Abstract
High-throughput technologies have led to advances in the recognition of disease pathways and their underlying mechanisms. To investigate the impact of micro-RNAs on the disease process in multiple sclerosis, a prototypic inflammatory neurological disorder, we examined cerebral white matter from patients with or without the disease by micro-RNA profiling, together with confirmatory reverse transcription–polymerase chain reaction analysis, immunoblotting and gas chromatography-mass spectrometry. These observations were verified using the in vivo multiple sclerosis model, experimental autoimmune encephalomyelitis. Brains of patients with or without multiple sclerosis demonstrated differential expression of multiple micro-RNAs, but expression of three neurosteroid synthesis enzyme-specific micro-RNAs (miR-338, miR-155 and miR-491) showed a bias towards induction in patients with multiple sclerosis (P < 0.05). Analysis of the neurosteroidogenic pathways targeted by micro-RNAs revealed suppression of enzyme transcript and protein levels in the white matter of patients with multiple sclerosis (P < 0.05). This was confirmed by firefly/Renilla luciferase micro-RNA target knockdown experiments (P < 0.05) and detection of specific micro-RNAs by in situ hybridization in the brains of patients with or without multiple sclerosis. Levels of important neurosteroids, including allopregnanolone, were suppressed in the white matter of patients with multiple sclerosis (P < 0.05). Induction of the murine micro-RNAs, miR-338 and miR-155, accompanied by diminished expression of neurosteroidogenic enzymes and allopregnanolone, was also observed in the brains of mice with experimental autoimmune encephalomyelitis (P < 0.05). Allopregnanolone treatment of the experimental autoimmune encephalomyelitis mouse model limited the associated neuropathology, including neuroinflammation, myelin and axonal injury and reduced neurobehavioral deficits (P < 0.05). These multi-platform studies point to impaired neurosteroidogenesis in both multiple sclerosis and experimental autoimmune encephalomyelitis. The findings also indicate that allopregnanolone and perhaps other neurosteroid-like compounds might represent potential biomarkers or therapies for multiple sclerosis.
Keywords: microRNA, multiple sclerosis, experimental autoimmune encephalomyelitis, neurosteroid
Introduction
Diseases of the CNS are frequently driven by complex alterations in the brain's gene expression programs, as evidenced by classical approaches as well as high-throughput systems of biological analyses (Chabas et al., 2001; Lock et al., 2002; Han et al., 2008; Noorbakhsh et al., 2009). Multiple sclerosis is a debilitating chronic inflammatory neurological disorder, which is characterized by infiltration of CNS white matter by myelin-reactive lymphocytes, causing myelin and axonal injury (Sospedra and Martin, 2005). The underlying pathogenic mechanisms of multiple sclerosis remain uncertain, but several candidate disease processes including aberrant innate or adaptive immune responses, combined with neurodegeneration have been posited (Trapp and Nave, 2008). Similar to other neurological diseases, transcriptomic, proteomic, lipidomic and metabolomic studies have revealed complex alterations of a variety of molecular networks in multiple sclerosis-derived tissues (Lock et al., 2002; Kanter et al., 2006; Han et al., 2008).
Micro-RNAs are small RNA molecules that exert essential roles in the regulation of gene expression through translational silencing or messenger RNA degradation (Ambros, 2001). While micro-RNA-mediated regulation of gene expression is involved in controlling spatiotemporal patterns of gene expression required for normal development and functioning of eukaryotic organisms (Carrington and Ambros, 2003; Ambros, 2004), perturbed expression of micro-RNAs has been implicated in different diseases (Kloosterman and Plasterk, 2006; Asli et al., 2008). Indeed, previous studies have pointed to the potential involvement of micro-RNAs in the pathogenesis of neurodegenerative disorders including Alzheimer's and Parkinson's diseases as well as HIV-associated dementia (Hebert and De Strooper, 2009; Weinberg and Wood, 2009; Noorbakhsh et al., 2010).
Herein, using high-throughput microarray micro-RNA profiling coupled with complementary methodologies, we investigated potential molecular perturbations in brain white matter from patients with multiple sclerosis and other disease controls. The working hypothesis was that specific changes in micro-RNA profiles in the white matter of patients with multiple sclerosis point to disordered homeostatic mechanisms in the CNS, leading to new perspectives on multiple sclerosis pathogenesis. We defined and verified the impact of specific micro-RNA alterations on multiple sclerosis pathogenesis, focusing on alterations in neurosteroidogenic machinery in the CNS. In particular, our multi-platform studies demonstrated the expression of the neuroprotective steroid allopregnanolone and related molecules in the white matter of patients with multiple sclerosis to be reduced. Moreover, treatment with allopregnanolone was found to be neuroprotective in different model systems of multiple sclerosis.
Materials and methods
Human brain tissue samples
Brain tissues were collected at autopsy with consent from age- and sex-matched groups of patients with or without multiple sclerosis for micro-RNA profiling, reverse transcription–polymerase chain reaction (PCR) analyses and neurosteroid measurements. Control subjects included 10 individuals (five males; mean age: 59 ± 6.1 years) who were diagnosed with cancer/leukaemia (n = 3), stroke, (n = 3), cardiac arrest (n = 2) and sepsis (n = 2). The group of patients with multiple sclerosis included 16 individuals (seven males, mean age 55 ± 7.8 years) who had been classified as relapsing-remitting (n = 3), primary progressive (n = 3) and secondary progressive (n = 10) and showed an estimated disability status score of 7–10 before death. Micro-RNA microarray analyses together with reverse transcription–PCR analyses of proinflammatory transcripts were performed in four specimens from patients without multiple sclerosis and seven specimens from patients with multiple sclerosis. Tissues from another set of patients with (n = 5) and without (n = 5) multiple sclerosis were used for independent confirmation of the expression levels of micro-RNAs by reverse transcription–PCR. Neurosteroid measurements were performed on all specimens from patients with (n = 16) and without multiple sclerosis (n = 10) including samples from the same patients used in the above molecular studies. All tissues for micro-RNA and neurosteroid analyses were collected from frontal lobe normal appearing white matter juxtaposed to lesions, based on proximal neuropathological analyses. Tissues were stored at −80°C at The Laboratory for Neurological Infection and Immunity Brain Bank at the University of Alberta, as previously reported (Noorbakhsh et al., 2005, Zhu et al., 2009).
Micro-RNA extraction and analysis
The Qiagen miRNeasy kit was used for extracting micro-RNA-containing total RNA from autopsy tissues, according to the manufacturer's instructions. Samples were analysed for RNA quality on a Bioanalyser2100 and RNA concentration was measured on a Nanodrop instrument. The samples were then labelled using the miRCURY™ Hy3™/Hy5™ power labelling kit and hybridized on the miRCURY™ LNA Array (v.11.0, Exiqon). Locked nucleic acid microRNA arrays make use of ‘locked nucleic acid’ technology for enhancing the efficiency and specificity of hybridization of short RNA sequences. All capture probes for the control spike-in oligonucleotides produced signals in the expected range, confirming successful labelling and hybridization. The quantified signals (background corrected) were normalized using the global LOWESS (LOcally WEighted Scatterplot Smoothing) regression algorithm. Micro-RNAs showing >1.5-fold changes between multiple sclerosis and control tissues were considered for further analysis. CIMminer (http://discover.nci.nih.gov/cimminer) was used to perform hierarchical clustering and generate heat maps for the differentially expressed micro-RNAs.
Real-time reverse transcription– polymerase chain reaction
Real-time reverse transcription–PCR using SYBR Green dye on a Bio-Rad i-Cycler (Richmond, CA) was used for confirmation of micro-RNA levels derived from microarray studies as well as gene expression analyses (Power et al., 2003). For micro-RNA analyses, the Qiagen miRNeasy kit was used for extracting micro-RNA-containing total RNA from autopsy tissues. Exiqon miRCURY locked nucleic acid primers sets were used for sequence-specific first strand complementary DNA synthesis, followed by reverse transcription–PCR. Geometric mean of threshold cycles of seven different small/ribosomal RNAs were used to normalize the micro-RNA expression data. For gene expression analyses, total RNA was extracted from cultured cells or brain tissue using TRIzol® (Invitrogen) followed by first strand complementary DNA synthesis using random hexamers (Roche). Oligonucleotide PCR primer sequences for human cytokines were published previously (Noorbakhsh et al., 2005; Ellestad et al., 2009). Oligonucleotide PCR primer sequences for human and mouse neurosteroidogenic enzymes as well as mouse cytokines are shown in Table 1. Oligonucleotide PCR primer sequences for mouse Akr1c/e isoforms real-time reverse transcription–PCR have previously been published (Vergnes et al., 2003). Gene expression data were normalized against glyceraldehyde-3-phosphate dehydrogenase messenger RNA levels.
Table 1.
Real-time reverse transcription–PCR primer sequences
| Gene name | Species | Primer | Sequence |
|---|---|---|---|
| 3βHSD1 | Human | Sense | GTA GCC GGG CCC AAC TCC T |
| Antisense | AAT CGG CTT CCT TCC CCA TAG A | ||
| 3βHSD2 | Human | Sense | GCC GGG CCC AAC TCC TAC AA |
| Antisense | CCG CCA GCA CAG CCT TCT CA | ||
| 5αReductase1 | Human | Sense | AAA TAC GCT GAA ATG GAG GTT GAA |
| Antisense | AGG GGG CTA TAG AAG GGA TTG TG | ||
| 5αReductase2 | Human | Sense | CCC CGG AAA ACT GGT ATG GCT ATT |
| Antisense | AGT TGC TTG GGG CTT CTG CTG TA | ||
| AKR1C1 | Human | Sense | GTA AAG CTT TAG AGG CCA C |
| Antisense | CAC CCA TGC TTC TTC TCG G | ||
| AKR1C2 | Human | Sense | GTA AAG CTC TAG AGG CCG T |
| Antisense | CAC CCA TGG TTC TTC TCG A | ||
| GAPDH | Human | Sense | AGC CTT CTC CAT GGT GGT GAA GAC |
| Antisense | CGG AGT CAA CGG ATT TGG TCG | ||
| MIP-1α | Mouse | Sense | CAC CAC TGC CCT TGC TGT TC |
| Antisense | GAT CTG CCG GTT TCT CTT AGT CA | ||
| IL-12p40 | Mouse | Sense | GGA AGC ACG GCA GCA GAA TA |
| Antisense | AAC TTG AGG GAG AAG TAG GAA TGG | ||
| F4/80 | Mouse | Sense | GCT GTG AGA TTG TGG AAG CA |
| Antisense | AGT TTG CCA TCC GGT TAC AG | ||
| CD3ε | Mouse | Sense | TCT CGG AAG TCG AGG ACA GT |
| Antisense | TTG AGG CTG GTG TGT AGC AG | ||
| 3βHSD1 | Mouse | Sense | TGC AGG GCC CAA CTC GTA |
| Antisense | TGC CCA GGC CAC ATT TTC | ||
| 3βHSD2 | Mouse | Sense | AGG GCA TCT CTG TTG TCA TCC AT |
| Antisense | TGC CTT CTC AGC CAT CTT TTT G | ||
| 5αReductase1 | Mouse | Sense | CAG GAA GGG CAA TGG GAG GGT GTT |
| Antisense | TGT CTG GGG GTC AAA GGG GTC TGC | ||
| 5αReductase2 | Mouse | Sense | CAT GCG GTT TAG CGT CGG TGT CT |
| Antisense | CCA AAG CGT AGC CCA TCC ATT CAA | ||
| AKR1C6 | Mouse | Sense | GCT GAT TGC CCT TCG CTA CC |
| Antisense | ACA TCT GGC ACA AGG CAC AAA AT | ||
| AKR1C14 | Mouse | Sense | CAC ATT GGG AAG TTC ACG AGA CA |
| Antisense | AAG CCA ACT GGA ATT CAA AAA CCT | ||
| GAPDH | Mouse | Sense | TTC ACC ACC ATG GAG AAG GC |
| Antisense | GGC ATG GAC TGT GGT CAT GA |
Experimental autoimmune encephalitis induction and treatments
C57BL/6 mice (10–12 weeks old) were used for experimental autoimmune encephalitis (EAE) induction with myelin oligodendrocyte glycoprotein (MOG)35–55 peptide as previously described (Ellestad et al., 2009). For allopregnanolone treatment, animals received daily intraperitoneal injections of allopregnanolone (10 mg/kg) after the onset of clinical symptoms until the end of the experiment. Control animals received daily injections of 30% β-cyclodexterin vehicle. Animals were assessed daily for disease severity for up to 30 days following immunization using a 0–5 point scoring scale (Tsutsui et al., 2004). For neuropathological analyses, CNS tissues were removed from a separate group of allopregnanolone/vehicle-treated animals at the peak of the disease (Days 16–17 post immunization). Measurement of the neurosteroid levels by gas chromatography–mass spectrometry as well as the measurement of neurosteroidogenic pathway enzyme transcripts and 3α-hydroxysteroid dehydrogenase protein levels were also performed on hindbrain tissues isolated from the animals at the peak of the disease. All experiments were approved by the University of Alberta Health Sciences Animal Care and Use Committee.
