Abstract
Purpose
KRAS wild-type status is an imperfect predictor of sensitivity to anti-EGFR monoclonal antibodies in colorectal cancer (CRC), motivating efforts to identify novel molecular aberrations driving RAS. This study aimed to build a quantitative readout of RAS pathway activity to: (1) uncover molecular surrogates of RAS activity specific to CRC; (2) improve the prediction of cetuximab response in patients; (3) suggest new treatment strategies.
Methods
A model of RAS pathway activity was trained in a large CRC dataset and validated in three independent CRC patient datasets. Novel molecular traits were inferred from the TCGA CRC data. The ability of the RAS model to predict resistance to cetuximab was tested in mouse xenografts and three independent patient cohorts. Drug sensitivity correlations between our model and large cell line compendiums were performed.
Results
The performance of the RAS model was remarkably robust across 3 validation datasets. (1) Our model confirmed the heterogeneity of the RAS phenotype in KRAS wild-type patients, and suggests novel molecular traits driving its phenotype (e.g. MED12 loss, GBXW7 mutation, MAP2K4 mutation). (2) It improved the prediction of response and progression free survival (HR=2.0; p<.01) to cetuximab compared to KRAS mutation (xenograft and patient cohorts). (3) Our model consistently predicted sensitivity to MEK inhibitors (p<.01) in 2 cell panel screens.
Conclusions
Modeling the RAS phenotype in CRC allows for the robust interrogation of RAS pathway activity across cell lines, xenografts, and patient cohorts. It demonstrates clinical utility in predicting response to anti-EGFR agents and MEK inhibitors.
Introduction
In the past decade, the management of metastatic colorectal cancer (CRC) patients has been profoundly improved by the introduction of anti-EGFR monoclonal antibodies (i.e. cetuximab, panitumumab)(1,2). The subsequent identification of KRAS mutation as a predictor of resistance to these agents(3) has resulted in a restriction of their regulatory approval to the subset of KRAS wild-type tumors. Consequently, virtually all patients with metastatic CRC are tested for KRAS mutation status and receive adapted anti-tumor strategies.
A growing body of evidence suggests that KRAS mutation status alone is not sufficient to predict the response to anti-EGFR monoclonal antibodies. First, not all KRAS wild-type tumors respond to therapy with anti-EGFR agents(2,4). Second, other molecular abnormalities such as BRAF, HRAS, NRAS, PIK3CA, P53, PTEN, or IGF1R have been implicated in the resistance to these agents(5–10). Finally, the impact of specific KRAS mutations like KRAS p.G13D on sensitivity to anti-EGFR monoclonal antibodies remains actively debated(11,12,13).
Several groups have attempted to improve the prediction of response to anti-EGFR agents using gene expression signatures(14–16), although none of these signatures has been independently validated in external datasets. The recent availability of multiple, large CRC datasets with coherent high-throughput molecular profiling - concomitant to the emergence of powerful modeling frameworks - provides the opportunity to interrogate RAS biology at a high resolution. The present study aims to develop a more precise measure of the RAS phenotype – defined as a model based assessment of RAS dependency using gene expression - in the CRC setting to improve existing therapeutic strategies and offer new treatment options for colorectal cancer patients.
Methods
Patient Cohorts
As training set, we used n=334 fresh frozen colorectal cancer tissues collected at the Koo Foundation Sun-Yat-Sen Cancer Center (KFSYSCC) from 2000-2004 and profiled on the Affymetrix U133 plus 2.0 platform. After RNA and microarray quality control procedures (Supplementary Materials), 322 samples were retained. Taqman real-time PCR was used for detection of mutations in KRAS codon 12 and 13 as previously described(17). QC analysis of the microarray data revealed 2 outliers, which were removed from further analysis. Following the intersection of all samples that had both microarray and KRAS mutation status, 290 samples were available for analysis.
As validation dataset, we used the following publicly available and previously published datasets: Gaedcke, J et al(18) (n=65 patients, GEO id: GSE20842), Khambata-Ford S et al(15), (n=68 patients; GEO id: GSE5851), TCGA (The Cancer Genome Atlas) CRC dataset(19) (n=206 patients; https://tcga-data.nci.nih.gov/tcga). Patient characteristics are described in Supplementary Table 1.
