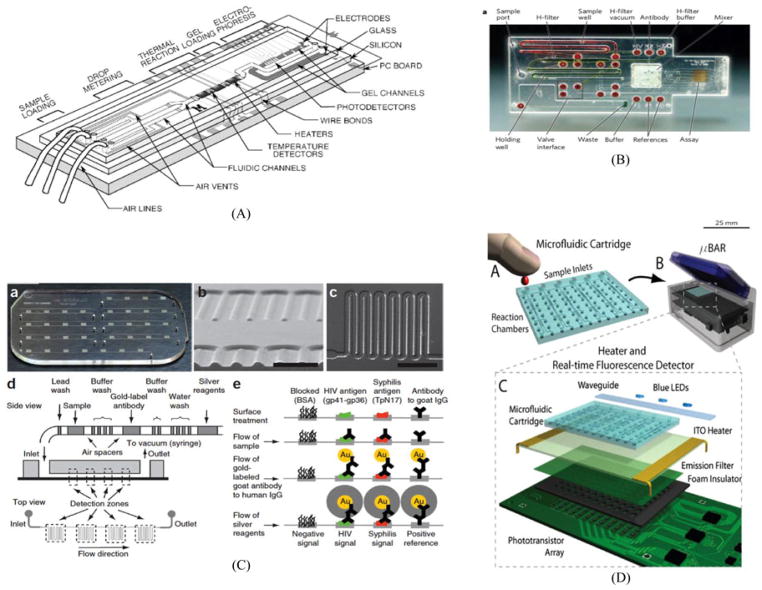
Abstract
Integrated molecular diagnostic systems (iMDx), which are automated, sensitive, specific, user-friendly, robust, rapid, easy-to-use, and portable, can revolutionize future medicine. This review will first focus on the components of sample extraction, preservation, and filtration necessary for all point-of-care devices to include for practical use. Subsequently, we will look for low-powered and precise methods for both sample amplification and signal transduction, going in-depth to the details behind their principles. The final field of total device integration and its application to the clinical field will also be addressed to discuss the practicality for future patient care. We envision that microfluidic systems hold the potential to breakthrough the number of problems brought into the field of medical diagnosis today.
Index Terms: Diagnostics, integration, microfluidics, point-of-care (POC)
I. Introduction
IN comparison to the commonly equipped hospital laboratory, a majority of on-site medical field ranging from international military zones to areas of rural and impoverished isolations have lagged behind the rest of the world in terms of health care. While many technological advances in the developed world have allowed the average hospital to successfully treat its patients, several key challenges oppose similar gains to the rest of the world. In an effort to alleviate the equipment burden, significant growth in the field of microfluidics as depicted by reviews such as Fiorini and Chiu has propelled the concept of “lab on a chip” as a viable alternative [1]. When in a developing area, the need for careful sample preparation and device design is critical to long lasting medical diagnostics. Crude samples, whether it be blood, urine, or saliva, must first be treated before the proper biomolecules can be detected. Such prerequisites call for buffer dilutions in addition to several other prediagnostic preparation steps for sample retrieval and conditioning. Methods of on-chip sample preparation, sample mixing, and cellular lysis have to preserve the biomolecules necessary, while at the same time create a simple, durable, and energy-efficient device design for any resource applications. Once preserved and filtered, sensitive, low-powered sample amplification and detection of desired components becomes another obstacle to a POC platform. Biomolecules of interest such as DNA, RNA, and proteins become primary methods of disease detection and individually dictate separate collection of treatment protocols. Although each biomolecule demands its own set of amplification steps, a device which collectively tests for all three allows for a series of confirmatory steps necessary for both a precise and accurate diagnosis. Integrating the entire diagnostic process on a single, easy-to-use device to effectively apply in a clinical setting plays the final lock in bridging the gap between the developed and developing world. A fully functional and efficient integrated molecular diagnostic system (iMDx) platform must be able to accurately detect the desired biomarkers while independently handling a biospecimen from retrieval to signal transduction. Creating an elegant but durable design incorporating all components of sample treatment and detection will offer the next step to solving clinical issues with a greater level of simplification for both a developed and developing setting. As of now, elegant reviews written by Vo-Dinh and Cullum, and Bashir et al. have already noted the timed development of biosensors and separate on-chip components aiding in medical diagnostics [2], [3]. This review paper focuses on the field of microfluidics to comfortably fulfill the necessary medical niche in low-resource settings, and will discuss critical criteria to achieve reliable and affordable diagnostic systems. Each section of the review will be equipped with a timeline portraying the series of developments leading up to possible integrations into an iMDx platform.