Immunofluorescence and histological analyses
Antibodies against Iba-1 (1:500; Wako), CD3 (1:100; Santa Cruz Biotechnology Inc.) and myelin basic protein (1:1,000; Sternberger Monoclonals), followed by appropriate secondary antibodies were used for immunofluorescent detection of macrophage/microglial, T lymphocyte and myelin immunoreactivity, as previously described (Noorbakhsh et al., 2006). For quantification, Iba-1 or CD3-immunopositive cells were counted using an upright fluorescent microscope (Axioskop2; Carl Zeiss MicroImaging Inc.) in spinal cord white matter. Quantitative analysis of myelin basic protein reactivity was performed using Scion Image software (Scion Corporation). Spinal cords were also stained with Bielschowsky's silver impregnation method. Each animal's whole ventral white matter area was scanned and photographed using light microscopy and the Spot imaging system (Diagnostic Instruments). The number of silver-positive axons was quantified per square millimeter using Scion Image software (Scion Corporation).
Cell cultures and treatments
Primary oligodendrocyte cultures were established from adult Sprague-Dawley rat brains and plated in polyornithine-coated chamber slides (Nunc) as described previously (Oh et al., 1999). Cells were pretreated with 10 nM allopregnanolone for 8 h before exposure to 100 ng/ml recombinant tumor necrosis factor-α (Roche Diagnostics) for 48 h. Cells were then fixed and immunostained with anti-2′,3′-cyclic-nucleotide 3′-phosphodiesterase (Millipore) antibody for quantitative immunodetection, as described previously (Noorbakhsh et al., 2005). Monocyte-derived macrophages were prepared from the blood of healthy donors as described before (Tsutsui et al., 2004). Differentiated macrophages were pretreated with different concentrations of allopregnanolone, followed by phorbol 12-myristate 13-acetate (50 ng/ml) treatment for 12 h. Treated macrophages were lysed in TRIzol® followed by RNA extraction. Allopregnanolone was purchased from Steraloids. For in vivo experiments, allopregnanolone was dissolved in 30% β-cyclodexterin (Sigma) at 1.25 and 10 mg/ml, respectively. For in vitro treatments, allopregnanolone was dissolved at 100 μM in dimethyl sulphoxide.
Micro-RNA transfection and target verification analyses
In order to verify the ability of the candidate micro-RNAs to knock down the expression of specific target genes, we used a commercial firefly luciferase-3′-untranslated region target clone assay system. Briefly, vectors encoding the firefly luciferase open reading frame fused with the 3′-untranslated region of AKR1C1 or AKR1C2 (Genecopoeia) were co-transfected into HEK293T cells along with vectors expressing immature micro-RNA sequences (Genecopoeia). A vector expressing a scrambled micro-RNA sequence was used as a control. Luciferase-3′-untranslated region target plasmids also encoded Renilla luciferase under the control of a second promoter as an internal control. HEK293T cells seeded in 96-well plates were transfected with a mixture of 25 ng/well of luciferase-3′-untranslated region target plasmid together with 50 ng/well of micro-RNA expression plasmid using Lipofectamine™ LTX (Invitrogen). Following 72 h of incubation at 37°C and 5% CO2, both firefly and Renilla luciferase activity were measured using the Dual-Glo dual luciferase assay system (Promega) according to the manufacturer's recommended protocols. Assay plates were read using a Bio-Tek Synergy HT plate reader (Bio-Tek instruments). To analyse the effect of micro-RNA overexpression on endogenous levels of AKR1C1/C2, human Huh7 hepatocyte cells seeded in 12-well plates were transfected with vectors expressing immature micro-RNA sequences (Genecopoeia) or a control scrambled micro-RNA, using Lipofectamine™ LTX (Invitrogen). Cells were lysed with TRIzol® after 48 h and used for RNA extraction and real-time reverse transcription–PCR analysis of gene expression.
Micro-RNA in situ hybridization
Micro-RNA in situ hybridization was performed on cryosections of white matter blocks from patients with multiple sclerosis and controls. Double digoxigenin-labelled locked nucleic acid probes (Exiqon) were used for hsa-miR-338-3p, as well as for U6snRNA and miR-159 as positive controls. A probe with a scrambled sequence was used as negative control. The hybridization process was performed according to the instructions provided by the probe manufacturer. In brief, after proteinase K treatment and paraformaldehyde fixation, sections were rinsed in phosphate buffered saline and pre-hybridized for 2 h in hybridization buffer (50% formamide, 5× saline sodium citrate buffer, 0.1% Tween, 50 μg/ml heparin, 500 μg/ml yeast RNA, pH 6). Slides were then hybridized with 20 nM of individual probes diluted in hybridization buffer at 42°C overnight. After hybridization, slides were rinsed in 2× saline sodium citrate buffer, followed by stringency washes (50% formamide, 2× saline sodium citrate buffer, three times for 30 min each) at hybridization temperature. Slides were then washed in phosphate buffered saline with Tween 20 several times, blocked in blocking buffer (2% sheep serum, 2 mg/ml bovine serum albumin in phosphate buffered saline with Tween 20) and immunostained with anti-digoxigenin alkaline phosphatase Fab fragments (1:2000, Roche Diagnostics) overnight. Nitroblue tetrazolium chloride/5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt solution was used for detecting the immunoreactivity (Roche Diagnostics).
Gas chromatography–mass spectrometry
Gas chromatography–mass spectrometry analysis was used to measure levels of several neurosteroids in the CNS tissue, as described previously (Ahboucha et al., 2008). Protein was precipitated by the addition of methanol followed by centrifugation. The supernatant was retained and the steroids isolated using solid-phase extraction with Oasis HLB cartridges (Waters). The samples were then derivatized with heptafluorobutyrylimidazole (Sigma-Aldrich) and the resultant derivatives analysed by gas chromatography combined with negative ion chemical ionization mass spectrometry (an Agilent 6890 gas chromatographer coupled to a 5973N mass selective detector). Standard curves were prepared for each steroid. Deuterated (d4) pregnenolone (provided by Dr R. H. Purdy) was carried throughout the entire procedure as internal standard. Cholesterol was analysed by gas chromatography combined with electron-impact mass spectrometry, with 5-α-cholestrol as an internal standard.
Immunoblotting
Immunoblot detection of 3α-hydroxysteroid dehydrogenase was performed using polyclonal rabbit anti-3α-hydroxysteroid dehydrogenase (1:1000 dilution), a generous gift from Synthia Mellon, UCSF. Following overnight incubation with primary antibody, blots were incubated with goat anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Chemicon International, 1:5000) for 1 h at room temperature. Peroxidase activity on the membrane was detected by chemiluminescence (Roche Diagnostics).
Cell proliferation and cytokine assays
T-cell proliferation and cytokine assays were performed using splenocytes derived from the spleens of MOG35–55-immunized mice, as described previously (Ellestad et al., 2009). Fluorochrome-conjugated antibodies against CD3 (145-2C11; BD Biosciences), CD4 (RM4-5), IFN-γ (XMG1.2), IL-17A (eBio17B7) and FoxP3 (FJK-16s) were obtained from eBioscience and used at the manufacturer's recommended concentrations. For intracellular staining, the FoxP3 staining buffer set (eBioscience) was used according to the manufacturer-supplied protocols. Stained cells were analysed on a BD FACSCanto flow cytometer using FACSDiva software for acquisition. Multicolor compensation was performed using BD Biosciences Compbeads. Cytometry data were analysed using FlowJo version 7.2.2 (Tree Star software).
Statistical analyses
Statistical analyses were performed using GraphPad InStat version 3.0 (GraphPad Software) for both parametric and non-parametric comparisons. Analysis was performed using the Mann–Whitney U test for neurobehavioral studies. ANOVA followed by Tukey–Kramer Multiple Comparisons test was used for statistical comparisons between more than two groups, as indicated in individual figure legends. Comparisons between two groups were performed using Mann–Whitney U test or unpaired Student's t-test, as indicated in figure legends. P < 0.05 were considered significant.
Results
Micro-RNA profiling shows differential expression of micro-RNAs in the brains of patients with multiple sclerosis
To investigate the extent of micro-RNA perturbations in multiple sclerosis as a means to discovering important pathways dysregulated in this disease, we investigated micro-RNA expression in the white matter of brains from patients with and without multiple sclerosis using micro-RNA microarrays, followed by reverse transcription–PCR confirmation and target verification. Brain-derived white matter from seven patients with multiple sclerosis and four patients without multiple sclerosis was examined initially and categorized based on the extent of inflammatory gene expression, including the transcript levels of CD3ε, CD8β, CD11c, MHC–II and FoxP3, which represent different populations of inflammatory cells, which infiltrate the CNS during disease, as well as pro- and anti-inflammatory cytokines, i.e. interleukin-1β, interleukin-6, interleukin-10, interleukin-12p35, interleukin-23p19 and transforming growth factor-β. The transcriptional analyses revealed a spectrum of inflammatory changes among different white matter tissues from patients with multiple sclerosis, distinguishing a group with lower levels of inflammatory transcripts (four patients) and a group with higher levels of inflammatory markers (three patients), as evidenced by hierarchical clustering of relative inflammatory transcript abundance (Supplementary Fig. 1). Given that whole tissue micro-RNA analyses would reflect the micro-RNA content of both parenchymal and infiltrating cells, in the present study we focused on the lower levels of inflammatory transcripts exhibited by patients with multiple sclerosis, which was indicative of less intense leucocyte infiltration. Indeed, this category was more likely to represent neural cell-specific micro-RNA alterations relevant to multiple sclerosis disease progression, including aspects of ongoing cellular damage or repair processes important in multiple sclerosis pathogenesis and disease progression.
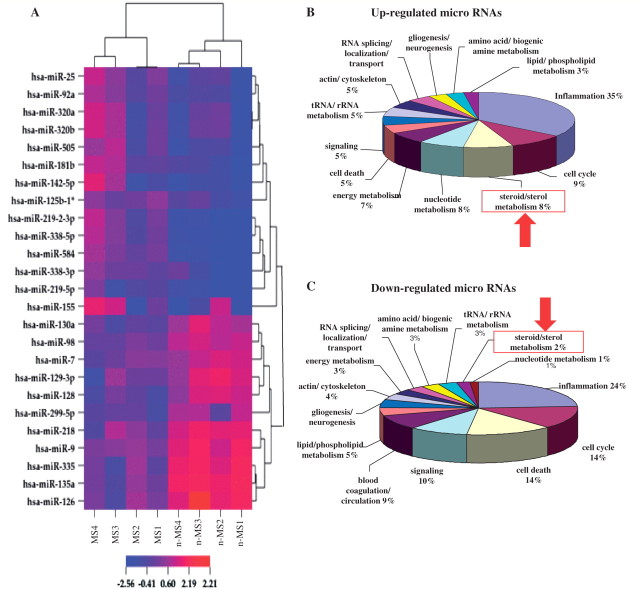
Microarray analyses of the micro-RNA pool derived from each brain specimen showed differential expression of multiple micro-RNAs distinguishing white matter from patients with and without multiple sclerosis (Fig. 1A, heatmap). Micro-RNAs showing >1.5-fold changes between tissues from patients with or without multiple sclerosis were considered for further analysis. Real-time reverse transcription–PCR analyses using micro-RNA-specific primers confirmed the microarray data (Supplementary Figs 2 and 3) although age- and sex-associated changes were not observed herein. Dysregulation of the majority of the micro-RNAs whose altered expression were observed in microarray analysis, but also confirmed by reverse transcription–PCR in the same set of samples was also independently confirmed in another set of brain white matter specimens from five patients with and five patients without multiple sclerosis, which were interrogated in terms of both neuroinflammation and micro-RNA levels by reverse transcription–PCR (Table 2). Thus, micro-RNA profiling by both array and reverse transcription–PCR in different multiple sclerosis and non-multiple sclerosis white matter sample sets revealed marked differences between clinical groups.
