Abstract
MicroRNAs (miRNAs) have emerged as biomarkers of metabolic status, etiological factors in complex disease, and promising drug targets. Recent reports suggest that miRNAs are critical regulators of pathways underlying the pathophysiology of type 2 diabetes. In this study, we demonstrate by deep sequencing and real-time quantitative PCR that hepatic levels of Foxa2 mRNA and miR-29 are elevated in a mouse model of diet-induced insulin resistance. We also show that Foxa2 and miR-29 are significantly upregulated in the livers of Zucker diabetic fatty (fa/fa) rats and that the levels of both returned to normal upon treatment with the insulin-sensitizing agent pioglitazone. We present evidence that miR-29 expression in human hepatoma cells is controlled in part by FOXA2, which is known to play a critical role in hepatic energy homeostasis. Moreover, we demonstrate that miR-29 fine-tunes FOXA2-mediated activation of key lipid metabolism genes, including PPARGC1A, HMGCS2, and ABHD5. These results suggest that miR-29 is an important regulatory factor in normal metabolism and may represent a novel therapeutic target in type 2 diabetes and related metabolic syndromes.
Introduction
Type 2 diabetes is characterized in part by resistance to insulin action in the liver and other metabolic tissues (1). MicroRNAs (miRNAs) are widely recognized as important regulators of a diverse array of biological processes (2), including metabolism (3). Recently, miRNAs have also emerged as stable plasma biomarkers of physiologic and metabolic status (4,5), etiological factors in complex disease (6), and promising therapeutic targets (7,8). miRNA-mediated gene regulation occurs principally at the posttranscriptional level and has been the subject of intense research over the past decade. Several miRNAs have been implicated in the pathobiology of a variety of metabolic disorders, including type 2 diabetes (9), cardiovascular disease (10), and obesity (11). Recently, we reported that miR-27b is a posttranscriptional regulatory hub in liver lipid metabolism and is altered in dyslipidemia (12). Another group of studies demonstrated that miR-33 modulates lipoprotein metabolism in mice (13–17) and nonhuman primates (18), as well as insulin signaling in cultured human hepatocytes (19). And, most recently, both miR-103/107 and miR-802 (20) were shown to regulate insulin sensitivity and glucose tolerance in mice (20,21). These findings strongly support the notion that miRNAs are critical players in pathways that underlie metabolic disease etiology, thus raising the possibility that miRNA-based therapy could be relevant for type 2 diabetes and related metabolic syndromes.
miR-29 has been demonstrated to be an important regulator of numerous biological processes, including neuronal maturation (22), fibrosis (23), hematopoiesis (24), replicative senescence (25), and immune response (26). Our recent in silico work identified miR-29 as the strongest candidate miRNA regulatory hub in the type 2 diabetes gene network (27). Other groups have shown that miR-29 is highly responsive to glucose and may regulate β-cell proliferation and insulin secretion (28,29). We sought to investigate miR-29 in the liver, which is a metabolic tissue of critical relevance to type 2 diabetes etiology.
In this study, we demonstrate that 1) hepatic miR-29 and Foxa2 mRNA are significantly upregulated in two different animal models of insulin resistance, 2) the insulin-sensitizing drug pioglitazone corrects hepatic miR-29 and Foxa2 levels in the Zucker diabetic fatty (ZDF) rat model of diabetes, 3) miR-29 levels in hepatocytes are controlled in part by the insulin-regulated transcription factor (TF) FOXA2, and 4) miR-29 fine-tunes FOXA2-mediated regulation of key lipid metabolism genes. Taken together, our findings implicate miR-29 as an important regulatory factor for lipid homeostasis and motivate future studies to investigate the utility of miR-29 as a tissue biomarker of type 2 diabetes drug efficacy, as well as a potential therapeutic target in metabolic syndromes.
Research Design and Methods
Animal Studies
Female C57BL/6J mice were from a University of North Carolina (UNC) at Chapel Hill colony and started at 4 weeks of age on high-fat diet (HFD) (D01060502, 45% kcal from fat) or matched low-fat diet (LFD) (D01060501, 10% kcal from fat) (Research Diets, New Brunswick, NJ). Livers were isolated after 16 weeks of diet and RNA was isolated using the Norgen Total RNA Purification Kit (Thorold, Ontario, Canada). Male ZDF rats (Charles Rivers Laboratories) were acclimated for 2 weeks and had access to a standard chow diet (LabDiet, St. Louis, MO). Four weeks of pioglitazone treatment (30 mg/kg/day) was started at 8 weeks of age. Blood was collected once a week during treatment to measure glucose levels. Livers were isolated at 12 weeks of age, and RNA was isolated using TRIzol.
Cell Culture
Huh7 cells (human hepatoma) were obtained from Dr. Stanley Lemon’s laboratory at UNC at Chapel Hill. Huh7 cells were maintained in 5 mmol/L glucose Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10% FBS, 2 mmol/L L-glutamine, 1 mmol/L Na-pyruvate, and 1× NEAA (Invitrogen, Grand Island, NY), in 100 mm of collagen 1–coated cell culture dishes (Becton Dickinson, Bedford, MA). For transfections, cells were split into 6-well collagen 1–coated plates (Becton Dickinson) to approximately 70–80% confluency, and allowed 24 h to adhere. All cells were cultured in a humidified incubator at 37°C and 5% CO2.
