ubiquitin chain-extending E2, E2S/UBE2S (Fig. 5C). In addition, the in vitro assay confirmed that the ZBR–RL region of Emi1 also inhibits ubiquitin chain elongation (Fig. 5D), as reported with the intact APC/C polyubiquitylation system, in combination with E2C [41,48].
3.6. The ZBR domain of Emi2 inhibits the ubiquitylation by the APC/C cullin-RING module combined with E2C
The ZBR–RL fragment of Emi2 interacts with the cullin–winged-helix subdomain of ANAPC2, ANAPC2CW, as described above (Fig. 3C; Supplementary Fig. S2C). The ANAPC2CW region is reportedly sufficient for E2C binding and E3 activity [20]. We therefore established an in vitro polyubiquitylation system using the minimum cullin-RING ligase (CRL) module of APC/C (mCRLA), which was prepared by the co-expression of ANAPC2CW and ANAPC11 in an E. coli cell-free protein synthesis system (Fig. 6A and B). The mCRLA exhibited the CRL E3 activity of APC/C to elongate ubiquitin chains, and Emi2 ZBR–RL significantly decreased the rate of mCRLA-mediated ubiquitin chain elongation (Fig. 6C; Supplementary Fig. S5A). Our mCRLA protein lacks eukaryotic posttranslational modifications, whereas most APC/C subunits, including ANAPC2, are phosphorylated in the cell [75]. Consequently, no eukaryotic posttranslational modification is required, for either ubiquitin chain elongation or its inhibition by Emi2 ZBR–RL. Thus, the mCRLA system was shown to include the target of Emi2 ZBR–RL, and was therefore employed hereafter for the in vitro ubiquitin polymerization inhibition assay.
Remarkably, the ZBR fragment without the C-terminal tail still exhibited the inhibitory activity against ubiquitin chain elongation (Fig. 6D; Supplementary Fig. S5A). In contrast, an Emi1 fragment corresponding to the ZBR domain without the C-terminal tail, consisting of the PZ region and the RL residues, did not inhibit ubiquitin chain elongation by the mCRLA system (Fig. 6E). Correspondingly, overexpression of the ZBR fragment of Emi1 did not impair mitosis under the same conditions (Fig. 3D and E).
The inhibitory activities of the Emi2 ZBR–RL and ZBR fragments against ubiquitin chain elongation were compared (Fig. 6F and G; Supplementary Fig. S5A). In comparison with the control, the levels of poly-ubiquitin chain production were reduced to 28% (78% reduction) and 58% (42% reduction) in the presence of the ZBR–RL and ZBR fragments, respectively. The inhibitory activity of the Emi2 ZBR fragment is increased only by about twofold, by fusion to the C-terminal tail. Hence, the properties of the ZBR fragments distinguish Emi2 from Emi1.
We then performed the in vitro ubiquitylation assay using the mCRLA system for the Emi2 ZBR mutants, K587M and K586M. Note that these two Lys residues do not have any corresponding residues in Emi1. While the K586M mutation only slightly reduced the inhibitory activity against ubiquitin chain elongation, the K587M mutation resulted in loss of function (Fig. 6H and I; Supplementary Fig. S5C). The HSQC spectra of the WT and mutant ZBR fragments displayed well-dispersed peaks characteristic of folded proteins, indicating that the two mutants are fully folded (Supplementary Fig. S6D). Thus, these residues, especially K587, within the Emi2 ZBR domain are responsible for the inhibition of the ubiquitylation by the mCRLA. We then examined the ubiquitylation sensitivity of the ZBR domain mutants in cells, by transfecting the AcGFP-Emi2 ZBR into HEK 293T cells, and found that the K587M mutant was more easily polyubiquitylated than the WT and the other mutants (Supplementary Fig. S6A–C). These results indicated that K587 is crucial for the inhibitory activity of the Emi2 ZBR domain against ubiquitin chain elongation. On the other hand, the K586M mutation has the largest influence on the cell division abnormalities induced by the ZBR fragment (Fig. 4A and B), although K586 contributes only slightly to the inhibition of ubiquitin chain elongation (Fig. 6H and I; Supplementary Fig. S5C, S6B, C). The side chain of K586 is oriented oppositely, as compared with that of K587 (Fig. 4E; Supplementary Fig. S6D). Furthermore, the K586M mutation impairs the Cdc20·APC/C interaction for both the ZBR and ZBR–RL fragments (Fig. 4C and D), and thus K586 contributes substantially to prevent Cdc20 re-association with the core APC/C (Fig. 4F).
