Abstract
A new database search algorithm has been developed for identification of intact cross-links in proteins and peptides from tandem mass spectrometric data. Using this algorithm, intact cross-links can be identified and characterized in proteins and peptides with high confidence. The algorithm was tested using BS3 (bis[sulfosuccinimidyl] suberate) cross-linked Cytochrome C. Five cross-links were identified and verified for spatial plausibility by comparison with its three-dimensional structure at optimized experimental conditions. The distributions of statistical scores for true and false positives and receiver operating characteristic analysis indicate that the algorithm is capable of discriminating true positive cross-linked peptide-spectrum matches from false ones. It has also been demonstrated that the MassMatrix database search engine is capable of searching for intact cross-links in complex Escherichia coli proteome samples cross-linked by BS3. The new algorithm in MassMatrix offers an additional approach for the discovery of cross-links in proteins and peptides from tandem mass spectrometric data.
Keywords: chemical cross-link, tandem mass spectrometry, database search, proteomics
Introduction
Three-dimensional (3D) protein structures and protein-protein interactions are of great importance to study the functions of proteins. Identification of cross-links in proteins can provide invaluable information regarding a protein’s structure, conformation, and interactions.1–5 Liquid chromatography coupled with tandem mass spectrometry (LC–MS/MS) has become one of the most widely used tools in MS based proteomics for protein ID and characterization.6 Given the high resolution and mass accuracy of modern mass spectrometers, identification of intact cross-links in proteins and peptides by use of LC–MS/MS has recently become feasible.7
Database search algorithms are the most widely used approach to identify peptides and proteins from tandem MS data.8 Due to the increased search space for searches with cross-links, false positives need to be controlled by use of validated scoring algorithms and decoy search strategy.9,10 Traditional database search programs, such as Mascot,11 SEQUEST,12 and OMSSA,13 cannot be used for analysis of cross-linked proteins/peptides. For this reason, several specialized methods have been developed specifically for this type of analysis.7,14–17 Unfortunately, these approaches do not use well-validated scoring algorithms, making false discovery rate determinations difficult. Furthermore, some of those programs lack user-friendly interface and require extensive user input during data analysis.
Here we describe a database search algorithm for identification of cross-links in proteins and peptides by use of tandem mass spectrometry. This new cross-link search algorithm is an extension of a validated database search engine with three probability-based scoring algorithms. For peptides and proteins without any cross-links or disulfide bonds, these scoring algorithms have been validated against Mascot, SEQUEST, OMSSA, and X!Tandem.18,19 It has been shown that for data with high mass accuracy, MassMatrix provided better sensitivity than Mascot, SEQUEST, X!Tandem, and OMSSA for a given specificity.19 The scoring algorithms have also been validated for peptides and proteins with disulfide bonds by use of peptide standards with known disulfide bonds and bovine pancreatic ribonuclease A (RNaseA).20 A user-friendly web interface is also available for the program (http://www.massmatrix.net) and the search form for peptides and proteins with cross-links and disulfide bonds is the same as that for peptides and proteins without any cross-links or disulfide bonds. Therefore, search for cross-links and disulfide bonds in MassMatrix by use of tandem MS data is as easy as that for peptides and proteins without any cross-links and disulfide bonds in MassMatrix search engine. The algorithm was tested using data sets collected on a LTQ-FT mass spectrometer for the tryptic digests of Cytochrome C cross-linked by BS3. Five cross-links were identified and verified for spatial plausibility by comparison with its 3D structure. It has also been demonstrated that MassMatrix database search engine is capable of searching for intact cross-links in complex Escherichia coli proteome samples cross-linked by BS3.
Experimental Section
Materials, Sample Preparation, and Mass Spectrometry
Horse heart Cytochrome C, was purchased from Sigma-Aldrich (St. Louis, MO) and the cross-linking reagent BS3 (bis[sulfosuccinimidyl] suberate) was purchased from Thermo Fisher Scientific (Rockford, IL). Horse heart Cytochrome C and the cross-linking reagent BS3 were prepared by dissolving in phosphate buffer solution (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4, pH 7.4). Cross-linking reactions were performed at various cross-linking reagent to protein molar ratios of 1:1, 2.5:1, 5:1, 10:1, 25:1, 50:1, and 100:1 and a final protein concentration of 0.12 mg/mL (0.01 mM) at room temperature at 4 °C for 60 min. Reactions were also performed at a cross-linking reagent to protein ratio of 10:1 and different protein concentrations of 0.06, 0.12, 0.6, and 2.4 mg/mL. Cross-linking reactions were then quenched by adding 1 M glycine (pH 9.0) solution. The cross-linked protein samples were purified by SDS-PAGE and the monomer bands were cut and digested by trypsin (Promega, Madison, WI) with a substrate to enzyme ratio of 50:1 at 37 °C overnight. Tryptic peptides were then extracted by 50% acetonitrile with 5% formic acid three times.
Escherichia coli cells (BL21) were cultured in LB broth using 200 rpm shaking speed at 37 °C until OD600 reached 1.0. Cells were harvested by centrifugation at 5000× g for 10 min and the pellet was saved and resuspended in phosphate buffer solution. Lysate was sonicated and cell debris was removed by centrifugation at 15000× g for 30 min at 4 °C. The supernatant was subjected to the in vitro cross-linking reaction. The Cross-linking reagent BS3 was added to cell lysate with a final concentration of 2 mM followed by incubation at 4 °C for 60 min. The cross-linking reaction was then quenched by adding Tris-HCl (pH 8.0) with the final concentration of 100 mM for 30 min at 4 °C. Cell lysate was separated by SDS-PAGE and the whole lane of gel was divided into multiple pieces according to molecular weight for the in-gel trypsin digestion. The tryptic peptides were collected from each gel piece and analyzed by nano-LC–MS/MS.
Nano-LC–MS/MS experiments were performed on a LTQ-FT mass spectrometer (Thermo Fisher Scientific, Inc., Waltham, MA) equipped with a nanoelectrospray ion source (New Objective, Inc., Woburn, MA) in positive ion mode. The enzyme digested cross-linked protein samples were injected onto a self-packed precolumn (150 µm I.D. × 20 mm, 5 µm, 200 Å). Chromatographic separation was performed on a self-packed reversed phase C18 nanocolumn (75 µm I.D. × 300 mm, 5 µm, 100 Å) by using 0.1% formic acid in water (mobile phase A) and 0.1% formic acid in 80% acetonitrile (mobile phase B). A linear gradient from 5 to 40% mobile phase B for 40 min at a flow rate of 300 nL/min was applied. A scan cycle was initiated with a full-scan survey MS spectrum (mass range of 300–2000 Da) performed on the FT-ICR mass spectrometer with resolution of 100 000 at 400 Da. The ten most abundant ions detected in this scan were subjected to a MS/MS experiment performed in the linear ion trap. Ion accumulation (Auto Gain Control target number) and maximal ion accumulation time for full-scan and MS/MS were set at 1 × 106 ions, 1000 ms and 5 × 104 ions, 200 ms. Ions were fragmented by use of CID (collision induced dissociation) with a normalized collision energy of 35%, activation Q of 0.3, and activation time of 30 ms.