Figure 1.
Microarray micro-RNA profiling reveals differential expression of multiple micro-RNAs in brains of patients with multiple sclerosis. Microarray analysis of micro-RNAs extracted from tissues of patients with and without multiple sclerosis revealed differential expression of multiple micro-RNAs in multiple sclerosis brains. (A) The heatmap represents only those micro-RNAs whose expression was altered by at least 1.5-fold with both microarray and real-time reverse transcription–PCR analyses. Gene ontology term enrichment analysis of predicted targets of differentially expressed micro-RNAs showed targeting of different biological processes by multiple sclerosis-induced (B) or multiple sclerosis-suppressed (C) micro-RNAs.
Table 2.
miRNA fold change in multiple sclerosis versus non-multiple sclerosis white matters
| miRNA ID | MS/non-MS ratio | P-value |
|---|---|---|
| Downregulated miRNAs (microarray and PCR) | ||
| hsa-mir-7 | 0.243 | 0.009 |
| hsa-mir-299-5p | 0.349 | 0.018 |
| hsa-miR-135a | 0.371 | 0.017 |
| hsa-mir-218 | 0.396 | 0.006 |
| hsa-miR-129-3p | 0.518 | 0.012 |
| hsa-miR-9 | 0.533 | 0.002 |
| hsa-mir-128 | 0.534 | 0.019 |
| hsa-miR-130A | 0.549 | 0.023 |
| hsa-mir-126 | 0.603 | 0.039 |
| hsa-miR-335 | 0.616 | 0.037 |
| hsa-miR-98 | 0.663 | 0.030 |
| miRNA ID | Fold change (MS/control) | P-value |
|---|---|---|
| Upregulated miRNAs (microarray and PCR) | ||
| hsa-miR-25 | 1.677 | 0.026 |
| hsa-miR-505* | 1.934 | 0.000 |
| hsa-miR-320b | 1.986 | 0.003 |
| hsa-miR-320a | 1.995 | 0.003 |
| hsa-miR-338-3p | 2.119 | 0.036 |
| hsa-miR-181b | 2.182 | 0.013 |
| hsa-mir-125b1 | 2.218 | 0.018 |
| hsa-miR-92a | 2.292 | 0.003 |
| hsa-mir-155 | 2.451 | 0.024 |
| hsa-miR-584 | 2.456 | 0.013 |
| hsa-miR-219-2-3p | 2.749 | 0.008 |
| hsa-miR-338-5p | 2.949 | 0.006 |
| hsa-miR-219-5p | 2.989 | 0.028 |
| hsa-miR-142-5p | 3.461 | 0.024 |
As shown in Fig. 1A, micro-RNAs (miRNAs) showing >1.5-fold change at microarray analysis were further analysed with real-time reverse transcription–PCR. The upper and lower panels show miRNAs with altered expression confirmed by PCR analysis, showing also a minimum 1.5-fold change at the PCR level. MS = multiple sclerosis.
Bioinformatics reveal differentially expressed micro-RNAs targeting multiple pathways in multiple sclerosis
Focusing on the ‘low-inflammation’ multiple sclerosis group, bioinformatic analyses were performed to gain insight into the functional importance of differential micro-RNA expression in brains from patients with multiple sclerosis. The predicted messenger RNA targets for each of the up- or downregulated micro-RNAs were determined using the Sanger micro-RNA (www.mirbase.org) and European Molecular Biology Laboratory microcosm databases. All predicted messenger RNA targets were analysed for over-represented functional classes using the DAVID bioinformatics Functional Annotation Chart tool (http://david.abcc.ncifcrf.gov). Gene families containing a minimum of five transcripts with a minimum ‘fold enrichment’ of two were considered for further analyses. Gene ontology term analyses of targeted gene families revealed the majority of differentially expressed micro-RNAs targeted genes involved in immune responses and inflammation (Fig. 1B and C). Among gene families targeted by multiple sclerosis up-regulated micro-RNAs (Up-micro-RNAs), genes involved in inflammation, cell cycle regulation, steroid biosynthesis and nucleotide metabolism ranked the highest (Fig. 1B). Gene families targeted by multiple sclerosis downregulated micro-RNAs were composed of inflammation, cell cycle, cell death and signaling-related genes (Fig. 1C). As discussed above, part of the multiple sclerosis upregulated micro-RNAs might be derived from infiltrating inflammatory cells, which were minimal in non-multiple sclerosis tissues, leading to the over-representation of these genes when analysing the targets of upregulated micro-RNAs. However, several multiple sclerosis-downregulated micro-RNAs also targeted immune response/inflammation-related genes, likely reflecting an intrinsic component of inflammation independent of immune cell infiltration. Of note, in the present analyses, all inflammation-related transcripts (pro- or anti-inflammatory/immunomodulatory) were included in the ‘inflammation’ group. Emergence of cell cycle-related genes as one of the major micro-RNA-targeted groups might also reflect the ongoing inflammatory, remyelinating or reactive gliogenic processes, which involve proliferation of immune or neural cells, respectively. Most interestingly, the genes involved in steroid biosynthesis were highly targeted by upregulated micro-RNAs with minimal targeting by downregulated micro-RNAs, which pointed to a loss of potentially protective/restorative molecules (Fig. 1B and C). In fact, steroid biosynthesis genes were the chief group of genes, which showed a significant ‘targeting bias’ towards upregulated micro-RNAs (Supplementary Fig. 4). Hence, the present bioinformatic analyses indicated neurosteroidogenesis might be a previously unrecognized, but plausible, molecular network disrupted in the pathogenesis of multiple sclerosis.
Neurosteroid biosynthetic machinery is targeted by multiple sclerosis-dysregulated micro-RNAs
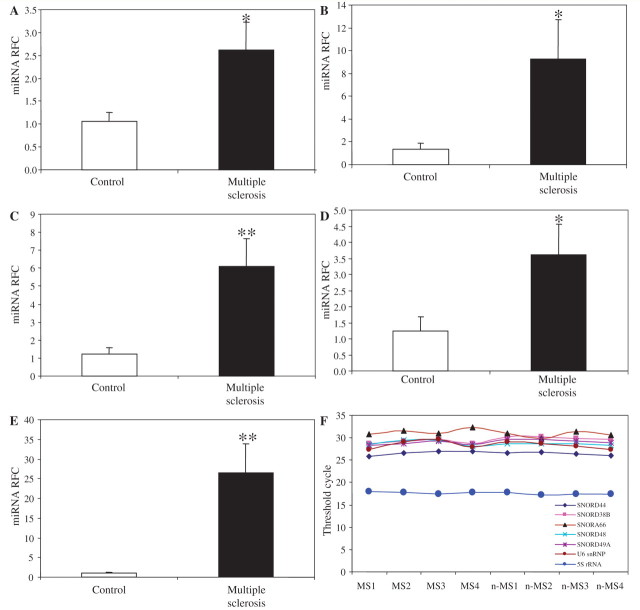
Steroids produced by prototypic steroidogenic organs, i.e. gonads and adrenals, were considered previously to be the chief source of steroid-like molecules found in brain and other neural tissues. However, pioneering studies in late 1980s indicated that neural cells and tissues were capable of producing steroids from cholesterol de novo (Hu et al.,1987; Goascogne et al., 1989). Indeed, it is now widely recognized the brain possesses the capability and complete enzymatic machinery for synthesizing steroids (Rupprecht and Holsboer, 1999; Mellon, 2007). Brain-derived steroids (termed ‘neurosteroids’) exert their effects independently of peripheral steroids and numerous studies have highlighted their role in neural cell function, as well as their involvement in several diseases (Le Melledo and Baker, 2002; Mellon and Griffin, 2002; Schumacher et al., 2003; Griffin et al., 2004; MacKenzie et al., 2007; Giatti et al., 2010). Because the gene ontology analyses of the predicted targets for multiple sclerosis-dysregulated micro-RNAs pointed to sterol/steroid biosynthesis as one of the principal targets of upregulated micro-RNAs, we explored the potential dysregulation in steroid biosynthetic machinery in the brains of patients with multiple sclerosis. Proteomic data derived from comprehensive mass spectrometric analysis of different types of multiple sclerosis lesions has been reported previously (Han et al., 2008). We interrogated this proteomic database to identify all proteins related to steroid biosynthesis with their associated spectral counts both in control tissues and multiple sclerosis lesions. Several steroid-related enzymes were detected in the database, which could be categorized into two functional groups; a group of alpha-ketoreductases and a group of 17-β-hydroxysteroid dehydrogenases. Most of the enzymes in both groups showed reduced spectral counts in multiple sclerosis lesions in the proteomic database (Supplementary Fig. 5). The largest and most extensively studied cluster of alpha-ketoreductases in the human genome consists of four genes (AKR1C1–4) located on chromosome 10 (10p15-p14), all encoding potent 3-α-hydroxysteroid dehydrogenase activity (Penning et al., 2000). Among the four isoforms, AKR1C2 followed by AKR1C1 showed the highest expression levels in the brain (Penning et al., 2000). AKR1C3 was also detected in the brain at lower levels, while AKR1C4 showed liver-specific expression (Penning et al., 2000). Given this similarity in terms of global changes observed with micro-RNA and proteomic approaches, in which steroid synthesis enzymes are targeted by upregulated micro-RNAs, while also showing reductions in the proteomic analyses, we then analysed the potential impact of micro-RNA changes on this group of enzymes more specifically. Focusing on AKR1C1 and -C2 major brain isoforms, we determined the expression levels of the micro-RNAs predicted to target these two isoforms. Examination of microarray data showed upregulation of micro-RNAs targeting both isoforms (miR-338-3p, miR-491-3p and miR-155), as well as AKR1C1-targeting miR-183 in brains of patients with multiple sclerosis, while only one AKR1C2-targeting micro-RNA (miR-342-3p) showed downregulation. To confirm the altered expression of these micro-RNAs in multiple sclerosis, we analysed their levels of expression in the independent set of autopsied white matter specimens (five patients with and five patients without multiple sclerosis) by real-time reverse transcription–PCR. The analysis of these new sets of tissues also showed similar changes for miR-155, miR-183, miR-338-3p, miR-338-5p and miR-491-3p (Fig. 2A–F), indicating these changes were common in and specific to white matter specimens from patients with multiple sclerosis. Of interest, when examining the expression of micro-RNAs which specifically target the AKR1C4 liver isoform, no induction was detected (data not shown), a finding which suggests a degree of specificity in the upregulation of micro-RNAs targeting brain-expressed isoforms.
Figure 2.
Independent confirmation of dysregulation of AKR1C-targeting micro-RNAs in a separate set of multiple sclerosis/non-multiple sclerosis brain tissues. Real-time reverse transcription–PCR analysis performed on an independent set of multiple sclerosis and non-multiple sclerosis samples showed significant induction of hsa-miR-155 (A), hsa-miR-183 (B), hsa-miR-338-3p (C), hsa-miR-338-5p (D), as well as hsa-miR-491-3p (E), confirming the results obtained from microarray analyses. All micro-RNA data were normalized against the geometric mean of seven different housekeeping small RNAs/snords, which showed consistent expression across all samples (F). Data are shown as mean ± SEM (multiple sclerosis, n = 5; non-multiple sclerosis n = 5; Student's t-test; *P < 0.05, **P < 0.01; miRNA = micro-RNA; MS = multiple sclerosis; nMS = non-multiple sclerosis controls; RFC = reduced folate carrier).