To assess the ability of our model to predict cetuximab response, we used the following datasets: Julien S et al (20) (n=54 mouse xenografts, n=19 patients; ArrayExpress id: E-MTAB-991), Khambata-Ford S et al(15) (n=68 patients; GEO id: GSE5851), and INSERM (n=85 patients; GEO id under process). Patient characteristics are described in Supplementary Table 2.
To assess the drug response of MEK inhibition, we use the following datasets: Barretina J, et al(21) (n=19 cell lines, http://www.broadinstitute.org/ccle/home), Garnett M et al(22), (n=15 cell lines, http://cancerrxgene.org) , Jürchott K et al(23) (n=12 cell lines; GEO id: GSE18232), and mouse xenografts (n=11; ArrayExpress id: E-MEXP-3557).
Bioinformatics and Statistical Analysis
Quality control analysis for outlier detection was performed on all data using principal component analysis (PCA). We used the penalized ElasticNet(24) regression model to predict KRAS mutation (codon 12 and 13). Optimal hyper-parameters (alpha and lambda in the ElasticNet) were selected using 5-fold cross-validation. Multiple bootstraps of the data (n=100) were used to assess over-fitting. Two-sided, non-parametric methods were used for all statistical evaluations. Kaplan-Meier estimates and log-rank test comparisons were used for survival analyses. Multivariate cox regression was used to infer association with progression hazard. To assess the predictive performance of our model, concordance index and proportion of the variance were computed. All statistical analysis was conducted using the R version 3.0 and Bioconductor 2.12. All microarray data was processed and normalized using RMA(25). RNA-seq normalization was performed using conditional quantile normalization on the unnormalized count data(26). Comprehensive information of our modeling and statistical approaches can be found in the Supplementary Methods.
Results
A Model of the RAS Phenotype in Colorectal Cancer
We first assessed the performance of four recently published gene expression signatures(14,16,27,28) to classify samples with KRAS mutation in several large human CRC datasets. Notably, these signatures were derived from several disease contexts, and were not specifically optimized for prediction of RAS within CRC. We observed that all these signatures performed poorly in distinguishing KRAS mutant from wild-type samples (Supplementary Figure 1A), underscoring the need of developing a specific RAS CRC model.
We trained a RAS model on an unpublished gene expression dataset (KFSYSCC, n=290) derived from primary colon and rectal tumor samples to discriminate KRAS mutation status from wild-type. The output of our predictive model is a continuous value between 0 and 1, which we refer to as the RAS Index Score or RIS, with higher values corresponding to a stronger RAS phenotype. Internal bootstrapping of the data demonstrates robust prediction performance (Supplementary Figure 1B, median AUC=0.81). The performance of our model was robust across all validation datasets (AUC= 0.78 - 0.90), regardless of the location of the tumor in the intestine (primary colon, primary rectum, and metastatic colorectal disease) or of the RNA profiling platform (Affymetrix, Agilent, RNAseq) (Supplementary Figure 1C, Supplementary Table 4). The significant genes comprising our model are described in Supplementary Table 3.
To assess the specificity of our model to CRC, we repeated this evaluation in 4 lung adenocarcinoma and 1 endometrial carcinoma datasets. Overall, we observed much lower AUC performance in lung and uterine tumors (AUC= 0.53 – 0.68) compared to CRC (Supplementary Table 3), suggesting that our RAS phenotype is specific to CRC and not generalizable to other disease types.