The ubiquitin chain-extending E2, E2S/UBE2S, is an auxiliary factor for APC/C required for recovery from abnormally stopped cell proliferation, but not for normal cell-cycle progression [76]. The E2S drives the elongation of K11-linked ubiquitin chains and enhances protein degradation by branched ubiquitin chains [77,78]. Recent studies reported that both E2C and E2S are required for rapid cyclin B degradation and exit from cytostatic arrest upon fertilization in Xenopus eggs [49]. The C-terminal RL residues of Emi2 competitively inhibits the binding of ubiquitin chain-extending E2, E2S/UBE2S to ANAPC10, and the RL residues of Emi1/2 compete with the tail of E2S for APC/C binding [48,49]. On the other hand, our results revealed that the ZBR–RL fragment of Emi2 interacts with ANAPC2, and clearly showed that the ZBR–RL fragments of Emi1 and Emi2 are available to inhibit the ubiquitin chain elongation by ANAPC2–11 and E2C (Fig. 5). In addition, E2C interacts with ANAPC2 [20,73], while E2S interacts with ANAPC10 [49]. Thus, it is possible that the Emi2·APC2–11 interaction inhibits APC2–11·E2C-mediated ubiquitylation, whereas the Emi2·APC10 interaction inhibits E2S-mediated ubiquitylation. On the other hand, Emi2 reportedly did not prevent the association of E2C with APC/C in the ubiquitylation reaction [47]. Moreover, our results showed that the ZBR–RL and ZBR fragments of Emi2 preferentially inhibit ubiquitin elongation (Figs. 5 and 6). Therefore, the Emi2 ZBR domain functions by interfering with the ubiquitin chain assembly reaction, rather than interrupting E2C binding to ANAPC2.
3.7. The key residues exerting the characteristics of the Emi2 ZBR domain
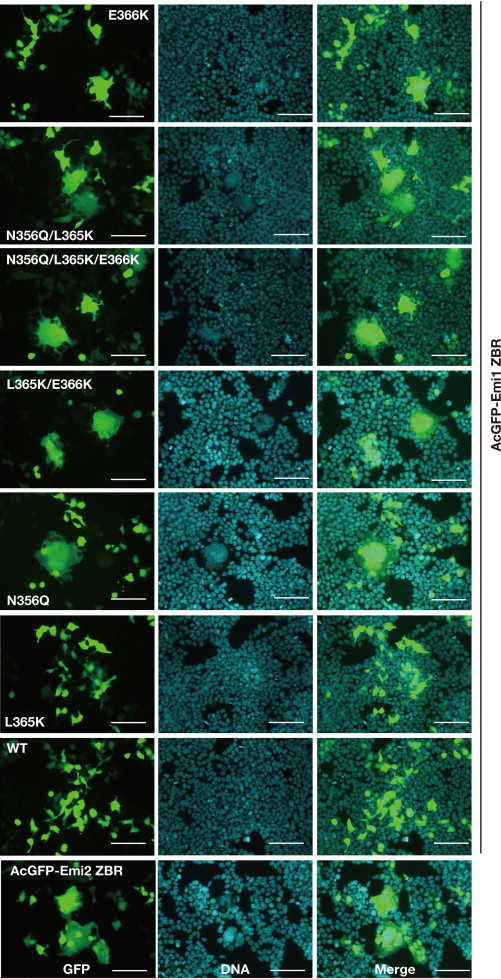
The functional residues of the ZBR domains were compared between Emi1 and Emi2 (Fig. 7A, B, and D). The ZBR residues K571, Q577, K586, K587, and C606 are highly conserved among the Emi2 orthologs from vertebrates (Fig. 3C; Supplementary Fig. S3A). In contrast, with the exception of K571, none of these residues are conserved in Emi1 or important for its function [41]. Thus, it seems that the roles of the ZBR domains are completely different between Emi2 and Emi1. Notably, the ZBR residues Q577, K586, and K587 (Fig. 7A) are highly conserved among the Emi2 orthologs from vertebrates. These residues are likely to be responsible for the Emi2-specific ZBR-domain functions. We therefore transplanted the Emi2-specific residues into the Emi1 ZBR fragment (Fig. 6C–E). These Emi1 ZBR mutants, except for N356Q (corresponding to Q577 of Emi2), gained the ubiquitylation inhibitory activity against the mCRLA system (Fig. 6E). These results confirmed that Q577 of Emi2 is not involved in the inhibition of ubiquitin chain elongation. Moreover, we examined the effects of these mutations on the mitotic phenotype of HEK293T cells, by a transfection assay with the expression vector encoding the AcGFP fused to the Emi1 ZBR-domain fragment. Mutations in the Emi1 ZBR fragment actually conferred the abnormal Emi2 ZBR-induced mitotic phenotype, in the order N356Q/L365K/E366K ≈ N356Q/L365K > N356Q ⩾ L365K/E366K >> E366K > L365K (Fig. 7E; Supplementary Fig. S7). We observed the Emi2-like abnormal mitotic phenotype in the Emi1 ZBR mutants with N356Q. In contrast, the L365K and E366K mutations hardly influenced cell division. These results indicate that the contribution of the APC/C activator separation to the cell-division inhibiting activity is superior to that of the ubiquitin elongation inhibition.
Fig. 7.
