Abstract
IP3Ks and IP6Ks each regulate specialized signaling activities by phosphorylating either InsP3 or InsP6 respectively; what are the molecular determinants of these different kinase activities? We address this question by determining the crystal structure of an enzymatic parallel to a “living fossil”: an Entamoeba histolytica hybrid IP6K/IP3K. Through molecular modeling and mutagenesis, we also extrapolated our findings to human IP6K2, which retains vestigial IP3K activity. Two structural elements, an α-helical pair and a rare, two-turn 310 helix, together forge a substrate-binding pocket with an open-clamshell geometry. InsP6 forms substantial contacts with both structural elements. Relative to InsP6, enzyme-bound InsP3 rotates 55° closer to the α-helices, which provide most of the protein’s interactions with InsP3. These non-overlapping substrate orientations are unprecedented for an inositol phosphate kinase. This arrangement also suggests an unusual evolutionary trajectory for a primordial kinase that could have favored efficient bi-functionality, prior to propagation of separate IP3Ks and IP6Ks.
Keywords: structure, kinases, specificity, inositol pyrophosphates, evolution
Introduction
Phosphate is a universal device for imposing specificity in cell signaling. The phosphate group’s very bulk establishes geometric constraints on ligand-protein and protein-protein interactions. Additionally, the phosphate’s negative charge at physiological pH also bestows specificity through ionic and hydrogen bonding to only certain amino acid residues. Signaling specificity is further enhanced through interactions that utilize multiple phosphates, such as those that are placed in various combinations around the six carbon inositol ring. The resulting molecules – inositol phosphates (InsPs) – each have a distinct three-dimensional phosphate pattern that can encode unique signaling properties. Within this family of signaling molecules there is a subgroup that possess functionally-significant, “high-energy” diphosphate groups: the inositol pyrophosphates (also known as diphosphoinositol polyphosphates) 1,2. These molecules, particularly InsP7 and InsP8, are now viewed as being of fundamental importance in all eukaryotes 2. Inositol pyrophosphates play pivotal roles in several aspects of cellular and organismic metabolic homeostasis 1,3-7, and they regulate interferon transcription in response to viral infection 8. Indeed, cell-signaling by the inositol pyrophosphates is considered to be among the most evolutionarily ancient functions of the entire inositol phosphate family 9. Thus, there is considerable interest in the kinases that synthesize InsP7 and InsP8.
For example, mammals express three isoforms of IP6Ks 10,11 that each add a second phosphate group to a pre-existing phosphate at the 5-position of the inositol ring. These kinases thereby convert InsP6 to 5-PP-Ins(1,2,3,4,6)P5(5-InsP7)12,13. The type 1 and type 2 isoforms (IP6K1 and IP6K2 respectively) have been the subject of a number of genetic studies with startling phenotypes. Silencing of IP6K1 in pancreatic β-cells inhibits insulin secretion 4. Gene disruption of murine IP6K1 increases the insulin sensitivity of target tissues 14, reduces spermatogenesis 14 and compromises hemostasis 15. Furthermore, in MEFs prepared from IP6K1−/− mice, DNA repair by homologous recombination is impaired 16, the epigenetic program is altered 17 and mitochondria are defective 3. A reduction in IP6K2 expression through an antisense strategy reduces apoptosis 18, while IP6K2−/− mice show increased susceptibility to carcinogen-induced squamous cell carcinoma 19. As IP6Ks have so many important functions, the determination of their structures is a key research goal: such information illuminates catalytic mechanisms, regulatory processes, and evolutionary relationships with other kinases, while also offering templates for the rational design of selective inhibitors as research tools or for therapeutic applications. However, IP6Ks have been stubbornly resistant to structural analysis for many years. Our goal has been to fill this significant void in our structural and molecular understanding, so as to further our insight into the entire inositol phosphate kinase family.
Entamoeba histolytica express an intriguing IP6K homologue (Genbank: XP_648490.2) that, secondary to its InsP6 kinase activity, also phosphorylates the 6-OH of Ins(1,4,5)P3 20 - an inositol phosphate multikinase (IPMK)-like activity 10,21,22. IPMK itself is positionally promiscuous in that it is a 3-, 5- and 6-kinase 23. Another non-specific inositol phosphate kinase is ITPK1, which phosphorylates the 1-, 5- and 6-positions around the inositol ring 24. Within the active sites of IPMK and ITPK1, the plane of the inositol rings in all of the alternate substrates are proposed to occupy the same spatial orientation, enabling each substrate to interact with a common set of protein-contacts 25-28. However, there has not previously been any direct structural confirmation of that hypothesis, because the published crystal structures of IPMK 26,28 and ITPK1 27,29 lack bound substrate. We therefore considered that structural analysis of the IP6K from E. histolytica could generate new ideas concerning specificity determinants of promiscuity within this enzyme family.
IP6Ks are members of a wider inositol phosphate kinase family (Pfam PF03770) that includes IPMKs and IP3Ks; these enzymes all share a PxxxDxKxG (“PDKG”) catalytic motif 10,21. Phylogenetic analysis 9 has led to the hypothesis that this kinase family arose from a primordial IP6K precursor. However, such an evolutionary pathway would be highly unusual, at least according to current thinking 30. The issue of concern is that the original substrate (InsP6) for the putative progenitor kinase is both larger and substantially more polar than the substrates for the descendant kinases: the InsP3 and InsP4 that are phosphorylated by IPMKs 26,28 and the InsP3 that is specifically phosphorylated at the 3-OH by IP3Ks 31,32. The traditional viewpoint is that improvements in the efficiency of catalysis of the smaller substrates (InsP3/InsP4) would evolve through compression of the active site 30. However, that would generally be expected to reduce activity against the larger InsP6 molecule - a “negative trade-off” - that would impede organismic fitness and thereby select against the emergence of the independent InsP3/InsP4 kinase activities 30. Such restrictions would seem inevitable if all substrates interacted with common structural elements. Nevertheless, the significant InsP3 kinase activity of E. histolytica IP6K 20 suggests it has somewhat overcome the constraints of negative trade-off; structural analysis could reveal how this was accomplished. We now describe several crystal complexes of the IP6K from E. histolytica, including those that contain either large (Ins(1,3,4,5,6)P5 / InsP6) or small (Ins(1,4,5)P3) substrates. Furthermore, we extrapolate our findings to mammalian IP6Ks by site-directed mutagenesis of the human IP6K2.
Results
Ins(1,4,5)P3 and InsP6 phosphorylation by EhIP6KA
In our study, a recently characterized IP6K expressed by E. histolytica 20 (Genbank: XP_648490.2) is referred to as EhIP6KA, in order to distinguish it from two other gene products of E. histolytica that are currently also annotated as IP6Ks in Genbank. HPLC of the InsP7 synthesized by EhIP6KA (solid line, Fig. 1A) found it to co-elute with the 5-InsP7 produced by HsIP6K2 12 (dotted line, Fig. 1A). A previous study 20 also concluded from HPLC that EhIP6KA synthesizes 5-InsP7. Nevertheless, current HPLC methodology alone does not completely resolve all possible InsP7 isomers 33,34. Furthermore, there have been reports that amoeba contain atypical isomers of inositol pyrophosphates 35. Considering that positional specificity is such a central feature of our study (see the Introduction), we employed NMR for definitive characterization of the InsP7 product 12,13.
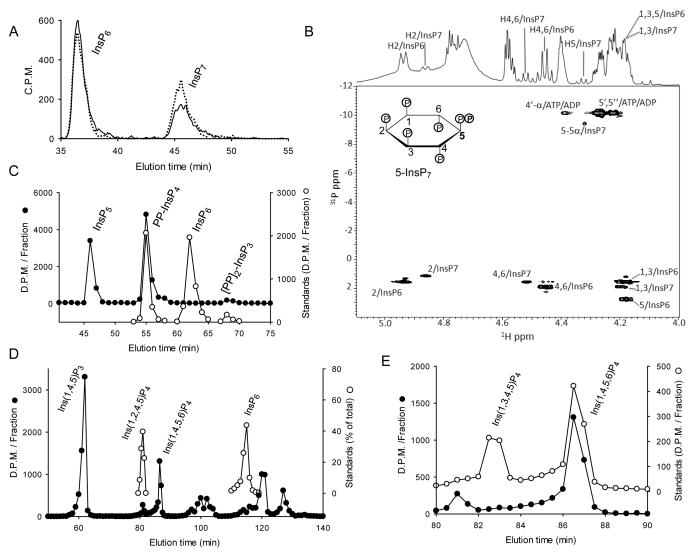
Fig. 1. HPLC and NMR analysis of the products of Ins(1,4,5)P3 and InsP6 phosphorylation by EhIP6KA.