Quantitative model uncovers new molecular traits associated with RAS phenotype
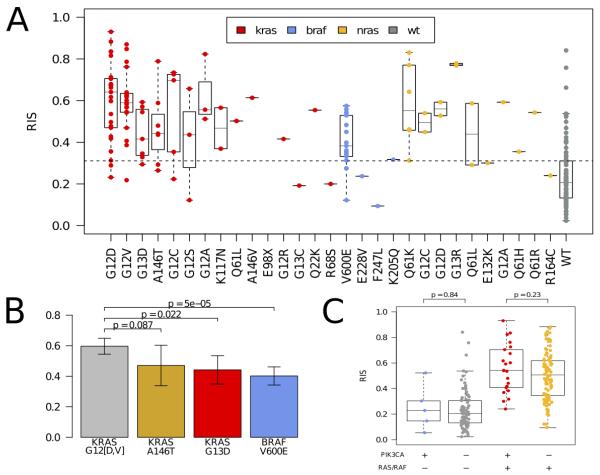
We investigated how molecular aberrations contribute to the RAS phenotype using the TCGA CRC cohort. Expectedly, KRAS, BRAF, and NRAS mutated samples exhibited a highly significant elevated RIS compared to wild-type samples, (p < 2e-04, Figure 1A, Supplementary Figure 3). Moreover, while we observed that the canonical activating KRAS mutations of codons 12, 13, 61, and 146 had a constitutively high RIS, there were large differences among them. The RIS of KRAS p.A146T and p.G13D samples was significantly lower than samples with a KRAS codon 12 mutations mutation (Figure 1B). A balanced bootstrap analysis in the KFSYSCC dataset confirmed that the lower RIS of KRAS p.G13D was not an artifact of the lower number of KRAS p.G13D samples relative to codon 12 mutations samples in the training data (Supplementary Figure 2). Interestingly, the KRAS p.K117N and p.Q22K mutants had a high RIS and may be novel (albeit rare) activating mutations (Figure 1A). While BRAF p.V600E showed in aggregate a higher RIS compared to wild-type, it had a significantly lower RIS than KRAS codon 12 mutations (p < 5e-05) and NRAS (p < 6.0e-3), (Figure 1B, Supplementary Figure 3). We observed no significant contribution of PIK3CA mutations toward RIS variation looking at PIK3CA mutations in aggregate (Figure 1C) or by PIK3CA exon (Supplementary Figure 4). Repeating this analysis using CRC cell lines from the CCLE dataset confirmed these results in vitro (Supplementary Figure 5).
Figure 1.
[A] Distribution of RAS phenotype as assessed by RIS according to amino acid change for KRAS (red), NRAS (yellow), and BRAF (blue) mutations in the TCGA colorectal cohort. Wild-type in this context is in relation to the KRAS, NRAS and BRAF genes. [B] Comparison of RIS distributions for KRAS p.A146T, KRAS p.G13D, and BRAF p.V600E with KRAS p.G12D/V. KRAS p.A146T and KRAS p.G13D show a lower RIS (p = .08 and .02, respectively, Mann-Whitney test) while BRAF p.V600E is significantly lower (p < 5e-05, Mann-Whitney test). [C] Assessment of RIS for samples with PIK3CA mutations (n=27). We observe that PIK3A mutant and BRAF/RAS wild-type samples do not have a significantly different RIS from the PIK3CA/RAS/RAF wild-type samples. Similarly, the double mutant samples do not have a significantly different RIS compared to the PIK3CA wild-type and RAS/RAF mutant samples. This suggests that PIK3CA mutations do not impact the RAS phenotype independent of RAS/RAF mutations.
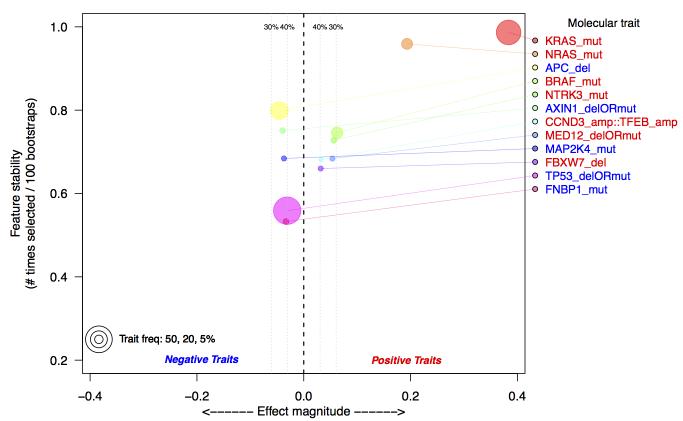
The distribution of RIS in samples annotated as RAS wild-type (i.e. KRAS, NRAS, and BRAF wild-type) reveals substantial heterogeneity (Figure 1A, Supplementary Figure 3). We hypothesize that this indicates the presence of alternative molecular aberrations that may contribute to the RAS phenotype. To investigate this, we incorporated somatic mutations, amplifications, and homozygous deletions from ~400 cancer genes (Sanger Cancer Census(29)) into a feature selection model to identify additional aberrations that could explain RIS. By training multiple bootstrapped models, we were able to observe the frequency an aberration appeared in a model. The most frequently selected genomic aberrations below our false discovery criteria (see Supplemental Methods) are shown in Figure 2. As a positive control, we observed that KRAS, NRAS, and BRAF are the 1st, 2nd and 4th ranked aberrations, respectively. Other aberrations associated with high RIS are NTRK3 mutation, CCND3/TFEB amplification, MED12 loss, and FBXW7 loss. Interestingly, MED12 loss has been recently shown to activate MEK/ERK through activation of TGFBR2 signaling, and may cause resistance to therapies acting upstream to MAPK/ERK(30). Among the aberrations associated with a low RIS is MAP2K4, a mitogen activating kinase implicated in JNK pathway signaling(31), and the tumor suppressor TP53. While TP53 deletions or mutations are not mutually exclusive with KRAS mutations (p > .5), we observe a significant, negative association between TP53 aberrations and RIS (p=.0071, Wilcoxon rank-sum test).