Transfection Studies
Huh7 cells were plated on collagen 1–coated 6-well plates (Becton Dickinson) 1 day before transfection. At ∼70–80% confluency, the cells were transfected with either 10 nmol/L miRIDIAN hsa-miR-29a mimic (Thermo Scientific, Waltham, MA), 10 nmol/L mmu-miR-29a-3p LNA inhibitor (Exiqon, Woburn, MA), or 100 nmol/L ON-TARGETplus human siRNA against FOXA2 (Thermo Scientific) using either DharmaFECT 4 (Thermo Scientific) or Lipofectamine 2000 (Life Technologies, Grand Island, NY) transfection reagent. A human FOXA2 open reading frame (ORF) expression vector containing FOXA2 transcript variant 1 in the pCMV6-XL5 plasmid (OriGene, Rockville, MD) was transfected (1 μg) using DharmaFECT Duo transfection reagent (Thermo Scientific). Forty-eight hours after transfection, total RNA was isolated from the cells using the Total RNA Purification Kit from Norgen.
Small RNA Sequencing Analysis
Total RNA was extracted from mouse liver tissue using the Norgen Total RNA Purification Kit. RNA quality was assessed by Agilent 2100 Bioanalyzer, and only very high-quality samples with a RNA Integrity Number (RIN) above 8.0 were considered further. Small RNA libraries (n = 2 for each of HFD-fed and LFD-fed mice) were generated using the Illumina TruSeq Small RNA library preparation kit. These libraries were then sequenced on the Illumina HiSeq 2000 platform (50 bp reads). miRNA and isomiR identification and quantitation were performed as described previously (27).
Gene Expression (RNA) Analysis
Total RNA was isolated from cultured Huh7 cells or mouse liver tissue using the Total RNA Purification Kit and subjected to DNase treatment using the TURBO DNA-free Kit (Applied Biosystems, Grand Island, NY). Complementary DNA was synthesized using either the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) or the High-Capacity RNA-to-cDNA Kit (Applied Biosystems), according to the manufacturer’s instructions. Real-time PCR amplification was performed using TaqMan miRNA or gene expression assays in TaqMan Universal PCR Master Mix (miRNA qPCR) or TaqMan Gene Expression Master Mix (gene expression qPCR) on a Bio-Rad CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Richmond, CA). Reactions were performed in triplicate using either U6 (miRNA expression) or RPS9 (gene expression) as the internal control. miRNA and mRNA levels were expressed as relative quantitative values (RQVs). All TaqMan assays used in this study were purchased from Applied Biosystems and include miR-29a (4427975, 002112), miR-29b (4427975, 000412), miR-29c (444087, 000587), miR-15a (4427975, 000389), U6 (4427975–001973), RPS9 (human—4331182, Hs02339424_g1; mouse—4331182, Mm00850060_s1), ABHD5 (4331182, Hs01104373_m1), HMGCS2 (4331182, Hs00985427_m1), G6PC (human—4331182, Hs00609178_m1; mouse—4331182, Mm00839363_m1), PPARGC1A (4331182, Hs01016719_m1), and FOXA2 (human—4331182, Hs00232764_m1; mouse—4331182, Mm01976556_s1; rat—4331182, Rn0145600_m1).
Western Blotting
Protein was isolated 48 to 72 h after transfection. RIPA Buffer (Sigma-Aldrich), supplemented with complete protease inhibitor (Roche, Indianapolis, IN), phosphatase inhibitor (Thermo Scientific), 100 mmol/L PMSF in 100% isopropanol, 0.1% β-mercaptoethanol (VWR International, Radnor, PA), and 1 mol/L DTT (Fisher Scientific, Pittsburgh, PA), was used to passively lyse adhered cells. The lysate was collected and flash frozen before clarification by centrifugation. Protein concentration was calculated using the Pierce Microplate BCA Protein Assay Kit—Reducing Agent Compatible (Thermo Scientific) and run on an Any kD Mini-PROTEAN TGX Precast Gel (Bio-Rad). Transfer was conducted using Bio-Rad Midi Transfer Packs on the Bio-Rad Trans-Blot Turbo Blotting System. The membranes were blocked in 5% nonfat dry milk (Sigma-Aldrich) in TBST and probed with 1:800 anti-ABHD5 antibody (Abnova, Cat. # H00051099-M01, Taiwan) in 4% BSA (Applied Biosystems, Cat. # AM2616) in TBST. Goat anti-rabbit secondary antibody (Abcam, Cat. # ab131366) was diluted 1:3,000 in 1% milk/TBST. β-Actin peroxidase (Sigma-Aldrich Cat. # A3854) or GAPDH (Cell Signaling Technology; Danvers, MA; Cat. # 8884s) diluted 1:40,000 in 1% milk/TBST were used as control subjects. Signal was detected using the Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare Life Sciences, Piscataway, NJ) following manufacturer instructions, and was exposed using the LI-COR C-DiGit Blot Scanner (LI-COR Biosciences, Lincoln, NE). Signal intensity was determined by densitometry using LI-COR Biosciences Image Studio Software.
Reporter Gene (Luciferase) Assays
Human embryonic kidney 293T (HEK293T) cells were plated at 1 × 105 cells/mL for 24 h prior to transient transfection using DharmaFECT 4 and 500 ng/mL MT01 Firefly/Renilla luciferase reporter with the entire 3′ UTR of HMGCS2 cloned immediately downstream of Firefly luciferase (GeneCopoeia). Cells were dual transfected with 10 nmol/L of miR-29a mimic for 48 h prior to cell lysis and dual luciferase assays (GeneCopoeia). Site-directed mutagenesis was completed using QuikChange II XL kits (Stratagene) and the following primers: Reverse 5′-agccgttgcaccgtcaggcacaggg-3′ and Forward 5′-ccctgtgcctgacggtgcaacggct-3′. A 3 base deletion was created in the middle of the predicted “seed” target site for miR-29 (…tgcctgacggtggtgcaacggctgatgga…).