A, HPLC analysis (Partisphere SAX, Gradient 1; see Experimental Procedures) of InsP6 phosphorylation by either EhIP6KA (57 ng, 20 min; solid line) or HsIP6K2 (7.6 ng, 20 min; dotted line). B, NMR analysis of the products of the reaction between InsP6 and ATP catalyzed by EhIP6KA; the Fig. depicts an overlay of the 1D 1H and 2D 1H/31P HSQC spectra. The acquisition parameters are given under Experimental Procedures. Panel C, HPLC analysis (Partisphere SAX, Gradient 2; see Experimental Procedures) of the phosphorylation of approx. 9000 D.P.M. [14C]Ins(1,3,4,5,6)P5 by EhIP6KA (6 μg, 20 min, 50 μl; filled circles). Also shown are the elution of [3H]-labeled internal standards of PP-InsP4 and [PP]2-InsP3, as well as InsP6 (open circles). Panels D and E depict representative HPLC assays (90 min; 200 μl) that contained approx. 10,000 D.P.M. [3H]Ins(1,4,5)P3 plus 2 μg EhIP6KA. After quenching and neutralization, products (filled circles) were analyzed by Q100 SAX HPLC (Gradient 3). Panel D includes the elution positions of standards of [3H]Ins(1,2,4,5)P4 and [3H]InsP6 (open circles), determined in parallel HPLC runs. Panel E zooms in on the InsP4 region of the chromatograph; elution of internal [14C] standards of Ins(1,3,4,5)P4 and Ins(1,4,5,6)P4 are also shown (open circles).
To prepare sufficient quantities of pure InsP7 for NMR analysis, we adopted a strategy that has proved useful in the past 12, namely, to analyze reaction mixtures (Fig. 1B). The 1H chemical shifts of the unreacted InsP6 and the product InsP7 were assigned by comparison to reported InsP6 chemical shifts 13 and from an analysis of the 2D 1H-1H COSY spectrum of the reaction mixture (Supplementary Table 1). The proton resonances from InsP6 and InsP7 are labeled in the 1D 1H spectrum (Fig. 1B). The integral from the InsP6 protons in the 4/6 positions is approximately 3 times larger than the corresponding integral arising from the PPInsP5 product, indicating that about 25% of the InsP6 was phosphorylated. To verify the assignments and differentiate the InsP6 and InsP7 resonances, the 1D 1H and 31P spectra were reacquired after spiking the sample with an additional 100 μl of 5 mM InsP6. The major difference between the InsP6 and InsP7 in the new 1H spectrum is that the resonance from H-5 is shifted to higher chemical shift and no longer overlaps the H-1,3 resonances as it does in InsP6 (see also Supplementary Table 1).The remainder of the 1H resonances exhibit less dramatic changes relative to the corresponding resonances in InsP6. The H-1,3 and H-4,6 resonances of InsP7 are equivalent due to the symmetry of molecule. These data are consistent with the site of phosphorylation being at the 5-position. With the 1H chemical shift assignments in hand, the 31P chemical shifts of InsP7 were assigned from a combined analysis of 2D 1H-31P HSQC and 31P-31P COSY spectra (Supplementary Table 1). The HSQC spectrum correlates the 1H shifts at each site to the 31P shifts of the phosphate on the same carbon, and the COSY spectrum correlates the 31P shifts of the two 31P nuclei in the diphosphate moiety. The H-2, H-1,3 and H-4,6 proton chemical shifts correlate to 31P chemical shifts from the monophosphate groups in InsP7, and the H-5 proton shift correlates to the 31P chemical shift of the α-phosphate of the only diphosphate group (Fig. 1B), so we conclude that the site of phosphorylation is the 5-position. Note the two other α-phosphate resonances arise from the ADP product and from unreacted ATP (Fig. 1B). The β-phosphate chemical shift of InsP7 is assigned from its correlation to the α-phosphate chemical shift in the 31P-31P COSY spectrum. Thus, our NMR data reinforce the conclusion reached by HPLC that EhIP6KA, like mammalian IP6Ks, converts InsP6 to 5-InsP7.
Mammalian IP6Ks also phosphorylate Ins(1,3,4,5,6)P5, yielding PP-InsP412,36. We found that EhIP6KA likewise phosphorylates Ins(1,3,4,5,6)P5, and the major product co-elutes with a PP-[3H]InsP4 standard 12,36; no InsP6 was formed (Fig. 1C). A small amount of [PP]2-InsP312,36 also accumulated (Fig. 1C). The first order rate constant for Ins(1,3,4,5,6)P5 phosphorylation (0.15 ± 0.006 s−1 mg−1, n=3) is 40-fold lower than that for InsP6(6.1 ± 0.64 s−1 mg−1, n=3). The rate of phosphorylation of Ins(1,4,5)P3, another EhIP6K substrate 20, was 0.028 ± 0.005 s−1 mg−1 (n=3), which is 220-fold slower than that for InsP6. HPLC indicates that Ins(1,4,5,6)P4 is the major InsP4 product formed from Ins(1,4,5)P3 (Fig. 1D,E and ref20). A new finding in our study is the accumulation of an additional InsP4 isomer that co-elutes with a standard of Ins(1,2,4,5)P4 (Fig. 1D). The phosphorylation of an inositol phosphate at the 2-position has only previously been observed for IP5K 26,37. We also confirmed that there were no pyrophosphorylated molecules associated with the InsP4 products, because they were not dephosphorylated upon their incubation with DIPP1 (Supplementary Fig. 1), an inositol pyrophosphate phosphatase that removes the β-phosphate from the diphosphate group 36,38.
Overall structure of EhIP6KA
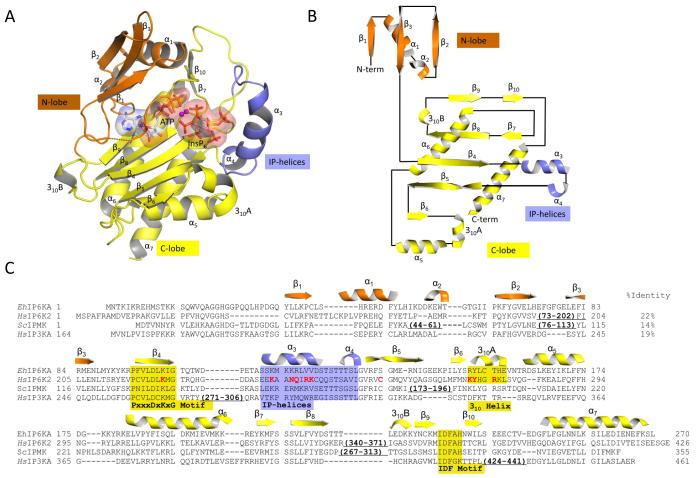
The structure of EhIP6KA (Fig. 2A,B,C; Table 1) was determined through two approaches. First, the structure of the fusion protein MBP-EhIP6KA (apo-enzyme; residues 32-270, space group P212121) was determined by a molecular replacement approach using MBP (1ez9) and IP3K (1w2c) as search models. That information was then used for further molecular replacement in order to solve the structures of crystal complexes that bound ATP plus either Ins(1,4,5)P3, Ins(1,3,4,5,6)P5, or InsP6 (residues 27-270, space group I4122), and a crystal of the apo-enzyme (residues 20-270, space group P21). In the latter, the N-terminal residues 20-29 were disordered. For each asymmetric unit, our data are consistent with one molecule of MBP-EhIP6KA in space group P212121, one molecule of substrate-bound EhIP6KA in space-group I4122, and two molecules of apo-EhIP6KA in space group P21. The latter crystal structure was analyzed with the Protein Interfaces, Surfaces and Assemblies service (PISA; https://www.ebi.ac.uk/pdbe/pisa/) which estimated an interface area of 814.7 Å2 and total gained free energy upon interface formation of −10.8 kcal/mol. It was also assigned a complexation significance score of zero, indicating that the apo structure is a monomer. Furthermore, gel filtration indicates that apo-EhIP6KA is a monomer in solution.
Fig. 2. Overall structure of EhIP6KA.
A, Ribbon-plot of EhIP6KA structure. ATP and InsP6 are shown as sticks within a transparent surface. Two Mg atoms are depicted as magenta balls. B, Topology diagram. C, A manual alignment of amino-acid sequences (EhIP6KA, XP-648490.2; HsIP6K2, NP_057375.2; ScIPMK, NP_010458.3; HsIP3KA, NP_002211.1), guided by the structural elements that have been observed in crystal structures, and in the case of HsIP6K2, secondary structural predictions. Outside of the conserved catalytic core of the HsIP6K2 are two significant insertions that are omitted from the alignment. The first of these (residues 73-202) includes a specialized HSP90-binding domain 54. The second insertion (residues 340-371) includes a CK2-regulated, ubiquitination motif 55. The secondary structural elements from EhIP6KA are depicted above its sequence and are color-coded orange for the N-lobe, yellow for the C-lobe and blue for the IP-helix. Structural elements that directly participate in substrate interactions are highlighted by shading. Residues in HsIP6K2 that were selected for mutagenesis are colored red. PDB codes for EhIP6KA are 4O4B, 4O4C, 4O4D, 4O4E, 4O4F.