Figure 2.
Top selected molecular aberrations that explain RIS in TCGA colorectal data. Aberrations are defined as either somatic mutations (mut), homozygous deletion (del) or amplifications (amp). Aberrations are identified using a multivariate Lasso model, regressed against the estimated RIS. Along the y-axis is the frequency with which an aberration is selected from 100 bootstraps of the data; the x-axis is the effect magnitude, computed as the average beta value from the Lasso model over all bootstrapped models. Estimated false discovery thresholds are shown as vertical lines, and only aberrations below a 40% false discovery threshold are plotted. The selection of KRAS and NRAS as the top selected features serves as a positive control.
RAS gene expression model improves prediction of response to cetuximab in KRAS wild-type patients
We investigated whether our model was able to predict response to cetuximab in a recently published panel of human CRC (n=19) and mouse xenografts (n=54)(20). The RAS model separated responders vs non-responders to cetuximab (p < .05) better than KRAS and/or BRAF mutation status (p > .1) both in the human specimens and mouse xenografts (Supplementary Figure 6).
Next, we applied our model to a 68 metastatic CRC patient cohort(32) treated with cetuximab monotherapy. Again, our RAS model discriminated patient with disease control (DC) (AUC=0.75) better than KRAS status alone (AUC=0.70). Using the median RIS as a threshold, our RAS model separated non-responders (p=3.19e-4, Fisher exact test) better than KRAS mutation status (p=3.3e-3), (Supplementary Figure 7). Importantly, the positive predictive value of our model for disease control was higher than the KRAS status alone: 64% (16/25 patients) versus 51% (19/37 patients), respectively.
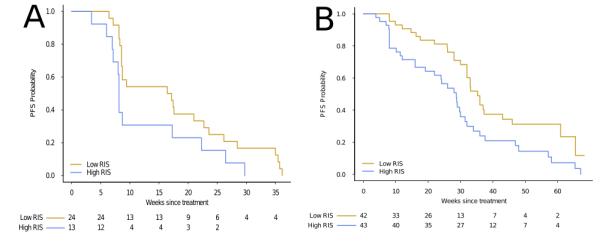
We next asked whether our RIS score could separate the patient cohort based on progression free survival (PFS) time. Dichotomizing the patient cohort on the median RIS value, we observed a significant separation of survival time with our model (HR=2.4, CI95% [1.37-4.23]; p=.001 log-rank test; c-index=.612 (se=.04)), as well as with the KRAS status alone (HR=2.3, CI95% [1.24-4.35]; p=.008 log-rank test; c-index=.56 (se=.038)). A multivariate cox regression analysis including KRAS mutation status and RIS revealed RIS to be a significant factor (HR=2.0, CI95% [1.03-3.83], p < .05) compared to the KRAS mutation variable (HR=1.56, CI95% [.76-3.2], p > .2)(Supplementary Table 5B). Finally, restricting our analysis to the KRAS wild-type patient cohort (n=37), we continued to observe a significant separation of progression free survival (HR=2.3, CI95% [1.2-4.9], p=.016 log-rank test; Figure 3A).
Figure 3.
Kaplan-Meier plots comparing progression free survival (PFS) in two KRAS wild-type, advanced CRC patient cohorts treated with cetuximab. For both cohorts, we dichotomize on the median of the estimated RIS. [A] KRAS wild-type patients (n=37) from the Khambata-Ford data set, separated by RIS (p=.016, log-rank test, HR=2.3, 1.2-4.9 95%CI). [B] KRAS wild-type patient (n=85) from blinded validation set. PFS is significantly lower in the high RIS patients compared to low RIS patient (p=.007 log-rank test, HR =2.0 CI95% [1.2-3.3]).