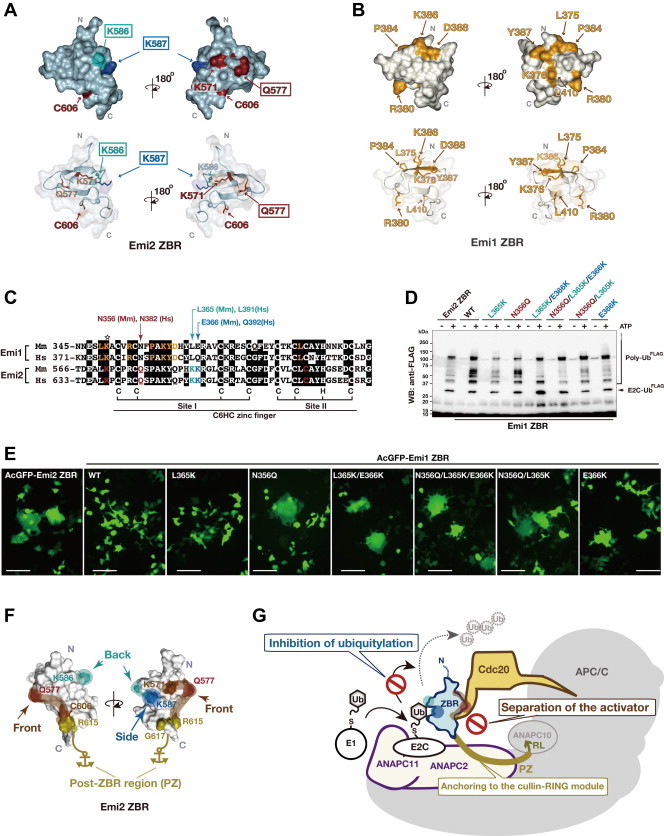
The key residues exerting the characteristic inhibitory activity of the Emi2 ZBR-domain against APC/C. (A and B) Comparison of the Emi2 and Emi1 ZBRs by surface mapping of the functional residues involved in the inhibition of the APC/C E3 activity. The images on the top show surface representations of Emi2 ZBR (A) and Emi1 ZBR (B), in the same orientation. The merged images of the ribbon diagram and the surface structure are displayed at the bottom, respectively. The Emi2 residues colored red-brown and cyan correspond to the putative Cdc20 separation mechanism illustrated in Fig. 4F. K587 (cobalt blue) is responsible for the inhibition of ubiquitin chain elongation (Fig. 5H and I; Supplementary Fig. S6). The Emi1 residues colored yellow-orange are related to the inhibition of the ubiquitin chain elongation by the APC/C-Cdh1 with E2C [41]. (C) Pairwise sequence alignment of the ZBR–RL regions from the mouse (Mm) and human (Hs) Emi/Erp family proteins. The amino acid sequence similarity between mouse and human: Emi1, 79.2% identical, Emi2, 86.3% identical (GENETYX). Arrows indicate the target residues within the Emi1 ZBR domain for the transplantation of Emi2-specific residues: N356Q of Mm Emi1 for Q577 of Mm Emi2; L365K of Mm Emi1 for K586 of Mm Emi2; E366K of Mm Emi1for K587 of Mm Emi2. The star indicates the functional Lys (K) residue conserved in the Emi1 and Emi2 ZBRs. (D) WB showing the effects of the transplantation of Emi2-specific residues into the Emi1 ZBR fragment on the mCRLA-mediated ubiquitin chain elongation. (E) Cellular phenotypes associated with the AcGFP-Emi1 ZBR mutants constructed by the transplantation of the Emi2-specific residues. Scale bar, 100 μm. Images of these transfectants stained with Hoechst 33342 for nuclear DNA are shown in Supplementary Fig. S7. (F) The functional residues of the Emi2 ZBR-domain surface, which are related to the inhibition of the APC/C activity. (G) Schematic representation of the inhibition mechanisms against APC/C-Cdc20 by the Emi2 ZBR domain and the post-ZBR (PZ) region. The C-terminal tail of Emi2 consists of the PZ region and the RL residues. The PZ region binds to ANAPC2 within the cullin-RING ligase module of APC/C (ANAPC2–11 subcomplex) while the RL residues bind to ANAPC10. The Emi2 ZBR domain inhibits the ubiquitin chain elongation by the ANAPC2–11 subcomplex combined with E2C. On the other hand, the Emi2 ZBR domain binds to the coactivator Cdc20 and impairs its association with the APC/C core complex, thereby turning off the E3 activity. The PZ region enhances the ZBR-mediated activities by anchoring it to the APC/C catalytic core complex. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Collectively, as shown in Fig. 7F, the putative Cdc20-dissociating residues of the Emi2 ZBR domain, K571, Q577, and C606, are clustered on one face (designated here as the “front face”) of the domain. On the other hand, the K587 residue, required for the inhibition of ubiquitin chain elongation, is located on the “side face”. The K586 residue, on the “back face”, is involved in the inhibition of ubiquitylation and responsible for preventing the re-association of APC/C·Cdc20. Consequently, the ZBR domain of Emi2 exerts the cell-division inhibiting activity through a combination of two mechanisms (Fig. 7G). In one, the ZBR domain binds to the coactivator Cdc20 and impairs its association with the APC/C core complex. In the other, the ZBR inhibits the ubiquitin chain elongation by the APC/C cullin-RING ligase (CRL) module, ANAPC2–11 complex, in combination with the ubiquitin chain initiating E2, E2C. In addition, we determined the solution structure of the Emi2 ZBR domain, and the post-ZBR (PZ) region interacts with the CRL module of APC/C.
4. Conclusions
The present finding of the two novel inhibitory mechanisms (separation of Cdc20 and inhibition of E2C) for the Emi2 ZBR domain will provide a solid foundation for future studies on the functions of Emi2 in in vivo systems, i.e., mammalian oocytes and early embryos. In particular, we identified the single mutations within the Emi2 ZBR domain that selectively impair each of the two new inhibitory mechanisms, which will be useful for distinguishing the multiple inhibitory mechanisms potentially mediated by Emi2. We hope that our findings will be shared and utilized among the scientific community.