Database Search and Search Parameters
The RAW data files collected on the mass spectrometer were converted to mzXML files by use of MassMatrix data conversion tools (version 1.3, http://www.massmatrix.net/download). Isotope distributions for the precursor ions of the MS/MS spectra were deconvoluted to obtain the charge states and monoisotopic m/z values of the precursor ions during the data conversion. The mzXML files for the Cytochrome C samples were searched against a limited custom protein database by use of the web-based MassMatrix search engine (version 2.3.4, http://www.massmatrix.net). The custom database was composed of the Cytochrome C protein sequence along with decoy sequences. The decoy sequences included a reversed Cytochrome C sequence and 20 randomized Cytochrome C sequences. Forty-one mzXML data files from the gel bands of the Escherichia coli samples from two replicate experiments were searched collectively against an Escherichia coli K-12 strain sequence database containing 4285 protein sequences (http://www.genome.wisc.edu/sequencing/k12.htm21). The search parameters in MassMatrix were set as follows: (i) enzyme: trypsin; (ii) missed cleavage: 2; (iii) modifications: variable iodoacetamide derivative of cysteine and variable oxidation of methionine; (iv) mass tolerances of 10 ppm and 0.6 Da for the precursor and product ions respectively; (v) maximum number of modifications allowed for each peptide: 2; (vi) peptide length: 6–40 amino acid residues; (vii) score thresholds of 5.3 and 1.3 for the pp and pptag scores respectively.
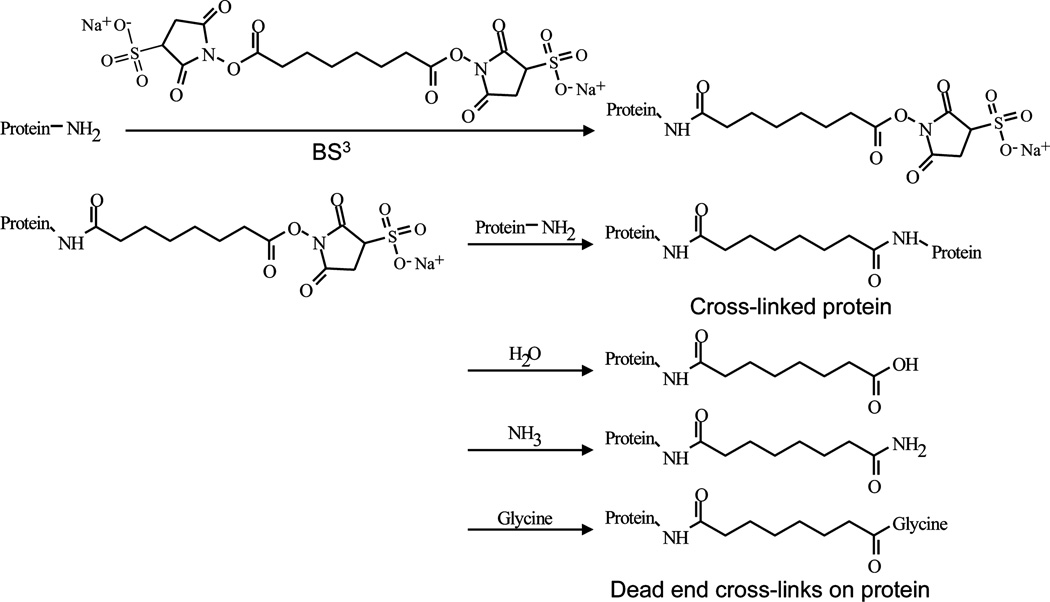
The structure and reaction of the cross-linking reagent are shown in Figure 1. The chemical formula of the cross-link between two lysine sites is specified as C8H10O2 with a monoisotopic mass of 138.068 Da in MassMatrix. Three additional variable modifications for lysine were also specified during the searches due to the dead-end cross-links (Figure 1). Cross-linked lysine was specified to be noncleavable by trypsin and the maximum number of cross-links allowed for each peptide was 2.
Figure 1.
Structure of cross-linking reagent BS3, its reaction with lysine, and the possible dead-end cross-links.
Results and Discussion
Search Algorithm
The classification of chemical cross-links in peptides and chemical cross-linked peptides is the same as that for disulfide bonds and disulfide-linked peptides as described previously.20 In brief, cross-links in peptides were classified into two types: interchain cross-links and intrachain cross-links. Peptides with more than 2 cross-links are difficult to characterize by tandem MS due to poor fragmentation and large size. Therefore, only peptides with up to 2 cross-links were considered in MassMatrix. Those peptides were classified into 4 types: type 1 peptides only have interchain cross-links; type 2 peptides only have intrachain cross-links; type 3 peptides are hybrids with both inter- and intrachain cross-links; and type 4 peptides have circular chains.20 Dead-end cross-links in peptides are considered as modifications to the peptides. Searching for modifications and cross-links in MassMatrix is performed in a single stage. Therefore, cross-linked peptides with various modifications and dead-end cross-links can be identified in MassMatrix.
The algorithm was developed based on the disulfide search algorithm in MassMatrix.20 In the disulfide search algorithm reported previously, only links between cysteine residues are searched by MassMatrix. In the cross-link search algorithm described herein, cross-links between any two amino acid residues are supported by MassMatrix. Potential link sites can be the same amino acid residues or two different amino acid residues. The disulfide search algorithm can be reproduced by a special case of the cross-link algorithm where the two link sites are both cysteines. The algorithm has three search modes. The first two search modes of the algorithm are the same as the exploratory and confirmatory search modes of the disulfide search algorithm described previously.20 In brief, in the exploratory search mode, all occurrences of A and B residues in the protein sequences are considered to be variable link sites, i.e. all those residues may or may not form cross-links of type A–B. During searching, MassMatrix will generate all possible combinations of cross-links by assuming that any two A and B residues are capable of forming a cross-link. In the confirmatory search mode, only the cross-links specified in the protein database by the user will be considered and searched against experimental data. Cross-links are specified in the sequences of a custom database. In the custom .FASTA or .BAS MassMatrix databases, cross-links are coded as “A($i)” and “B($i)”, where i is the index number of the specified cross-link, and A and B are the two related link sites. Each link has two related link sites. A new search mode, called semiexploratory search mode, has been added to the algorithm. In this mode, a limited exploratory search of cross-links will be performed between the amino acid residues labeled by “($)” or “($x)” in the database where x is any number.
During searching, proteins are digested in silico based on the specified proteolysis reagents with consideration given to the presence of cross-links generated from either the exploratory/semiexploratory search mode or as input by the user in confirmatory search mode. These hypothetical peptides are then fragmented using the appropriate fragmentation model and scored against the experimental tandem MS data.