To confirm these findings, we evaluated whether our model could predict PFS in a larger, blinded dataset consisting of KRAS wild-type, advanced diseased patients (n=85) treated with cetuximab(8). Cox regression analysis revealed a significant association between PFS and RIS (HR=5.7, CI95% [2.0-15.85]; p=7.8e-04; c-index=.639 (se=.043)) (Supplementary Table 6) treating RIS as a continuous variable. When dichotomized on the median RIS, patients with high RAS phenotype exhibited a longer PFS (35.3 weeks) as compared to low RIS (28.0 weeks) (HR =2.0 CI95% [1.2-3.3] p=6.4e-03) (Figure 3B).
High RAS Phenotype Predicts MEK Sensitivity
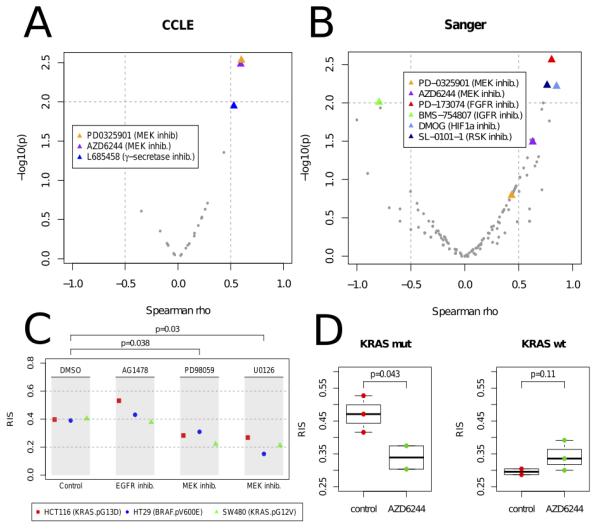
To investigate which therapies should be applied to patients exhibiting a high RAS phenotype, we applied our model to the 19 CRC cell lines with drug response data available from CCLE(21) and computed the correlation between RIS and the drug sensitivity values measured for 24 recent anti-cancer drugs. Interestingly, we observed a significant correlation between RIS and drug sensitivity measurements for the two MEK inhibitors (PD-0325901 & Selumitinib, p < 0.003, Spearman rank test), (Figure 4A). The mutation status of KRAS and/or BRAF did not predict MEK response (p > .2).
Figure 4.
Volcano plots of Spearman correlation (x-axis) and significance (y-axis) evaluated between drug sensitivity (AUC) and RAS index score (RIS) in the [A] Cancer Cell-Line Encyclopedia (CCLE, 24 cancer drugs) and [B] Sanger Cell Line drug panel (130 drugs). Significant drug associations (p < .01) are highlighted. [C] RIS measured in 3 CRC cell lines (HCT116, HT29 and SW480) following treatment with anti-EGFR drug AG1478, and MEK inhibitors PD098059 and U0126. Reduction of RIS is observed in the cell lines treated with MEK (p < .05). As a negative control, the EGFR treated cell lines exhibited no reduction of RIS. [D] Xenograft CRC model showing distribution of RIS pre- (red) and post (green) treatment with MEK inhibitor Selumitinib (AZD6244) (n=10). RIS reduction is observed in the KRAS mutant cell line (HCT116) treated with MEK inhibitor, whereas the KRAS wild-type treated cell line (HKH2) shows a modest increase in RIS.
To expand the drug sensitivity analysis, we tested the RIS association with the cell line drug sensitivity data available from the Sanger dataset(22) with 130 drugs profiled (Figure 4B). We confirmed the association of the MEK inhibitor AZD6244 (Selumitinib) with the RAS phenotype (Spearman rho=0.629, p=0.032). Moreover, other significant associations included the FGFR inhibitor PD-173074 (R=0.804,p=0.0027), IGF1R inhibitor BMS-754807 (R=-0.794,p=0.0060), the HIF1A drug DMOG (R=0.85,p=0.0060) and the p90 ribosomal S6 kinases inhibitor (R=0.76,p=0.0058).
To corroborate the association between the RAS phenotype and response to MEK inhibition, we applied our model to a gene expression panel derived 3 CRC cell lines profiled pre and post MEK inhibition(23) (SW480, HCT116, and HT29) (Figure 4C). We observed that the MEK inhibitor consistently reduced the RIS in all of these cell lines (p=0.038, PD098059; p=0.030, U0126), (Figure 4C). We observed similar findings in a xenograft panel of HCT116 (KRAS p.G13D) and HKH2 (KRAS wild-type) cell lines (n=10) treated with the MEK inhibitor Selumitinib (Figure 4D).