Neurosteroidogenic enzyme expression is regulated by particular micro-RNAs
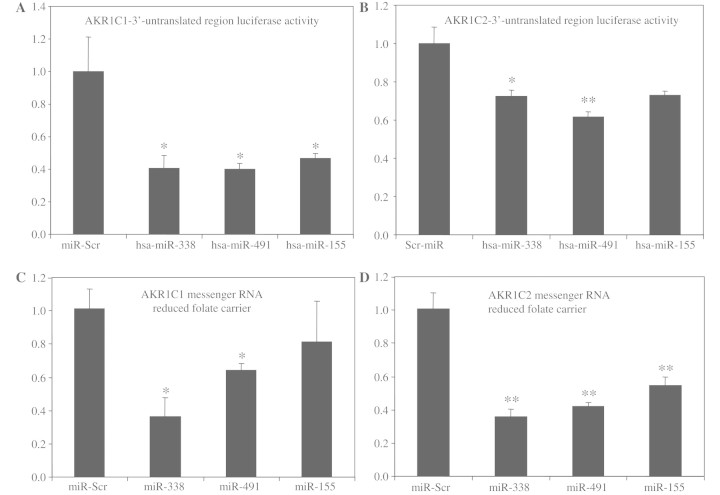
The interaction of the micro-RNAs predicted to target AKR1C1 and AKR1C1C2 isoforms with the 3′-untranslated regions of these enzymes were validated experimentally using a luciferase-3′-untranslated region assay. HEK293T cells were co-transfected with vectors encoding the firefly luciferase open reading frame fused to the 3′-untranslated region of either AKR1C1 or AKR1C2 and plasmids expressing the immature forms of hsa-miR-338, -491 or -155 or a plasmid expressing a scrambled micro-RNA sequence. Quantification of luciferase activity after co-transfection revealed significant suppression of luciferase activity for cells co-transfected with AKR1C1-3′-untranslated region and all three micro-RNA-expressing vectors compared with cells expressing the scrambled micro-RNA sequence (Fig. 3A). The same results were obtained for AKR1C2-3′-untranslated region with miR-338 and -491, while miR-155 showed non-significant suppression (Fig. 3B). The effects of micro-RNAs on gene expression have been shown to involve both messenger RNA degradation and translational inhibition, but recent findings indicate that the major effects of micro-RNAs may be due to lowered messenger RNA levels (Guo et al., 2010). To investigate whether altered expression of micro-RNAs might affect the endogenous expression levels of each enzyme isoform, cells expressing high levels of AKR1C1 or AKR1C2 (Huh7 hepatocyte cells) were transfected with vectors expressing different immature micro-RNA sequences. Consistent with target prediction analyses and luciferase assays, overexpression of miR-338 and miR-491 suppressed the expression levels of AKR1C1, while miR-155 showed a non-significant reduction, compared with cells transfected with a vector expressing a scrambled micro-RNA sequence (Fig. 3C). Moreover, transfection with all three micro-RNA clones suppressed endogenous AKR1C2 levels in Huh7 cells (Fig. 3D).
Figure 3.
Luciferase-3′-untranslated region and overexpression assays verify the interaction of select micro-RNAs with AKR1C1 and AKR1C2 transcripts. Co-transfection of HEK293T cells with vectors encoding firefly luciferase, ligated to the 3′-untranslated region of AKR1C1 (A) or AKR1C2 (B), together with vectors expressing the immature forms of hsa-miR-338, -491 or -155, individually showed significant suppression of luciferase activity, compared with cells transfected with a vector expressing a scrambled micro-RNA sequence. Transfection of Huh7 cells with vectors expressing immature micro-RNA sequences showed significant suppression of AKR1C1 transcripts after miR-338 and miR-491 transfection, compared with cells transfected with a vector expressing a scrambled micro-RNA sequence (C). Moreover, AKR1C2 transcript levels were significantly suppressed in cells overexpressing miR-338, -491, or -155 immature sequences (D), compared with scrambled-miR-expressing cells. Firefly luciferase levels were normalized against Renilla luciferase, which was expressed as an internal control in the target vectors (*P < 0.05, **P < 0.01, Tukey–Kramer Multiple Comparisons Test).
As mentioned, both brain parenchymal cells and infiltrating inflammatory cells might harbour micro-RNA species which exhibit upregulation in multiple sclerosis. Focusing on miR-338-3p, which exerted the greatest reduction in endogenous AKR1C1 and AKR1C2 levels, we performed in situ hybridization on brain tissues from patients with and without multiple sclerosis using micro-RNA-specific locked nucleic acid digoxigenin-labelled probes. Probes specific for the widely expressed miR-159 and U6snRNA were used as positive controls, whereas a scrambled micro-RNA sequence was used as the negative control. As expected, in situ hybridization analyses of brain sections with miR-159/U6snRNA probes revealed substantial reactivity on neural cells on all brain sections (Supplementary Fig. 6A and B). Of interest, in situ hybridization analyses of brains with a probe specific for the AKR1C-targeting miR-338-3p disclosed expression of this micro-RNA sequence on brain parenchymal cells rather than on perivascular infiltrating cells (Supplementary Fig. 6C and D). In situ hybridization using a scrambled control micro-RNA probe did not demonstrate any reactivity on any of the tissue sections (Supplementary Fig. 6E and F). Overall, these data indicated that select micro-RNA species, which were upregulated in neural cells of patients with multiple sclerosis targeted specific enzyme isoforms involved in the synthesis of neurosteroids, which might lead to pathogenic alterations in neurosteroidogenesis during the course of the disease.
Neurosteroid biosynthetic machinery is suppressed in multiple sclerosis
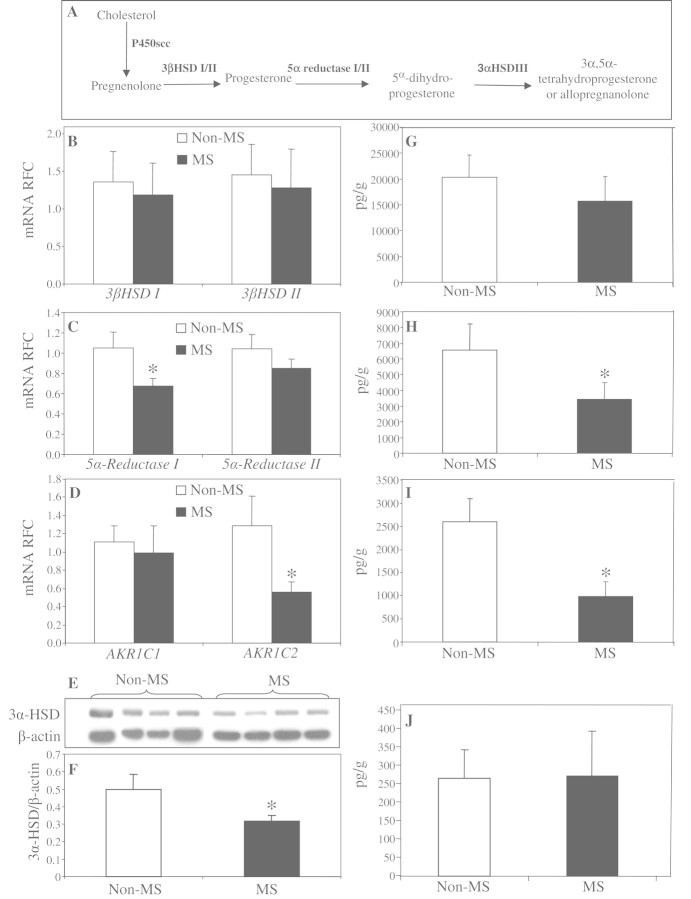
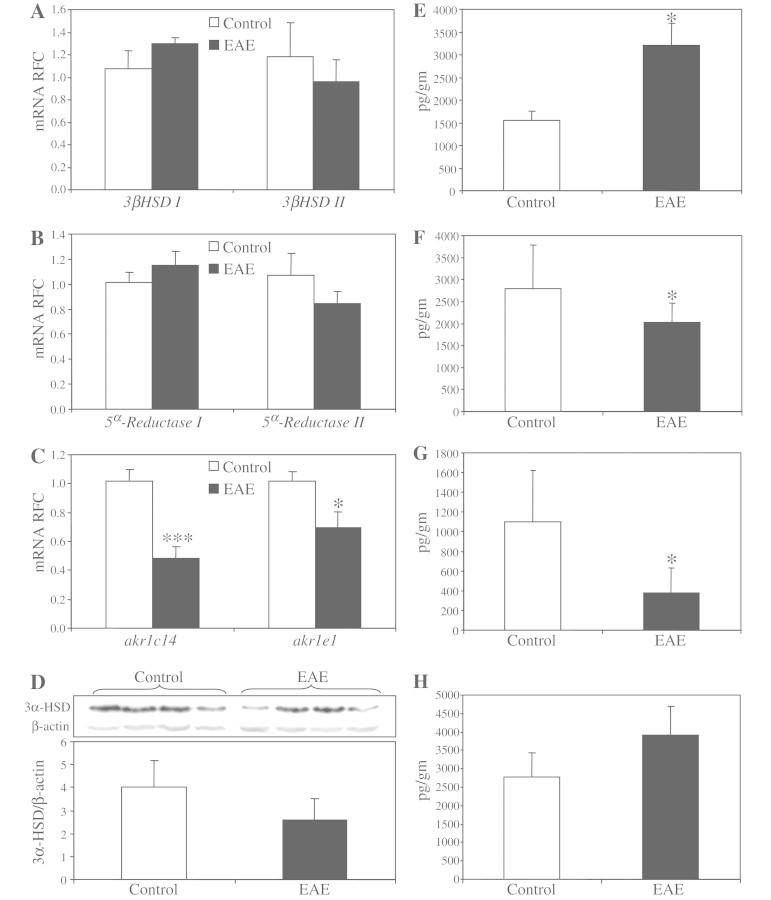
By encoding the majority of the brain's 3α-hydroxysteroid dehydrogenase activity, AKR1C1 and AKR1C2 (alternatively called 20α3α-hydroxysteroid dehydrogenase and 3α-hydroxysteroid dehydrogenase III, respectively), are essential for neurosteroid biosynthesis in the brain. Preceded by 3β-hydroxysteroid dehydrogenase and 5α-reductase in the metabolic pathway, AKR1C1 and AKR1C2 catalyze the final step in the reduction of progesterone to tetrahydroprogesterone and its major 3α,5α isoform, known as allopregnanolone (Fig. 4A), a neurosteroid with neuroprotective and neurotrophic properties (Charalampopoulos et al., 2008; Wang et al., 2008a). Using real-time reverse transcription–PCR, we analysed the transcript levels of the neurosteroidogenic enzymes involved in all three steps of allopregnanolone biosynthesis starting from pregnenolone. Transcriptional analyses did not show a significant difference in transcripts of 3β-hydroxysteroid dehydrogenase isoforms in the white matter of patients with or without multiple sclerosis (Fig. 4B), whereas there was a significant downregulation of 5α-reductase isoform 1 transcripts (Fig. 4C) in patients with multiple sclerosis. Transcriptional analysis of AKR1C isoforms showed a non-significant reduction for the less-abundant AKR1C1 isoform with substantially suppressed levels of AKR1C2 transcripts in the brains of patients with multiple sclerosis (Fig. 4D). Consistent with the messenger RNA data, western blot analyses using an antibody against 3α-hydroxysteroid dehydrogenase showed significantly lower immunoreactivity in multiple sclerosis (Fig. 4E and F). These findings recapitulated our previous data, which had showed the upregulation of AKR1C-targeting micro-RNAs in multiple sclerosis.