Table 1.
Data collection and structure refinement statistics
| PDB Accession IDs | 4O4B | 4O4C | 4O4D | 4O4E | 4O4F |
| EhIP6K1 residues | MBP-32-270 | 20-270 | 27-270 | 27-270 | 27-270 |
| Ligand | Apo | Apo | ATP- Ins(1,4,5)P3 |
ATP-Ins(1,3,4,5,6)P5 | ATP-InsP6 |
| Data collection | |||||
| Space group | P212121 | P21 | I4122 | I4122 | I4122 |
| Cell dimensions a,b, c (Å), β (°) |
117.63 51.98 117.33 |
43.42 128.37 52.53, β=114.3° |
102.74 102.74 111.74 |
103.00 103.00 110.98 | 102.23 102.23 109.49 |
| Resolution (Å)* | 50-1.8 (1.83) | 50-1.9 (1.93) | 50-2.1 (2.14) | 50-1.9 (1.93) | 50-1.7 (1.73) |
| Rsym * | 0.057 (0.457) | 0.078 (0.235) | 0.086 (0.551) | 0.049 (0.527) | 0.053 (0.540) |
| I/σI* | 39.6 (3.1) | 25.0 (3.6) | 29.1 (3.4) | 42.9 (2.6 | 56.7 (2.4) |
| Completeness (%)* | 99.4 (99.4) | 98.6 (93.3) | 99.4 (100.0) | 99.3 (93.9) | 96.4 (71.6) |
| Redundancy * | 5.0 (4.8) | 3.7 (3.0) | 6.7 (6.2) | 8.1 (6.3) | 9.8 (5.2) |
| Refinement | |||||
| Resolution(Å)* | 1.8 (1.84) | 2.2 (2.27) | 2.1 (2.16) | 1.9 (1.95) | 1.7 (1.74) |
| No. reflections | 64104 | 26476 | 16743 | 22265 | 29589 |
| Rwork * | 19.6 (27.7) | 22.9 (24.3) | 17.3 (20.2) | 18.3 (25.4) | 19.1(28.4) |
| Rfree * | 22.1 (34.1) | 28.6 (28.2) | 22.7 (25.2) | 21.9 (29.5) | 22.2(36.2) |
| No. atoms | |||||
| Protein | 5010 | 4039 | 2080 | 2084 | 2075 |
| Ligand/ion | 57 | 65 | 69 | ||
| Solvent | 654 | 328 | 198 | 200 | 200 |
| B-factors (Å2) | |||||
| Protein | 16.8 | 19.8 | 30.1 | 23.8 | 23.7 |
| Ligand/ion | 52.6 | 36.9 | 39.3 | ||
| Solvent | 47.1 | 29.7 | 46.3 | 41.5 | 42.1 |
| R.m.s. deviations | |||||
| Bond length(Å) | 0.006 | 0.007 | 0.012 | 0.011 | 0.01 |
| Bond Angle (°) | 1.10 | 1.18 | 1.71 | 1.73 | 1.83 |
The numbers in parentheses are for the highest resolution shell.
Analysis of the overall fold of EhIP6KA (Fig. 2A,B) offers the first description of some architectural elements that are shared with IPMKs and IP3Ks, while also revealing some molecular differences that may contribute to individual catalytic preferences. Among conserved elements, we identified domains in EhIP6KA that are similar to the so-called N- and C-domains that comprise the ATP-binding sites in both IPMK 26,28 and HsIP3KA 31,32. First, an N-terminal lobe in EhIP6KA can be designated from residues 28-91, which consist of three antiparallel β-strands; the β1 and β2 strands are connected by two short helices α1 and α2. Second, we can designate a C-terminal lobe (residues 92-102 and 132-270), which is an α+β fold with five, central antiparallel β-strands including β4-6, as well as β8 and β9 (connected by a 3-residue 310 helix), a pair of small antiparallel β-strands (β7 and β10), and three α–helices (α5-α7). Also in the C-lobe of EhIP6KA, between the β6 strand and the α5 helix, is an unusually long (6-residue), two-turn 310 helix (310A; Fig. 2A) that is not present in either IPMK 26,28 or HsIP3KA 31,32. A 310 helix of this length occurs relatively rarely because, being thinner and more tightly wound that classical α-helices, it would usually be too unstable 39,40. However, as we describe below, the highly-specialized nature and orientation of the amino-acid side chains in this 310 helix not only stabilize this structural element, but also make it functionally integral to substrate binding by EhIP6KA.
Description of the nucleotide binding region of EhIP6KA
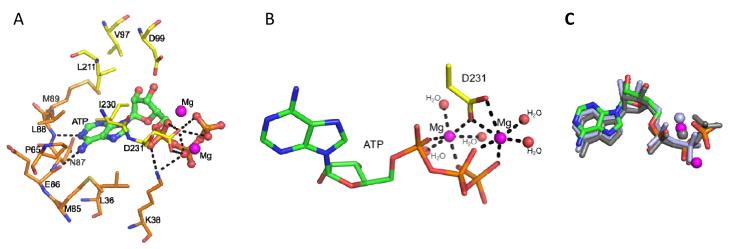
We observed that ATP is sandwiched between the N- and C-lobes of EhIP6KA (Fig. 2A); nucleotide binding did not alter protein conformation. Within a conserved IDF tripeptide (Fig. 2C) is Asp231, which co-ordinates with two Mg ions and makes polar contacts with the nucleotide phosphates (Fig. 3A,B). The orientation of the nucleotide in EhIP6KA is superimposable upon its position in the other kinases that share the PDKG motif, except for the more flexible γ-phosphate of ATP (Fig. 3C; Supplementary Fig. 2), which may contribute to the unusually low affinity of IP6Ks for this nucleotide 41. The adenosine moiety of ATP is docked in a hydrophobic pocket in EhIP6KA that is surrounded by residues Leu36, Pro65, Met85, Asn87, Met89 from the N-lobe and Leu211 and Ile230 from the C-lobe (Fig. 3A,4A). The N1 and N6 atoms of adenine are hydrogen bonded with the amide nitrogen and carbonyl oxygen of Leu88 and Glu86 respectively, which anchors the adenosine in the hydrophobic pocket (Fig. 3A,4A). The ATP-ribose group is loosely confined by several Van der Waals contacts with Val97, Asp99, Leu211, and Ile230. Lys38 forms two hydrogen bonds with oxygen atoms from the α- and β-phosphates (Fig. 3A,4A).
Fig. 3. Nucleotide binding by EhIP6KA.
A, ATP is depicted as a stick and ball model. Two Mg atoms are depicted as magenta spheres. Polar contacts are shown with dashed lines. Amino acids are shown as stick. B, Metal coordination. Two Mg atoms are depicted as magenta spheres. Water molecules are depicted as red spheres. The structure of ATP and Asp231are shown as stick models. Polar contacts to coordinate with Mg atoms are shown with dashed lines. C, The orientation of the EhIP6KA-bound nucleotide (green for carbon, red for oxygen, blue for nitrogen and orange for phosphorus atoms) and Mg atoms (magenta spheres) are superimposed upon that for HsIP3KA (grey stick represent AMPPNP (ATP analog); grey spheres represent Mg), and ScIPMK (light blue stick and spheres represent ADP and Mg).
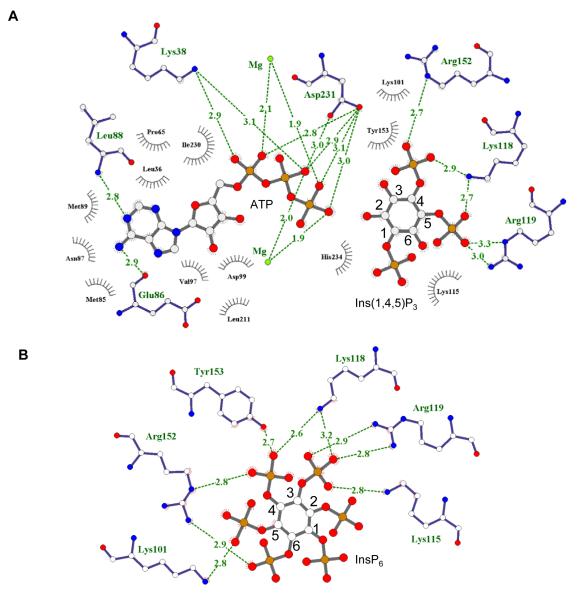
Fig. 4. Ligplots showing interactions of EhIP6KA with ATP and inositol phosphates.
Ligplots are shown for A, ATP and Ins(1,4,5)P3 and B, InsP6. Hydrogen bonds are shown in green dashed lines; bond distances are denoted in Angstroms. Residues that make hydrophobic interactions are depicted as grey eyelashes (cutoff distance is 3.9 Å). Atoms are shown white for carbon, red for oxygen, blue for nitrogen and orange for phosphorus.