CID (collision induced dissociation) and ETD (electron transfer dissociation) fragmentation methods are currently supported in MassMatrix. Cross-linked peptides may also possess multiple cross-linked peptide chains. In MassMatrix fragmentation model, each chain undergoes fragmentation independently. Only product ions created from the rupture of a single bond are considered and internal fragments are not searched. Therefore, when one chain undergoes fragmentation to create product ions, other chain(s), if any, will be intact and considered as a modification to the cross-linked amino acid residue on the first chain. Product ions with neutral losses of H2O and NH3 due to residues with hydroxyl and amino groups are also considered in the CID fragmentation model of Mass-Matrix.22 Similar to peptides with disulfide bonds,20 type 1 peptides are the preferred configuration for sequencing by tandem mass spectrometry due to the fact that their fragmentation allows for good sequence coverage. Other peptide types can suffer from lower sequence coverage due to blind spots in the product ion series.20
The scoring models involved in the cross-link search algorithm are the same as those used for peptides without an cross-links and those with disulfide bonds as described previously.18–20,23,24 These models have been validated by use of large tandem MS data sets collected on various mass spectrometers against large protein databases and tandem MS data sets from proteins and peptides with intact disulfide bonds. Three independent statistical scores, pp, pp2, and pptag, from the scoring models are mainly used to evaluate quality of peptide-spectrum matches in MassMatrix. Among these three scores, pptag is the best standard to discriminate true matches from false ones18 and will be used for the discussion herein.
Database search results from MassMatrix are reported in html files. Protein and peptide match lists can also be exported to tables presented in .CSV text file format in MassMatrix (version 2.3.4 or later). Cross-link assignments are generated using a posthoc analysis program, XMapper (http://www.massmatrix.net/xmapper), from the MassMatrix html search results. The identified cross-linked peptides are assigned to the cross-links in the proteins to give cross-link identifications. A cross-link is scored in XMapper using the following equation
| (1) |
where N is the number of peptides assigned to the cross-link, np is the number of spectral matches for peptide p with the cross-link, pp, pp2 and pptag are the statistical scores for a spectral match.18,23 This score algorithm was derived from the validated protein score algorithm of MassMatrix search engine.18 The cross-link identifications are reported by XMapper in a .CSV data file and a heat map figure for each protein match.
Validation of the Cross-Link Search Algorithm in MassMatrix
The cross-link search algorithm in MassMatrix was first tested using a data set for a tryptic digest of Cytochrome C cross-linked by BS3 on a LTQ-FT mass spectrometer. The Cross-linking reagent to protein ratio was 25:1 and the final protein concentration was 0.12 mg/mL. The tandem MS data set contained 6982 MS/MS spectra. The data set was searched against a limited database containing the Cytochrome C protein sequence, a reversed Cytochrome C sequence and twenty randomized Cytochrome C sequences. The summary of the search space is listed in Table 1. The search space of the protein database in terms of number of theoretically calculated peptides was dramatically increased when cross-links were considered. 2.10 × 106 peptides were calculated from the database and the total search time was 55 s. A total of 760 peptide-spectrum matches were identified for the Cytochrome C of which 459 were cross-linked peptide-spectrum matches and the overall sequence coverage was 96%. The complete list of true positive peptide-spectrum matches with cross-links is provided in Supplementary Table 1 (Supporting Information).
Table 1.
Summary of the Search Space for a Tryptic Digest of BS3 Cross-Linked Cytochrome C against a Limited Database Containing the Cytochrome C Sequence and 21 Decoy Sequencesa
| sequences | # of peptides without cross-links |
# of cross-linked peptides |
total search time |
|---|---|---|---|
| Cytochrome C | 1.28 × 103 | 9.26 × 104 | 55 s |
| 21 Decoy Sequences | 2.48 × 104 | 1.98 × 106 |
The search was performed on a server with AMD Athlon 64 3800+ CPU processors in serial mode and a single CPU core was used. All peptides with and without modifications and/or dead-end cross-links were considered.
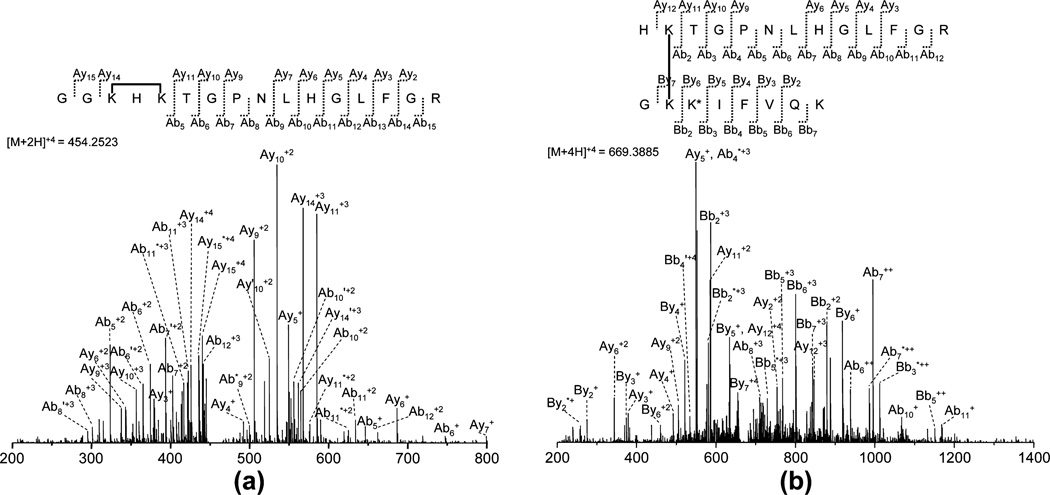
Two representative spectra for cross-linked peptides are shown in Figure 2. It can be seen that the majority of the product ions of cross-linked peptides are the ions created from the rupture of a single bond and those with neutral losses of small molecules. Product ions from internal fragments are ignorable in CID fragmentation. Therefore, the CID fragmentation model in MassMatrix that only considers product ions from the rupture of a single bond and neutral losses is appropriate for cross-linked peptides.
Figure 2.
Representative MS/MS spectra obtained for two cross-linked peptides with (a) an intrachain cross-link and (b) an interchain cross-link in the search of tryptic digest of cross-linked Cytochrome C by BS3. Product ions from neutral loss are labeled by * (loss of ammonia) and ′ (loss of water). The lysine residue with a dead-end cross-link is denoted as K*.
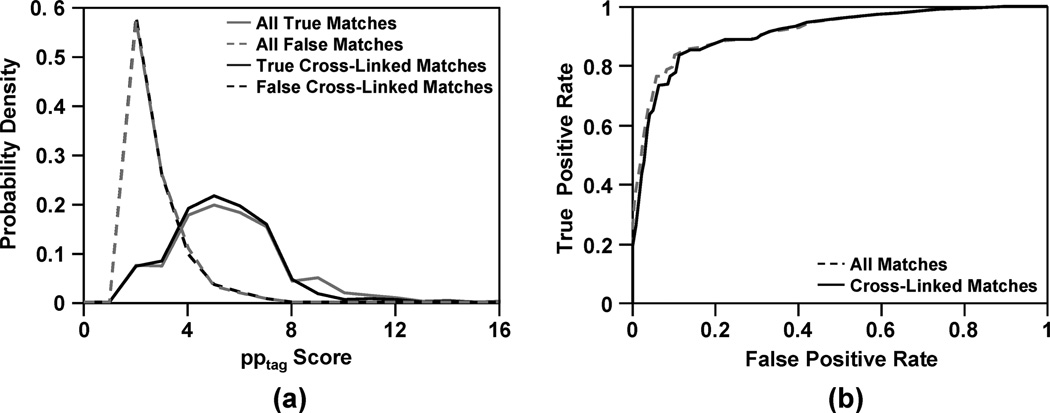
Figure 3 shows the pptag score distributions for TPs and FPs, and the receiver operating characteristic (ROC) analysis for the peptide-spectrum matches identified in MassMatrix. Because the search space for the decoy sequences was much larger than that for the target Cytochrome C sequence, peptide matches from the target Cytochrome C were considered true positives (TPs) and those from the decoy sequences were considered false positives (FPs).9 True positive rate (TPR) and false positive rate (FPR) are calculated by TP/(TP + FN) and FP/(FP + TN), respectively, for a certain threshold, where FN is number of false negatives (TPs under the threshold) and TN is number of true negatives (FPs under the threshold). It can be seen that the scoring model was able to discriminate TPs from FPs for peptides with and without cross-links with small overlap between the two distributions. ROC analysis also indicates that the algorithm performs well for both peptides with and without cross-links. Area under the curve (AUC) for the ROC curves indicates sensitivity and specificity. An ideal algorithm with an AUC equal to 1.0 achieves 100% sensitivity and specificity. It can be seen from Figure 3b that the AUC for peptides with and without cross-links are 0.91 and 0.92 respectively. This indicates that MassMatrix has good sensitivity and specificity for both types of peptides with and without cross-links and is able to identify majority of the peptide-spectrum matches at a low false positive rate.