Discussion
KRAS mutation status is recognized as a key determinant of resistance to anti-EGFR antibodies in colorectal cancer. The increasing number of targeted agents that impact the EGFR/RAS/MEK signaling pathway - and the failure of these therapies to translate seamlessly between disease types - provides strong motivation to assess the activity of this pathway in a context dependent manner, i.e. lung cancer vs. colorectal cancer. Herein, we provide a robust quantitative readout of the RAS phenotype that is specific to colorectal cancer. Using this model, we were able to (i) unravel novel molecular aberrations that contribute to the RAS phenotype (e.g. KRAS specific amino acid changes, NRAS, and BRAF); (ii) improve the prediction of response and survival to cetuximab treatment in KRAS wild-type patients; (iii) and uncover targeted strategies matched to the patient’s RAS phenotype (e.g. MEK inhibitors).
The specificity of KRAS mutation to predict resistance to anti-EGFR therapy is very high while the wild-type status of KRAS has a low sensitivity anti EGFR response(2,4). Our model improves the discrimination of KRAS wild-type patients that are likely to benefit from cetuximab therapy both in terms of progression free survival and response rate. Particularly, our model demonstrated a significant PFS hazard ratio in two independent datasets: HR=2.0 (1.2-3.3) in the INSERM patient cohort (p < .05) and HR=2.3 (1.2-4.9) (p < .05) in the KRAS wild-type patients from the Khambata-Ford dataset. In the latter dataset, we also observed that 64% of the patients with KRAS wild-type and low RIS exhibited partial response or stable disease as compared to only 51% of the patients with KRAS wild-type only. The fact that most of the patients in these two datasets received conventional chemotherapy plus cetuximab may have influenced our models. However, our results are consistent with other studies describing an overlap between RAS- and cetuximab- gene expression models(33), thus confirming the central role of KRAS in the prediction of response to cetuximab.
Interestingly, our model not only identifies wild-type KRAS patients not suitable for cetuximab, but also uncovers potential alternative treatment strategies. In that regard, we highlight the sensitivity of MEK inhibitors in colorectal cell lines exhibiting a high RAS phenotype. Moreover, we have shown that high RAS phenotype is reversed by MEK inhibition, both in vitro and in xenograft models. Although this information has great potential for changing clinical practice, these warrant confirmation in large prospective cohorts.
Finally, our RAS model has allowed us to probe the relative potency of specific oncogenic mutations contributing to the RAS phenotype. Particularly, we found that KRAS p.G13D mutated samples had a lower RAS phenotype than p.G12D or p.G12V mutant samples. This is consistent with the recent literature describing a different molecular phenotype of the KRAS p.G13D mutation (34,35) as well as suggesting a possible benefit of cetuximab for patients harbouring this specific trait (12,36). Second, BRAF mutated samples also exhibited a lower RAS phenotype as compared to KRAS p.G12D and p.G12V mutated samples, suggesting a smaller effect in the activation of RAS downstream signaling. This is corroborated in a recent large phase III trial that first line metastatic patients harboring in BRAF mutations in their tumors could benefit from a combination of chemotherapy plus anti-EGFR therapy(13),(37) but remains a matter of debate for second or additional lines of treatment(38). We did not observe any relationship between specific PIK3CA mutations and the RAS phenotype as measured by RIS, aligning with recent data suggesting an absence of effect of these mutations in predicting cetuximab resistance(39). Further functional studies are warranted to confirm these findings.
In conclusion, we have produced a powerful molecular classifier of the RAS phenotype specific to the CRC setting. We provide strong evidence by translating efficiently our tools across various platforms (Affymetrix, Agilent, RNAseq) and datasets (cell line and xenograft panels, and patient cohorts). Importantly, our classifier improves the identification of KRAS wild-type patients likely to benefit from anti-EGFR monoclonal antibodies. Furthermore, our study suggests the application of MEK inhibitors to patients with a high RAS phenotype. These findings have strong potential for practice changes at the bedside, but require prospective validation in patient cohorts.
Supplementary Material
Translational Relevance.
Modeling the RAS phenotype in CRC allows for the robust interrogation of RAS molecular dependencies across cell lines, xenografts, and patient cohorts. It demonstrates clinical utility in predicting response to anti-EGFR antibodies and MEK inhibitors.