Figure 4.
Neurosteroid biosynthetic machinery and neurosteroid levels are dysregulated in multiple sclerosis. Schematic representation of the enzymatic reactions involved in allopregnanolone biosynthesis (A). Analysed by real-time reverse transcription–PCR, transcript levels of different isoforms of 3β-hydroxysteroid dehydrogenase (HSD) were not changed in the brains of patients with multiple sclerosis (B). However, transcript levels of 5α-reductase 1 (C), as well as AKR1C2 (D) were significantly lower in patients with multiple sclerosis compared with control subjects. Western blot analysis also showed reduced protein levels of 3α-HSD in multiple sclerosis (E and F). Gas chromatography–mass spectrometry analysis disclosed unaltered levels of pregnenolone in the brains of patients with multiple sclerosis (G), whereas the levels of dehydroepiandrosterone (H) and allopregnanolone (I) were significantly reduced in these subjects compared with controls. The levels of the minor 3α5β-tetrahydroprogesterone isoform were not changed in brain (J). Transcript levels are represented as relative fold change ± SEM. Neurosteroid levels are shown as mean ± SEM. Non-multiple sclerosis control subjects included 10 individuals (five males; mean age: 59 ± 6.1 years) and the multiple sclerosis group included 16 patients (seven males, mean age 55 ± 7.8 years; Student's t-test; *P < 0.05; mRNA = messenger RNA; MS = multiple sclerosis; nMS = non-multiple sclerosis controls; RFC = reduced folate carrier).
Given these enzymatic alterations, white matter neurosteroid levels were investigated by gas chromatography–mass spectrometry in specimens of white matter from patients with and without multiple sclerosis. The levels of the chief neurosteroid precursor, pregnenolone, did not differ between clinical groups (Fig. 4G). However, a significant reduction was observed in the levels of allopregnanolone, as well as another neurosteroid, dehydroepiandrosterone, in specimens of multiple sclerosis white matter compared with controls (Fig. 4H and I). These changes appeared to be specific to allopregnanolone, as the other isoforms of tetrahydroprogesterone, i.e. 3α,5β-tetrahydroprogesterone (Fig. 4J) and 3β,5α-tetrahydroprogesterone (data not shown), were not altered in multiple sclerosis. Overall, these findings pointed to micro-RNA-mediated alterations in neurosteroidogenic machinery which were associated with reduced levels of specific neurosteroids in the white matter of patients with multiple sclerosis.
Allopregnanolone exerts neuroprotective and immunomodulatory effects in vitro
Allopregnanolone is known to exert neuroprotective effects and regulate neural cell physiology and development through binding to γ-aminobutyric acid (GABA)A receptors expressed on neural cells (Lambert et al., 2003; Barbaccia, 2004). We investigated the potential neuroprotective effects of allopregnanolone using primary cultures of rat oligodendrocytes. In vitro treatment of oligodendrocytes with allopregnanolone prevented tumour necrosis factor-α-induced oligodendrocytotoxicity, suggesting a protective role for allopregnanolone in inflammation-mediated neural cell death (Fig. 5A–C). Moreover, our data indicated that treatment of oligodendrocytes with allopregnanolone enhanced the expression of oligodendrocyte maturation markers while simultaneously diminishing the expression of markers of premature oligodendrocytes, thereby promoting oligodendrocyte differentiation (F. Noorbakhsh and C. Power, unpublished results).
Figure 5.
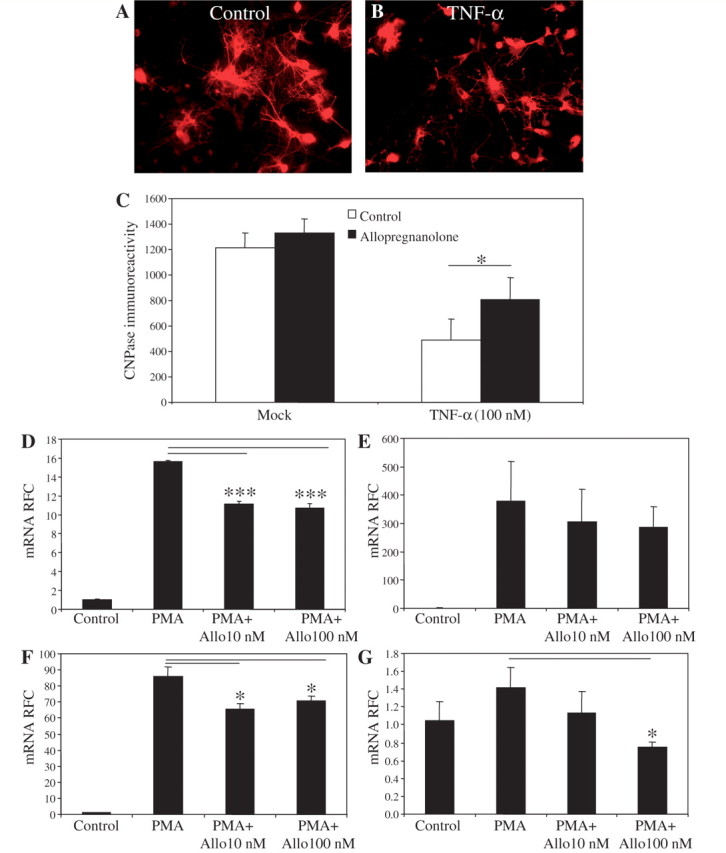
Allopregnanolone protects oligodendrocytes and regulates proinflammatory gene expression. Cultured primary oligodendrocytes (A) were treated with recombinant tumour necrosis factor-α (B), which reduced their viability, as quantified by 2', 3'-cyclic nucleotide 3'-phospho-diesterase immunoreactivity (C). However, pretreatment with allopregnanolone prevented tumour necrosis factor (TNF)-α-induced oligodendrocytotoxicity (C). Allopregnanolone treatment of human monocyte-derived macrophages diminished phorbol ester myristate-induced expression of proinflammatory mediators, TNF-α (D), IL-8, (E) IL-1β; (F) and IDO (G). Values are shown as mean ± SEM. (Original magnification ×200; Tukey–Kramer Multiple Comparisons Test; *P < 0.05, ***P < 0.001). Experiments were performed in five (oligodendrocyte cultures) and three replicates (monocyte-derived macrophage cultures) and repeated at least twice. mRNA = messenger RNA; PMA = phorbol ester myristate; RFC = reduced folate carrier.
In addition to the nervous system, recent findings have shown that the immune system also possesses the capacity to generate and respond to GABA (Bhat and Steinman, 2009). Monocytoid cells express functional GABAA receptors, and their immune activity can be modulated through treatment with known GABAA receptor agonists, including topiramate and gabaculine (Bhat and Steinman, 2009). Using human monocyte-derived macrophage cultures, we investigated the impact of allopregnanolone treatment on macrophage-derived cytokine production. In vitro treatment of primary monocyte-derived macrophage cultures with allopregnanolone diminished PMA-induced expression of the proinflammatory genes TNF-α (Fig. 5D), IL-8 (Fig. 5F) and IDO (Fig. 5G), all recognized to contribute to the pathogenesis of multiple sclerosis. While allopregnanolone-mediated neuroprotective effects point to the adverse impact of reduced brain allopregnanolone levels in neurological disease, allopregnanolone regulation of innate immune cell gene expression also highlights its ability to modulate neuroinflammation and ensuing neurodegeneration.
Neurosteroid biosynthetic machinery is reduced in experimental autoimmune encaphalitis
There is a relative conservation of the enzymes involved in neurosteroid synthesis machinery between humans and mice. However, with regard to murine homologues of human AKR1C enzymes, there is some divergence between humans and mice. The mouse genome contains a cluster of nine transcriptionally active alpha-keto-reductases on chromosome 13, i.e. akr1c6, c12-14, c18-21 and the more distantly related akr1e1 (Vergnes et al., 2003). Previous studies have analysed the tissue-specific expression of these enzymes, showing the expression of akr1c12-14, akr1c18-19 and akr1e1 in murine brain tissue (Vergnes et al., 2003). Although mouse akr1c/e isoforms show divergence from their human homologues, when analysed for their targeting with murine micro-RNA sequences, substantial overlap exists with the micro-RNAs targeting human isoforms, with mmu-miR-338 targeting several isoforms and mmu-miR-155 and mmu-miR-183 targeting individual isoforms of the enzyme in the mouse (data not shown). Considering these similarities, we extended the micro-RNA expression analysis to the MOG35–55-induced EAE model, using real-time reverse transcription–PCR for individual micro-RNAs. Reverse transcription–PCR analyses of mouse hindbrain tissue derived from EAE and control animals at the peak of the disease (Days 16–17 post immunization) showed upregulation of mmu-miR-155 and mmu-miR-338-3p in EAE hindbrains compared with controls (Supplementary Fig. 7A and C). Mmu-miR-338-5p was not altered in EAE, whereas analysis of mmu-miR-183 and -491 revealed a non-significant increase in EAE compared with controls (Supplementary Fig. 7B–F). Importantly, murine akr1c/e isoforms are also predicted to be targeted by multiple other micro-RNAs, which are either not conserved between human and mouse or do not show dysregulation in multiple sclerosis brains and thus were not included in these analyses. Nevertheless, the present studies showed a remarkable degree of similarity between disease-induced micro-RNA changes in human and mouse systems.
We then investigated the transcript levels of neurosteroidogenic enzymes involved in allopregnanolone production in the CNS of MOG-induced EAE animals. Similar to transcriptional analysis of multiple sclerosis tissues, the CNS levels of 3β-hydroxysteroid dehydrogenase isoforms were not different between EAE and control animals (Fig. 6A). Likewise, the levels of 5-α reductase isoforms did not differ between EAE and control mice in CNS tissues (Fig. 6B). As mentioned above, 3α-hydroxysteroid dehydrogenase activity is encoded by multiple enzyme isoforms in the mouse genome, among which six isoforms have been shown to be expressed in the CNS. Using isoform-specific primers, we first examined the relative expression levels of different enzyme isoforms in the mouse CNS. Average threshold cycles of real-time reverse transcription–PCR analyses showed that akr1e1, followed by akr1c14, were the two most abundant enzyme isoforms in the brain (Supplementary Fig. 8). Comparison of the transcript levels for each enzyme isoform showed significant suppression of the two most abundant isoforms, akr1e1 and akr1c14, in EAE hindbrains compared with controls (Fig. 6C). Western blot analysis of protein levels revealed a reduction of 3α-hydroxysteroid dehydrogenase immunoreactivity in protein lysate derived from EAE brains, although this did not reach statistical significance (Fig. 6D).
Figure 6.
Neurosteroid biosynthetic machinery and neurosteroid levels are dysregulated in experimental autoimmune encephalomyelitis. Similar to multiple sclerosis brains, real-time reverse transcription–PCR did not show any differences for the transcript levels of 3β-hydroxysteroid dehydrogenase in the hindbrains of experimental autoimmune encephalomyelitis mice (A). Likewise, the transcript levels of 5α-reductase 1 were not changed in multiple sclerosis brains, while 5α-reductase 2 showed a non-significant reduction in experimental autoimmune encephalomyelitis hindbrains (B). In contrast, the two most abundant isoforms of akr1c/e, i.e. akr1e1 and akr1c14, were significantly suppressed in experimental autoimmune encephalomyelitis hindbrains (C). Western blot analysis of EAE hindbrains displayed reduced 3α-hydroxysteroid dehydrogenase (3α-HSD) immunoreactivity in protein lysates derived from EAE tissues (D). Gas chromatography–mass spectrometry of neurosteroid levels showed enhanced levels of pregnenolone (E) in EAE-derived CNS tissues. However, the levels of dehydroepiandrosterone (F) and allopregnanolone (G) were significantly suppressed in EAE hindbrains, whereas the levels of the 3β,5α-tetrahydroprogesterone (THP) isoform were unchanged (H). Transcript levels are represented as relative fold change mean ± SEM. All tissues were extracted from the animals at the peak of the disease (Days 16–17 post immunization). Neurosteroid levels are shown as mean ± SEM (EAE, n = 9 and controls, n = 9; Student's t-test; *P < 0.05, ***P < 0.005; mRNA = messenger RNA; RFC = reduced folate carrier).