Figure 3.
(a) Distributions of pptag scores for true and false positive peptide-spectrum matches and (b) ROC curves of the search of a tryptic digest of cross-linked Cytochrome C by BS3. In ROC curve, a value toward the top of the graph indicates higher sensitivity and a value to the left indicates higher specificity.
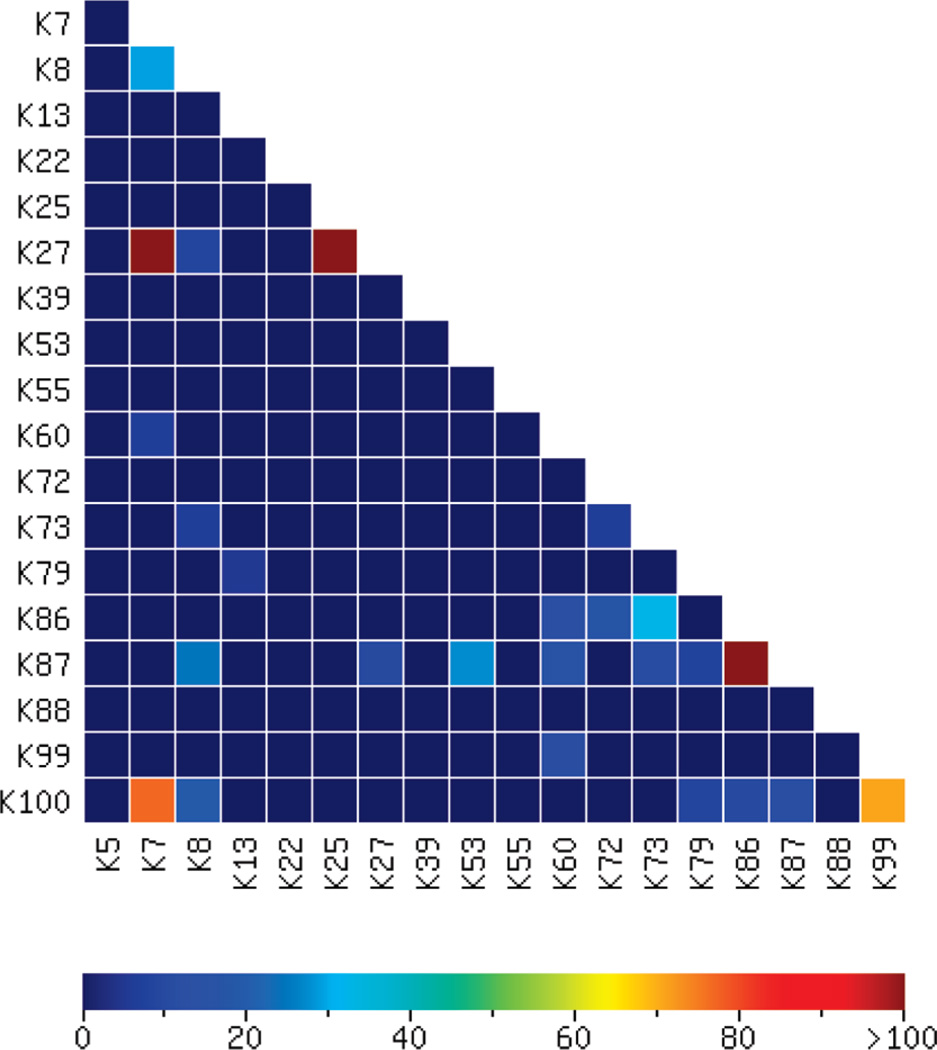
Cytochrome C contains 19 lysine residues and can potentially form 171 cross-links between any two lysine residues. 459 cross-linked peptide-spectrum matches for Cytochrome C were assigned to 25 cross-links as listed in Supplementary Table 2 (Supporting Information). The quality of an identified cross-link is evaluated by the score calculated from eq 1 and the scores of all identified cross-links for Cytochrome C are mapped in a heat map using XMapper as shown in Figure 4. A majority of the cross-links are background cross-links of relatively low occurrence and low scores in MassMatrix and XMapper (cells in light blue or cyan in Figure 4). These background cross-links are the interprotein cross-links formed on proteins with structure altered during the cross-linking experiment. They are biologically irrelevant and represent the noise present in cross-link determination.
Figure 4.
Cross-links in the BS3 cross-linked Cytochrome C mapped by tandem MS experiment and subsequent database search in MassMatrix. Each cell in the heat map represents a cross-link between two lysine residues. Confidence of the identification of each cross-link is indicated by its score displayed in the heat map. The cross-linking reagent to protein ratio was 25:1 and the final protein concentration was 0.12 mg/mL.