Conflict of interest
The authors have no conflicts of interest to declare.
Acknowledgements
The authors thank A.C.F. Perry, RIKEN Center for Developmental Biology, for the cDNA clones encoding mouse Emi1, Emi2, and Cdc20, and Y. Hayashizaki, RIKEN Omics Science Center, for the FANTOM clone (1110015A16) encoding E2C. The authors also thank the Support Unit for Bio-Material Analysis, especially K. Otsuki and M. Usui for the mass spectrometry analysis, at RIKEN BSI Research Resources Center, and H. Fujimoto for the FACS analysis at the RIKEN Research Center for Allergy and Immunology. We would also like to thank the members of the RIKEN Systems and Structural Biology Center, particularly T. Harada, Y. Fujikura, S. Watanabe, and T. Kigawa for protein preparation, H. Shimizu for advice on protein purification methods, Y. Terazawa and T. Fujimoto for technical assistance with the baculovirus-insect cell expression system, K. Tsuganezawa and A. Tanaka for providing the LSM 510 system, and E.C. Morishita and S. Kusano for their helpful comments on the manuscript. This work was supported by the RIKEN Special Postdoctoral Researcher Program to S.S., JSPS KAKENHI Grant Number 20710178 to S.S., the Targeted Proteins Research Program, and the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan to S.Y.
Contributor Information
Mikako Shirouzu, Email: mikako.shirouzu@riken.jp.
Shigeyuki Yokoyama, Email: yokoyama@riken.jp.
Appendix A. Supplementary data
Supplementary Figure 1.
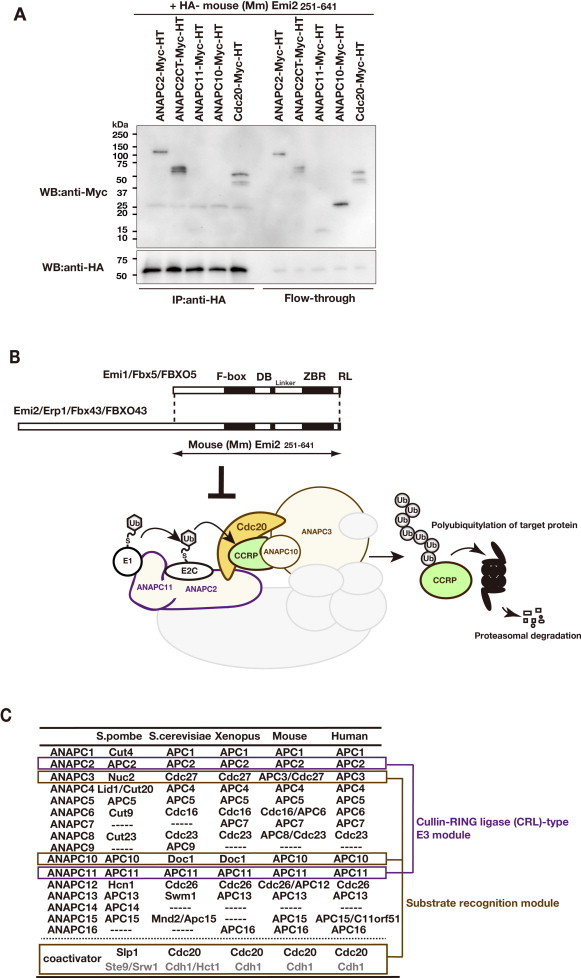
The major components of the Emi2-mediated APC/C inhibition system, related to Fig. 2. (A) Co-IP assay using the HA-tagged F-box–DB–ZBR–RL fragment of Emi2 (residues 251–641 of mouse [Mm] Emi2) and Myc-tagged APC/C subunits: ANAPC2, ANAPC2-CT (C-terminus; residues 337–837 of mouse ANAPC2, corresponding to the cullin-homology region), ANAPC11, ANAPC10, and Cdc20. The recombinant proteins were produced in the conventional in vitro rabbit reticulocyte transcription/translation system. Here, the C-terminally tagged ANAPC10 did not co-immunoprecipitate with Emi2, whereas the N-terminally tagged ANAPC10 co-immunoprecipitated with Emi2 [49]. (B) Schematic representation of the polyubiquitylation reaction by the substrate-recognition catalytic module of APC/C-Cdc20. Cyclin B1 (CycB1) is the target cell-cycle regulatory protein (CCRP) of APC/C-Cdc20. The Emi2 C-terminal CSF active region, which shares high amino acid sequence similarity with its homolog, Emi1, functions as an APC/C inhibitor. (C) The core subunits and coactivators of APC/C. The cullin and RING subunits, ANAPC2 and ANAPC11, are outlined in purple, and the subunits of the substrate-recognition module are outlined in light brown (see also panel B). ANAPC3/Cdc27, ANAPC10/Doc1, and a coactivator constitute the substrate-recognition module.
Supplementary Figure 2.