Given the present dysregulation of the enzymes involved in allopregnanolone biosynthesis, we then investigated whether the levels of neurosteroids were altered in tissues of the CNS in EAE mice. Unlike patients with multiple sclerosis, in which brain pregnenolone levels were unchanged, hindbrains from EAE animals showed enhanced levels of pregnenolone (Fig. 6E). Similar to the specimens of multiple sclerosis white matter, neurosteroid measurements disclosed decreased levels of dehydroepiandrosterone (Fig. 6F) as well as allopregnanolone (Fig. 6G) in EAE hindbrains. Again, the changes in allopregnanolone levels were specific to the 3α5α isoform, as the levels of 3α5β isoform of tetrahydroprogesterone were unchanged in the hindbrains of EAE animals (Fig. 6H). Thus, these findings indicated that the neurosteroidogenic machinery was disrupted in CNS during EAE, leading to diminished expression of the enzymes involved in allopregnanolone biosynthesis, coupled with reduced allopregnanolone levels in the CNS. Moreover, the observations of reduced neurosteroid levels were associated with enhanced levels of murine micro-RNAs that were predicted to target the transcripts of akr1c/e enzymes. Overall, these observations point to similar underlying mechanisms, whereby the same neuroprotective pathway is perturbed in both the human and mouse CNS during neuroinflammatory disease processes such as multiple sclerosis and EAE.
Allopregnanolone treatment diminishes neuroinflammation and disease severity in experimental autoimmune encephalitis
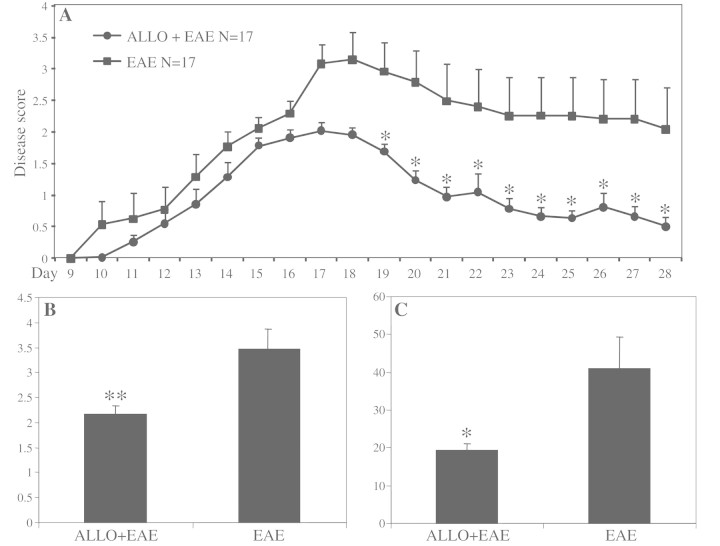
In view of the downregulation of allopregnanolone-synthesizing enzyme transcripts associated with lower levels of neurosteroids in multiple sclerosis and EAE CNS tissues, we investigated whether treatment of EAE mice with allopregnanolone affected disease severity. MOG35–55-induced EAE mice received daily intraperitoneal injections of allopregnanolone (10 mg/kg) or vehicle after signs of the EAE onset. Neuropathological analyses showed preserved spinal cord myelin in allopregnanolone-treated EAE animals compared with vehicle-treated animals with EAE (Fig. 7A), a finding that was confirmed by quantification of myelin basic protein-immunoreactive areas in several spinal cord sections from EAE mice (Fig. 7B). Allopregnanolone treatment also reduced immunoreactivity of the monocytoid cell marker, Iba-1 (Fig. 7A) as well as the lymphocytic marker, CD3ε (Fig. 7A), in EAE mouse-derived lumbar spinal cords. Indeed, quantification of the Iba-1 and CD3ε immunoreactive cells showed significant reductions in the numbers of immunopositive cells per surface unit of spinal cord (Fig. 7C and D) for allopregnanolone-treated EAE mice compared with vehicle-treated animals. Moreover, silver staining of lumbar-sacral spinal cords exhibited less axonal injury in EAE mice receiving allopregnanolone treatment (Fig. 7A), which was quantified by counting the number of silver-stained axons in the lumbar spinal cord (Fig. 7E). These data indicated that allopregnanolone treatment suppressed neuroinflammation together with preventing myelin and axonal injury in EAE.
Figure 7.
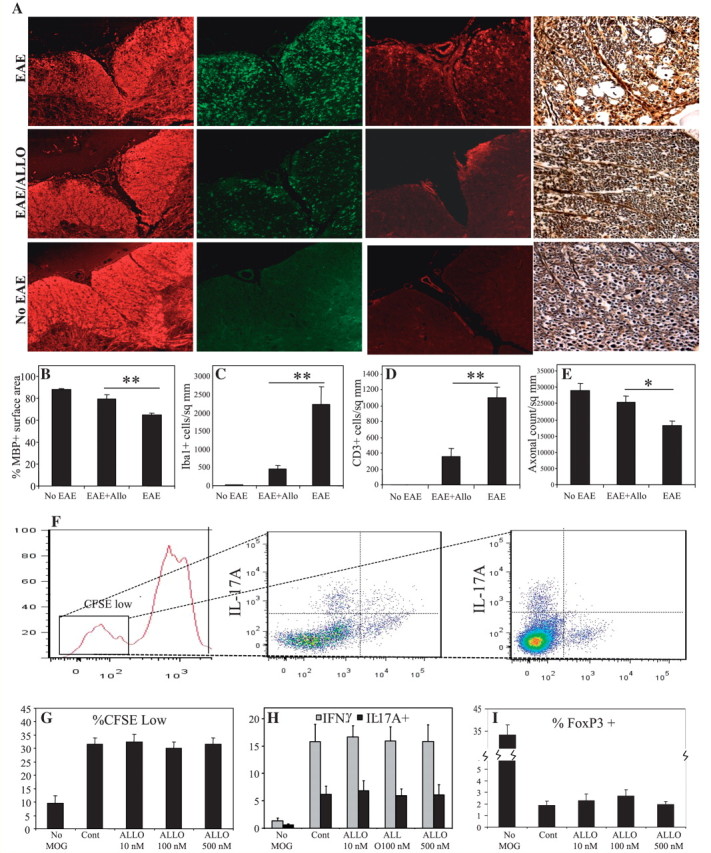
Allopregnanolone treatment suppresses neuroinflammation, demyelination and axonal injury in mice affected by EAE. MOG-immunized mice receiving daily intraperitoneal injections of allopregnanolone (ALLO; 10 mg/kg) after the onset of neurological symptoms were euthanized at the peak of the disease and hindbrain tissues were isolated. Analysis of myelin basic protein immunoreactivity showed preserved myelin in allopregnanolone-treated experimental autoimmune encephalomyelitis mice compared with vehicle-treated animals (A, 1st column, B). Likewise, immunoreactivity of the monocytoid cell marker, Iba-1 (A, 2nd column, C), and the lymphocyte marker, CD3ε (A, 3rd column, D), were lower in the spinal cords of EAE mice treated with allopregnanolone compared with vehicle-treated experimental autoimmune encephalomyelitis animals. Bielchowsky (silver) staining revealed loss of axons in vehicle-treated EAE mice, which was less severe in allopregnanolone-treated animals (A, 4th column, E). Moreover, allopregnanolone treatment of splenocyte cultures derived from MOG-immunized mice, did not affect the percentage of proliferated [carboxyfluorescein succinimidyl ester (CFSE)low] CD4+ T-cells (F and G). Likewise, allopregnanolone treatment did not change the proportion of interferon (IFN)-γ or interleukin (IL)-17A immunopositive CFSElow CD4+ T-cells (H), while the proportion of FoxP3+ T regulatory cells showed a non-significant increase (I). Values are shown as mean ± SEM. (Original magnification ×200, Tukey–Kramer Multiple Comparisons test; *P < 0.05, **P < 0.01).
To investigate whether the effects of allopregnanolone on EAE neuropathology are due to modulation of MOG-reactive lymphocyte responses, we established splenocyte cultures from spleens of MOG35–55-induced EAE mice. Treatment of splenocyte cultures with different concentrations of allopregnanolone (up to 500 nM), in the presence of MOG, did not affect antigen-specific proliferation of CD4+ T-cells as measured by carboxyfluorescein succinimidyl ester fluorescence (Fig. 7F and G). Likewise, intracellular immunostaining did not reveal any changes in interferon-γ or interleukin-17 immunoreactivity in MOG-treated T-cells (Fig. 7H) with a non-significant increase in the number of FoxP3+ T regulatory cells (Fig. 7I). These data indicated that the effects of allopregnanolone were likely mediated through direct neuroprotection together with modulation of monocytoid cell activity without affecting antigen-specific T-cell responses.
In accordance with neuropathological data, neurobehavioural analyses showed significantly diminished disease severity after allopregnanolone treatment (Fig. 8A). This was associated with reduced maximum disease scores as well as cumulative scores in allopregnanolone-treated animals (Fig. 8B and C).
Figure 8.
Allopregnanolone treatment diminishes neurobehavioural deficits in mice affected by EAE. MOG-immunized mice received daily intraperitoneal injections of allopregnanolone (ALLO; 10 mg/kg) after the onset of neurological symptoms until the end of scoring period. Consistent with neuropathological findings, allopregnanolone treatment significantly diminished clinical disease severity in mice over the course of the disease (A). Treatment of experimental autoimmune encephalomyelitis mice with allopregnanolone also significantly reduced the maximum score reached by each animal during the disease period (B) as well as the total clinical score (sum of scores) (C). Disease scores are shown as mean ± SEM (n = 17 per group of animals for EAE experiments, Mann–Whitney U test; *P < 0.05, **P < 0.01).
Discussion
Herein, we report that profiling of multiple sclerosis white matter by high-throughput microarray analyses and related multiplatform technologies pointed to the induction of several micro-RNAs involved in neurosteroidogenesis; this was supported by suppression of neurosteroid synthesis in multiple sclerosis. These studies are the first report of perturbed neurosteroidogenesis in multiple sclerosis and the related model, EAE, which also showed improved outcomes in terms of neurobehavioural deficits, neuropathology and neuromolecular changes with neurosteroid (allopregnanolone) replacement (Supplementary Fig. 9). Gene ontology analyses disclosed the potential targeting of multiple biological processes by differentially expressed micro-RNAs, based on arrays composed of 1000 human micro-RNAs, many of which have prior well-known roles in multiple sclerosis pathogenesis. However, steroid biosynthetic machinery exhibited the most robust targeting bias towards upregulated micro-RNAs in white matter of patients with multiple sclerosis. This observation was complemented by suppressed expression of critical neurosteroidogenic enzymes (3α–hydroxysteroid dehydrogenase isoforms, targeted by miR-338, -155 and -491), evident by reverse transcription–PCR, in situ hybridization, western blotting and reduced neurosteroid levels in multiple sclerosis white matter measured by gas chromatography–mass spectrometry. We also verified the putative targets, specific neurosteroidogenic enzymes, for each of the identified micro-RNAs in the present studies using the firefly-Renilla assay. Similar reductions in neurosteroidogenic enzymes and neurosteroids were observed in EAE CNS tissues, and moreover, treatment of EAE animals with allopregnanolone (depleted in both multiple sclerosis and EAE), ameliorated the disease severity and neuroinflammation. These findings underscore the potential value of high-throughput analyses for revealing new therapeutic targets in multiple sclerosis. Moreover, the present studies also suggest that the neurosteroid allopregnanolone, or perhaps closely related compounds, might represent unique therapeutic options for people with multiple sclerosis (Supplementary Fig. 9).