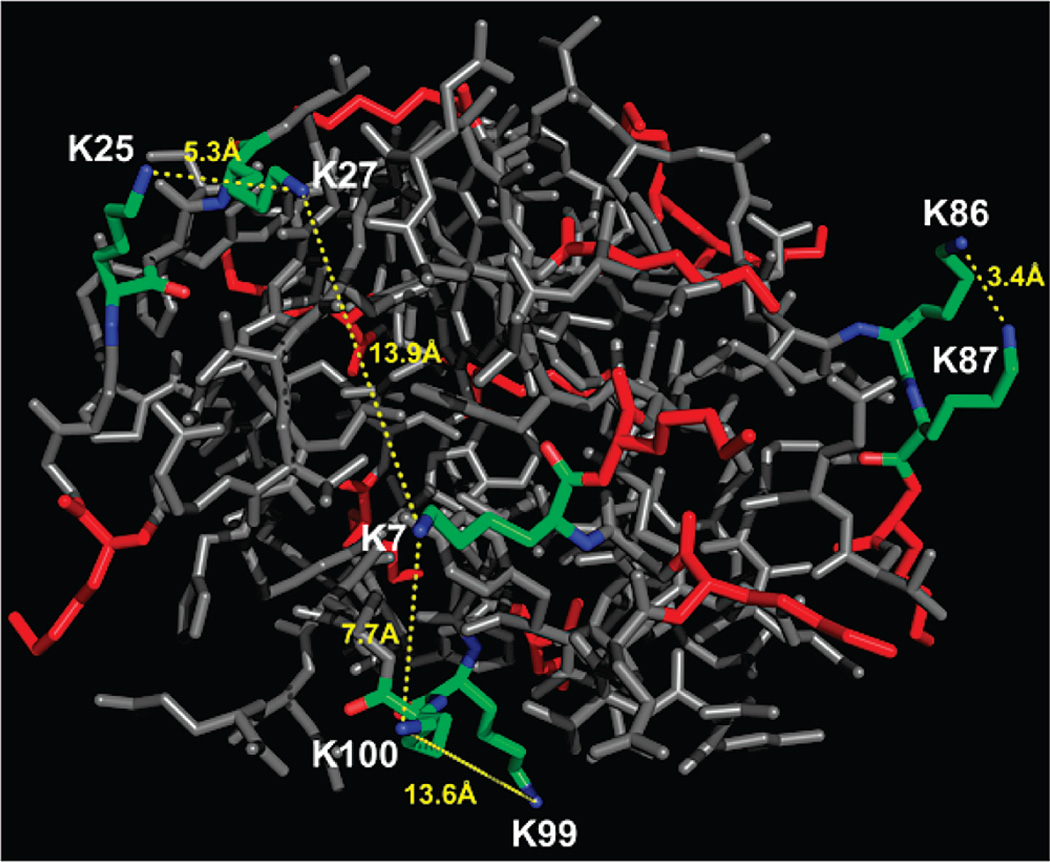
The cross-links formed on the Cytochrome C protein due to its representative 3D structure (PDB code: 1HRC) were present at higher occurrence and higher abundance. Those cross-links were identified in MassMatrix with significantly higher scores than the background cross-links. As shown in Figure 4, five nonbackground cross-links (cells in red or brown), K7–K27, K25–K27, K86–K87, K7–K100, and K99–K100, for Cytochrome C were identified with scores of 160.57, 160.45, 110.4, 76.87, and 70.8, respectively. These cross-links were further verified by comparison with the 3D structure of Cytochrome C as shown in Figure 5. The detailed results for the five nonbackground cross-links are listed in Table 2 and representative MS/MS spectra with matched peaks highlighted and a table of theoretical masses calculated from the MassMatrix fragmentation model for peptide matches with nonbackground cross-links are shown in Supplementary Figure 1 (Supporting Information). The distances of the two lysine residues in the 3D structure for cross-links K25–K27, K86–K87, and K7–K100 are 5.3 Å, 3.4 Å, and 7.7 Å, respectively. They are shorter than the length of the cross-link, which is 12.0 Å. For cross-links K7–K27 and K99–K100, the distances between the two link-sites are 13.9 Å and 13.6 Å, respectively. They are slightly longer than the length of the cross-link but reasonable given the differences between crystal and solution-phase structures.
Figure 5.
Evaluation of spatial plausibility for Cytochrome C cross-links identified by MassMatrix (PDB code: 1HRC).25
Table 2.
Validated Cross-Links and Their Assigned Peptide Matches As Identified by MassMatrix for the BS3 Cross-Linked Cytochrome C at a Cross-Linking Reagent to Protein Ratio of 25:1 and a Final Protein Concentration of 0.12 mg/mL
| Cross-link | Score | Distance(Å) | Cross-linked Peptide | |
|---|---|---|---|---|
| K7–K27 | 160.57 | 13.9 | [23:38]–[6.8] | |
| [23:38]–[6.13] | ||||
| [26:38]–[6.8] | ||||
| [26:38]–[6.13] | ||||
| K25–K27 | 160.45 | 5.3 | [23:38] | |
| K86–K87 | 110.40 | 3.4 | [80:88] | |
| K7–K100 | 76.87 | 7.7 | [6:13]–[100:104] | |
| [92–104]–[6:8] | ||||
| [100–104]–[6:8] | ||||
| K99–K100 | 70.80 | 13.6 | [92–104] | |
The scores of the background cross-links were at a similar level to that of the false positive cross-links identified for decoy proteins. Therefore, the background cross-links can be controlled by the target-decoy search strategy.9,10 False discovery rate (FDR) at a certain threshold for cross-link identification can be calculated by FPcross-link/(TPcross-link + FPcross-link), where TPcross-link and FPcross-link are the numbers of true and false cross-link identifications above the threshold. All five nonbackground cross-links survived the false positive control and all of the background cross-links were filter at FDR of 5%.
Effect of Different Cross-Linking Conditions
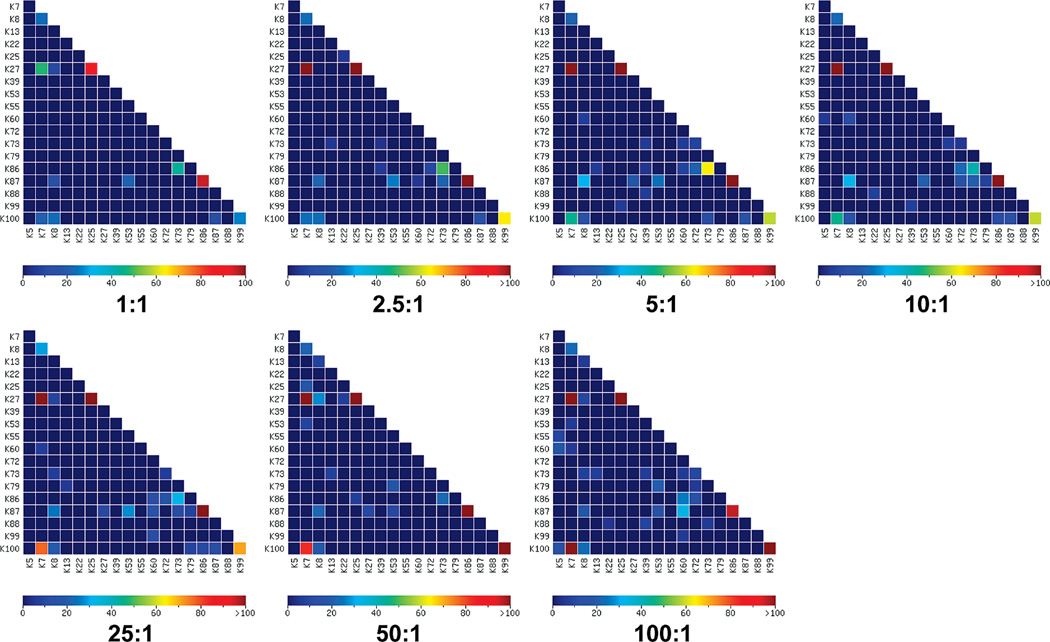
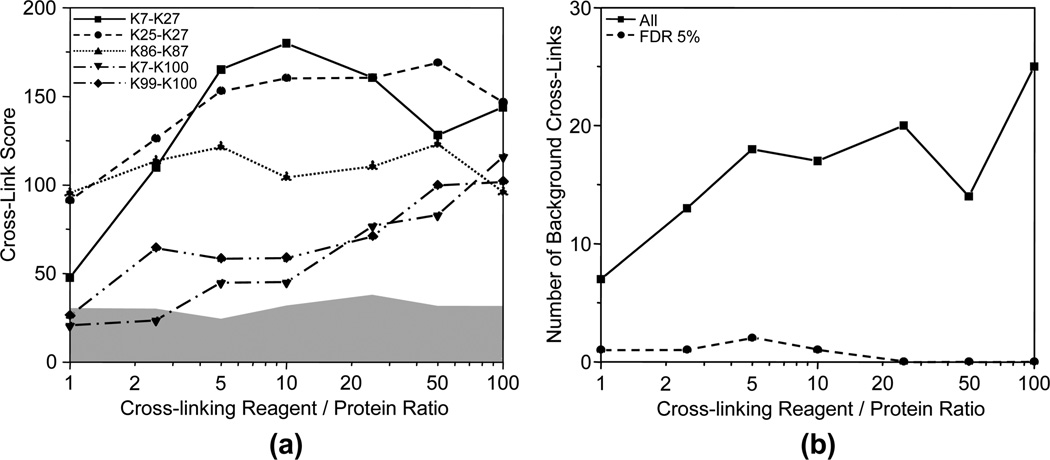
The effect of cross-linking reagent to protein ratio on the cross-linking experiment was evaluated by the Cytochrome C samples cross-linked by BS3 at different cross-linking reagent to protein ratios of 1:1, 2.5:1, 5:1, 10:1, 25:1, 50:1, and 100:1 with a final protein concentration of 0.12 mg/mL. The heat maps of the cross-links identified in these samples are shown in Figure 6. All five nonbackground cross-links were identified when the ratio is equal to or bigger than 25:1. However, two or more nonbackground cross-links were not identified with scores well above those of background cross-links when the ratio