Bioinformatics
Chromatin immunoprecipitation sequencing (ChIP-seq) data for Foxa2 in mouse liver and islet were published previously (30). Chromatin occupancy sites based on these ChIP-seq data were obtained directly from the lead author of the study (Brad Hoffman, University of British Columbia). Candidate Foxa2 target genes in mouse liver and islet were assembled by cross-referencing the chromosomal locations of Foxa2 occupancy sites with gene promoter regions (defined as windows 5 kb upstream of transcription start sites as annotated in the RefSeq database). The miR-29ab promoter region was identified as recently described (31). Target site prediction for miR-29 was performed with TargetScan 6.2 (downloaded from http://www.targetscan.org). Statistical enrichment of predicted miR-29 target sites among Foxa2 target genes in the mouse liver and islet was assessed according to our recently published method, mirHub (27), using the “non-network” mode and requiring a predicted target site to be conserved among at least three mammalian species including mouse.
Results
Hepatic miR-29 Is Upregulated in Animal Models of Insulin Resistance and Is Corrected by Treatment With the Insulin-Sensitizing Drug Pioglitazone
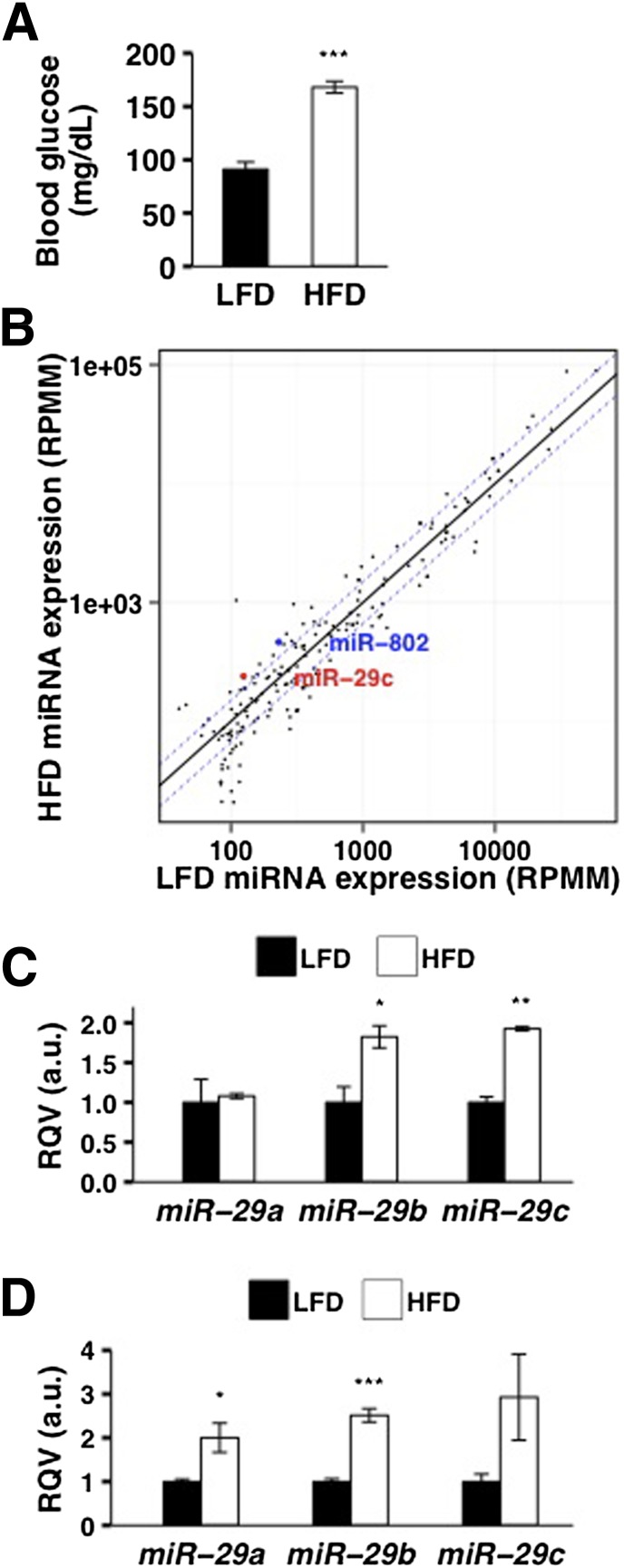
To determine if hepatic miR-29 levels are altered in the insulin-resistant state, we investigated two different animal models of metabolic dysfunction. First, we studied female C57BL/6J mice placed on a 16-week HFD (45% kcal from fat), which resulted in significantly elevated (∼1.8-fold, P < 0.001) fasting blood glucose levels relative to age-, sex-, and strain-matched mice on LFD (10% kcal from fat) (Fig. 1A). We performed deep sequencing of liver small RNAs and found that miR-29b (∼1.8-fold, P < 0.05) and miR-29c (∼1.9-fold, P < 0.001) were significantly elevated in HFD-fed mice (Fig. 1B and C, Supplementary Table 1), matching the fold increase in miR-802 (Fig. 1B, Supplementary Table 1), which was previously identified as a critical mediator of obesity-induced glucose intolerance (20). To validate this finding, we performed real-time quantitative PCR (RT-qPCR) and confirmed that hepatic levels of miR-29 were significantly increased in HFD-fed mice (Fig. 1D).
Figure 1.
Hepatic miR-29 levels are upregulated in diet-induced insulin resistance in mice. A: Fasting blood glucose levels of C57BL/6J female mice on HFD for 16 weeks (n = 3) and matched LFD for 16 weeks (n = 3) are shown. B: miRNA expression levels (RPMM, reads per million mapped reads) based on deep sequencing analysis of small RNAs from the livers of HFD-fed (n = 2) and LFD-fed (n = 2) C57BL/6J female mice are shown. Each circle represents a miRNA that is expressed at RPMM >100 in at least one murine liver sample. Dashed blue lines represent 1.5-fold difference in expression between HFD-fed and LFD-fed mice. C: Relative levels (based on sequencing) of miR-29a, miR-29b, and miR-29c in the livers of HFD-fed (n = 2) and LFD-fed (n = 2) C57BL/6J female mice are shown. D: Relative levels (based on RT-qPCR) of miR-29a, miR-29b, and miR-29c in the livers of HFD-fed (n = 3) and LFD-fed (n = 3) C57BL/6J female mice are shown. P values were calculated according to the one-tailed unpaired Student t test. a.u., arbitrary unit. *P < 0.05; **P < 0.01; ***P < 0.001.