Structural analysis of the Emi2 ZBR–RL and ZBR regions, related to Fig. 3. (A) The 2D [1H, 15N]-HSQC spectra of the Emi2 ZBR and ZBR–RL fragments. (B) Stereo view of the backbone traces for the 20 conformers of the solution structure of the ZBR domain of Emi2 (mouse Emi2; residues 566–617). (C) The 2D [1H, 15N]-HSQC spectra of the ZBR-RL fragment (mouse Emi2; residues 547–641), showing selected amide shift changes in the titration of the ANAPC2CW fragment (mouse ANAPC2 cullin-Winged-helix subdomain; residues 512–837). (D) Schematic diagrams showing the domain architectures of the E3 ubiquitin ligase subfamily, RING-between-RING fingers (RBR) proteins and the Emi/Erp family (Emi1 and Emi2). ZF, zinc-finger domain; NZF, Npl4-type ZF domain; UBA, ubiquitin-associated domain; UBL, ubiquitin-like domain; RBR, RING-between-RING domain; NLS, nuclear localization signal; RWD, domain found in RING finger-containing proteins, WD repeat-containing proteins, and DEAD-like helicases; POTRA, polypeptide transport-associated domain; CUL7L and CUL1L, Cullin 7- and Cullin 1-like domain; DOC, Doc1/ANAPC10-like domain.
Supplementary Figure 3.
Additional information about the mutants of the Emi2 ZBR–RL and ZBR fragments, related to Fig. 4. (A) Multiple sequence alignment of the C6HC-type zinc finger domain of RBR proteins and Emi2 homologs from different organisms (The domain architectures of the Emi/Erp family and the RBR protein family are shown in Supplementary Fig.S2D). Species abbreviations are the same as in Fig. 3C. Green characters indicate highly conserved residues in Emi2 and its somatic paralog, Emi1. Cyan characters indicate highly conserved residues among Emi2 orthologues. Zinc-binding sites are shown in black boxes; conserved residues among the RBR and Emi/Erp proteins are shown in shaded boxes. Arrows indicate single amino acid substitutions introduced within the ZBR domain for phenotype analysis. Black arrows indicate the ZBR domain mutations that rescued the Emi2 ZBR–RL fragment (residues 547–641 of mouse Emi2)-induced mitotic defects (see also Fig. 4A). The E568A, L570V, Y582P, and S592A mutants of AcGFP-Emi2 ZBR–RL were poorly expressed and/or formed aggregates, indicating denatured protein structures. (B) The cell phenotype was not influenced by the expression of P579A (indicated by an asterisk), although its expression was higher than that of the WT. (C) A binding analysis of the core APC/C to the ZBR mutants of AcGFP-Emi2 ZBR–RL and ZBR in the cells. The cell extracts from the indicated HEK293T transfectants were used for co-IP with an anti-GFP antibody. Western blots (WB) of the immunoprecipitates (IP) and cell lysates were probed with the indicated antibodies. M, mouse monoclonal antibodies; R, rabbit polyclonal antibodies.
Supplementary Figure 4.
In vitro ubiquitylation assay using core enzymatic reaction components for polyubiquitylation by APC/C, related to Fig. 5. (A) In vitro polyubiquitin chain formation by the fundamental ubiquitylation system of APC/C. (B) and (C) Purified recombinant proteins used in the in vitro ubiquitylation assay. (B) APC/C-related catalytic components: UbFLAG (N-terminal FLAG-tagged ubiquitin), E1 (GST-tagged ubiquitin-activating enzyme), E2C (UBE2C/UbcH10), and E3 (full-length APC/C subunit-2 and -11 subcomplex, ANAPC2–11; co-expressed in the baculovirus-insect cell system). (C) Emi2 fragments synthesized in an E. coli cell-free system in the presence of Zn2+ ions (the band with an asterisk is the putative degradation product of Emi2499–641). (D) Zinc (Zn)-dependent folding of the Emi2 C-terminal fragments containing the ZBR domain (see also Fig. 5C). The requirement of Zn2+ ions for protein synthesis and solubility was assayed in cell-free synthesis reactions with (+) or without (−) 50 μM ZnCl2. Total and supernatant (Sup) fractions were analyzed by SDS-PAGE followed by CBB staining. (E) Effect of the addition of an Emi2 C-terminal fragment on E1-catalyzed ubiquitin activation. The DB–ZBR–RL fragment of Emi2 did not impair the ubiquitin activation by E1.
Supplementary Figure 5.
Ubiquitylation inhibitory activities of Emi2 ZBR–RL and ZBR fragments, related to Fig. 6. (A) Replicates of in vitro ubiquitylation assays to compare the abilities of the Emi2 ZBR–RL and ZBR fragments to inhibit polyubiquitin chain assembly, related to Fig. 6G. Ubiquitin adducts containing ubiquitin dimers and partially ubiquitylated Emi2 fragments were occasionally observed (asterisks). (B) The WB with the anti-E2C antibody shows that the auto-ubiquitylation of E2C was hardly affected by the Emi1 and Emi2 ZBR fragments, related to Fig. 6E. The white line indicates the junction between two membranes in simultaneous WB detection (original data shown at the bottom). Upper panel with high contrast shows these membranes arranged at angles in the same direction. (C) Replicates of the in vitro ubiquitylation assay using the mCRLA system, to determine the effects of ZBR mutations on in vitro polyubiquitin chain assembly. (D) The 2D [1H, 15N]-HSQC spectra of the Emi2 ZBR mutants, K586M and K587M.
Supplementary Figure 6.
