Abstract
Metastasis is a complex process that propagates cells from the primary or initial site of the cancer occurrence to distant parts of the body. Cancer cells break from the cancer site and circulate through the bloodstream or lymph vessels, allowing them to reach nearly all parts of the body. These circulating tumour cells (CTCs) contain specialized metastasis-initiating cells (MICs) that reside in the biological heterogeneous primary tumour. Researchers have hypothesized that metastasis of renal cell carcinoma is initiated by circulation of MICs in patients’ blood and bone marrow. Based on the cancer stem/progenitor cell concept of carcinogenesis, understanding the molecular phenotypes of metastasis-initiating cells (MICs) in renal cancer could play a vital role in developing strategies for therapeutic interventions in renal cancer. Existence of MICs among CTCs in renal carcinoma has not been proven in large scale. However, some studies have reported that specialized markers are found on the surface of circulating cells from the primary tumour. In mice, MICs have been isolated from CTCs using such markers, which have then been transplanted into xenograft model to show whether they give rise to metastasis in different organs. Considering these findings, in this review we have attempted to summarize the studies connected with MICs and their gene expression profiles that are responsible for metastasis in renal cancer.
Keywords: Circulating tumour cells, metastasis-initiating cells, renal cell carcinoma, tumour-initiating cells
INTRODUCTION
Renal Cell Carcinoma (RCC)
Renal cell carcinoma (RCC) is the most common epithelial malignancy of human adult kidneys, accounting for 3% of all neoplasms. At 40%, RCC has the highest mortality [1]. The most important prognostic factor in RCC is metastatic dissemination of disease. Approximately 30% of patients with RCC will be diagnosed with metastatic disease, and 60% of these patients will die from aggressive disease and metastases [2]. Within Europe, 42,000 patients are diagnosed with RCC, and 25,000 of these die each year.
Immunotherapies such as interferon (INF)-α and interleukin (IL)-2 have been used for decades in metastatic RCC treatment but have provided positive effects in only 10 – 20% of cases [3,4]. For the past ten years, clinical studies have used targeted molecular therapies such as orally administered sorafenib, sunitinib, pazopanib and tivozanib (receptor tyrosine kinase inhibitors), as well as mammalian target of rapamycin (mTOR) inhibitors (everolimus and temsirolimus) for treating patients with RCC. First-, second- and third-line treatments have shown some positive results. However, continuous treatments with these drugs are associated with a high incidence of toxic effects and resistance [5,6]. The targets of these agents are the signalling pathways responsible for angiogenesis and cancer-cell proliferation. The clinical response is very low, and five-year survival of patients with metastatic RCC is only 10% [7-9].
Tumour-Initiating Cells (TICs) in Renal Cell Carcinoma (RCC)
Increasing evidence shows the existence of small populations of tumour-initiating cells (TICs) or cancer stem cells (CSCs) that reside in the vicinity of the heterogeneous mass of the tumour. TICs have been identified in melanoma [19] and many solid tumours, including brain [10], breast [11], prostate [12], ovarian [13], colon [14], gastric [15], pancreatic [16], head and neck [17] and liver cancers [18]. Like normal stem cells, these specialized tumour regenerating cells share common properties and can be characterized by their ability to self-renew and their capacity to form serially transplantable tumours in immune-deficient mice. On the basis of different protein expression on the cell surface, cells such as CD133, CD105, NCAM, selecting side population (SP) can be easily isolated and identified through aldehyde dehydrogenase 1 (ALDH1) and rhodamine 123 (Rh123) dye activity [20-23] (Fig. 1). During conventional treatments such as radiation and chemotherapy, these cells are not targeted and are responsible for resistance. Afterward, treatment failure results in patient relapse. Currently, there are no efficient treatments for TICs. However, a number of pre-clinical trials have been performed using different therapeutic approaches to halt the proliferation of TICs. These therapies include induction of cell differentiation and blockage of TIC maintenance pathways [24].
Fig. (1).