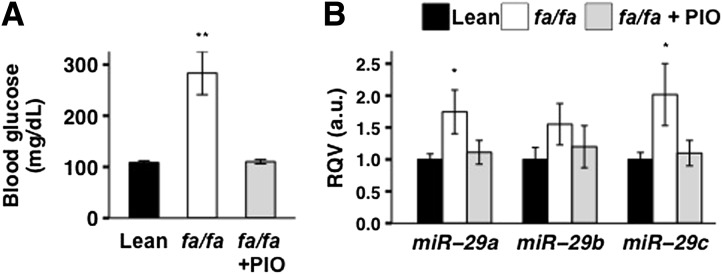
Next, we examined the ZDF fa/fa rat model, which closely mimics human adult-onset diabetes (32). We showed that, as expected, fasting blood glucose levels were significantly elevated (∼2.5-fold, P < 0.005) in 12-week-old male obese fa/fa rats compared with age- and sex-matched lean healthy littermates (Fig. 2A). We then demonstrated by RT-qPCR that hepatic miR-29a and miR-29c levels were significantly (P < 0.05) higher for the fa/fa rats compared with the lean littermate control subjects (Fig. 2B). Strikingly, we also observed that treatment with the insulin-sensitizing drug pioglitazone for 4 weeks, which markedly improved glycemia (Fig. 2A), reduced hepatic miR-29 expression to levels comparable to those of the lean control subjects (Fig. 2B).
Figure 2.
Hepatic miR-29 levels are elevated in a rat model of diabetes and corrected by treatment with the insulin-sensitizing drug pioglitazone (PIO). Fasting blood glucose levels (A) and relative hepatic levels (based on RT-qPCR) of miR-29a, miR-29b, and miR-29c (B) in 12-week-old healthy male rats (n = 8), ZDF fa/fa male littermates (n = 11), and pioglitazone-treated (4 weeks) ZDF fa/fa male rats (n = 6) are shown. P values were calculated according to the one-tailed unpaired Student t test. a.u., arbitrary unit. *P < 0.05; **P < 0.01.
Hepatic miR-29 Expression Is Controlled in Part by the Insulin-Regulated TF FOXA2
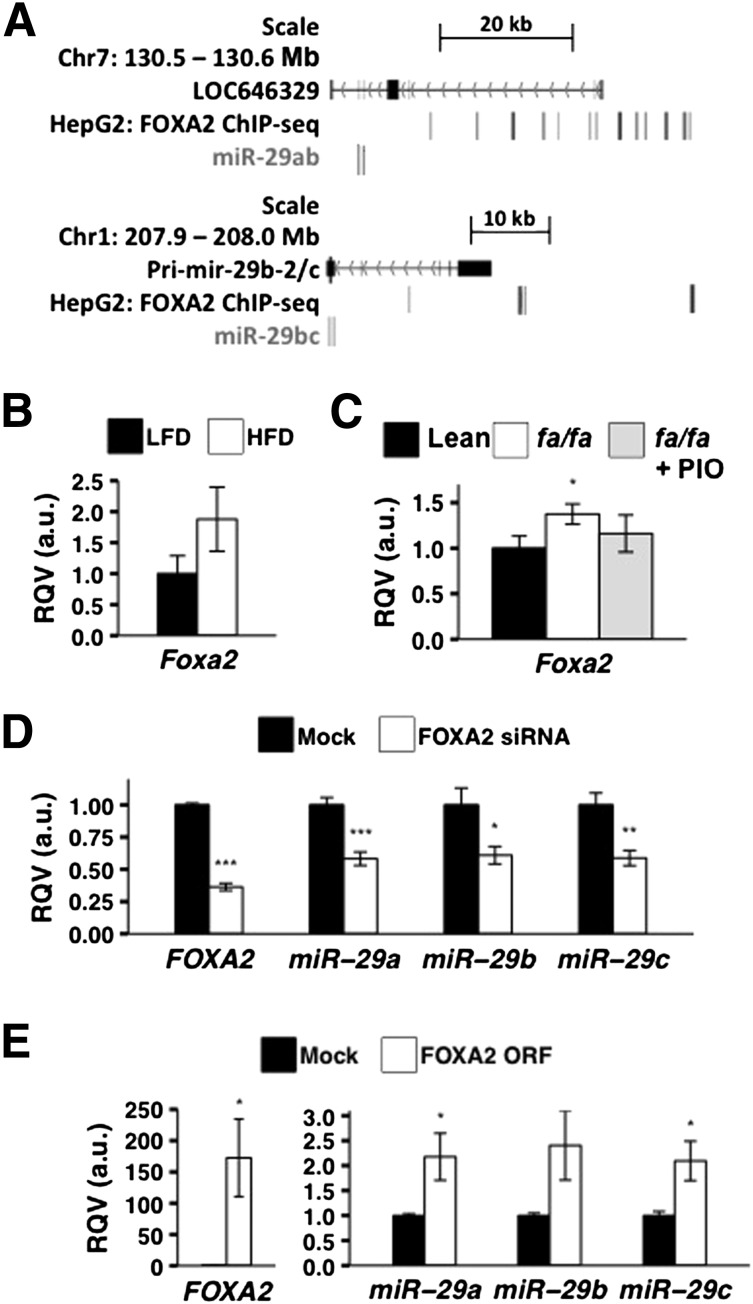
To investigate the molecular mechanism(s) that mediates the upregulation of miR-29 in insulin resistance, we sought to identify hepatic TFs regulated by insulin signaling that could be involved in the control of miR-29 expression. First, we identified the transcription start sites (TSS) of miR-29a/b-1 (chromosome 7) and miR-29b-2/c (chromosome 1) in human hepatoma cells (HepG2) by analyzing chromatin data from ENCODE, using our previously described bioinformatic pipeline (31). This strategy revealed that the most proximal active TSS for miR-29a/b-1 is ∼36.5 kb upstream of the mature miR-29a sequence; for miR-29b-2/c, it is ∼20 kb upstream of the mature miR-29c sequence. We scanned these regions for areas of open chromatin and TF occupancy in HepG2 cells, as determined by ENCODE, and found >10 binding sites for FOXA2 at the miR-29a/b-1 locus and 4 binding sites at the miR-29b-2/c locus (Fig. 3A). FOXA2 is negatively regulated by insulin (33,34) and opposes insulin action (35) by promoting hepatic lipid catabolism and fatty acid oxidation (36). To further support the finding in HepG2 cells, we mined a recently published TF ChIP-seq data set from adult mouse liver (30) and detected Foxa2 chromatin occupancy at the mouse miR-29 promoter regions (data not shown).