is smaller than 25:1. Figure 7a shows the dependence of the scores of the five nonbackground cross-links on the cross-linking reagent to protein ratio. It further confirms that the cross-link scores increase with the increase of the ratio for four nonbackground cross-links, i.e. K7–K27, K25–K27, K7–K100, and K99–100. The score of cross-link K86–K87 was independent of the ratio in a range of 1:1 to 100:1 at a protein concentration of 0.12 mg/ mL. Figure 7b shows the numbers of all the background cross-links and those at FDR of 5%. The number of total background cross-links increases with the increase of ratios when the ratio is smaller than 5:1. This trend becomes much less significant when the ratio is bigger than 5:1. However, the number of background cross-links at FDR of 5% shows no significant dependence on the ratio and stays low for all ratios. This indicates that the increased number of background cross-links at high ratios can be controlled at FDR of 5%. All background cross-links can be filtered at FDR of 5% when the cross-linking reagent to protein ratio is bigger than 25:1. In summary, high cross-linking reagent to protein ratio favors the cross-link determination in a ratio range of 1:1 to 25:1 at a final protein concentration of 0.12 mg/mL. The scores of nonbackground cross-links become saturated and the benefit of high ratios becomes less significant when the ratio is larger than 25:1.
Figure 6.
Heat maps of identified cross-links for Cytochrome C cross-linked by BS3 at different cross-linking reagent to protein standard ratios of 1:1, 2.5:1, 5:1, 10:1, 25:1, 50:1, and 100:1 with a final protein concentration of 0.12 mg/mL.
Figure 7.
(a) Scores of the five nonbackground cross-links, and (b) numbers of all background cross-links (-■-) and the background cross-links at FDR of 5% (-●-) for Cytochrome C cross-linked by BS3 at different cross-linking reagent to protein standard ratios of 1:1, 2.5:1, 5:1, 10:1, 25:1, 50:1, and 100:1 with a final protein concentration of 0.12 mg/mL. The gray area represents the score level of the false positive cross-links.
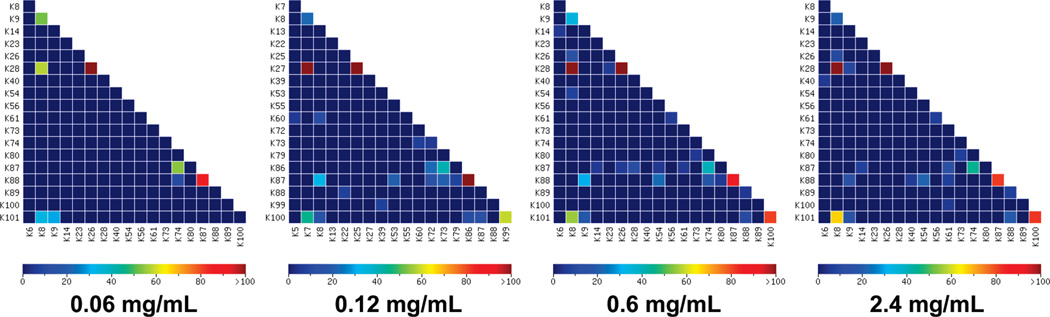
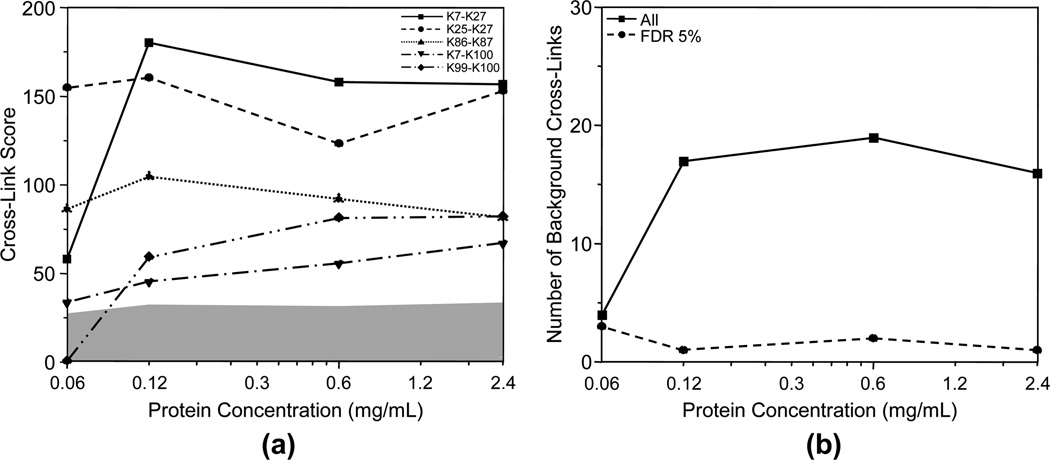
The effect of protein concentration on the cross-linking experiment was evaluated by the Cytochrome C samples cross-linked by BS3 at a cross-linking reagent to protein ratio of 10:1 with various protein concentrations of 0.06, 0.12, 0.60, and 2.4 mg/mL. The heat maps of the cross-links identified in the four samples with different protein concentrations are shown in Figure 8. It can be seen that the samples with higher protein concentration have higher scores for the five nonbackground cross-links. This improvement becomes much less significant when the protein concentration is higher than 0.60 mg/mL. This is confirmed by the dependence of the scores of the nonbackground cross-links on the protein concentration as shown in Figure 9a. The number of total background cross-links increases with the increase of protein concentration when the concentration is lower than 0.12 mg/mL. However, the number of background cross-links at FDR of 5% is independent of the protein concentration and stays low for all protein concentrations. This indicates that the increased number of background cross-links at high protein concentration can still be controlled and does not negatively affect the results. In summary, high protein concentration favors the cross-link determination experiment and this benefit becomes insignificant when the protein concentration is higher than 0.12 mg/ mL at a cross-linking reagent to protein ratio of 10:1.