Characterization of the Emi2 ZBR mutants, related to Fig. 6H. (A) Morphological properties of HEK293T cells expressing the indicated mutants of AcGFP-Emi2 ZBR at 3 days post-transfection, related to panel B. Scale bar, 50 μm. (B–C) The effects of the indicated ZBR mutants on the in cell ubiquitylation sensitivities. The FK2 antibody recognizes ubiquitin conjugates, but not free ubiquitin. (D) Ribbon diagrams with selected side chains drawn in stick representations. The Emi2 ZBR domain (blue silver) is superimposed on the Emi1 ZBR domain (ivory white). The color-codes of the side chains correspond to those in Fig 7A and B.
Supplementary Figure 7.
Cellular phenotypes associated with the AcGFP-Emi1 ZBR mutants constructed by the transplantation of Emi2-specific residues, related to Fig. 7E. Scale bar, 100 μm.
Supplementary Table.
References
- 1.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Weissman A.M., Shabek N., Ciechanover A. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 2011;12:605–620. doi: 10.1038/nrm3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deshaies R.J., Joazeiro C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 4.Peters J.M. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan M., Morgan D.O. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- 6.Pfleger C.M., Lee E., Kirschner M.W. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- 8.Primorac I., Musacchio A. Panta rhei: the APC/C at steady state. J. Cell Biol. 2013;201:177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwab M., Neutzner M., Mocker D., Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 2001;20:5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol. Cell Biol. 2001;21:5190–5199. doi: 10.1128/MCB.21.15.5190-5199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vodermaier H.C., Gieffers C., Maurer-Stroh S., Eisenhaber F., Peters J.M. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr. Biol. 2003;13:1459–1468. doi: 10.1016/s0960-9822(03)00581-5. [DOI] [PubMed] [Google Scholar]
- 12.Thornton B.R., Ng T.M., Matyskiela M.E., Carroll C.W., Morgan D.O., Toczyski D.P. An architectural map of the anaphase-promoting complex. Genes Dev. 2006;20:449–460. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimata Y., Baxter J.E., Fry A.M., Yamano H. A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Mol. Cell. 2008;32:576–583. doi: 10.1016/j.molcel.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 14.Izawa D., Pines J. Mad2 and the APC/C compete for the same site on Cdc20 to ensure proper chromosome segregation. J. Cell Biol. 2012;199:27–37. doi: 10.1083/jcb.201205170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraft C., Vodermaier H.C., Maurer-Stroh S., Eisenhaber F., Peters J.M. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol. Cell. 2005;18:543–553. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 16.Passmore L.A., Barford D. Coactivator functions in a stoichiometric complex with anaphase-promoting complex/cyclosome to mediate substrate recognition. EMBO Rep. 2005;6:873–878. doi: 10.1038/sj.embor.7400482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol. Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Chao W.C., Kulkarni K., Zhang Z., Kong E.H., Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 19.Tian W., Li B., Warrington R., Tomchick D.R., Yu H., Luo X. Structural analysis of human Cdc20 supports multisite degron recognition by APC/C. Proc. Natl. Acad. Sci. U.S.A. 2012;109:18419–18424. doi: 10.1073/pnas.1213438109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Z., Li B., Bharadwaj R., Zhu H., Ozkan E., Hakala K., Deisenhofer J., Yu H. APC2 Cullin protein and APC11 RING protein comprise the minimal ubiquitin ligase module of the anaphase-promoting complex. Mol. Biol. Cell. 2001;12:3839–3851. doi: 10.1091/mbc.12.12.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J., Sheung J., Dong G., Coquilla C., Daniel-Issakani S., Payan D.G. High-throughput screening for inhibitors of the e3 ubiquitin ligase APC. Methods Enzymol. 2005;399:740–754. doi: 10.1016/S0076-6879(05)99049-6. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt A., Rauh N.R., Nigg E.A., Mayer T.U. Cytostatic factor: an activity that puts the cell cycle on hold. J. Cell Sci. 2006;119:1213–1218. doi: 10.1242/jcs.02919. [DOI] [PubMed] [Google Scholar]
- 23.Pesin J.A., Orr-Weaver T.L. Regulation of APC/C activators in mitosis and meiosis. Annu. Rev. Cell Dev. Biol. 2008;24:475–499. doi: 10.1146/annurev.cellbio.041408.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H., Lee D.J., Oh S.P., Park H.D., Nam H.H., Kim J.M., Lim D.S. Mouse emi1 has an essential function in mitotic progression during early embryogenesis. Mol. Cell Biol. 2006;26:5373–5381. doi: 10.1128/MCB.00043-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J., Grimison B., Lewellyn A.L., Maller J.L. The anaphase-promoting complex/cyclosome inhibitor Emi2 is essential for meiotic but not mitotic cell cycles. J. Biol. Chem. 2006;281:34736–34741. doi: 10.1074/jbc.M606607200. [DOI] [PubMed] [Google Scholar]