The Molecular determinants of Inositol Phosphate binding
We have obtained crystal structures of EhIP6KA in complex with three different substrates: Ins(1,4,5)P3, Ins(1,3,4,5,6)P5, and InsP6 (Fig. 4A,B, 5A,B,C,D, Supplementary Fig. 3). Simulated annealing omit-maps and 2Fo-Fc maps show clear density of the substrate’s inositol ring; the individual phosphate groups and their specific positions can each be identified (Supplementary Fig. 3).
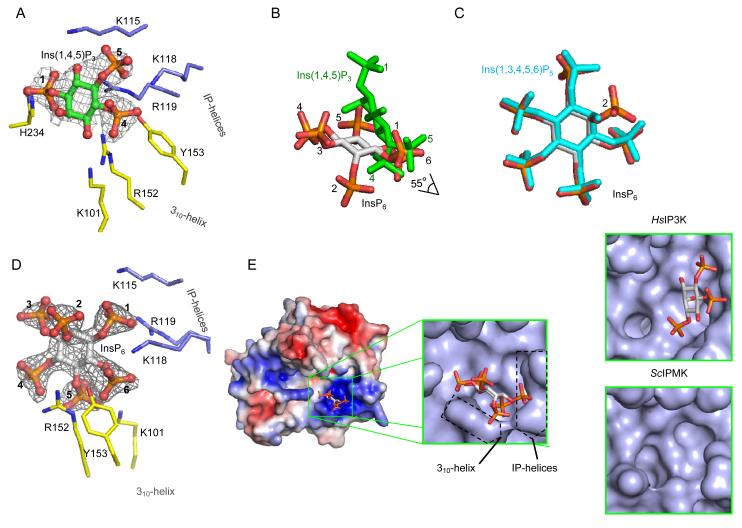
Fig. 5. Inositol phosphate binding site for EhIP6KA.
A, Binding of Ins(1,4,5)P3 (stick and ball model; green for carbon, red for oxygen and orange for phosphorus atoms. The phosphate groups are numbered). The refined 2Fo-Fc electron density map is contoured at 1.3 σ. B, Overlay of InsP6 (grey stick model with orange and red phosphate groups) and Ins(1,4,5)P3 (green stick). C, Overlay of InsP6 and Ins(1,3,4,5,6)P5 (cyan stick). D, Binding of InsP6 (stick and ball model; green for carbon, red for oxygen and orange for phosphorus atoms). The phosphate groups are numbered. Amino acids are shown as stick models. The refined 2Fo-Fc electron density map is contoured at 1.3 σ. E, Electrostatic surface plot with blue and red coloration to respectively indicate positive and negative electrostatic potentials at physiological pH. In the magnified surface representation, the positions of the 310 helix and IP-helices are highlighted. Bound InsP6 is depicted as a stick model. Also shown are surface representation of Ins(1,4,5)P3 in the active site of HsIP3K (data from 32) and the active site of ScIPMK (data from 26; crystals did not contain substrate).
The binding site for InsP6, which lies in a shallow depression in the enzyme’s surface, can be likened to an “open-clamshell”. One jaw is represented by the IP helices, the other jaw comprises the 310A helix; note that neither HsIP3KA nor IPMK possess an equivalent to the 310A helix (Fig. 5E). The IP helices and the 310 helix are not directly connected, but they approach within sufficient proximity of each other to provide a metaphorical “hinge” to the clamshell. Here, the 1- and 6-phosphates of InsP6 make contacts with Lys115, Lys118 and Arg119 in the α-3/α-4 helical pair (Fig. 4B, 5D). Nevertheless, the plane of the inositol ring of InsP6 runs parallel to the contour of the 310A helix (Fig. 5E), enabling the 4- and 6-phosphates to interact with Arg152 and Tyr153 (Fig. 4B,5D). These contacts appear to be stabilized by cation-pi interactions between Arg152 and Tyr153, which in turn are facilitated by the specific architecture of the 310 helix (Supplementary Fig. 4).
Ins(1,3,4,5,6)P5 is also a substrate for EhIP6K (Fig. 1C,D). Indeed, Ins(1,3,4,5,6)P5 and InsP6 occupy a similar orientation within the active site (Fig. 5C), indicating how both can be phosphorylated at the same position. IP6Ks can also phosphorylate 1-InsP7 13. Assuming that InsP6 and 1-InsP7 also occupy similar orientations within the active site, molecular modeling indicates that the latter’s β-phosphate projects into the bulk phase and should not greatly influence substrate binding.
While it is the 5-phosphate group on InsP6 that is diphosphorylated by EhIP6KA (Fig. 1B), it is either the 2- or the 6-OH of Ins(1,4,5)P3 that are phosphorylated (Fig. 1D,E). Positional promiscuity in other inositol phosphate kinases has been rationalized by proposing alternative substrate-binding modes, in which the plane of the inositol ring occupies the same spatial orientation relative to the active site, enabling each substrate to share the same protein-contact points 25-28. This is not the case for EhIP6KA. Instead, a unique adaptation of EhIP6KA lies in the nature of its contacts with Ins(1,4,5)P3 that tilt the plane of the inositol ring by 55° relative to that of InsP6 (Fig. 5B). Thus, near the clamshell hinge, the 5-phosphate of Ins(1,4,5)P3 somewhat overlaps the space that would be occupied by the 1-phosphate of InsP6 (Fig. 5B). However, at the open end of the clamshell, the 1-phosphate of Ins(1,4,5)P3 is positioned 7.5 Å away from the location of the 3-phosphate of InsP6 (Fig. 5B). In fact, steric restrictions prevent InsP6 from occupying the same space as Ins(1,4,5)P3 in the substrate-binding pocket.
The 310 helix makes only limited contact with Ins(1,4,5)P3: the 4-phosphate has just one hydrogen bond with Arg152 and there are weak Van der Waals interactions with Tyr153 (Fig. 4A,5A). Instead, the plane of the inositol ring in Ins(1,4,5)P3 is tilted closer to the IP helices, which are what this substrate mainly interacts with. Lys118 and Arg119 make contacts with the 4- and 5-phosphates of Ins(1,4,5)P3 (Fig. 4A, 5A). The 1-phosphate, 6-hydroxyl and 5-phosphate of Ins(1,4,5)P3 also have Van der Waals interactions with Lys115 (Fig. 4A). The 2-OH of Ins(1,4,5)P3 is positioned 3.7 Å from the γ-phosphate of ATP (Supplementary Fig. 5), which is close enough to permit an in-line phosphoryl-transfer reaction 42. Indeed, our HPLC analysis revealed Ins(1,2,4,5)P4 to be a product of Ins(1,4,5)P3 phosphorylation (Fig. 1D,E).
Furthermore, we remodeled the orientation of the inositol ring in enzyme-bound Ins(1,4,5)P3, by flipping it 180° across its 1/4 axis, whereupon the 6-OH of Ins(1,4,5)P3 was then positioned 2.9 Å from the γ-phosphate of ATP (Supplementary Fig. 5). This rationalizes the 6-kinase activity against Ins(1,4,5)P3 by EhIP6KA (Fig. 1D,E). Thus, Ins(1,4,5)P3 can be held in the active site of EhIP6KA in two alternative, productive orientations, yielding either Ins(1,2,4,5)P4 or Ins(1,4,5,6)P4 as products.
Another interesting observation is that, within the ligand-binding pocket of EhIP6KA, the orientation of the plane of the inositol ring of Ins(1,4,5)P3 shows a remarkable degree of spatial conservation with that for HsIP3KA (Fig. 5A,E, Supplementary Fig. 5 and ref 32); one inositol ring is simply flipped 180° across its 2,3/5,6 axis relative to the other so that, in HsIP3KA, the 3-OH that is phosphorylated lies 4.6 Å from the γ-phosphate of ATP (Supplementary Fig. 5), which is within the distance that permits an in-line, phospho-transfer reaction 42. Thus, our data suggest that EhIP6KA be considered an enzymatic parallel to a “living fossil”, that illuminates the potential nature of a primordial InsP6/InsP3 kinase. In this putative protein, the open-clamshell topology of the active site provides a partial diversification of specificity determinants for Ins(1,4,5)P3 and InsP6, prior to its propagation into separate enzymes that could specialize in either of the two catalytic activities. We used mutagenesis to pursue this idea (see below).