Figure 3.
FOXA2 regulates miR-29 expression. A: FOXA2 occupancy in HepG2 at the miR-29ab and miR-29b-2/c genomic loci are shown (based on ENCODE ChIP-seq data). LOC646329 represents the putative primary transcript of miR-29a and miR-29b-1 on chromosome 7 and Pri-miR-29b-2 represents the putative primary transcript of miR-29b-2 and miR-29c on chromosome 1. B: Relative levels of Foxa2 mRNA in the livers of C57BL/6J female mice on HFD for 16 weeks (n = 3) and matched LFD for 16 weeks (n = 3) are shown. C: Relative levels of Foxa2 mRNA in the livers of 16-week-old healthy male rats (n = 8), ZDF fa/fa male littermates (n = 11), and pioglitazone-treated (PIO) (4 weeks) ZDF fa/fa male rats (n = 6) are shown. D: Effects of FOXA2-siRNA treatment (100 nmol/L) in Huh7 cells on FOXA2 (mock, n = 14; siRNA, n = 13), miR-29a (mock, n = 12; siRNA, n = 12), and miR-29b and miR-29c (mock, n = 10; siRNA, n = 8) expression levels are shown. E: Effects of FOXA2 open reading frame (ORF) overexpression plasmid (1 μg) in Huh7 cells on FOXA2 (mock, n = 6; plasmid, n = 5), miR-29a (mock, n = 6; plasmid, n = 5), miR-29b (mock, n = 5; plasmid, n = 4), and miR-29c (mock, n = 6; plasmid, n = 5) are shown. All transfections were conducted in triplicate and results were validated by at least two independent experiments. P values were calculated according to the two-tailed unpaired Student t test. a.u., arbitrary unit. *P < 0.05; **P < 0.01; ***P < 0.001.
We next performed RT-qPCR and observed that hepatic Foxa2 mRNA levels were increased in both HFD-fed mice (Fig. 3B) and diabetic fa/fa rats (P < 0.05) (Fig. 3C). Moreover, as with miR-29 (Fig. 3D), hepatic Foxa2 expression in the fa/fa rats returned to that of the lean control subjects upon treatment with pioglitazone (Fig. 3C). To more directly evaluate the potential for FOXA2 to regulate hepatic miR-29 levels, we performed small interfering RNA (siRNA)–mediated knockdown of FOXA2 in Huh7 cells. After 48 h of siRNA treatment, FOXA2 mRNA was significantly reduced (P < 10−12) (Fig. 3D). Under these conditions, we observed an almost twofold downregulation of miR-29a (P < 0.001), miR-29b (P < 0.05), and miR-29c (P < 0.01) (Fig. 3D). We also transiently transfected Huh7 cells with a FOXA2 expression vector (1 μg), which led to an ∼172-fold upregulation in FOXA2 mRNA levels (P < 0.05) and a concomitant more than twofold increase in miR-29a (P < 0.05), miR-29b (P = 0.06), and miR-29c (P < 0.05). Collectively, these data suggest that the insulin-regulated FOXA2 is a transcriptional activator of miR-29.