While micro-RNAs are well known to participate in fine-tuning gene expression programs underlying specific processes including neuronal differentiation, synaptogenesis and synaptic plasticity in the nervous system, their role in neurological disorders is increasingly being appreciated. Several micro-RNA expression profiling studies performed on tissues derived from patients affected by neurodegenerative disorders have implicated micro-RNAs in the disease process. Studies of brain tissues from individuals with Alzheimer's disease show dysregulation of micro-RNAs in different brain regions, with amyloid precursor protein processing pathways and innate immune mechanisms representing the two major targets of altered micro-RNAs (Hebert et al., 2008, Wang et al., 2008b; Roshan et al., 2009). Likewise, studies performed on spinocerebellar ataxia tissues revealed several micro-RNAs which control the expression of disease-associated proteins (Johnson et al., 2008). Studies of Parkinson's disease have led to the identification of a micro-RNA that regulates the maturation and function of midbrain dopaminergic neurons, which is downregulated in brains of patients with Parkinson's disease (Kim et al., 2007). Until recently, studies exploring the role of micro-RNAs in multiple sclerosis were limited to three repeats performed on patient blood. A study by Otaegui et al. (2009) identified micro-RNAs whose altered expression in peripheral blood mononuclear cells was associated with relapse (miR-18b and miR-599) or remission (miR-96) of disease. Likewise, a study by Keller et al. (2009) identified a set of micro-RNAs, including miR-145, in the blood of patients with multiple sclerosis, which could accurately differentiate patients with relapsing–remitting multiple sclerosis from healthy controls. Another study by Du et al. (2009) detected upregulation of miR-326 in peripheral blood leucocytes of patients with multiple sclerosis and identified its role as a key regulator of Th17 differentiation and multiple sclerosis/EAE disease severity. More recently, the results of micro-RNA profiling of multiple sclerosis brain tissue using quantitative reverse transcription–PCR analyses of 365 human micro-RNAs were reported, which showed the targeting of the regulatory CD47 protein by three upregulated micro-RNAs in multiple sclerosis, leading to enhanced macrophage activity and myelin phagocytosis (Junker et al., 2009). Interestingly, the chief multiple sclerosis-induced micro-RNAs in this latter report, which included miR-155, showed localization in astrocytes. Hence, our studies confirm this previous observation, but also extend the understanding of micro-RNA-mediated effects in the CNS during the course of multiple sclerosis, pointing to a novel pathogenic mechanism, amenable to therapeutic interventions.
Although focal demyelinating lesions are typically considered to be the hallmark of multiple sclerosis pathology, imaging, molecular and histopathological studies have also shown that the so-called ‘normal-appearing white matter’, identified by its normal appearance in conventional MRI images or histochemically stained white matter sections, contain inflammation and axonal injury (Filippi and Rocca, 2005). Quantitative pathological studies have reported axonal loss in the normal appearing white matter of patients with multiple sclerosis (Evangelou et al., 2000) and focal clusters of activated microglia in the absence of demyelination and leucocyte infiltration. Moreover, these neuropathological changes in the normal appearing white matter of patients with multiple sclerosis often precede the appearance of gadolinium-enhancing lesions by months or years (van der Valk and Amor, 2009). Likewise, quantitative structural MRI techniques also indicate the presence of normal appearing white matter abnormalities from the onset of disease (Miller et al., 2003). Moreover, molecular studies have shown altered gene expression in normal appearing white matter, with components of cellular immune responses being equally dysregulated in normal appearing white matter and proximal demyelinating lesions (Lindberg et al., 2004), all supporting the notion of widespread white matter disease in multiple sclerosis. Given these circumstances, we deliberately selected normal appearing white matter proximate to lesions in order to gain insight into molecular abnormalities present in this region, which more likely demonstrate brain-intrinsic pathways involved in neuropathogenesis independent of infiltrating leucocytes.
Neurosteroids exert diverse effects on neural cell function and survival in the brain. The mechanisms involved include genomic actions mediated by nuclear steroid receptors and more rapid non-genomic effects mediated by their actions as allosteric modulators of at neurotransmitter receptors (Mellon and Griffin, 2002). Nonetheless, neurosteroids are also capable of interacting with cell surface neurotransmitter receptors to modulate neural cell physiology as recognized over the last decade (Sedlacek et al., 2008). In particular, GABAA receptors and the modulation of their activity by neurochemicals, e.g. allopregnanolone, have been extensively investigated in the context of neurodegenerative disorders (Mellon and Griffin, 2002; Lambert et al., 2003). Mediating the effects of the chief inhibitory neurotransmitter in the CNS, GABAA receptors represent one of the most elaborate neurotransmitter receptor structures, harboring multiple binding sites for allosteric modulators and neuroactive compounds, e.g. benzodiazepines, barbiturates, ethanol, inhaled anesthetics and neuroactive steroids (Johnston, 1996; Hosie et al., 2007). GABAA receptors are widely expressed on neurons and glial cells in the nervous system, modulating neurogenesis, neuronal survival, migration and synaptogenesis (Ge et al., 2007; Mellon, 2007). Allopregnanolone has been shown to promote neurogenesis in both rodent and human neuroprogenitor cells, likely through binding the GABAA receptor (Wang et al., 2005, 2008b), which complements its putative regenerative properties in the aged brain and Alzheimer's disease (Brinton and Wang, 2006). Moreover, allopregnanolone promotes myelination and myelin protein synthesis in peripheral nerves (Martini et al., 2003) as well as in the CNS (Ghoumari et al., 2003). Of note, these effects are independent of the widely recognized effects of progesterone in promoting neural cell survival, neurogenesis and myelination mediated through the nuclear receptor for progesterone (Sakamoto et al., 2002; Ghoumari et al., 2003, 2005; Zhang et al., 2010). In addition to neural cells, GABAA receptors are also expressed on resident and circulating monocytoid cells of the immune system and they have been suggested to ameliorate inflammation by modulating the activity of antigen-presenting cells (Alam et al., 2006; Bhat et al., 2010). Herein, allopregnanolone treatment of human monocytes diminished proinflammatory gene expression. However, allopregnanolone did not influence antigen-specific lymphocyte proliferation and differentiation. This latter observation indicated that, unlike glucocorticoids frequently used in the treatment of autoimmune disorders, allopregnanolone likely lacks direct immunosuppressive effects; hence, this and related molecules might provide new therapeutic options, devoid of the side-effects of common immunosuppressive therapies, for treatment of multiple sclerosis and neuroinflammatory disorders. Of interest, ganaxolone, a methylated synthetic analogue of allopregnanolone, has previously been evaluated and shown to be safe, tolerable and therapeutically promising in several clinical trials for intractable epilepsy (Laxer et al., 2000, Pieribone et al., 2007). Future studies of micro-RNAs and other non-coding small RNA molecules in multiple sclerosis will likely highlight new families of disrupted genes, perhaps yielding other therapeutic targets and new diagnostic options for multiple sclerosis and related neuroinflammatory disorders.
Funding
Canadian Institutes for Health Research and Alberta Heritage Foundation for Medical Research Fellowships (to F.N.); Canadian Institutes for Health Research (C.P. and G.B.); Multiple Sclerosis Society of Canada (C.P.).
Supplementary material
Supplementary material is available at Brain online.
Acknowledgements
The authors thank Gail Rauw and Ingrid Catz for expert technical assistance and Synthia Mellon for providing reagents. C.P. and G.B. hold Canada Research Chairs (Tier 1) in Neurological Infection and Immunity, and Neurochemistry and Drug Development, respectively. C.P. is an Alberta Heritage Foundation for Medical Research Senior Scholar.
Glossary
Abbreviations
- EAE
experimental autoimmune encephalitis
- GABA
γ-aminobutyric acid
- MOG
myelin oligodendrocyte glycoprotein
- PCR
polymerase chain reaction
References
- Ahboucha S, Butterworth RF, Pomier-Layrargues G, Vincent C, Hassoun Z, Baker GB. Neuroactive steroids and fatigue severity in patients with primary biliary cirrhosis and hepatitis C. Neurogastroenterol Motil. 2008;20:671–9. doi: 10.1111/j.1365-2982.2007.01080.x. [DOI] [PubMed] [Google Scholar]
- Alam S, Laughton DL, Walding A, Wolstenholme AJ. Human peripheral blood mononuclear cells express GABAA receptor subunits. Mol Immunol. 2006;43:1432–42. doi: 10.1016/j.molimm.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;28 doi: 10.1016/s0092-8674(01)00616-x. 107: 823–6. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Asli NS, Pitulescu ME, Kessel M. MicroRNAs in organogenesis and disease. Curr Mol Med. 2008;8:698–710. doi: 10.2174/156652408786733739. [DOI] [PubMed] [Google Scholar]
- Barbaccia ML. Neurosteroidogenesis: relevance to neurosteroid actions in brain and modulation by psychotropic drugs. Crit Rev Neurobiol. 2004;16:67–74. doi: 10.1615/critrevneurobiol.v16.i12.70. [DOI] [PubMed] [Google Scholar]
- Bhat R, Axtell R, Mitra A, Miranda M, Lock C, Tsien RW, et al. Inhibitory role for GABA in autoimmune inflammation. Proc Natl Acad Sci USA. 2010;107:2580–5. doi: 10.1073/pnas.0915139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat R, Steinman L. Innate and adaptive autoimmunity directed to the central nervous system. Neuron. 2009;64:123–32. doi: 10.1016/j.neuron.2009.09.015. [DOI] [PubMed] [Google Scholar]
- Brinton RD, Wang JM. Therapeutic potential of neurogenesis for prevention and recovery from Alzheimer’s disease: allopregnanolone as a proof of concept neurogenic agent. Curr Alzheimer Res. 2006;3:185–90. doi: 10.2174/156720506777632817. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–8. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chabas D, Baranzini SE, Mitchell D, Bernard CC, Rittling SR, Denhardt DT, et al. The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science. 2001;294:1731–5. doi: 10.1126/science.1062960. [DOI] [PubMed] [Google Scholar]
- Charalampopoulos I, Remboutsika E, Margioris AN, Gravanis A. Neurosteroids as modulators of neurogenesis and neuronal survival. Trends Endocrinol Metab. 2008;19:300–7. doi: 10.1016/j.tem.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, et al. MicroRNA miR-326 regulates T(H)-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–9. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- Ellestad KK, Tsutsui S, Noorbakhsh F, Warren KG, Yong VW, Pittman QJ, et al. Early life exposure to lipopolysaccharide suppresses experimental autoimmune encephalomyelitis by promoting tolerogenic dendritic cells and regulatory T cells. J Immunol. 2009;183:298–309. doi: 10.4049/jimmunol.0803576. [DOI] [PubMed] [Google Scholar]
- Evangelou N, Esiri MM, Smith S, Palace J, Matthews PM. Quantitative pathological evidence for axonal loss in normal appearing white matter in multiple sclerosis. Ann Neurol. 2000;47:391–5. [PubMed] [Google Scholar]
- Filippi M, Rocca MA. MRI evidence for multiple sclerosis as a diffuse disease of the central nervous system. J Neurol. 2005;252(Suppl 5):v16–24. doi: 10.1007/s00415-005-5004-5. [DOI] [PubMed] [Google Scholar]