Molecular modeling and mutagenesis of a human IP6K
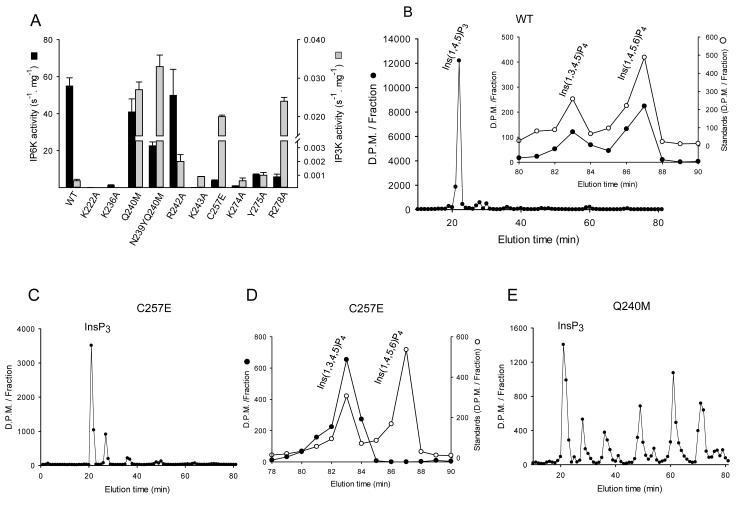
So as to consolidate our structural work with EhIP6KA, and also to extrapolate it to mammalian IP6Ks, we turned to HsIP6K2 as a model for mutagenic studies. Interestingly, we found that HsIP6K2 harbors a vestigial Ins(1,4,5)P3 kinase activity, some five orders of magnitude weaker than the InsP6 activity (Fig. 6A). The products of Ins(1,4,5)P3 phosphorylation are Ins(1,3,4,5)P4 and Ins(1,4,5,6)P4 (Fig. 6B). Although the Ins(1,4,5)P3 kinase activity was relatively low, it was sufficient to permit us to pursue the specificity determinants for each substrate.
Fig. 6. HPLC analysis of the kinase activities of wild-type and mutant IP6Ks.
A, InsP6 kinase (black bars) and Ins(1,4,5)P3 kinase (grey bars) activities of wild-type HsIP6K2 and the indicated mutants (means ± standard errors, n ≥3). Panels B shows representative HPLC analysis (Partisphere SAX; Gradient 2) of Ins(1,4,5)P3 kinase activities of wild-type HsIP6K2 (8 μg, 7 hr). The inset to panel B shows HPLC analysis (Q100 SAX; Gradient 3) of the InsP4 products formed by the wild-type enzyme (closed circles) together with the elution of internal standards of [14C]-Ins(1,3,4,5)P4 and [14C]-Ins(1,4,5,6)P4 (open circles). Panel C shows a representative HPLC analysis (Partisphere SAX; Gradient 2) of Ins(1,4,5)P3 kinase activity of a Cys257Glu mutant of HsIP6K2 (5 μg, 1 hr). Panel D shows a representative HPLC analysis (Q100 SAX; Gradient 3) of the InsP4 products formed by a Cys257Glu mutant of HsIP6K2 (5 μg, 45 min). Panel E shows a representative HPLC analysis of Ins(1,4,5)P3 kinase activity of a Gln240Met mutant of HsIP6K2 (11 μg, 7 hr).
For a positive control in the mutagenic experiments, we prepared a Lys222Ala mutant of HsIP6K2. This residue (equivalent to Lys101 in EhIP6KA; Fig. 2C, 4B, 5A,5D), lies within the conserved PxxxDxKxG catalytic center (Fig. 2C). A HsIP6K2 construct in which this residue was mutated to Ala has previously been used as a kinase-impaired control when studying the effects of over-expression of wild-type enzyme in intact cells 43. Nevertheless, to our knowledge, the degree to which this mutation inhibits recombinant enzyme activity has not previously been published. We found that the K222A mutant of HsIP6K2 exhibits negligible catalytic activity (Fig. 6A).
Our structural analysis of EhIP6KA indicates that Lys115 in the IP-helices of EhIP6KA has multiple interactions with both substrates (Fig. 4,5). We mutated the corresponding Lys236 in HsIP6K2 (Fig. 2C) to Ala; catalytic activity against both substrates was strongly reduced (Fig. 6A). Lys118 and Arg119 are two other residues in the IP-helices of EhIP6KA that make contact with both substrates (Fig. 4,5). These residues are not precisely replicated in HsIP6K2, according to both our sequence alignments (Fig. 2C) and our molecular model of the mammalian kinase (Supplementary Fig. 6A). Instead, our modeling indicates that Lys243 in HsIP6K2 has functionally significant interactions with the 6-phosphate of InsP6 (Supplementary Fig. 6A). Indeed, an Lys243Ala mutation in HsIP6K2 decreased InsP6 kinase activity 600-fold (Fig. 6A) without affecting Ins(1,4,5)P3 phosphorylation. These data speak to the specialization of the InsP6 kinase activity of HsIP6K2.
We next used mutagenesis to test our hypothesis drawn from structural data that the 310A helix in IP6Ks is particularly important for binding InsP6. Arg152 and Tyr153 in the 310A helix of EhIP6KA align with Lys274 and Tyr275 in the equivalent region of HsIP6K2 (Fig. 2C). We found that Lys274Ala and Tyr275Ala mutants of HsIP6K2 exhibited a decrease in InsP6 kinase activities of 55- and 8-fold respectively; the fact that these two mutations did not affect Ins(1,4,5)P3 kinase activities (Fig. 6A) confirms our conclusion that they play only a minor role in binding Ins(1,4,5)P3. Interestingly, the 310A helix of HsIP6K2 additionally contains an Arg residue at position 278 for which there is no equivalent in EhIP6KA (Fig. 2C). Molecular modeling of HsIP6K2 suggests Arg278 might contribute positive charge to the substrate binding pocket, and/or stabilize the 310 helix through cation-pi interactions with Tyr274 (Supplementary Fig. 6B). Consistent with these ideas, an Arg278Ala mutant of HsIP6K2 exhibited a 9-fold reduction in InsP6 kinase activity compared to wild-type enzyme (Fig. 6A). Interestingly, the Arg278Ala mutant also showed a dramatic increase in Ins(1,4,5)P3 kinase activity (Fig. 6A). It is possible Arg278 has a negative impact on Ins(1,4,5)P3 phosphorylation by attracting it into a non-productive orientation.
The electronegative Glu333 in the C-lobe of HsIP3KA 32 (Supplementary Fig. 6C) is replaced by Ser136 in EhIP6KA and Cys257 in HsIP6K2 (Fig. 2C). Structural modeling of this region of HsIP6K2 (Supplementary Fig. 6B,C) indicates a Cys257Glu replacement in HsIP6K2 would cause electrostatic repulsion of InsP6 but not Ins(1,4,5)P3. Indeed, we prepared a Cys257Glu mutant of HsIP6K2 and it showed 14-fold lower InsP6 kinase activity than wild-type enzyme (Fig. 6A). Moreover, this mutant showed a 33-fold increase in the rate of phosphorylation of Ins(1,4,5)P3 (Fig. 6A), specifically at the 3-OH group (Fig. 6C,D). These results indicate the importance of Glu333 in the evolution of catalytic specificity of HsIP3KA.
Finally, a hydrophobic patch in the IP-helices in HsIP3KA and ScIPMK (Tyr315/Met316 in HsIP3KA, Met151 in ScIPMK; Fig. 2C), against which the inositol ring is proposed to rest, is hypothesized to enhance Ins(1,4,5)P3 binding 32. This hydrophobic patch is missing from both EhIP6KA and HsIP6K2 (Fig. 2C). We introduced HsIP3KA-like, hydrophobic residues into the corresponding region of the IP-helices of HsIP6K2 by preparing a single Gln240Met mutant and a tandem Asn239Tyr/Gln240Met mutant. These two mutant enzymes both exhibited a 50-fold increase in the rate of Ins(1,4,5)P3 phosphorylation (Fig. 6A). Moreover, the variety of products that were formed was significantly increased (compare Figs. 6B and 6E), perhaps by favoring multiple ligand-binding modes. Arg242Ala is another mutation that increases the hydrophobicity of the IP-helices; this elicited a 3-fold increase in Ins(1,4,5)P3 phosphorylation (Fig. 6A). Significantly, InsP6 kinase activity of HsIP6K2 was not substantially affected by these mutations to either Asn239, Gln240 or Arg242 (Fig. 6A). These mutagenic data confirm the structural information that indicate InsP6 and Ins(1,4,5)P3 have non-overlapping orientations in the substrate-binding site. InsP6 cannot interact with the synthetic hydrophobic patch since the plane its inositol ring is tilted away from the plane of the IP helices, unlike that of Ins(1,4,5)P3 (Fig. 5B,E). These mutagenic experiments also offer evolutionary insight by providing proof-of-principle of the nature of mutations to a progenitor kinase that could have increased Ins(1,4,5)P3 kinase activity without negative trade off from a compromised InsP6 kinase activity.