Acknowledgments
Funding source: Justin Guinney, Charles Ferté, Erich Huang, and Jonathan Derry were supported by the grant U54CA149237 from the Integrative Cancer Biology Program of the National Cancer Institute . Pierre Laurent-Puig was supported by the Programme Biointellligence. Charles Ferté was supported by the Bettencourt Schueller Foundation.
Footnotes
Disclosure of interest: Pierre Laurent-Puig received honoraria from Pfizer, Merck-serono, Amgen, Astra-Zeneca, Sanofi, Roche honoraria; Jonathan Dry, Robert McEwen,, Claus Bendtsen, Kevin Hudson, and Brian Dougherty are employees of Astra-Zeneca.
References
- 1.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. The New England Journal of Medicine. Massachusetts Medical Society. 2004:337–45. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, Köhne C-H, Hitre E, Zaluski J, Chang Chien C-R, Makhson A, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. The New England Journal of Medicine. Massachusetts Medical Society. 2009;360:1408–17. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 3.Lièvre A, Bachet J-B, Boige V, Cayre A, Le Corre D, Buc E, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. Journal of Clinical Oncology. 2008;26:374–9. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 4.Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, De Braud F, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. Journal of Clinical Oncology. 2009:663–71. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- 5.De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. The lancet oncology. Vol. 12. Elsevier Ltd; 2011. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer; pp. 594–603. [DOI] [PubMed] [Google Scholar]
- 6.Mao C, Yang ZY, Hu XF, Chen Q, Tang JL. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Annals of oncology official journal of the European Society for Medical Oncology ESMO. 2011 doi: 10.1093/annonc/mdr464. [DOI] [PubMed] [Google Scholar]
- 7.Oden-Gangloff A, Di Fiore F, Bibeau F, Lamy A, Bougeard G, Charbonnier F, et al. TP53 mutations predict disease control in metastatic colorectal cancer treated with cetuximab-based chemotherapy. British Journal of Cancer. Nature Publishing Group. 2009;100:1330–5. doi: 10.1038/sj.bjc.6605008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet J-B, Lecomte T, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. Journal of Clinical Oncology. Am Soc Clin Oncol. 2009;27:5924–30. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 9.Huang F, Xu L-A, Khambata-Ford S. Correlation between gene expression of IGF-1R pathway markers and cetuximab benefit in metastatic colorectal cancer. Clinical Cancer Research. 2012;18:1156–66. doi: 10.1158/1078-0432.CCR-11-1135. [DOI] [PubMed] [Google Scholar]
- 10.Van Cutsem E, Köhne C-H, Láng I, Folprecht G, Nowacki MP, Cascinu S, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. Journal of Clinical Oncology. American Society of Clinical Oncology. 2011:2011–9. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 11.De Roock W, Di Nicolantonio F, et al. JDJ ASsociation of kras p.g13d mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA: The Journal of the American Medical Association. 2010;304:1812–20. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 12.Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E. Association of KRAS G13D Tumor Mutations With Outcome in Patients With Metastatic Colorectal Cancer Treated With First-Line Chemotherapy With or Without Cetuximab. Journal of Clinical Oncology. 2012;30:3570–7. doi: 10.1200/JCO.2012.42.2592. [DOI] [PubMed] [Google Scholar]
- 13.Peeters M, Douillard J-Y, Van Cutsem E, Siena S, Zhang K, Williams R, et al. Mutant KRAS Codon 12 and 13 Alleles in Patients With Metastatic Colorectal Cancer: Assessment As Prognostic and Predictive Biomarkers of Response to Panitumumab. Journal of Clinical Oncology. 2013;31:759–65. doi: 10.1200/JCO.2012.45.1492. [DOI] [PubMed] [Google Scholar]
- 14.Loboda A, Nebozhyn M, Klinghoffer R, Frazier J, Chastain M, Arthur W, et al. A gene expression signature of RAS pathway dependence predicts response to PI3K and RAS pathway inhibitors and expands the population of RAS pathway activated tumors. BMC medical genomics. BioMed Central. 2010;3:26. doi: 10.1186/1755-8794-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. Journal of Clinical Oncology. 2007;25:3230–7. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 16.Baker JB, Dutta D, Watson D, Maddala T, Munneke BM, Shak S, et al. Tumour gene expression predicts response to cetuximab in patients with KRAS wild-type metastatic colorectal cancer. Br J Cancer. Cancer Research UK. 2011;104:488–95. doi: 10.1038/sj.bjc.6606054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Roock W, Piessevaux H, De Schutter J, Janssens M, De Hertogh G, Personeni N, et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Annals of oncology official journal of the European Society for Medical Oncology ESMO. 2008;19:508–15. doi: 10.1093/annonc/mdm496. [DOI] [PubMed] [Google Scholar]