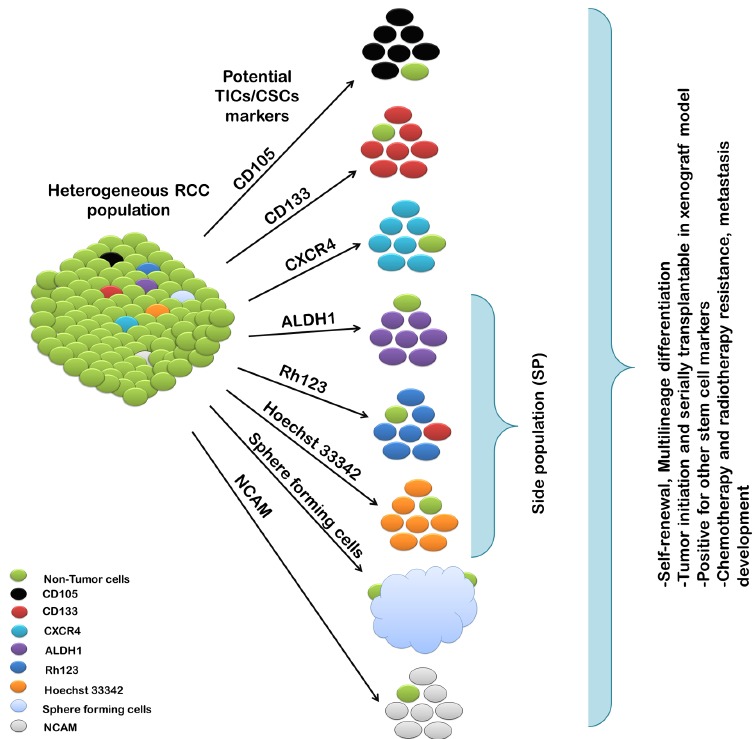
Diagram showing enrichment methods for tumour initiating cells (TICs) in renal cell carcinoma (RCC) and TIC characteristics.
TICs have been identified and isolated from RCC patients and cell lines using mesenchymal stem cell marker CD105 (endoglin) [25,26]. To evaluate the tumorigenicity of the isolated cells, CD105+ (1x106) and CD105- (1x106) cells were transplanted subcutaneously into severely compromised immunodeficient (SCID) mice [25]. CD105+ populations showed induced tumours in 100% of cases (10/10). In contrast, 10% tumour incidence (1/10) was observed with CD105- cells. These cells appear to be important in RCC since angiogenesis is critical for tumour growth and development of metastasis, and CD105 is a membrane glycoprotein that is highly expressed in activated endothelial cells associated with angiogenesis. CD105 protein is part of a receptor for TGF-B1 and TGF-B3 that promotes cell proliferation and differentiation. Moreover, endoglin expression was inversely correlated with Tumour-Node-Metastasis (T-N-M) [27]. Thus, endoglin could be a marker of tumour neovascularization and provide prognostic information in RCC development.
Others have also identified TICs using Hoechst 33342 dye as epithelial side population (SP) in malignant RCC human kidney tissue [28]. SP population cells demonstrated greater potential of colony-forming efficiency. They were enriched for high proliferation in culture conditions and stem cell-like characteristics. Additionally, spheres derived from culturing single cells of RCC cell lines were enriched for cancer stem-like cells [29]. These sphere-forming cells were able to self-renew in in vivo and in vitro models and presented higher expression of ‘stemness’ genes and resistance to chemotherapeutic agents and radiotherapy compared to monolayer-adherent cells. Recently, Huang et al. [30] reported cancer stem cell-like cells with SP phenotype using Hoechst 33342 dye in human primary RCC cell lines. Less than 5% SP population has been reported in all studied RCC cell lines. Sorted SP populations have high expression of ABCB1, ABCG2 and ABCC1 proteins and are resistant to chemotherapy and radiotherapy. However, pre-treatment of SP cells with verapamil, an ABC transporter inhibitor, reversed drug resistance, thereby demonstrating that the ABC transporter could be responsible for drug resistance [30,31]. Moreover, cancer-testis antigen and HSP40 family member DNAJB8 were important in maintaining the TIC/CSC phenotype of the SP population in RCC cells, and overexpression of DNAJB8 increased the percentage of SP cells [32].
CD133 has been investigated as a putative stem cell marker in many solid tumours, including RCC [21,33]. A small population of CD133+ cells have been found in RCC and characterized as renal resident progenitor cells [26, 34]. CD133+ cells were able to differentiate into endothelial and epithelial cells. Therefore, a low number of CD133+ cells in RCC might be an early event in stem cell differentiation and possibly in malignant transformation. In addition, when undifferentiated CD133+ cells from RCC were subcutaneously transplanted alone into SCID in mice, the mice did not give rise to tumour formation, suggesting CD133+ are not TICs. However, when co-transplantated with renal carcinoma cells, CD133+ progenitors fostered tumour engraftment, growth and development. The tumours formed in vivo demonstrated that CD133+ derived endothelial cells were able to form functional vessels enriched in human HLA class I connected with the mouse vasculature. This showed that CD133 can contribute to the growth factor stimulation necessary for angiogenesis [34]. The expression level of CD133 in RCC patients biopsies were not correlated with clinical pathology and prognostic significance [35].
The chemokine receptor CXCR4 is a putative stem cell marker, and its increased expression has been recently reported in renal carcinoma [36,37]. Two RCC cell lines derived from the primary and metastatic site were used to demonstrate that high expression of CXCR4 is associated with a more tumorigenic cell line [38]. RCC cell lines derived from the metastatic site were found enriched in CXCR4+ cells and capable of forming larger spheres in vitro, generating more tumours in mice when compared with CXCR4- cells [24]. These results demonstrated that CXCR4+ cells as a TIC subpopulation the differences in CXCR4 expression reveal differences in the TICs content in RCC cell lines. Moreover, CXCR4+ cells exhibited greater resistance to tyrosine kinase inhibitors and expressed other ‘stemness’ associated genes. Surgical biopsies of the RCC patients confirmed that the high expression of CXCR4 has significant prognostic value and therapeutic importance [35].