Discussion
In this study we describe the crystal structure of a bifunctional IP6K/IP3K from E. histoloytica (EhIP6KA), and we extrapolate our findings to a human IP6K with mutagenic and biochemical experiments. The most important, new ideas that we draw from this information are as follows: first, the geometry of the substrate binding site is revealed to exhibit a relatively spacious, open clamshell geometry; one jaw is represented by a pair of α-helices, the other jaw is an unusually-long 310 helix. Second, we show that InsP3 and InsP6 have non-overlapping orientations within the substrate pocket. Thus, relatively large inositol phosphates (Ins(1,3,4,5,6)P5 / InsP6) form contacts with both the α-helices and the 310 helix. In contrast, the smaller InsP3 molecule is rotated 55° closer to the α-helices; the latter provide most of the protein’s interactions with InsP3. Third, we describe an unusual evolutionary trajectory that could have favored efficient bi-functionality in a primordial kinase, prior to propagation of separate, dedicated IP3Ks and IP6Ks.
Previous ideas concerning how specialized enzymes diverged from promiscuous progenitors are based on the paradigm that the native catalytic activity should generally involve a substrate that is smaller than the emerging substrate, without a change in its degree of polarity 30. For the putative primordial IP6K 9, the original, preferred substrate (InsP6) would have been both larger and substantially more polar than the InsP3 substrates for the emerging kinases (IPMK and IP3K). Usually, the evolution of increased catalytic activity against smaller substrates depends upon compression of the active site 30. That development would be expected to reduce activity against the larger InsP6 molecule - a “negative trade-off” - that would impede organismic fitness and thereby select against the emergence of the InsP3/InsP4 kinase activities 30. However, our substrate-bound structures of EhIP6KA uncover a partial separation of the binding sites for Ins(1,4,5)P3 and InsP6. It is because of these non-overlapping substrate orientations that the insertion of a hydrophobic patch into the α-helices can enhance IP3K activity without affecting IP6K activity: an avoidance of negative trade-off which would have favored efficient bi-functionality. It appears that E. histolytica may be a particularly appropriate model for gaining insight into evolution of a hybrid IP6K/IP3K, since current annotations of this organism’s genome indicate that it does not encode a specialized Ins(1,4,5)P3 3-kinase. Thus, we conclude that EhIP6KA is the enzymatic equivalent of a “living fossil” that is suggestive of the nature of a primordial IP6K prior to gene duplication. After the latter event had occurred, it would appear that evolutionary gain of function of Ins(1,4,5)P3 phosphorylation would then have involved atrophy of the 310 helix and consolidation of the role of the IP helices in substrate binding (Fig. 5E). In HsIP3KA, the IP helices have been replaced with a distinct, highly-structured 63-residue inositol-phosphate binding domain: four helices with greater structural constraints that limit substrate selectivity to Ins(1,4,5)P3 alone 31,32. The 310 helix has also been lost from IPMK (Fig. 5E), but the substrate-binding pocket remains relatively spacious, thereby retaining a degree of positional promiscuity.
Each of the inositol phosphate kinases that share the PDKG motif - IP6Ks, IPMKs and IP3Ks - are specialized to synthesize unique inositol phosphates with differing biological actions. Our molecular understanding of these catalytic specializations has been incomplete, since among these proteins, only HsIP3KA has previously been co-crystallized with substrate 32. In contrast, the published crystal structures of IPMK 26,28 lacked bound substrate. Thus, our first atomic-level description of a substrate-bound IP6K fills a significant void in our structural and molecular understanding of the determinants of catalytic activity in the entire PDKG-motif kinase family. This information is also relevant to IP5K - previously thought to be the only inositol phosphate kinase capable of phosphorylating the 2-OH. Nevertheless, in crystals of substrate-bound IP5K, the orientation of the axial 2-OH of Ins(1,3,4,5,6)P5, relative to ATP, is very similar to the orientation of the equatorial 3-OH of Ins(1,4,5)P3, relative to ATP, in crystals of HsIP3KA 44. Furthermore, the current study shows that the inositol ring of Ins(1,4,5)P3 occupies very similar relative positions in the active sites of EhIP6KA and HsIP3KA (Fig. 5; supplementary Fig. 5). This spatial conservation of substrate binding between these different inositol phosphate kinases is also matched by spatial conservation of some key active-site residues 45. Such observations help to rationalize our observation that EhIP6KA phosphorylates Ins(1,4,5)P3 at the 2-position (Fig. 1D,E; Supplementary Fig. 5). Finally, our IP6K structure provides the missing template necessary for the rational design of inhibitors that might selectively target each of the different kinases that contain the PDKG motif, both for research tools and therapeutic applications.
Materials and Methods
In this study we use EhIP6KA as an abbreviation for the E. histolytica IP6K with Genbank accession number XP_648490.2, so as to distinguish it from two other gene products of that are also annotated as IP6Ks. A previous study 20 named this enzyme as EhIPK1. A codon-optimized cDNA for expression in E coli of EhIP6KA was synthesized by Genscript Inc (see Supplementary Methods). The Gateway expression system (Invitrogen) was used to subclone EhIP6KA into the pDest-566 vector. This vector encodes a 6×His tag, a maltose binding protein tag and TEV protease cleavage site at the N terminus. ArcticExpress™ (DE3) competent cells (Stratagene) were transformed with the resultant plasmid. An overnight culture of E. coli cells carrying the pDest-566-EhIP6KA was inoculated into 2× yeast extract tryptone medium at pH 7.5 and grown at 37 °C to A595 = 0.7. Isopropyl β-d-thiogalactopyranoside (0.1 mM) was then added and cultures were continued at 15 °C for 1 day. The cells were disrupted (Constant Systems Ltd) at 20 KPsi. Recombinant EhIP6KA proteins were purified with a Ni-NTA agarose column (Qiagen) followed by a HiTrap™ Q FF column (GE Healthcare). The bound proteins were released by TEV protease in the presence of 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1.5 mM DTT, 10 mM ATP, 20 mM MgCl2, then applied to a HiTrap™ Q FF column. In the case of the MBP fusion protein, this TEV cleavage step was omitted. As a final step, a Superdex™ 200 gel filtration column (GE Healthcare) was used with a running buffer of 50 mM NaCl and 20 mM Tris-HCl, pH 7.5.
Human IP6K2 cDNA was ordered from American Type Culture Collection (I.M.A.G.E. Clone ID 6162048, ATCC number 10437631). The Gateway expression system (Invitrogen) was used to subclone the kinase into the pDest-566 vector. Lemo21(DE3) Competent E. coli cells (New England Biolabs) were transformed with the resultant plasmid. An overnight culture of E. coli cells carrying the pDest-566-HsIP6K2 was inoculated into 2× yeast extract tryptone medium supplemented with 0.6 mM L-rhamnose at pH 7.5 and grown at 37 °C to A595 = 0.7. Isopropyl β-D-thiogalactopyranoside (0.1 mM) was then added and cultures were continued at 14 °C for 2 days. The cells were disrupted using a constant cell disruption system (Constant Systems LTD) under 20 KPsi. Recombinant HsIP6K2 was purified with a Ni-NTA agarose column (Qiagen) followed by a HiTrap™ Heparin HP column (GE Healthcare), TEV protease cleavage and a HiTrap™ Q FF column (GE Healthcare). As a final step, a Superdex™ 200 gel filtration column (GE Healthcare) was used with a running buffer of 150 mM NaCl and 20 mM Tris-HCl, pH 7.5. Mutant proteins were purified similarly. Recombinant DIPP1 was expressed and purified as previously described 46 with an additional, heparin-affinity purification step. The purity of all proteins was estimated to be >95% as judged by SDS-PAGE. The purified proteins were concentrated and stored at −80 °C.
Sample preparation for NMR analysis
Full length EhIP6KA (1-3 mg) was incubated in 3 ml medium containing 150 mM NaCl, 20 mM Tris-HCl (pH 7.0), 2 mM MgCl2, 3mM ATP, and 3mM InsP6 at 37 °C for one day and then 25 °C for three days. The reaction sample was centrifuged and filtered to remove protein, and then vacuum dried at 45 °C for 30 min.