Figure 8.
Heat maps of identified cross-links for Cytochrome C cross-linked by BS3 at a cross-linking reagent to protein ratio of 10:1 and different protein concentrations of 0.06, 0.12, 0.6, and 2.4 mg/mL.
Figure 9.
(a) Scores of the five nonbackground cross-links, and (b) numbers of all background cross-links (-■-) and the background cross-links at FDR of 5% (-●-) for Cytochrome C cross-linked by BS3 at a cross-linking reagent to protein ratio of 10:1 and different protein concentrations of 0.06, 0.12, 0.6, and 2.4 mg/mL. The gray area represents the score level of the false positive cross-links.
Cross-Link Search of A Complex Proteome Sample
A protein database can create much more theoretical cross-linked peptides than those without any cross-links (Table 1). The search space of a protein database in terms of number of theoretically calculated peptides can be increased by up to thousands of times when cross-links are considered. Due to the dramatically increased search space, searching tandem mass spectrometric data from complex proteome samples for cross-links against large protein databases is very challenging. It requires enormous computational resources and takes significantly longer time than those searches without considering cross-links. In order to make it practical to search cross-links in complex proteome samples, a staged search strategy is introduced in MassMatrix. Using this strategy, the search is performed in two stages to reduce the search space and time. In the first stage, the tandem mass spectrometric data set is searched against the large protein database without considering any cross-links. In the second stage, protein matches with significant scores from the first stage search will be searched for cross-links using the tandem mass spectrometric data. This strategy is valid given an assumption that the proteins with intact cross-links in the complex proteome samples can always create a certain number of peptides without any cross-links after enzymatic digestion. Those peptides without any cross-links can be used to identify the proteins in the first stage and the second stage search for cross-links can be focused on a limited number of proteins to reduce the search space and time. However, a staged database search violates the assumption used in the target-decoy search strategy. Therefore, the target-decoy search strategy cannot be used to estimate and control false positive rates in the staged database searches of cross-links. In future, with the advance of new computational hardware and the optimization of the search algorithm in MassMatrix, nonstaged search for cross-links in complex proteome samples against a large protein database will become feasible and the target-decoy search strategy can be used to estimate and control false positive rates.
Database search of cross-links in complex proteome samples using the staged search strategy is evaluated using the Escherichia coli proteome in vitro cross-linked by BS3. The samples from two replicate experiments were preseparated by SDS-PAGE and 41 bands were cut and in-gel digested with trypsin and analyzed by LC–MS/MS on a LTQ-FT mass spectrometer. The 41 tandem mass spectrometric data sets containing 341,613 MS/MS spectra in total were searched collectively as a whole against an Escherichia coli K-12 strain protein database containing 4,285 protein sequences. 3.76 × 107 peptides were calculated from the database in the staged search strategy and the total search time was 36.70 min. The complete list of identified protein matches in the search is provided in Supplementary Table 3 (Supporting Information) and the list of the identified cross-linked peptides is provided in Supplementary Table 4 (Supporting Information).
A total of 51 992 peptide-spectrum matches were identified for the collective search, of which only 3393 were cross-linked matches. This indicates that the cross-linked complex proteome sample was dominated by the peptides without any across-links due to the limited efficiency of cross-linking experiment. The high complexity of the sample, even after prefractionation by SDS-PAGE, further made the identification of cross-linked peptides and cross-links even more challenging. Among 456 identified significant protein matches only 59 proteins were identified with one or more significant cross-links with a score higher than 20. In general, these identified cross-links had lower scores than those identified in the previous Cytochrome C samples. The detailed results for the top 20 cross-links identified in the search are listed in Table 3. Among them, 12 cross-links from 9 proteins were verified for spatial plausibility by comparison with the published 3D structures. One cross-link had a distance of 16.62 Å longer than the cross-link length. This might be due to the difference between crystal and solution-phase structures of the protein. Another cross-link with a distance of 73.8 Å was not spatially plausible. However, this cross-link had a score lower than all other verified cross-links and was ranked 19th among all top 20 identified cross-links. Therefore, this cross-link might be a false discovery or background cross-link. The rest of 6 cross-links have no available structural data. In summary, only a limited number of cross-links can be identified in complex proteome samples using LC–MS/MS due to the dominating noncross-linked peptides and high sample complexity. This limitation is general to the cross-link determination using LC–MS/MS, not necessarily specific to the database search algorithm described herein. Therefore, it is preferable that the proteome samples are purified and/or enriched for cross-linked peptides.
Table 3.
List of Top 20 Cross-Links Identified by MassMatrix in the Complex Escherichia coli Proteome Sample Cross-Linked by BS3
| protein | Uni_Prot | PDB | cross-link | score | ranka | distance (Å) |
|---|---|---|---|---|---|---|
| 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase (dapD)2 | DAPD_ECOLI | – | K259–K263 | 44.73 | 15 | –b |
| Chaperone Hsp70; DNA biosynthesis; autoregulated heat shock proteins (dnaK) | DNAK_ECOLI | 2KHO | K245–K246 | 81.01 | 3 | 9.64 |
| K299–K304 | 47.61 | 11 | 10.12 | |||
| K246–K304 | 43.34 | 17 | 9.23 | |||
| K155–K562 | 41.54 | 19 | 73.8 | |||
| Glyceraldehyde-3-phosphate dehydrogenase A (gapA) | G3P1_ECOLI | 1DC5(dimer) | K213–K217 | 84.93 | 1 | 7.84e |
| Phosphoglyceromutase 1 (gpmA) | GPMA_ECOLI | 1E58 | K248–K250 | 46.09 | 14 | –d |
| Adenylate kinase activity; pleiotropic effects on glycerol-3-phosphate acyltransferase activity (adk) | KAD_ECOLI | 4AKE(dimer) | K157–K166 | 81.40 | 2 | 8.77e |
| N-acetylneuraminate lyase (aldolase); catabolism of sialic acid (nanA) | NANA_ECOLI | 1NAL(tetramer) | K71–K73 | 49.78 | 10 | 8.97e |
| Phosphoglucomutase (pgm) | PGM_ECOLI | – | K529–K533 | 55.60 | 7 | –b |
| PTS enzyme IIAB mannose-specific (manX) | PTNAB_ECOLI | 2JZH | K320–K323 | 62.57 | 5 | 12.98 |
| 50S ribosomal subunit protein L7/L12 (rplL) | RL7_ECOLI | 1CTF | K82–K85 | 46.89 | 12 | 8.69 |
| Putative enzyme (vacB) | RNR_ECOLI | – | K764–K816 | 51.36 | 9 | –b |
| 30S ribosomal subunit protein S3 (rpsC) | RS3_ECOLI | 2AVY(chain C) | K108–K147 | 74.21 | 4 | 16.62 |
| K79–K80 | 46.58 | 13 | 10.22 | |||
| 30S ribosomal subunit protein S4 (rpsD) | RS4_ECOLI | 2AVY(chain D) | K151–K156 | 57.30 | 6 | 6.45 |
| K150–K156 | 41.96 | 18 | 13.77 | |||
| 30S ribosomal subunit protein S19 (rpsS) | RS19_ECOLI | 2AVY(chain S) | K28–K29 | 43.59 | 16 | 5.96 |
| Membrane spanning protein required for outer membrane integrity (tolA) | TOLA_ECOLI | 1TOL | K82–K170 | 39.71 | 20 | –c |
| Orf hypothetical protein (yjgF) | YJGF_ECOLI | 1QU9 | K128–K131 | 53.58 | 8 | –c |
Ranking is based on cross-link scores.