The ability of stem cells to efflux dye such as Rhodamine 123 (Rh123) can be used to analyse and isolate these cells with progenitor characteristics [39,40]. Based on Rh123 staining, RCC cells were characterized as Rh123high and Rh123low cells in a recent study [39]. Serially transplanted Rh123high cells into SCID mice were able to form tumours in all cases (12/12 cases). However, no visible tumours were observed for Rh123low cells (1/12 cases) [39]. In addition, the Rh123high cells showed higher differentiating potential and increased survival capability against radiotherapy compared to the Rh123low cells. This finding indicates that TICs might exist in Rh123high populations in RCC cells, which is opposite the results reported in other cancers [41,42]. This could explain the different biological characteristics of RCC compared to other cancer tissues.
Other markers have been analysed in RCC. Pode-Shakked et al. [22] observed neural cell adhesion molecule- NCAM as a putative marker for malignant renal stem/ progenitor in Wilms’ tumour. However, this marker has not been investigated in RCC. The intracellular aldehyde dehydrogenase 1 (ALDH1) functions to catalyse the oxidation of aldehyde and maintain cellular homeostasis. Recent studies have shown that normal and cancer cells with high ALDH1 activity have great potential to function as TICs in metastatic RCC cell lines [43]. ALDH1 activity analysis revealed that SP populations from metastatic RCC cell lines have higher levels of activity for ALDH1 compared to NSP (non-side population). ALDH1+ cells also showed higher sphere-forming ability, self-renewal, tumorigenic and expressed higher mRNA levels of stem cell associated genes [43].
In summary, CD105, CXCR4 and ALDH1 activity could be suitable targets for identifying tumour- initiating cell markers in RCC [24, 25, 43]. (See Table 1.)
Table 1. Tumour-initiating cell (TIC) markers and function in RCC.
| TICs’ Identification Markers | Function in RCC | References |
|---|---|---|
| CD105 (Endoglin) |
Proliferation and differentiation in endothelial cells | Bussolati et al. [25] |
| CD133 (Prominin-1) |
Angiogenesis | Bruno et al. [34] |
| CXCR4 (Chemokine receptor type 4) |
Maintaining TICs and spread metastasis | Gassenmaier et al. [24] |
| ALDH1 activity | Maintain cellular homeostasis | Ueda et al. [43] |
| Hoechst 33342 staining | ABC activity | Addle et al. [28] |
| Rhodamine 123 staining | ABC activity | Lu et al. [39] |
| Sphere formation | Tumour initiation | Zhong et al. [29] |
Metastasis-Initiating Cells (MICs) as Circulating Tumour Cells (CTCs) in RCC
High cancer-associated mortality is mostly due to tumour metastasis. The term metastasis is used to indicate the successful dissemination of tumour cells from the primary tumour site to other parts of the body. Recent findings indicate that circulating tumour cells (CTCs) could contribute to metastasis [44]. The discovery of novel biomarkers for identifying CTCs in many human cancers has provided a novel way of understanding tumorigenesis at the cellular level. The mechanism of the formation of metastasis is comprised of three events: (1) A group of cancer cells detach from the primary tumour. (2) The CTSs circulate to distant parts of the body via the bloodstream or lymphatic system and adapt to the new environment. (3) The cells then settle in the new location to proliferate and colonize, giving rise to a metastatic tumour. However, this process is highly inefficient, as numerous disseminated cells die during migration, and some remain dormant for several years [44]. Because metastasis occurs through CTCs, examining these cells could lead to future research directions and help realize the cells’ clinical potential in RCC treatment and diagnosis.
The clinical relevance of disseminated CTCs in the blood stream of RCC patients remains a controversial subject, as the biology of these cells has been poorly understood [45]. Investigations in patients with different tumours have assessed the prognostic value of disseminated epithelial cells in bone marrow and peripheral blood detected by immunocytochemistry using anti-cytokeratin antibodies [46-48]. Only a few studies in RCC patients have demonstrated the possibility of cytokeratin-positive (CK+) cells as markers for CTCs investigated independently in peripheral blood and bone marrow [49,50]. An increase in the number of CK+ cells in RCC patients correlates with advanced tumour stage in RCC [51]. Bluemke et al. found two types of CTCs in the peripheral blood of RCC patients: cytokeratin (CK+) expressing cells and cells without cytokeratin expression, which are large with tumour-like morphology [49]. Only CK+ cells statistically correlated with the poor overall survival of RCC patients. Similar results have been reported in earlier studies [52,53].