miR-29 Fine-tunes FOXA2-Mediated Regulation of Key Hepatic Lipid Metabolism Genes
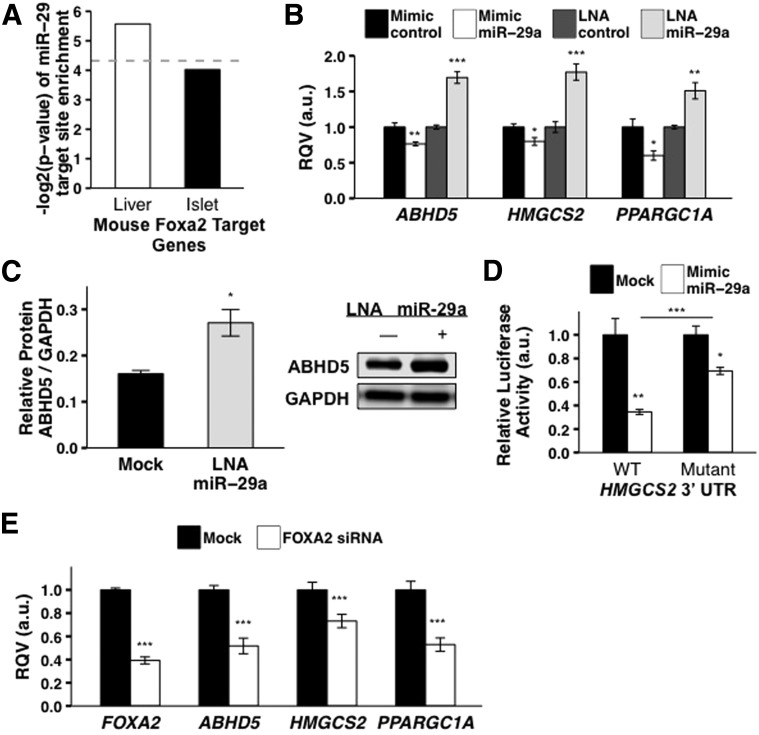
Recent studies of gene regulatory networks indicate that coordinated regulation by TFs and miRNAs confers robustness against environmental fluctuation (37). We assessed the extent to which miR-29 is predicted to regulate Foxa2 gene targets in the liver. First, we assembled a list of high-confidence hepatic target genes for Foxa2 from a published ChIP-seq study in mouse liver (Research Design and Methods). We then determined, using our previously published method mirHub (27), that the Foxa2 mouse liver target gene set was significantly enriched for predicted miR-29 target sites (Fig. 4A). Notably, we did not observe this enrichment among Foxa2 target genes in the mouse pancreatic islet, in which the Foxa2 regulatory network is rewired relative to the liver (30) (Fig. 4A). To evaluate further the predicted FOXA2–miR-29 feed-forward circuit, we experimentally tested three specific instances of the circuit with the genes HMGCS2, ABHD5, and PPARGC1A, which encode proteins that activate the enzymatic breakdown of fat in the liver (38,39). The mRNA levels of all three genes in Huh7 cells were significantly (P < 0.01) increased by the miR-29a locked nucleic acid (LNA) inhibitor and significantly (P < 0.05) reduced by the miR-29a mimic (Fig. 4B). Consistent with this observation, the protein levels of ABHD5 were also significantly (P < 0.05) upregulated by the miR-29a LNA inhibitor after 48 h (Fig. 4C). Also, to determine if miR-29 regulation of HMGCS2 is mediated through the 3′ UTR, we performed a reporter gene assay (Research Design and Methods). Overexpression of the miR-29a mimic (100 nmol/L) in HEK293T cells significantly reduced (P < 0.01, ∼65% loss) relative Firefly luciferase activity when the HMGCS2 3′ UTR was inserted downstream of the Firefly reporter gene (Fig. 4D). Moreover, targeted deletion (3 bp) of the predicted miR-29 target site in the HMGCS2 3′ UTR substantially mitigated the repressive effect of miR-29 on Firefly activity (Fig. 4D). Finally, siRNA-mediated knockdown of FOXA2 led to a significant (P < 0.01) decrease in the expression of HMGCS2, ABHD5, and PPARGC1A (Fig. 4E). The latter observation suggests that FOXA2 is the primary driver of the expression levels of its target genes, whereas miR-29 serves as a feed-forward negative modulator (Fig. 5).
Figure 4.
miR-29 fine-tunes FOXA2-mediated regulation of key lipid metabolism genes. A: Significant enrichment of predicted miR-29 target sites among FOXA2-bound genes in mouse liver but not in mouse islet is shown. Dashed line reflects P = 0.05. B: Effects of the miR-29a mimic (10 nmol/L) and the miR-29a LNA inhibitor (10 nmol/L) in Huh7 cells on the mRNA levels of FOXA2-bound genes HMGCS2 (mimic, n = 7; LNA, n = 7), ABHD5 (mimic, n = 8; LNA, n = 6), and PPARGC1A (mimic, n = 7; LNA, n = 7) are shown. C: Effect of the miR-29a inhibitor (LNA, 10 nmol/L) in Huh7 cells on protein levels of ABHD5 is shown (mock, n = 3; LNA, n = 3). D: Effects of the miR-29a mimic (100 nmol/L) in HEK293T cells on the relative activity of Firefly reporter constructs containing either wild-type or mutated HMGCS2 3′ UTR are shown. Firefly activity was normalized to Renilla activity. E: Effects of FOXA2-siRNA treatment (100 nmol/L) in Huh7 cells on mRNA levels of HMGCS2 (mock, n = 10; siRNA, n = 9), ABHD5 (mock, n = 10; siRNA, n = 10), and PPARGC1A (mock, n = 8; siRNA, n = 4) are shown. P values were calculated according to the two-tailed unpaired Student t test. a.u., arbitrary unit. *P < 0.05; **P < 0.01; ***P < 0.001.
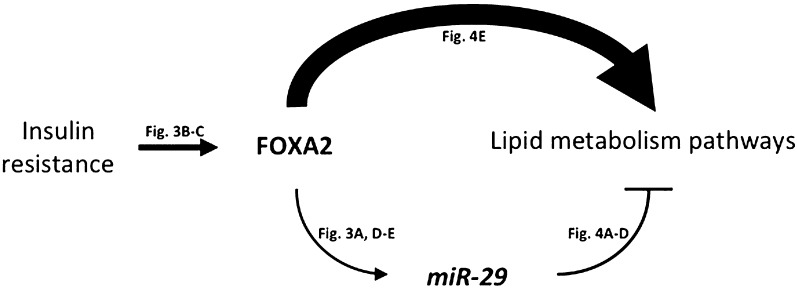
Figure 5.
Schematic of the FOXA2:miR-29 regulatory circuit in the liver. A possible model of FOXA2:miR-29 circuitry in the liver is shown. In the insulin-resistant state, FOXA2 activity is upregulated, which in turn elevates miR-29 levels. FOXA2 drives the expression of genes involved in lipid metabolism, and miR-29 acts as a feed-forward fine-tuner of many of the same genes.
Discussion
This study leveraged in vivo, in vitro, and in silico analyses to uncover a role for miR-29 as a potentially critical regulator of hepatic metabolic pathways. A prior study suggested that miR-29 is significantly elevated in the livers of the diabetic mouse model db/db (40); however, to our knowledge, this result had not been validated in other models. We showed in this study that liver miR-29 levels are elevated in two different animal models of metabolic dysfunction and, notably, are returned to normal levels upon treatment with an insulin sensitizer, pioglitazone, in the ZDF fa/fa rat. This finding signals the possibility that miRNAs could serve as tissue biomarkers of drug efficacy in type 2 diabetes.