No structure data available.
Partial structure data available but not including the identified cross-linkedlysine residues.
Coordinates of the required atoms for distance measurement are missing in PDB.
Shorted distance is used for a cross-link if the protein forms a dimer or tetramer
Conclusions
A new database search algorithm has been developed to identify intact cross-links in proteins and peptides by use of tandem MS data. The search algorithm is based on the validated statistical scoring models in MassMatrix and has been incorporated in MassMatrix for automated database search of intact cross-links in proteins and peptides from tandem MS data. The algorithm was tested using data sets collected on a LTQ-FT mass spectrometer for the tryptic digests of Cytochrome C cross-linked by BS3 at different experimental conditions. Five cross-links were identified by MassMatrix and their spatial plausibility was verified by comparison with the published three-dimensional structure of Cytochrome C. Cross-linking experiments at different cross-linking reagent to protein ratio and protein concentrations were also performed in this study. High cross-liking reagent to protein ratio favors the cross-link determination in ratio of 1:1 to 25:1 at a protein concentration of 0.12 mg/mL. This benefit of high ratios becomes less significant when the ratio is bigger than 25:1. In addition, protein concentration also has positive effect on the cross-link determination experiment when the protein concentration is lower than 0.12 mg/mL. The distributions of statistical scores for true and false positives and receiver operating characteristic analysis indicate that the algorithm is capable of discriminating true positive cross-linked peptide-spectrum matches from false ones. It has also been demonstrated that MassMatrix database search engine is capable of searching for intact cross-links in complex Escherichia coli proteome samples cross-linked by BS3.
Supplementary Material
Acknowledgment
We appreciate Dr. Mei-I Su for discussion and Mr. Chi-Shuen Chu and Ms. Ying-Shiuan Li for assistance in experiment setup. The study was funded by The Searle Funds at the Chicago Community Trust to the Chicago Biomedical Consortium, University of Illinois at Chicago, The Genomics Research Center and Institute of Biological Chemistry of Academia Sinica, The Ohio State University, National Institutes of Health CA107106 and CA101956 and the Leukemia and Lymphoma Society.
Footnotes
Supporting Information Available: The complete lists of true positive peptide-spectrum matches with cross-links and all cross-links identified by MassMatrix in the search of tryptic digest of cross-linked Cytochrome C by BS3 are provided in Supplementary Table 1 and Supplementary Table 2. Supplementary Figure 1 provides tandem mass spectra with matched peaks highlighted and a table of theoretical masses calculated from the MassMatrix fragmentation model for each peptide match with nonbackground cross-links in Cytochrome C. The complete lists of protein matches and significant cross-links identified in the collective search of the Escherichia coli proteome sample cross-linked by BS3 are provided in Supplementary Table 3 and Supplementary Table 4. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sinz A. Chemical cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes. J. Mass Spectrom. 2003;38:1225–1237. doi: 10.1002/jms.559. [DOI] [PubMed] [Google Scholar]
- 2.Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 3.Borch J, Jorgensen TJD, Roepstorff P. Mass spectrometric analysis of protein interactions. Curr. Opin. Chem. Biol. 2005;9:509–516. doi: 10.1016/j.cbpa.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 4.Back JW, de Jong L, Muijsers AO, de Koster CG. Chemical cross-linking and mass spectrometry for protein structural modeling. J. Mol. Biol. 2003;331:303–313. doi: 10.1016/s0022-2836(03)00721-6. [DOI] [PubMed] [Google Scholar]
- 5.Trakselis MA, Alley SC, Ishmael FT. Identification and mapping of protein-protein interactions by a combination of cross-linking, cleavage, and proteomics. Bioconjugate Chem. 2005;16(4):741–750. doi: 10.1021/bc050043a. [DOI] [PubMed] [Google Scholar]
- 6.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 7.Rinner O, Seebacher J, Walzthoeni T, Mueller LN, Beck M, Schmidt A, Mueller M, Aebersold R. Identification of cross-linked peptides from large sequence databases. Nat. Methods. 2008;5(4):315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadygov RG, Cociorva DC, Yates JR. Large-scale database searching using tandem mass spectra: Looking up the answer in the back of the book. Nat. Methods. 2004;1(3):195–202. doi: 10.1038/nmeth725. [DOI] [PubMed] [Google Scholar]
- 9.Huttlin EL, Hegeman AD, Harms AC, Sussman MR. Prediction of error associated with false-positive rate determinantion for peptide identification in large-scale proteomics experiments using a combined reversed and forward peptide sequence database strategy. J. Proteome Res. 2007;6:392–398. doi: 10.1021/pr0603194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 11.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence database using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 12.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 13.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J. Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 14.Schilling B, Row RH, Gibson BW, Guo X, Young MM. MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 2003;14:834–850. doi: 10.1016/S1044-0305(03)00327-1. [DOI] [PubMed] [Google Scholar]
- 15.Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell. Proteomics. 2007;6(12):2200–2211. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen T, Thaysen-Andersen M, Larsen N, Jorgensen FS, Houen G, Hojrup P. Determination of protein conformation by isotopically labelled cross-linking and dedicated software: Application to the chaperone, calreticulin. Int. J. Mass Spectrom. 2007;268:217–226. [Google Scholar]
- 17.Lee YJ, Lackner LL, Nunnari J, Phinney BS. Shotgun cross-linking analysis for studying quaternary and tertiary protein structures. J. Proteome Res. 2007;6(10):3908–3917. doi: 10.1021/pr070234i. [DOI] [PubMed] [Google Scholar]
- 18.Xu H, Freitas MA. Monte Carlo simulation based algorithms for analysis of shotgun proteomic data. J. Proteome Res. 2008;7(7):2605–2615. doi: 10.1021/pr800002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, Freitas MA. MassMatrix: A database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9(6):1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H, Zhang L, Freitas MA. Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine. J. Proteome Res. 2008;7(1):138–144. doi: 10.1021/pr070363z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blattner FR, Plunkett Gr, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 22.Paizs B, Suhai S. Fragmentation pathways of protonated peptides. Mass Spectrom. Rev. 2005;24:508–548. doi: 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]
- 23.Xu H, Freitas MA. A Mass Accuracy Sensitive Probability Based Scoring Algorithm for Database Searching of Tandem Mass Spectrometry Data. BMC Bioinform. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu H, Wang L, Sallans L, Freitas MA. A Hierarchical MS2/ MS3 Database Search Algorithm for Automated Analysis of Phosphopeptide Tandem Mass Spectra. Proteomics. 2009;9(7):1763–1770. doi: 10.1002/pmic.200800282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bushnell GW, Louie GV, Brayer GD. High-resolution three-dimensional structure of horse heart cytochrome c. J. Mol. Biol. 1990;214(2):585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.