In one study conducted with non-metastatic RCC patients, no prognostic relevance was found for disseminated cytokeratin-positive (CK+) cells in the bone marrow [54]. However, in another study, Buchner et al. [50] found the prognostic values of disseminated CK+ cells in the bone marrow of patients with metastatic RCC indicated that these cells played an important role in tumour spread in metastatic RCC. The researchers also found that immunocytochemical detection of these cells could be useful in the assessment of clinical outcomes in patients. However, further characterization of the cells is necessary to evaluate their malignant potential as a relevant therapeutic target for novel systematic therapy. Recently, El-Heiliebi et al. used a new blood filtration technique to distinguish circulating non-hematologic cells (CNHCs) as CTCs in the blood of RCC patients based on cytomorphological criteria [55].
Gene Expression Pattern in the Development of Metastasis RCC
Gene expression analysis has been widely adopted in clinical and laboratory research to discern genomic background and identify genes that might serve as a prognostic biomarker in metastatic RCC. Identifying such genes could serve to delineate novel targets for the invention of specific anti-cancer drugs. In a recent study, gene expression profiling was performed by comparing microarrays on samples from metastatic and primary RCC patients with other patients’ normal kidney tissue [56]. There were 95 gene sets significantly up-regulated in metastatic RCC. The majority of up-regulated genes in these sets were those responsible for DNA replication, cell cycle control, apoptosis and cell mortality. For instance, genes from the minichromosome maintenance gene family (MCM2, MCM4, MCM6 and MCM7), AURKA-coding for a kinase, involved cell cycle regulation and tumour metastasis progression, as well as FEN1 in DNA replication. Other researchers used a different approach to analyse the gene expression profiles based on stepwise progression and metastasis in RCC [57]. Samples from three different progressive sites of RCC were compared—Kidney tissue (N), through early tumour stage (T1) to distant metastasis (M). Caveolin 1, annexin A4 and lysyl oxidase were continuously deregulated on progression, indicating that these genes might be important in driving the tumour toward increased malignancy and metastasis [57].
In a study of 58 cases, Vasselli et al. identified a gene signature between the primary tumour and one with metastasis RCC [58]. Forty-five genes were consistently up- or down-regulated and associated with survival using the Cox proportional hazards model. In addition, vascular cell adhesion molecule 1 (VCAM-1) was revealed to be the most likely biomarker predicting survival of RCC patients. The higher expression for the VCAM-1 gene was significantly associated with longer survival compared to low VCAM-1 expression. Others have reported higher expression of CYYR1 and LDB2 genes associated with disease-free survival with higher expression in tumours that metastasized after 24 months compared to tumours that metastasized earlier [59]. In a different approach, two cell lines derived from a matched primary tumour and adrenal metastasis from the same RCC patients were analysed using similar technology [60]. EGFR, cadherin-6 and vimentin expression were higher in the metastatic cell line. Furthermore, those highly expressed genes were associated with growth factors, cell division, signal transduction and cell-cell adhesion function. Bockhorn et al. analysed the gene expression of cells shedding from two RCC cell lines with different metastatic potential [61]. Their analysis revealed that 23 deregulated genes were involved in metastasis. However, only caveolin-1, CD44 and α3-integrin expression were down regulated in cells shed from both cell lines. These expressions might be important in faster migration of the tumour cells and give shed cells a metastatic advantage.
Molecular differences between tumour subtypes of RCC have been largely unknown. In a study of 112 RCC and normal kidney samples, researchers distinguished the gene expression pattern between three major types of RCC: clear cell RCC (ccRCC), papillary RCC (pRCC) and chromophobe RCC (chRCC) [62]. Gene expression was widely correlated with RCC subtype. Down regulation of genes was reported in PRCC and chRCC compared with ccRCC. These genes were responsible for metabolism (GAPD), angiogenesis (ANGPTL4 and VEGF), cell adhesion (COL3A1 and FN1) and immune response (IGHG3 and HLA-DRBI). Up-regulation was reported in genes for osteopontin (SPP1) and retinol binding protein 4 (RBP4). Moreover, primary tumour gene expression delineated 12 such genes that were associated with tumour metastasis formation and patient survival. Among them, the most significantly deregulated genes were the human high-mobility group gene (HMGA1), mitochondrial dienoyl-CoA reductase (DECR1), GTP-binding protein (RAGB) and genes belonging to the gene families COL5A1, SLC13A3, SLC29A2, IGFBP3 and GUCY2C [62]. In summary, these finding could serve to discriminate clinically meaningful sub classification of RCC and expression pattern approaches to describe metastasis formation in various RCC sub-types.