- Ge S, Pradhan DA, Ming GL, Song H. GABA sets the tempo for activity-dependent adult neurogenesis. Trends Neurosci. 2007;30:1–8. doi: 10.1016/j.tins.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Ghoumari AM, Baulieu EE, Schumacher M. Progesterone increases oligodendroglial cell proliferation in rat cerebellar slice cultures. Neuroscience. 2005;135:47–58. doi: 10.1016/j.neuroscience.2005.05.023. [DOI] [PubMed] [Google Scholar]
- Ghoumari AM, Ibanez C, El-Etr M, Leclerc P, Eychenne B, O’Malley BW, et al. Progesterone and its metabolites increase myelin basic protein expression in organotypic slice cultures of rat cerebellum. J Neurochem. 2003;86:848–59. doi: 10.1046/j.1471-4159.2003.01881.x. [DOI] [PubMed] [Google Scholar]
- Giatti S, D’Intino G, Maschi O, Pesaresi M, Garcia-Segura LM, Calza L, et al. Acute experimental autoimmune encephalomyelitis induces sex dimorphic changes in neuroactive steroid levels. Neurochem Int. 2010;56:118–27. doi: 10.1016/j.neuint.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Goascogne C, Gouezou M, Robel P, Defaye G, Chambaz E, Waterman MR, et al. The cholesterol side-chain cleavage complex in human brain white matter. J Neuroendocrinol. 1989;1:153–6. doi: 10.1111/j.1365-2826.1989.tb00095.x. [DOI] [PubMed] [Google Scholar]
- Griffin LD, Gong W, Verot L, Mellon SH. Niemann-Pick type C disease involves disrupted neurosteroidogenesis and responds to allopregnanolone. Nat Med. 2004;10:704–11. doi: 10.1038/nm1073. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MH, Hwang SI, Roy DB, Lundgren DH, Price JV, Ousman SS, et al. Proteomic analysis of active multiple sclerosis lesions reveals therapeutic targets. Nature. 2008;451:1076–81. doi: 10.1038/nature06559. [DOI] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci USA. 2008;105:6415–20. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie AM, Wilkins ME, Smart TG. Neurosteroid binding sites on GABA(A) receptors. Pharmacol Ther. 2007;116:7–19. doi: 10.1016/j.pharmthera.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Hu ZY, Bourreau E, Jung-Testas I, Robel P, Baulieu EE. Neurosteroids: oligodendrocyte mitochondria convert cholesterol to pregnenolone. Proc Natl Acad Sci USA. 1987;84:8215–9. doi: 10.1073/pnas.84.23.8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington's disease. Neurobiol Dis. 2008;29:438–45. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Johnston GA. GABAA receptor pharmacology. Pharmacol Ther. 1996;69:173–98. doi: 10.1016/0163-7258(95)02043-8. [DOI] [PubMed] [Google Scholar]
- Junker A, Krumbholz M, Eisele S, Mohan H, Augstein F, Bittner R, et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132(Pt 12):3342–52. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- Kanter JL, Narayana S, Ho PP, Catz I, Warren KG, Sobel RA, et al. Lipid microarrays identify key mediators of autoimmune brain inflammation. Nat Med. 2006;12:138–43. doi: 10.1038/nm1344. [DOI] [PubMed] [Google Scholar]
- Keller A, Leidinger P, Lange J, Borries A, Schroers H, Scheffler M, et al. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One. 2009;4:e7440. doi: 10.1371/journal.pone.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–4. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–50. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Lambert JJ, Belelli D, Peden DR, Vardy AW, Peters JA. Neurosteroid modulation of GABAA receptors. Prog Neurobiol. 2003;71:67–80. doi: 10.1016/j.pneurobio.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Laxer K, Blum D, Abou-Khalil BW, Morrell MJ, Lee DA, Data JL, et al. Assessment of ganaxolone's anticonvulsant activity using a randomized, double-blind, presurgical trial design. Ganaxolone Presurgical Study Group. Epilepsia. 2000;41:1187–94. doi: 10.1111/j.1528-1157.2000.tb00324.x. [DOI] [PubMed] [Google Scholar]
- Le Melledo JM, Baker GB. Neuroactive steroids and anxiety disorders. J Psychiatry Neurosci. 2002;27:161–5. [PMC free article] [PubMed] [Google Scholar]
- Lindberg RL, De Groot CJ, Certa U, Ravid R, Hoffmann F, Kappos L, et al. Multiple sclerosis as a generalized CNS disease–comparative microarray analysis of normal appearing white matter and lesions in secondary progressive MS. J Neuroimmunol. 2004;152:154–67. doi: 10.1016/j.jneuroim.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Lock C, Hermans G, Pedotti R, Brendolan A, Schadt E, Garren H, et al. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat Med. 2002;8:500–8. doi: 10.1038/nm0502-500. [DOI] [PubMed] [Google Scholar]
- MacKenzie EM, Odontiadis J, Le Melledo JM, Prior TI, Baker GB. The relevance of neuroactive steroids in schizophrenia, depression, and anxiety disorders. Cell Mol Neurobiol. 2007;27:541–74. doi: 10.1007/s10571-006-9086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini L, Magnaghi V, Melcangi RC. Actions of progesterone and its 5alpha-reduced metabolites on the major proteins of the myelin of the peripheral nervous system. Steroids. 2003;68:825–9. doi: 10.1016/s0039-128x(03)00134-x. [DOI] [PubMed] [Google Scholar]
- Mellon SH. Neurosteroid regulation of central nervous system development. Pharmacol Ther. 2007;116:107–24. doi: 10.1016/j.pharmthera.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon SH, Griffin LD. Neurosteroids: biochemistry and clinical significance. Trends Endocrinol Metab. 2002;13:35–43. doi: 10.1016/s1043-2760(01)00503-3. [DOI] [PubMed] [Google Scholar]
- Miller DH, Thompson AJ, Filippi M. Magnetic resonance studies of abnormalities in the normal appearing white matter and grey matter in multiple sclerosis. J Neurol. 2003;250:1407–19. doi: 10.1007/s00415-003-0243-9. [DOI] [PubMed] [Google Scholar]
- Noorbakhsh F, Overall CM, Power C. Deciphering complex mechanisms in neurodegenerative diseases: the advent of systems biology. Trends Neurosci. 2009;32:88–100. doi: 10.1016/j.tins.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Noorbakhsh F, Ramachandran R, Barsby N, Ellestad KK, Leblanc A, Dickie P, et al. MicroRNA profiling reveals new aspects of HIV neurodegeneration: caspase-6 regulates astrocyte survival. FASEB J. 2010;24:1799–812. doi: 10.1096/fj.09-147819. [DOI] [PubMed] [Google Scholar]
- Noorbakhsh F, Tsutsui S, Vergnolle N, Boven LA, Shariat N, Vodjgani M, et al. Proteinase-activated receptor 2 modulates neuroinflammation in experimental autoimmune encephalomyelitis and multiple sclerosis. J Exp Med. 2006;203:425–35. doi: 10.1084/jem.20052148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorbakhsh F, Vergnolle N, McArthur JC, Silva C, Vodjgani M, Andrade-Gordon P, et al. Proteinase-activated receptor-2 induction by neuroinflammation prevents neuronal death during HIV infection. J Immunol. 2005;174:7320–9. doi: 10.4049/jimmunol.174.11.7320. [DOI] [PubMed] [Google Scholar]
- Oh LY, Larsen PH, Krekoski CA, Edwards DR, Donovan F, Werb Z, et al. Matrix metalloproteinase-9/gelatinase B is required for process outgrowth by oligodendrocytes. J Neurosci. 1999;19:8464–75. doi: 10.1523/JNEUROSCI.19-19-08464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaegui D, Baranzini SE, Armananzas R, Calvo B, Munoz-Culla M, Khankhanian P, et al. Differential micro-RNA expression in PBMC from multiple sclerosis patients. PLoS One. 2009;4:e6309. doi: 10.1371/journal.pone.0006309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penning TM, Burczynski ME, Jez JM, Hung CF, Lin HK, Ma H, et al. Human 3alpha-hydroxysteroid dehydrogenase isoforms (AKR1C1-AKR1C4) of the aldo-keto reductase superfamily: functional plasticity and tissue distribution reveals roles in the inactivation and formation of male and female sex hormones. Biochem J. 2000;351(Pt 1):67–77. doi: 10.1042/0264-6021:3510067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieribone VA, Tsai J, Soufflet C, Rey E, Shaw K, Giller E, et al. Clinical evaluation of ganaxolone in pediatric and adolescent patients with refractory epilepsy. Epilepsia. 2007;48:1870–4. doi: 10.1111/j.1528-1167.2007.01182.x. [DOI] [PubMed] [Google Scholar]
- Power C, Henry S, Del Bigio MR, Larsen PH, Corbett D, Imai Y, et al. Intracerebral hemorrhage induces macrophage activation and matrix metalloproteinases. Ann Neurol. 2003;53:731–42. doi: 10.1002/ana.10553. [DOI] [PubMed] [Google Scholar]
- Roshan R, Ghosh T, Scaria V, Pillai B. MicroRNAs: novel therapeutic targets in neurodegenerative diseases. Drug Discov Today. 2009;14:1123–9. doi: 10.1016/j.drudis.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Rupprecht R, Holsboer F. Neuroactive steroids: mechanisms of action and neuropsychopharmacological perspectives. Trends Neurosci. 1999;22:410–6. doi: 10.1016/s0166-2236(99)01399-5. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Ukena K, Tsutsui K. Dendritic spine formation in response to progesterone synthesized de novo in the developing Purkinje cell in rats. Neurosci Lett. 2002;322:111–5. doi: 10.1016/s0304-3940(02)00077-0. [DOI] [PubMed] [Google Scholar]
- Schumacher M, Weill-Engerer S, Liere P, Robert F, Franklin RJ, Garcia-Segura LM, et al. Steroid hormones and neurosteroids in normal and pathological aging of the nervous system. Prog Neurobiol. 2003;71:3–29. doi: 10.1016/j.pneurobio.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Sedlacek M, Korinek M, Petrovic M, Cais O, Adamusova E, Chodounska H, et al. Neurosteroid modulation of ionotropic glutamate receptors and excitatory synaptic transmission. Physiol Res. 2008;57(Suppl 3):S49–57. doi: 10.33549/physiolres.931600. [DOI] [PubMed] [Google Scholar]
- Sospedra M, Martin R. Immunology of multiple sclerosis. Annu Rev Immunol. 2005;23:683–747. doi: 10.1146/annurev.immunol.23.021704.115707. [DOI] [PubMed] [Google Scholar]
- Trapp BD, Nave KA. Multiple sclerosis: an immune or neurodegenerative disorder? Annu Rev Neurosci. 2008;31:247–69. doi: 10.1146/annurev.neuro.30.051606.094313. [DOI] [PubMed] [Google Scholar]
- Tsutsui S, Schnermann J, Noorbakhsh F, Henry S, Yong VW, Winston BW, et al. A1 adenosine receptor upregulation and activation attenuates neuroinflammation and demyelination in a model of multiple sclerosis. J Neurosci. 2004;24:1521–9. doi: 10.1523/JNEUROSCI.4271-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Valk P, Amor S. Preactive lesions in multiple sclerosis. Curr Opin Neurol. 2009;22:207–13. doi: 10.1097/WCO.0b013e32832b4c76. [DOI] [PubMed] [Google Scholar]
- Vergnes L, Phan J, Stolz A, Reue K. A cluster of eight hydroxysteroid dehydrogenase genes belonging to the aldo-keto reductase supergene family on mouse chromosome 13. J Lipid Res. 2003;44:503–11. doi: 10.1194/jlr.M200399-JLR200. [DOI] [PubMed] [Google Scholar]
- Wang JM, Johnston PB, Ball BG, Brinton RD. The neurosteroid allopregnanolone promotes proliferation of rodent and human neural progenitor cells and regulates cell-cycle gene and protein expression. J Neurosci. 2005;25:4706–18. doi: 10.1523/JNEUROSCI.4520-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JM, Liu L, Irwin RW, Chen S, Brinton RD. Regenerative potential of allopregnanolone. Brain Res Rev. 2008;57:398–409. doi: 10.1016/j.brainresrev.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, et al. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–23. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg MS, Wood MJ. Short non-coding RNA biology and neurodegenerative disorders: novel disease targets and therapeutics. Hum Mol Genet. 2009;18:R27–39. doi: 10.1093/hmg/ddp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yang R, Zhou R, Li L, Sokabe M, Chen L. Progesterone promotes the survival of newborn neurons in the dentate gyrus of adult male mice. Hippocampus. 2010;20:402–12. doi: 10.1002/hipo.20642. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Vergote D, Pardo C, Noorbakhsh F, McArthur JC, Hollenberg MD, et al. CXCR3 activation by lentivirus infection suppresses neuronal autophagy: neuroprotective effects of antiretroviral therapy. FASEB J. 2009;23:2928–41. doi: 10.1096/fj.08-128819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.