- 26.Shoji S. Mammalian Emi2 mediates cytostatic arrest and transduces the signal for meiotic exit via Cdc20. EMBO J. 2006;25:834–845. doi: 10.1038/sj.emboj.7600953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machida Y.J., Dutta A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 2007;21:184–194. doi: 10.1101/gad.1495007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu J.Y., Reimann J.D., Sorensen C.S., Lukas J., Jackson P.K. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1) Nat. Cell Biol. 2002;4:358–366. doi: 10.1038/ncb785. [DOI] [PubMed] [Google Scholar]
- 29.Verschuren E.W., Ban K.H., Masek M.A., Lehman N.L., Jackson P.K. Loss of Emi1-dependent anaphase-promoting complex/cyclosome inhibition deregulates E2F target expression and elicits DNA damage-induced senescence. Mol. Cell Biol. 2007;27:7955–7965. doi: 10.1128/MCB.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perrard M.H., Chassaing E., Montillet G., Sabido O., Durand P. Cytostatic factor proteins are present in male meiotic cells and beta-nerve growth factor increases mos levels in rat late spermatocytes. PLoS One. 2009;4:e7237. doi: 10.1371/journal.pone.0007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tischer T., Hormanseder E., Mayer T.U. The APC/C inhibitor XErp1/Emi2 is essential for Xenopus early embryonic divisions. Science. 2012;338:520–524. doi: 10.1126/science.1228394. [DOI] [PubMed] [Google Scholar]
- 32.Hormanseder E., Tischer T., Mayer T.U. Modulation of cell cycle control during oocyte-to-embryo transitions. EMBO J. 2013;32:2191–2203. doi: 10.1038/emboj.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tunquist B.J., Maller J.L. Under arrest: cytostatic factor (CSF)-mediated metaphase arrest in vertebrate eggs. Genes Dev. 2003;17:683–710. doi: 10.1101/gad.1071303. [DOI] [PubMed] [Google Scholar]
- 34.Masui Y., Markert C.L. Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes. J. Exp. Zool. 1971;177:129–145. doi: 10.1002/jez.1401770202. [DOI] [PubMed] [Google Scholar]
- 35.Lorca T., Cruzalegui F.H., Fesquet D., Cavadore J.C., Mery J., Means A., Doree M. Calmodulin-dependent protein kinase II mediates inactivation of MPF and CSF upon fertilization of Xenopus eggs. Nature. 1993;366:270–273. doi: 10.1038/366270a0. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto T.M., Iwabuchi M., Ohsumi K., Kishimoto T. APC/C-Cdc20-mediated degradation of cyclin B participates in CSF arrest in unfertilized Xenopus eggs. Dev. Biol. 2005;279:345–355. doi: 10.1016/j.ydbio.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt A., Duncan P.I., Rauh N.R., Sauer G., Fry A.M., Nigg E.A., Mayer T.U. Xenopus polo-like kinase Plx1 regulates XErp1, a novel inhibitor of APC/C activity. Genes Dev. 2005;19:502–513. doi: 10.1101/gad.320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tung J.J., Hansen D.V., Ban K.H., Loktev A.V., Summers M.K., Adler J.R., 3rd, Jackson P.K. A role for the anaphase-promoting complex inhibitor Emi2/XErp1, a homolog of early mitotic inhibitor 1, in cytostatic factor arrest of Xenopus eggs. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4318–4323. doi: 10.1073/pnas.0501108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inoue D., Ohe M., Kanemori Y., Nobui T., Sagata N. A direct link of the Mos-MAPK pathway to Erp1/Emi2 in meiotic arrest of Xenopus laevis eggs. Nature. 2007;446:1100–1104. doi: 10.1038/nature05688. [DOI] [PubMed] [Google Scholar]
- 40.Nishiyama T., Ohsumi K., Kishimoto T. Phosphorylation of Erp1 by p90rsk is required for cytostatic factor arrest in Xenopus laevis eggs. Nature. 2007;446:1096–1099. doi: 10.1038/nature05696. [DOI] [PubMed] [Google Scholar]
- 41.Frye J.J. Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nat. Struct. Mol. Biol. 2013;20:827–835. doi: 10.1038/nsmb.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohe M. Emi2 inhibition of the anaphase-promoting complex/cyclosome absolutely requires Emi2 binding via the C-terminal RL tail. Mol. Biol. Cell. 2010;21:905–913. doi: 10.1091/mbc.E09-11-0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Isoda M. Dynamic regulation of Emi2 by Emi2-bound Cdk1/Plk1/CK1 and PP2A-B56 in meiotic arrest of Xenopus eggs. Dev. Cell. 2011;21:506–519. doi: 10.1016/j.devcel.2011.06.029. [DOI] [PubMed] [Google Scholar]
- 44.Kipreos E.T., Pagano M. The F-box protein family. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-5-reviews3002. REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reimann J.D., Gardner B.E., Margottin-Goguet F., Jackson P.K. Emi1 regulates the anaphase-promoting complex by a different mechanism than Mad2 proteins. Genes Dev. 2001;15:3278–3285. doi: 10.1101/gad.945701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller J.J., Summers M.K., Hansen D.V., Nachury M.V., Lehman N.L., Loktev A., Jackson P.K. Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes Dev. 2006;20:2410–2420. doi: 10.1101/gad.1454006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang W., Wu J.Q., Chen C., Yang C.S., Guo J.Y., Freel C.D., Kornbluth S. Emi2-mediated inhibition of E2-substrate ubiquitin transfer by the anaphase-promoting complex/cyclosome through a D-box-independent mechanism. Mol. Biol. Cell. 2010;21:2589–2597. doi: 10.1091/mbc.E09-08-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W., Kirschner M.W. Emi1 preferentially inhibits ubiquitin chain elongation by the anaphase-promoting complex. Nat. Cell Biol. 2013;15:797–806. doi: 10.1038/ncb2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sako K., Suzuki K., Isoda M., Yoshikai S., Senoo C., Nakajo N., Ohe M., Sagata N. Emi2 mediates meiotic MII arrest by competitively inhibiting the binding of Ube2S to the APC/C. Nat. Commun. 