Genome expression analysis has been widely implicated in cancer research. Extracting relevant biological insight from such genome data sets remains a major challenge. In recent years, the Gene Set Enrichment Analysis (GSEA) method has been purposed to incorporate knowledge from databases with gene expression data set [63]. Maruschke et al. used the GSEA approach to analyse the expression profile in ccRCC by comparing poorly and well-differentiated tumour tissue samples [64]. Using comparative analysis, the researchers selected 16 expression gene sets out of 120. Genes in these sets were involved in cell motility, signalling, proliferation and metastasis and were gene products in the building of extracellular matrix and as cell surface markers.
conclusion
In conclusion, this short review of the current literature has shown that the activities of CD105, CXCR4 and ALDH1 represent the most prominent markers for identifying tumour-initiating cells in RCC. These cells are also associated with metastasis initiation and progression. In addition, cells expressing these markers share many common characteristics of the TIC/CSC phenotype, having strong proliferation potential, self-renewal ability, resistance against conventional therapy and the formation of tumours in in vivo studies. Gene expression profiling helps to define TICs and might help develop targeted therapeutics with refined diagnostic data for RCC cure. However, current described gene expression studies cannot be employed into clinical application. Therefore, additional confirmation is needed, and future studies might be helpful to identify relevance with the clinical response in RCC.
Acknowledgements
This research was supported by the Military Institute of Medicine statutory founding 1/1744 (101). CS, AMC was supported by the National Science Centre UMO-2011/01/B/ NZ5/02822 and 2011/01/B/NZ4/01602 projects. CS, AMC, IMK were supported by the Foundation for Polish Science TEAM project TEAM/2010-6/8. AMC was also supported by the Ministry of Science and Higher Education “Juventus” grant. The authors acknowledge the support of the Scribendi, Inc. for professional editing and proofreading of this manuscript.
Footnotes
The authors indicate no potential conflict of interest.
References
- 1.Chow T.F., Youssef Y.M., Lianidou E., Romaschin A.D., Honey R.J., Stewart R., et al. Differential expression profiling of microRNAs and their potential involvement in renal cell carcinoma pathogenesis. Clin. Biochem. 2010;43(1-2):150–158. doi: 10.1016/j.clinbiochem.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 2.Ljungberg B. Prognostic factors in renal cell carcinoma. Urologe (Ausg. A) 2004;43(Suppl. 3):S119–S120. doi: 10.1007/s00120-004-0594-6. [DOI] [PubMed] [Google Scholar]
- 3.Leibovich B.C., Han K.R., Bui M.H., Pantuck A.J., Dorey F.J., Figlin R.A., et al. Scoring algorithm to predict survival after nephrectomy and immunotherapy in patients with metastatic renal cell carcinoma: a stratification tool for prospective clinical trials. Cancer. 2003;98(12):2566–2575. doi: 10.1002/cncr.11851. [DOI] [PubMed] [Google Scholar]
- 4.Escudier B., Eisen T., Stadler W.M., Szczylik C., Oudard S., Siebels M., et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N. Engl. J. Med. 2007;356(2):125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 5.Buczek M., Escudier B., Bartnik E., Szczylik C., Czarnecka A. Resistance to tyrosine kinase inhibitors in clear cell renal cell carcinoma: From the patient's bed to molecular mechanisms. Biochim. Biophys. Acta. 2013 doi: 10.1016/j.bbcan.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Czarnecka A.M., Solarek W. The activity of tyrosine kinase inhibitors on clear cell renal cell carcinoma tumor initiating cells in hypoxic microenvironment.; 2012. [Google Scholar]
- 7.Motzer R.J., Hutson T.E., Cella D., Reeves J., Hawkins R., Guo J., et al. Pazopanib versus sunitinib in metastatic renal-cell carcinoma. N. Engl. J. Med. 2013;369(8):722–731. doi: 10.1056/NEJMoa1303989. [DOI] [PubMed] [Google Scholar]