The sample was prepared by taking up the reaction mixture in D2O to a volume of 600 ml and adjusting the pH to 6.2 using NaOH and HCl. In addition, sodium CDTA was added to a concentration of 20 mM to alleviate broadening of the 31P resonances due to chemical exchange of the phosphate groups with the high concentration of Mg2+ in the sample. All 1H and 31P chemical shifts were reference to ~5 mM internal DSS or to external 85% phosphoric acid, respectively. All NMR experiments were carried out on at 25 °C Varian Inova 600 MHz NMR spectrometers, equipped with either a 5 mm broad band NMR probe tuned to 31P on the observe channel and 1H on the decoupler channel, or a Varian 5 mm 1H{15N,13C} Cold Probe (Agilent, Santa Clara CA). The 1D 1H spectra were acquired with sweep widths of 12 ppm, with an acquisition time of 1 second, using a 1 second relaxation delay and with presaturation of the residual water resonance during the relaxation delay. The 1D 31P spectrum was obtained using a 50 ppm sweep width, with an acquisition time of 1.6 seconds, using a 1 second relaxation delay and with 1H decoupling during acquisition. The 2D 1H-31P HSQC spectrum was acquired with sweep widths of 12 ppm and 25 ppm in the 1H and 31P dimensions, respectively, with 142.3 ms and 42 ms acquisition times in these dimensions and with 1H decoupling during the acquisition. The 1/4J delay in the HSQC experiment was set to 31.25 ms, corresponding to a 1H-31P scalar coupling of 8 Hz. The 1H-1H and 31P-31P COSY spectra were acquired using gradient selected absolute value mode COSY experiments. The proton COSY experiment was acquired using 12 ppm sweep widths in both dimensions, with acquisition times of 342 ms and 71 ms in the directly and indirectly detected dimensions, respectively, and with a 1 second relaxation delay, during which time presaturation of the residual water signal was applied. The phosphorus COSY experiment was acquired using 25 ppm sweep widths in both dimensions, with acquisition times of 512 ms and 32 ms in the directly and indirectly detected dimensions, respectively, and with a 1 second relaxation delay. The 31P-31P COSY spectrum was also acquired with 1H-decoupling in both dimensions. All spectra were processed using Varian VnmrJ 2.2D software (Agilent, Santa Clara CA) on the Varian Inova spectrometer.
Enzyme Assays and HPLC analysis
Kinase assays were performed by incubating enzyme preparations with either [3H]Ins(1,4,5)P3 (American Radiolabeled Chemicals, Saint Louis, MO), [14C]-Ins(1,3,4,5,6)P5 (isolated from [14C]-labeled parotid acinar cells 47) or [3H]InsP6 (Perkin-Elmer; Waltham MA) at 37 °C in buffer containing 150 mM KCl, 20 mM HEPES pH 7.0, 12 mM MgSO4, 10 mM ATP, 1 mM EDTA. After either 20 min (InsP6 phosphorylation) or 0.5 - 7 hr (Ins(1,4,5)P3 phosphorylation) reactions were acid-quenched and neutralized 48, and analyzed by HPLC using either a 4.6 × 125 mm Partisphere 5 μm SAX column or a Q100 SAX column. The Partisphere column was eluted with one of two ammonium phosphate elution protocols: Gradient 1 48 or Gradient 2 49. The Q100 column was eluted with a gradient generated from Buffer A (1 mM Na2EDTA) and Buffer B (Buffer A plus 2M NH4H2PO4 pH 3.9 with H3PO4) in the following manner: 0 - 5 min, 0% B; 5 - 100 min, 0 - 42% B, 110 - 140 min, 42 - 70 % B (Gradient 3). Radioactivity was measured either with an in-line counter, or after collecting 1 ml or 0.5 ml fractions. Other radiolabeled inositol phosphates used for HPLC standards included [3H]Ins(1,2,4,5)P4 (prepared from [3H]Ins(1,3,4,5)P4 as described in 50), [14C]-Ins(1,3,4,5)P4 and [14C]-Ins(1,4,5,6)P4 (isolated from carbachol-stimulated, [14C]-labeled parotid acinar cells 47), PP-[3H]InsP4 and [PP]2-[3H]InsP3 (both prepared as described in 36).
Crystallization
The MBP fusion protein of EhIP6KA (residues 32-270) was crystallized by hanging drop vapor diffusion against a well buffer of 12% (w/v) PEG 3350, 50 mM NaH2PO4, pH 5.5 at 18 °C. The protein concentration was > 60 mg/ml. The core catalytic domain of EhIP6KA (residues 20-270) was crystallized by hanging drop vapor diffusion for one week against a well buffer of 8% (w/v) PEG 3350, 100 mM Na3Citrate, pH 5.2 at 25 °C, followed by dilution microseeding (Seed Bead™ kit, Hampton Research) for two weeks with a well buffer of 8% (w/v) PEG 3350, 100 mM Na3Citrate, pH 5.2, 8% ethylene glycol at 25 °C. The crystals were displayed as a thin plate and were anisotropic. Crystals of EhIP6KA (residues 27-270) emerged under 0.4 M NaH2PO4 in the presence of 10 mM ATP, plus either 10 mM InsP6, 5 mM Ins(1,3,4,5,6)P5 or 5 mM Ins(1,4,5)P3/20 mM MgCl2. To increase occupancy of inositol phosphates and remove the effect of phosphate ions, the crystals were further soaked under 22% (w/v) PEG 3350, 10 mM MgCl2, 10 mM ATP, 0.1 M sodium acetate, pH 5.2 and 20 mM InsP6, 10 mM Ins(1,3,4,5,6)P5 or 10 mM Ins(1,4,5)P3 for 3 days. Cryosolvent was prepared by adding 33% ethylene glycol into the soaking buffer.
Data Collection, Structure Determination and Refinement
Diffraction data were collected using APS beamlines 22-ID and 22-BM. All data were processed with the program HKL2000 51. Initial phases for the structure of MBP-EhIPK1 were determined by molecular replacement using MBP (1ez9) and HsIP3KA (1w2c) as searching models. Then the first structure was manually rebuilt with COOT 52 and refined with REFMAC from the CCP4 package 53. The other crystals structures were determined either by molecular replacement if the space group was different, or by using rigid body and direct Fourier synthesis, and refined with the equivalent and expanded test sets. A Cartesian simulated-annealing protocol was performed in the first round of refinement. Ligand topology and parameter files were prepared using the incorporated PRODRG program from the CCP4 package 53. The molecular graphics representations were prepared with the program PyMol (Schrödinger, LLC).
Supplementary Material
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. We thank T. M. Hall and L. C. Pedersen for discussions and critical reading of the manuscript, the NIEHS Collaborative crystallography group, the Advanced Photon Source (APS) Southeast Regional Collaborative Access Team (SER-CAT) 22-ID and 22-BM beam lines for assistance with crystallographic data collection.
Footnotes
Author contributions H.W., E.F.D. and S.B.S. performed experiments. All authors designed experiments and wrote the paper.
Conflict of Interest Statement The authors declare that there is no conflict of interest.
References
- 1.Shears SB. Diphosphoinositol polyphosphates: metabolic messengers? Mol. Pharmacol. 2009;76:236–252. doi: 10.1124/mol.109.055897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson MS, Livermore TM, Saiardi A. Inositol pyrophosphates: between signalling and metabolism. Biochem. J. 2013;452:369–379. doi: 10.1042/BJ20130118. [DOI] [PubMed] [Google Scholar]
- 3.Szijgyarto Z, Garedew A, Azevedo C, Saiardi A. Influence of inositol pyrophosphates on cellular energy dynamics. Science. 2011;334:802–805. doi: 10.1126/science.1211908. [DOI] [PubMed] [Google Scholar]
- 4.Illies C, et al. Inositol pyrophosphates determine exocytic capacity. Science. 2007;318:1299–1302. doi: 10.1126/science.1146824. [DOI] [PubMed] [Google Scholar]
- 5.Chakraborty A, et al. Inositol pyrophosphates inhibit AKT signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee YS, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gokhale NA, Zaremba A, Janoshazi AK, Weaver JD, Shears SB. PPIP5K1 Modulates Ligand Competition Between Diphosphoinositol Polyphosphates and PtdIns(3,4,5 P3 for Polyphosphoinositide-Binding Domains. Biochem. J. 2013;453:413–426. doi: 10.1042/BJ20121528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pulloor NK, et al. Human Genome-Wide RNAi Screen Identifies an Essential Role for Inositol Pyrophosphates in Type-I Interferon Response. PLoS Pathog. 2014;10:e1003981. doi: 10.1371/journal.ppat.1003981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett M, Onnebo SM, Azevedo C, Saiardi A. Inositol pyrophosphates: metabolism and signaling. Cell. Mol. Life Sci. 2006;63:552–564. doi: 10.1007/s00018-005-5446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saiardi A, Erdjument-Bromage H, Snowman A, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- 11.Saiardi A, Nagata E, Luo HR, Snowman AM, Snyder SH. Identification and characterization of a novel inositol hexakisphosphate kinase. J. Biol. Chem. 2001;276:39179–39185. doi: 10.1074/jbc.M106842200. [DOI] [PubMed] [Google Scholar]
- 12.Draskovic P, et al. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem. Biol. 2008;15:274–286. doi: 10.1016/j.chembiol.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 13.Lin H, et al. Structural analysis and detection of biological inositol pyrophosphates reveals that the VIP/PPIP5K family are 1/3-kinases. J. Biol. Chem. 2009;284:1863–1872. doi: 10.1074/jbc.M805686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhandari R, Juluri KR, Resnick AC, Snyder SH. Gene deletion of inositol hexakisphosphate kinase 1 reveals inositol pyrophosphate regulation of insulin secretion, growth, and spermiogenesis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2439–2353. doi: 10.1073/pnas.0712227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh S, et al. Inositol hexakisphosphate kinase 1 maintains hemostasis in mice by regulating platelet polyphosphate levels. Blood. 2013;122:1478–1486. doi: 10.1182/blood-2013-01-481549. [DOI] [PubMed] [Google Scholar]