- 18.Gaedcke J, Grade M, Jung K, Camps J, Jo P, Emons G, et al. Mutated KRAS results in overexpression of DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes chromosomes cancer. 2010;49:1024–34. doi: 10.1002/gcc.20811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muzny DM, Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. Nature Publishing Group. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julien S, Merino-Trigo A, Lacroix L, Pocard M, Goere D, Mariani P, et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clinical Cancer Research. 2012 doi: 10.1158/1078-0432.CCR-12-0372. [DOI] [PubMed] [Google Scholar]
- 21.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–307. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. Nature Publishing Group. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jürchott K, Kuban R-J, Krech T, Blüthgen N, Stein U, Walther W, et al. Cheung VG, editor. Identification of Y-Box Binding Protein 1 As a Core Regulator of MEK/ERK Pathway-Dependent Gene Signatures in Colorectal Cancer Cells. PLoS Genetics. Public Library of Science. 2010;6:19. doi: 10.1371/journal.pgen.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society - Series B: Statistical Methodology. Wiley Online Library. 2005;67:301–20. [Google Scholar]
- 25.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. Biometrika Trust. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 26.Hansen KD, Irizarry RA, Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012:1–13. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. Nature Publishing Group. 2006;439:353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 28.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. Elsevier Ltd. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nature Reviews Cancer. Nature Publishing Group. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knijnenburg T, Schlicker A, Roepman P, Mcdermott U, Huang S, Ho M, et al. MED12 Controls the Response to Multiple Cancer Drugs through Regulation of TGF-b Receptor Signaling. Cell. 2012;151:937–50. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. AAAS. 2002;298:1911–2. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 32.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. Journal of Clinical Oncology. 2007:3230–7. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 33.De Reyniès A, Boige V, Milano G, Faivre J, Laurent-Puig P. KRAS Mutation Signature in Colorectal Tumors Significantly Overlaps With the Cetuximab Response Signature. Journal of Clinical Oncology. 2008;26:2228–30. doi: 10.1200/JCO.2007.15.9186. [DOI] [PubMed] [Google Scholar]
- 34.Imamura Y, Morikawa T, Liao X, Lochhead P, Kuchiba A, Yamauchi M, et al. Specific Mutations in KRAS Codons 12 and 13, and Patient Prognosis in 1075 BRAF Wild-Type Colorectal Cancers. Clinical Cancer Research. 2012 doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C-C, Er T-K, Liu Y-Y, Hwang J-K, Barrio MJ, Rodrigo M, et al. Computational analysis of KRAS mutations: implications for different effects on the KRAS p.G12D and p.G13D mutations. PloS one. 2013;8:e55793. doi: 10.1371/journal.pone.0055793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. Jama The Journal Of The American Medical Association. Am Med Assoc. 2010;304:1812–20. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 37.Van Cutsem E, Köhne C-H, Láng I, Folprecht G, Nowacki MP, Cascinu S, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. Journal of Clinical Oncology. American Society of Clinical Oncology. 2011:2011–9. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 38.Seymour MT, Brown SR, Middleton G, Maughan T, Richman S, Gwyther S, Lowe C, Seligmann JF, Wadsley J, Maisey N, Chau I, Hill M, Dawson L, Falk S, O’Callaghan A, Benstead K, Chambers P, Oliver A, Marshall H, Napp VQP. Panitumumab and irinotecan versus irinotecan alone for patients with KRAS wild-type, fluorouracil-resistant advanced colorectal cancer (PICCOLO): a prospectively stratified randomised trial. Lancet Oncology. 2013;14:749–59. doi: 10.1016/S1470-2045(13)70163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jonker D, Karapetis C, O’Callaghan C. BRAF, PIK3CA, and PTEN status and benefit from cetuximab (CET) in the treatment of advanced colorectal cancer (CRC): Results from NCIC CTG/AGITG CO.17. J Clin Oncol. 2012;30:2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.