- 8.Sternberg C.N., Hawkins R.E., Wagstaff J., Salman P., Mardiak J., Barrios C.H., et al. A randomised, double-blind phase III study of pazopanib in patients with advanced and/or metastatic renal cell carcinoma: final overall survival results and safety update. Eur. J. Cancer. 2013;49(6):1287–1296. doi: 10.1016/j.ejca.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 9.Rini B.I., Escudier B., Tomczak P., Kaprin A., Szczylik C., Hutson T.E., et al. Comparative effectiveness of axitinib versus sorafenib in advanced renal cell carcinoma (AXIS): a randomised phase 3 trial. Lancet. 2011;378(9807):1931–1939. doi: 10.1016/S0140-6736(11)61613-9. [DOI] [PubMed] [Google Scholar]
- 10.Singh S.K., Clarke I.D., Terasaki M., Bonn V.E., Hawkins C., Squire J., et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 11.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins A.T., Berry P.A., Hyde C., Stower M.J., Maitland N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65(23):10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 13.Bapat S.A., Mali A.M., Koppikar C.B., Kurrey N.K. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005;65(8):3025–3029. doi: 10.1158/0008-5472.CAN-04-3931. [DOI] [PubMed] [Google Scholar]
- 14.Ricci-Vitiani L., Lombardi D.G., Pilozzi E., Biffoni M., Todaro M., Peschle C., et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 15.Haraguchi N., Utsunomiya T., Inoue H., Tanaka F., Mimori K., Barnard G.F., et al. Characterization of a side population of cancer cells from human gastrointestinal system. Stem Cells. 2006;24(3):506–513. doi: 10.1634/stemcells.2005-0282. [DOI] [PubMed] [Google Scholar]
- 16.Li C., Heidt D.G., Dalerba P., Burant C.F., Zhang L., Adsay V., et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 17.Prince M.E., Sivanandan R., Kaczorowski A., Wolf G.T., Kaplan M.J., Dalerba P., et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl. Acad. Sci. USA. 2007;104(3):973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z.F., Ngai P., Ho D.W., Yu W.C., Ng M.N., Lau C.K., et al. Identification of local and circulating cancer stem cells in human liver cancer. Hepatology. 2008;47(3):919–928. doi: 10.1002/hep.22082. [DOI] [PubMed] [Google Scholar]
- 19.Fang D., Nguyen T.K., Leishear K., Finko R., Kulp A.N., Hotz S., et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65(20):9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 20.Baccelli I., Trumpp A. The evolving concept of cancer and metastasis stem cells. J. Cell Biol. 2012;198(3):281–293. doi: 10.1083/jcb.201202014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bussolati B, Brossa A, Camussi G. Resident stem cells and renal carcinoma. 2011. [DOI] [PMC free article] [PubMed]
- 22.Pode-Shakked N., Metsuyanim S., Rom-Gross E., Mor Y., Fridman E., Goldstein I., et al. Developmental tumourigenesis: NCAM as a putative marker for the malignant renal stem/progenitor cell population. J. Cell. Mol. Med. 2009;13(8B):1792–1808. doi: 10.1111/j.1582-4934.2008.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan M.I., Czarnecka A.M., Krol M., Zdanowski R., Sobocinska A., Lewicki S., et al. 2013. [Google Scholar]
- 24.Gassenmaier M., Chen D., Buchner A., Henkel L., Schiemann M., Mack B., et al. CXC Chemokine Receptor 4 is Essential for Maintenance of Renal cell Carcinoma-Initiating Cells and Predicts Metastasis. Stem Cells. 2013;31(8):1467–1476. doi: 10.1002/stem.1407. [DOI] [PubMed] [Google Scholar]
- 25.Bussolati B, Bruno S, Grange C, Ferrando U, Camussi G. Identification of a tumor-initiating stem cell population in human renal carcinomas. 2008. [DOI] [PubMed]
- 26.Khan M.I., Czarnecka A.M., Szczylik C. 2012. [Google Scholar]
- 27.Sandlund J., Hedberg Y., Bergh A., Grankvist K., Ljungberg B., Rasmuson T. Endoglin (CD105) expression in human renal cell carcinoma. BJU Int. 2006;97(4):706–710. doi: 10.1111/j.1464-410X.2006.06006.x. [DOI] [PubMed] [Google Scholar]
- 28.Addla S.K., Brown M.D., Hart C.A., Ramani V.A., Clarke N.W. Characterization of the Hoechst 33342 side population from normal and malignant human renal epithelial cells. Am. J. Physiol. Renal Physiol. 2008;295(3):F680–F687. doi: 10.1152/ajprenal.90286.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong Y., Guan K., Guo S., Zhou C., Wang D., Ma W., et al. Spheres derived from the human SK-RC-42 renal cell carcinoma cell line are enriched in cancer stem cells. Cancer Lett. 2010;299(2):150–160. doi: 10.1016/j.canlet.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Huang B., Huang Y.J., Yao Z.J., Chen X., Guo S.J., Mao X.P., et al. Cancer stem cell-like side population cells in clear cell renal cell carcinoma cell line 769P. PLoS One. 2013;8(7):e68293. doi: 10.1371/journal.pone.0068293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golebiewska A., Brons N.H., Bjerkvig R., Niclou S.P. Critical appraisal of the side population assay in stem cell and cancer stem cell research. Cell Stem Cell. 2011;8(2):136–147. doi: 10.1016/j.stem.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Nishizawa S., Hirohashi Y., Torigoe T., Takahashi A., Tamura Y., Mori T., et al. HSP DNAJB8 controls tumor-initiating ability in renal cancer stem-like cells. Cancer Res. 2012;72(11):2844–2854. doi: 10.1158/0008-5472.CAN-11-3062. [DOI] [PubMed] [Google Scholar]