- 16.Jadav RS, Chanduri MV, Sengupta S, Bhandari R. Inositol Pyrophosphate Synthesis by Inositol Hexakisphosphate Kinase 1 is Required for Homologous Recombination Repair. J. Biol. Chem. 2012;288:3312–3321. doi: 10.1074/jbc.M112.396556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burton A, Azevedo C, Andreassi C, Riccio A, Saiardi A. Inositol pyrophosphates regulate JMJD2C-dependent histone demethylation. Proc. Natl. Acad. Sci. U. S. A. 2013;110:18970–18975. doi: 10.1073/pnas.1309699110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison BH, Bauer JA, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 mediates growth suppressive and apoptotic effects of interferon-b in ovarian carcinoma cells. J. Biol. Chem. 2001;276:24965–24970. doi: 10.1074/jbc.M101161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison BH, et al. Gene deletion of inositol hexakisphosphate kinase 2 predisposes to aerodigestive tract carcinoma. Oncogene. 2009;28:2383–2392. doi: 10.1038/onc.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loser B, et al. A novel Entamoeba histolytica inositol phosphate kinase catalyzes the formation of 5PP-Ins(1,2,3,4,6)P(5) Mol. Biochem. Parasitol. 2011 doi: 10.1016/j.molbiopara.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 22.Saiardi A, Caffrey JJ, Snyder SH, Shears SB. Inositol Polyphosphate Multikinase (ArgRIII) Determines Nuclear mRNA Export in Saccharomyces cerevisiae. FEBS Lett. 2000;468:28–32. doi: 10.1016/s0014-5793(00)01194-7. [DOI] [PubMed] [Google Scholar]
- 23.Chang S-C, Miller AL, Feng Y, Wente SR, Majerus PW. The human homologue of the rat inositol phosphate multikinase is an inositol 1,3,4,6-tetrakisphosphate 5-kinase. J. Biol. Chem. 2002;277:43836–43843. doi: 10.1074/jbc.M206134200. [DOI] [PubMed] [Google Scholar]
- 24.Yang X, Shears SB. Multitasking in Signal Transduction by a Promiscuous Human Ins(3,4,5,6)P4 1-Kinase/Ins(1,3,4)P3 5/6-Kinase. Biochem. J. 2000;351:551–555. [PMC free article] [PubMed] [Google Scholar]
- 25.Shears SB. How versatile are inositol phosphate kinases? Biochem. J. 2004;377:265–280. doi: 10.1042/BJ20031428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes W, Jogl G. Crystal structure of inositol phosphate multikinase 2 and implications for substrate specificity. J. Biol. Chem. 2006;281:38109–38116. doi: 10.1074/jbc.M606883200. [DOI] [PubMed] [Google Scholar]
- 27.Miller GJ, Wilson MP, Majerus PW, Hurley JH. Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase. Mol Cell. 2005;18:201–212. doi: 10.1016/j.molcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Endo-Streeter S, Tsui MK, Odom AR, Block J, York JD. Structural studies and protein engineering of inositol phosphate multikinase. J. Biol. Chem. 2012;287:35360–35369. doi: 10.1074/jbc.M112.365031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chamberlain PP, et al. Integration of inositol phosphate signaling pathways via human ITPK1. J. Biol. Chem. 2007;282:28117–28125. doi: 10.1074/jbc.M703121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khersonsky O, Tawfik DS. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 31.Miller GJ, Hurley JH. Crystal structure of the catalytic core of inositol 1,4,5-trisphosphate 3-kinase. Mol Cell. 2004;15:703–711. doi: 10.1016/j.molcel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez B, et al. Structure of a human inositol 1,4,5-trisphosphate 3-kinase; substrate binding reveals why it is not a phosphoinositide 3-kinase. Mol Cell. 2004;15:689–701. doi: 10.1016/j.molcel.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Albert C, et al. Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem. J. 1997;327:553–560. doi: 10.1042/bj3270553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kilari RS, Weaver JD, Shears SB, Safrany ST. Understanding inositol pyrophosphate metabolism and function: Kinetic characterization of the DIPPs. FEBS Lett. 2013;587:3464–3470. doi: 10.1016/j.febslet.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barker CJ, Illies C, Gaboardi GC, Berggren PO. Inositol pyrophosphates: structure, enzymology and function. Cell. Mol. Life Sci. 2009;66:3851–3871. doi: 10.1007/s00018-009-0115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saiardi A, Caffrey JJ, Snyder SH, Shears SB. The Inositol Hexakisphosphate Kinase Family: Catalytic Flexibility, and Function in Yeast Vacuole Biogenesis. J. Biol. Chem. 2000;275:24686–24692. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- 37.Verbsky JW, Wilson MP, Kisseleva MV, Majerus PW, Wente SR. The synthesis of inositol hexakisphosphate: Characterization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. J. Biol. Chem. 2002;277:31857–31862. doi: 10.1074/jbc.M205682200. [DOI] [PubMed] [Google Scholar]
- 38.Safrany ST, et al. A novel context for the “MutT” module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 1998;17:6599–6607. doi: 10.1093/emboj/17.22.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vieira-Pires RS, Morais-Cabral JH. 3(10) helices in channels and other membrane proteins. J. Gen. Physiol. 2010;136:585–592. doi: 10.1085/jgp.201010508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pal L, Basu G. Novel protein structural motifs containing two-turn and longer 3(10)-helices. Protein Eng. 1999;12:811–814. doi: 10.1093/protein/12.10.811. [DOI] [PubMed] [Google Scholar]
- 41.Padmanabhan U, Dollins DE, Fridy PC, York JD, Downes CP. Characterization of a selective inhibitor of inositol hexakisphosphate kinases: Use in defining biological roles and metabolic relationships of inositol pyrophosphates. J. Biol. Chem. 2009;284:10571–10582. doi: 10.1074/jbc.M900752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mildvan AS. Mechanisms of Signaling and Related Enzymes. Proteins. 1997;29:401–416. [PubMed] [Google Scholar]
- 43.Nagata E, et al. Inositol hexakisphosphate kinase-2, a physiologic mediator of cell death. J. Biol. Chem. 2005;280:1634–1640. doi: 10.1074/jbc.M409416200. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez B, Banos-Sanz JI, Villate M, Brearley CA, Sanz-Aparicio J. Inositol 1,3,4,5,6-pentakisphosphate 2-kinase is a distant IPK member with a singular inositide binding site for axial 2-OH recognition. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9608–9613. doi: 10.1073/pnas.0912979107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H, Falck JR, Hall TM, Shears SB. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat. Chem. Biol. 2012;8:111–116. doi: 10.1038/nchembio.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang X, Safrany ST, Shears SB. Site-directed mutagenesis of DIPP, a dual specificity MutT/Nudix-type hydrolase that attacks diadenosine polyphosphates and diphosphoinositol polyphosphates. J. Biol. Chem. 1999;274:35434–35440. doi: 10.1074/jbc.274.50.35434. [DOI] [PubMed] [Google Scholar]
- 47.Hughes PJ, Hughes AR, Putney JW, Jr., Shears SB. The regulation of the phosphorylation of inositol (1,3,4)-trisphosphate in cell free preparations and its relevance to the formation of inositol (1,3,4,6)-tetrakisphosphate in agonist-stimulated rat parotid acinar cells. J. Biol. Chem. 1989;264:19871–19878. [PubMed] [Google Scholar]
- 48.Weaver JD, Wang H, Shears SB. The kinetic properties of a human PPIP5K reveal that its kinase activities are protected against the consequences of a deteriorating cellular bioenergetic environment. Biosci. Rep. 2013;33:228–241. doi: 10.1042/BSR20120115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Safrany ST, Shears SB. Turnover of bis-diphosphoinositol tetrakisphosphate in a smooth muscle cell line is regulated by b2- adrenergic receptors through a cAMP-mediated, A-kinase-independent mechanism. EMBO J. 1998;17:1710–1716. doi: 10.1093/emboj/17.6.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brearley CA, Hanke DE. Metabolic evidence for the order of addition of individual phosphate esters to the myo-inositol moiety of inositol hexakisphosphate in the duckweed Spirodela polrhiza L. Biochem. J. 1996;314:227–233. doi: 10.1042/bj3140227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 54.Chakraborty A, et al. HSP90 regulates cell survival via inositol hexakisphosphate kinase-2. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1134–1139. doi: 10.1073/pnas.0711168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chakraborty A, et al. Casein kinase-2 mediates cell survival through phosphorylation and degradation of inositol hexakisphosphate kinase-2. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2205–2209. doi: 10.1073/pnas.1019381108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.