Two recent miRNA profiling studies reported that type 2 diabetes might be associated with reduced levels of plasma miR-29 (41,42). We observe in this study that miR-29 is elevated in the liver of animals with insulin resistance and diabetes. The apparent inverse correlation between plasma and liver miR-29 levels in type 2 diabetes is intriguing. It is now widely appreciated that miRNAs are stably present in circulation and are transported by a variety of different types of extracellular vehicles (EVs), including exosomes and high-density lipoproteins (43–47). Several studies have shown that numerous cell types secrete miRNAs, which can then be loaded onto EVs and delivered to recipient cells with functional integrity (48–50). However, the mechanisms that regulate intercellular miRNA transfer remain poorly characterized and represent a nascent but promising topic of research. Progress in this area will be critical for understanding why liver miR-29 is elevated but plasma miR-29 is reduced in type 2 diabetes. miR-29 is highly expressed in numerous metabolic tissues, including the pancreatic islet (27,51), and the relative contribution of each of these tissues to circulating miR-29 remains to be determined and merits further investigation.
We also demonstrated in this study that hepatic miR-29 expression is likely controlled at least in part by the insulin-regulated TF, FOXA2, which contains >10 ChIP-seq–derived binding sites in human hepatoma cells at the miR-29ab genomic locus on chromosome 7 and four binding sites at the miR-29bc genomic locus on chromosome 1. The evaluation of the combinatorial effect of these binding sites on miR-29 transcription is not trivial; however, it certainly warrants further investigation to more definitively establish direct FOXA2-mediated regulation of miR-29. Moreover, future studies in vivo should establish the extent to which FOXA2 controls miR-29 during hepatic insulin resistance.
Finally, we showed that miR-29 serves as a dampener of FOXA2-mediated activation of key lipid metabolism genes. For example, FOXA2 transcriptionally activates HMGCS2, which in turn is directly repressed by miR-29. It has been postulated that such TF:miRNA regulatory circuits, termed incoherent feed-forward loops, are likely important for noise buffering of gene expression (37,52,53). Further detailed studies in vivo may help elucidate the physiological importance of the FOXA2:miR-29 regulatory circuit in lipid homeostasis.
Overall, this study strongly suggests that miR-29 merits further investigation as a candidate biomarker of metabolic status and drug efficacy, an etiological factor in type 2 diabetes, and a potentially important therapeutic target for a range of metabolic disorders.
Supplementary Material
Article Information
Acknowledgments. The authors would like to thank Michael Erdos of the National Institutes of Health (NIH), Samir Kelada and Jeanette Baran-Gale of UNC at Chapel Hill, and Jonathan Haldeman of Duke University for their helpful suggestions regarding the study and the manuscript; Stanley Lemon of UNC at Chapel Hill for generously sharing Huh7 cells; and Brad Hoffman of the University of British Columbia for sharing the chromatin occupancy sites for FOXA2 in mouse liver and islet based on a previously published study.
Funding. This work was supported in part by an R00 grant (DK-091318-02) from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)/NIH (awarded to P.S.); a pilot and feasibility grant (NIDDK/NIH P30-DK-056350) from the UNC Nutrition Obesity Research Center (awarded to P.S.); a UNC Genetics and Molecular Biology T32 training grant (GM-007092-39) from the National Institute of General Medical Sciences (NIGMS)/NIH and UNC IMSD Fellowship through the Education Program grant (R25-GM-055336-13) from the NIGMS/NIH (awarded to B.C.E.P.); an R01 grant (RHL-109650) from the National Heart, Lung, and Blood Institute(NHLBI)/NIH (awarded to S.B.B.); a K99 grant (DK-100539) from NIDDK/NIH (awarded to J.M.); a K22 grant (K22-HL-113039) from NHLBI/NIH (awarded to K.C.V.); and a grant from the German Federal Ministry of Education and Research awarded to the German Center for Diabetes Research (P.S.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. C.L.K. and B.C.E.P. researched data and edited the manuscript. E.E.F., C.B., J.M., S.R.L., S.D., V.T., S.T., and S.B.B. researched data. P.K.L. contributed to discussion. K.C.V. researched data and contributed to the discussion. P.S. researched data, contributed to discussion, and wrote the manuscript. P.S. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db13-1015/-/DC1.
C.L.K. and B.C.E.P. contributed equally to this work.