- 33.Czarnecka A.M., Matak D., Solarek W., Khan M.I., Szczylik C. Hypoxia response regulates clear cell renal cell carcinoma tumor initiating cells.; 2013. [Google Scholar]
- 34.Bruno S., Bussolati B., Grange C., Collino F., Graziano M.E., Ferrando U., et al. CD133+ renal progenitor cells contribute to tumor angiogenesis. Am. J. Pathol. 2006;169(6):2223–2235. doi: 10.2353/ajpath.2006.060498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D'Alterio C., Cindolo L., Portella L., Polimeno M., Consales C., Riccio A., et al. Differential role of CD133 and CXCR4 in renal cell carcinoma. Cell Cycle. 2010;9(22):4492–4500. doi: 10.4161/cc.9.22.13680. [DOI] [PubMed] [Google Scholar]
- 36.Zagzag D., Krishnamachary B., Yee H., Okuyama H., Chiriboga L., Ali M.A., et al. Stromal cell-derived factor-1alpha and CXCR4 expression in hemangioblastoma and clear cell-renal cell carcinoma: von Hippel-Lindau loss-of-function induces expression of a ligand and its receptor. Cancer Res. 2005;65(14):6178–6188. doi: 10.1158/0008-5472.CAN-04-4406. [DOI] [PubMed] [Google Scholar]
- 37.Gelmini S., Mangoni M., Serio M., Romagnani P., Lazzeri E. The critical role of SDF-1/CXCR4 axis in cancer and cancer stem cells metastasis. J. Endocrinol. Invest. 2008;31(9):809–819. doi: 10.1007/BF03349262. [DOI] [PubMed] [Google Scholar]
- 38.Djafarzadeh R., Noessner E., Engelmann H., Schendel D.J., Notohamiprodjo M., von Luettichau I., et al. GPI-anchored TIMP-1 treatment renders renal cell carcinoma sensitive to FAS-meditated killing. Oncogene. 2006;25(10):1496–1508. doi: 10.1038/sj.onc.1209188. [DOI] [PubMed] [Google Scholar]
- 39.Lu J., Cui Y., Zhu J., He J., Zhou G., Yue Z. Biological characteristics of Rh123 stem-like cells in a side population of 786-O renal carcinoma cells. Oncol. Lett. 2013;5(6):1903–1908. doi: 10.3892/ol.2013.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Challen G.A., Little M.H. A side order of stem cells: the SP phenotype. Stem Cells. 2006;24(1):3–12. doi: 10.1634/stemcells.2005-0116. [DOI] [PubMed] [Google Scholar]
- 41.Wagner-Souza K., Diamond H.R., Ornellas M.H., Gomes B.E., Almeida-Oliveira A., Abdelhay E., et al. Rhodamine 123 efflux in human subpopulations of hematopoietic stem cells: comparison between bone marrow, umbilical cord blood and mobilized peripheral blood CD34+ cells. Int. J. Mol. Med. 2008;22(2):237–242. [PubMed] [Google Scholar]
- 42.Touil Y., Zuliani T., Wolowczuk I., Kuranda K., Prochazkova J., Andrieux J., et al. The PI3K/AKT signaling pathway controls the quiescence of the low-Rhodamine123-retention cell compartment enriched for melanoma stem cell activity. Stem Cells. 2013;31(4):641–651. doi: 10.1002/stem.1333. [DOI] [PubMed] [Google Scholar]
- 43.Ueda K., Ogasawara S., Akiba J., Nakayama M., Todoroki K., Sanada S., et al. Aldehyde dehydrogenase 1 identifies cells with cancer stem cell-like properties in a human renal cell carcinoma cell line. PLoS One. 2013;8(10):e75463. doi: 10.1371/journal.pone.0075463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacob K., Sollier C., Jabado N. Circulating tumor cells: detection, molecular profiling and future prospects. Expert Rev. Proteomics. 2007;4(6):741–756. doi: 10.1586/14789450.4.6.741. [DOI] [PubMed] [Google Scholar]
- 45.Gradilone A., Iacovelli R., Cortesi E., Raimondi C., Gianni W., Nicolazzo C., et al. Circulating tumor cells and “suspicious objects” evaluated through CellSearch(R) in metastatic renal cell carcinoma. Anticancer Res. 2011;31(12):4219–4221. [PubMed] [Google Scholar]
- 46.Braun S., Pantel K., Muller P., Janni W., Hepp F., Kentenich C.R., et al. Cytokeratin-positive cells in the bone marrow and survival of patients with stage I, II, or III breast cancer. N. Engl. J. Med. 2000;342(8):525–533. doi: 10.1056/NEJM200002243420801. [DOI] [PubMed] [Google Scholar]