2014;5:3667. doi: 10.1038/ncomms4667. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki T., Suzuki E., Yoshida N., Kubo A., Li H., Okuda E., Amanai M., Perry A.C. Mouse Emi2 as a distinctive regulatory hub in second meiotic metaphase. Development. 2010;137:3281–3291. doi: 10.1242/dev.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernhardt M.L., Kong B.Y., Kim A.M., O’Halloran T.V., Woodruff T.K. A zinc-dependent mechanism regulates meiotic progression in mammalian oocytes. Biol. Reprod. 2012;86:114. doi: 10.1095/biolreprod.111.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamano H. EMI1, a three-in-one ubiquitylation inhibitor. Nat. Struct. Mol. Biol. 2013;20:773–774. doi: 10.1038/nsmb.2626. [DOI] [PubMed] [Google Scholar]
- 53.Reimann J.D., Freed E., Hsu J.Y., Kramer E.R., Peters J.M., Jackson P.K. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 54.Kigawa T. Cell-free protein production system with the E. coli crude extract for determination of protein folds. Methods Mol. Biol. 2010;607:101–111. doi: 10.1007/978-1-60327-331-2_10. [DOI] [PubMed] [Google Scholar]
- 55.Lee M.S., Palmer A.G., 3rd, Wright P.E. Relationship between 1H and 13C NMR chemical shifts and the secondary and tertiary structure of a zinc finger peptide. J. Biomol. NMR. 1992;2:307–322. doi: 10.1007/BF01874810. [DOI] [PubMed] [Google Scholar]
- 56.Clore G.M., Gronenborn A.M. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol. 1994;239:349–363. doi: 10.1016/s0076-6879(94)39013-4. [DOI] [PubMed] [Google Scholar]
- 57.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 58.Kobayashi N., Iwahara J., Koshiba S., Tomizawa T., Tochio N., Guntert P., Kigawa T., Yokoyama S. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J. Biomol. NMR. 2007;39:31–52. doi: 10.1007/s10858-007-9175-5. [DOI] [PubMed] [Google Scholar]
- 59.Johnson B.A., Blevins R.A. NMR view: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 60.Herrmann T., Guntert P., Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 61.Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 62.Laskowski R.A., Rullmann J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 63.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 64.Case D.A. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsuda K. Structural basis for the dual RNA-recognition modes of human Tra2-beta RRM. Nucleic Acids Res. 2011;39:1538–1553. doi: 10.1093/nar/gkq854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Acquaviva C., Herzog F., Kraft C., Pines J. The anaphase promoting complex/cyclosome is recruited to centromeres by the spindle assembly checkpoint. Nat. Cell Biol. 2004;6:892–898. doi: 10.1038/ncb1167. [DOI] [PubMed] [Google Scholar]
- 67.Sakaue-Sawano A., Ohtawa K., Hama H., Kawano M., Ogawa M., Miyawaki A. Tracing the silhouette of individual cells in S/G2/M phases with fluorescence. Chem. Biol. 2008;15:1243–1248. doi: 10.1016/j.chembiol.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 68.Schreiber A. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Z., Yang J., Kong E.H., Chao W.C., Morris E.P., da Fonseca P.C., Barford D. Recombinant expression, reconstitution and structure of human anaphase-promoting complex (APC/C) Biochem. J. 2013;449:365–371. doi: 10.1042/BJ20121374. [DOI] [PubMed] [Google Scholar]
- 70.Wu Q. A role for Cdc2- and PP2A-mediated regulation of Emi2 in the maintenance of CSF arrest. Curr. Biol. 2007;17:213–224. doi: 10.1016/j.cub.2006.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Osaka F., Seino H., Seno T., Yamao F. A ubiquitin-conjugating enzyme in fission yeast that is essential for the onset of anaphase in mitosis. Mol. Cell Biol. 1997;17:3388–3397. doi: 10.1128/mcb.17.6.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mathe E., Kraft C., Giet R., Deak P., Peters J.M., Glover D.M. The E2-C vihar is required for the correct spatiotemporal proteolysis of cyclin B and itself undergoes cyclical degradation. Curr. Biol. 2004;14:1723–1733. doi: 10.1016/j.cub.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 73.Summers M.K., Pan B., Mukhyala K., Jackson P.K. The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol. Cell. 2008;31:544–556. doi: 10.1016/j.molcel.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kirkpatrick D.S., Hathaway N.A., Hanna J., Elsasser S., Rush J., Finley D., King R.W., Gygi S.P. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 75.Kraft C., Herzog F., Gieffers C., Mechtler K., Hagting A., Pines J., Peters J.M. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garnett M.J., Mansfeld J., Godwin C., Matsusaka T., Wu J., Russell P., Pines J., Venkitaraman A.R. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat. Cell Biol. 2009;11:1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meyer H.J., Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu T., Merbl Y., Huo Y., Gallop J.L., Tzur A., Kirschner M.W. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table.