References
- 1.Kadowaki T. Insights into insulin resistance and type 2 diabetes from knockout mouse models. J Clin Invest 2000;106:459–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim W, Kyung Lee E. Post-transcriptional regulation in metabolic diseases. RNA Biol 2012;9:772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karolina DS, Tavintharan S, Armugam A, et al. Circulating miRNA profiles in patients with metabolic syndrome. J Clin Endocrinol Metab 2012;97:E2271–E2276 [DOI] [PubMed] [Google Scholar]
- 6.Couzin J. MicroRNAs make big impression in disease after disease. Science 2008;319:1782–1784 [DOI] [PubMed] [Google Scholar]
- 7.van Rooij E, Purcell AL, Levin AA. Developing microRNA therapeutics. Circ Res 2012;110:496–507 [DOI] [PubMed] [Google Scholar]
- 8.Jackson AL, Levin AA. Developing microRNA therapeutics: approaching the unique complexities. Nucleic Acid Ther 2012;22:213–225 [DOI] [PubMed]
- 9.Fernandez-Valverde SL, Taft RJ, Mattick JS. MicroRNAs in β-cell biology, insulin resistance, diabetes and its complications. Diabetes 2011;60:1825–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest 2013;123:11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams MD, Mitchell GM. MicroRNAs in insulin resistance and obesity. Exp Diabetes Res 2012;2012:484696 [DOI] [PMC free article] [PubMed]
- 12.Vickers KC, Shoucri BM, Levin MG, et al. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology 2013;57:533–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rayner KJ, Suárez Y, Dávalos A, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010;328:1570–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Najafi-Shoushtari SH, Kristo F, Li Y, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010;328:1566–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marquart TJ, Allen RM, Ory DS, Baldán A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A 2010;107:12228–12232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horie T, Ono K, Horiguchi M, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A 2010;107:17321–17326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rayner KJ, Sheedy FJ, Esau CC, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest 2011;121:2921–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rayner KJ, Esau CC, Hussain FN, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature 2011;478:404–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dávalos A, Goedeke L, Smibert P, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A 2011;108:9232–9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornfeld JW, Baitzel C, Könner AC, et al. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature 2013;494:111–115 [DOI] [PubMed] [Google Scholar]
- 21.Trajkovski M, Hausser J, Soutschek J, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011;474:649–653 [DOI] [PubMed] [Google Scholar]
- 22.Kole AJ, Swahari V, Hammond SM, Deshmukh M. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev 2011;25:125–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A 2008;105:13027–13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han YC, Park CY, Bhagat G, et al. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med 2010;207:475–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez I, Cazalla D, Almstead LL, Steitz JA, DiMaio D. miR-29 and miR-30 regulate B-Myb expression during cellular senescence. Proc Natl Acad Sci U S A 2011;108:522–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma F, Xu S, Liu X, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol 2011;12:861–869 [DOI] [PubMed] [Google Scholar]
- 27.Baran-Gale J, Fannin EE, Kurtz CL, Sethupathy P. Beta cell 5′-shifted isomiRs are candidate regulatory hubs in type 2 diabetes. PLoS One 2013;8:e73240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bagge A, Clausen TR, Larsen S, et al. MicroRNA-29a is up-regulated in beta-cells by glucose and decreases glucose-stimulated insulin secretion. Biochem Biophys Res Commun 2012;426:266–272 [DOI] [PubMed] [Google Scholar]
- 29.Pullen TJ, da Silva Xavier G, Kelsey G, Rutter GA. miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1). Mol Cell Biol 2011;31:3182–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman BG, Robertson G, Zavaglia B, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res 2010;20:1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sethupathy P. Illuminating microRNA Transcription from the Epigenome. Curr Genomics 2013;14:68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yokoi N, Hoshino M, Hidaka S, et al. A novel rat model of type 2 diabetes: the Zucker fatty diabetes mellitus ZFDM rat. J Diabetes Res 2013;2013:103731 [DOI] [PMC free article] [PubMed]
- 33.Wolfrum C, Besser D, Luca E, Stoffel M. Insulin regulates the activity of forkhead transcription factor Hnf-3beta/Foxa-2 by Akt-mediated phosphorylation and nuclear/cytosolic localization. Proc Natl Acad Sci U S A 2003;100:11624–11629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon JC, Puigserver P, Chen G, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 2001;413:131–138 [DOI] [PubMed] [Google Scholar]
- 35.Puigserver P, Rodgers JT. Foxa2, a novel transcriptional regulator of insulin sensitivity. Nat Med 2006;12:38–39 [DOI] [PubMed] [Google Scholar]
- 36.Wolfrum C, Stoffel M. Coactivation of Foxa2 through Pgc-1beta promotes liver fatty acid oxidation and triglyceride/VLDL secretion. Cell Metab 2006;3:99–110 [DOI] [PubMed] [Google Scholar]
- 37.Osella M, Bosia C, Corá D, Caselle M. The role of incoherent microRNA-mediated feedforward loops in noise buffering. PLOS Comput Biol 2011;7:e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lord CC, Betters JL, Ivanova PT, et al. CGI-58/ABHD5-derived signaling lipids regulate systemic inflammation and insulin action. Diabetes 2012;61:355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vilà-Brau A, De Sousa-Coelho AL, Mayordomo C, Haro D, Marrero PF. Human HMGCS2 regulates mitochondrial fatty acid oxidation and FGF21 expression in HepG2 cell line. J Biol Chem 2011;286:20423–20430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandey AK, Verma G, Vig S, Srivastava S, Srivastava AK, Datta M. miR-29a levels are elevated in the db/db mice liver and its overexpression leads to attenuation of insulin action on PEPCK gene expression in HepG2 cells. Mol Cell Endocrinol 2011;332:125–133 [DOI] [PubMed] [Google Scholar]
- 41.Zampetaki A, Kiechl S, Drozdov I, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010;107:810–817 [DOI] [PubMed] [Google Scholar]
- 42.Kong L, Zhu J, Han W, et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol 2011;48:61–69 [DOI] [PubMed] [Google Scholar]
- 43.Boon RA, Vickers KC. Intercellular transport of microRNAs. Arterioscler Thromb Vasc Biol 2013;33:186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol 2012;23:91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miRNAs: cell-cell communication function? Front Genet 2013;4:119 [DOI] [PMC free article] [PubMed]
- 46.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108:5003–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res 2012;110:483–495 [DOI] [PubMed] [Google Scholar]
- 48.Montecalvo A, Larregina AT, Shufesky WJ, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012;119:756–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Comm 2011;2:282 [DOI] [PMC free article] [PubMed]
- 50.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van de Bunt M, Gaulton KJ, Parts L, et al. The miRNA profile of human pancreatic islets and beta-cells and relationship to type 2 diabetes pathogenesis. PLoS One 2013;8:e55272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell 2007;26:753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siciliano V, Garzilli I, Fracassi C, Criscuolo S, Ventre S, di Bernardo D. MiRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat Comm 2013;4:2364 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.