- 47.Partridge M, Brakenhoff R, Phillips E, Ali K, Francis R, Hooper R, et al. Detection of rare disseminated tumor cells identifies head and neck cancer patients at risk of treatment failure. 2003. [PubMed]
- 48.Wollenberg B., Walz A., Kolbow K., Pauli C., Chaubal S., Andratschke M. Clinical relevance of circulating tumour cells in the bone marrow of patients with SCCHN. Onkologie. 2004;27(4):358–362. doi: 10.1159/000079088. [DOI] [PubMed] [Google Scholar]
- 49.Bluemke K, Bilkenroth U, Meye A, Fuessel S, Lautenschlaeger C, Goebel S, et al. Detection of circulating tumor cells in peripheral blood of patients with renal cell carcinoma correlates with prognosis. 2009. [DOI] [PubMed]
- 50.Buchner A., Riesenberg R., Kotter I., Hofstetter A., Stief C., Oberneder R. Frequency and prognostic relevance of disseminated tumor cells in bone marrow of patients with metastatic renal cell carcinoma. Cancer. 2006;106(7):1514–1520. doi: 10.1002/cncr.21775. [DOI] [PubMed] [Google Scholar]
- 51.Bilkenroth U, Taubert H, Riemann D, Rebmann U, Heynemann H, Meye A. Detection and enrichment of disseminated renal carcinoma cells from peripheral blood by immunomagnetic cell separation. 2001. [DOI] [PubMed]
- 52.Ashida S, Okuda H, Chikazawa M, Tanimura M, Sugita O, Yamamoto Y, et al. Detection of circulating cancer cells with von hippel-lindau gene mutation in peripheral blood of patients with renal cell carcinoma. 2000. [PubMed]
- 53.Shimazui T., Yoshikawa K., Uemura H., Kawamoto R., Kawai K., Uchida K., et al. Detection of cadherin-6 mRNA by nested RT-PCR as a potential marker for circulating cancer cells in renal cell carcinoma. Int. J. Oncol. 2003;23(4):1049–1054. [PubMed] [Google Scholar]
- 54.Buchner A., Riesenberg R., Kotter I., Crispin A., Hofstetter A., Oberneder R. Detection and prognostic value of cytokeratin positive tumor cells in bone marrow of patients with renal cell carcinoma. J. Urol. 2003;170(5):1747–1751. doi: 10.1097/01.ju.0000091877.49439.cf. [DOI] [PubMed] [Google Scholar]
- 55.El-Heliebi A., Kroneis T., Zohrer E., Haybaeck J., Fischereder K., Kampel-Kettner K., et al. Are morphological criteria sufficient for the identification of circulating tumor cells in renal cancer? J. Transl. Med. 2013;11(1):214. doi: 10.1186/1479-5876-11-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maruschke M, Hakenberg OW, Koczan D, Zimmermann W, Stief CG, Buchner A. Expression profiling of metastatic renal cell carcinoma using gene set enrichment analysis. 2013. [DOI] [PubMed]
- 57.Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD, et al. Gene signatures of progression and metastasis in renal cell cancer. 2005. [DOI] [PubMed]
- 58.Vasselli J.R., Shih J.H., Iyengar S.R., Maranchie J., Riss J., Worrell R., et al. Predicting survival in patients with metastatic kidney cancer by gene-expression profiling in the primary tumor. Proc. Natl. Acad. Sci. USA. 2003;100(12):6958–6963. doi: 10.1073/pnas.1131754100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wozniak M.B., Le Calvez-Kelm F., Abedi-Ardekani B., Byrnes G., Durand G., Carreira C., et al. Integrative genome-wide gene expression profiling of clear cell renal cell carcinoma in Czech Republic and in the United States. PLoS One. 2013;8(3):e57886. doi: 10.1371/journal.pone.0057886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohno Y., Izumi M., Tachibana M., Kawamura T., Yoshioka K., Aoyagi T., et al. Characterization and gene expression analysis of novel matched primary and metastatic renal cell carcinoma cell lines. Oncol. Rep. 2008;20(3):501–509. [PubMed] [Google Scholar]
- 61.Bockhorn M., Roberge S., Sousa C., Jain R.K., Munn L.L. Differential gene expression in metastasizing cells shed from kidney tumors. Cancer Res. 2004;64(7):2469–2473. doi: 10.1158/0008-5472.can-03-0256. [DOI] [PubMed] [Google Scholar]
- 62.Sultmann H, von Heydebreck A, Huber W, Kuner R, Buness A, Vogt M, et al. Gene expression in kidney cancer is associated with cytogenetic abnormalities, metastasis formation, and patient survival. 2005. [PubMed]
- 63.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maruschke M., Reuter D., Koczan D., Hakenberg O.W., Thiesen H.J. Gene expression analysis in clear cell renal cell carcinoma using gene set enrichment analysis for biostatistical management. BJU Int. 2011;108(2 Pt 2):E29–E35. doi: 10.1111/j.1464-410X.2010.09794.x. [DOI] [PubMed] [Google Scholar]