II. Sample Preservation and Preparation
A. Sample Preservation
Sample/reagent preservation is a critical component for POC platforms in limited resource settings. In order to ensure a proper diagnostic performance, any necessary POC reagents or collected sample biospecimens need to be preserved during storage or transport for downstream assays. Integration of sample/reagent storage and downstream assays on a microfluidic system would significantly relieve demands from end-users, thereby offering a very viable solution in enabling an entire sample-to-answer platform in limited resource settings. Within the past decade, numerous strides have been made in developing and advancing on-chip sample preparation techniques for potential use in an iMDX platform (Fig. 1).
Fig. 1.
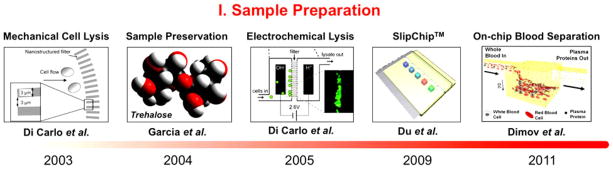
Chronicle development of on-chip sample preparation system: microfabricated nanobarb for mechanical cell lysis (2003) [37], sample preservation utilizing a naturally reoccurring organic compound (2004) [8], on-chip electrochemical integration for membrane disruption (2005) [31], innovative method for soft sample mixing through a SlipChip system (2009) [48], and gravity-driven fluid dynamic blood–plasma separation (2011) [19]. Reproduced from [8], [19], [31], [37], and [48] with permission.
A bioinspired material for sample preservation has recently received much attention, using a trehalose sugar-based glassification found in nature. The material has been modeled by tardigrades, known as water bears, and brine shrimp going through a phenomenon known as anhydrobiosis [4] as an active response to adverse environments to survive in extreme dehydration and elevated temperatures for over 120 years. This survival mechanism arrests their metabolic activity and turns each organism into a “tun” state after complete dehydration [5]. The trehalose within these anhydrobiotic organisms protects important biological elements (e.g., DNA, RNA, proteins, membranes, and cellular systems) from harsh environments by the formation of a glass-like shell, securely “shrink-wrapping” DNA and protecting it against degradation. It has a higher glass transition temperature than that of other general sugars, enabling glassification at an elevated temperature to ensure the use of trehalose only in extreme cases [6]. Protective effects of trehalose can be accounted for by two stabilizing mechanisms; the stabilization of membranes and lipid assemblies at very low levels of hydration, and the stabilization of biological macromolecules in the folded state under conditions that would normally promote their denaturation [7]–[10]. The unique properties of trehalose make it a sufficient candidate for long-term on-chip dehydration preservation at ambient temperatures. Other components such as sucrose and dextran have also been shown to complement the trehalose preservation of biomolecules. The Yager laboratory at the University of Washington showed a good example utilizing trehalose in a microfluidic system by performing a controlled release of dry reagents with temporal and spatial distribution upon rehydration [6], [8]. The lab developed a final formulation which consisted of 5% sucrose, 5% trehalose, and 1% BSA. Their experimentation showed the utility of storing reagents separately and then combining them immediately prior to use in a simple POC platform.
In an effort to advance the trehalose, Biomatrica, the Biostabiliity Company, has developed multiple commercial products based on the trehalose technique to stabilize and preserve DNA/RNA/enzymes/proteins either in purified form, whole blood, saliva, or tissues at room temperatures for up to 12 years. The use of these products are reflected in a series of publications including forensic applications [11], [12], sample preservation after collection [13], and storage of oligonucleotides [14].
An alternative material for sample/reagent preservation is sol–gel. Park et al. have developed a novel formulation of nanoporous sol–gel microarrays with physical properties applicable for protein immobilization and RNA–protein interaction in a microfluidic format [15]. The sol–gel system is capable of holding an extreme level of sensitivity (femtogram scale), due to the large amounts of active proteins that can be preserved in a 3-D dual-nanoporous sol–gel matrix. Sol–gel technology functions perfectly without the need for affinity capture tags or recombinant proteins, enabling the entrapment of proteins in their native state.
A method of freeze-drying, also known as lyophilization, has also been used as a method for sample and reagent storage in a wide variety of settings. Although lyophilizaiton does provide long-term stability to reagents, both freezing and drying steps have been known to add an additional stress to designated reagents. Research in lyophilization protocols have led to numerous lyoprotective effects through the addition of certain protective compounds beforehand, and continue to show great promise in the future [16], [17].
B. Sample Preparation
Sample preparations are typically performed in laboratory settings with time consuming, multistep, and labor-intensive processes. In order to fulfill the requisites for a fully functional POC platform, the capability of an autonomous, efficient, and multiplexed sample preparation method is vital for any selected downstream assays. Unfortunately, due to the diverse source of biospecimens, each with their respective source of detectable biomolecules, the development of both a wholesome and simple sample preparation technique becomes a challenge to implement in device designs.
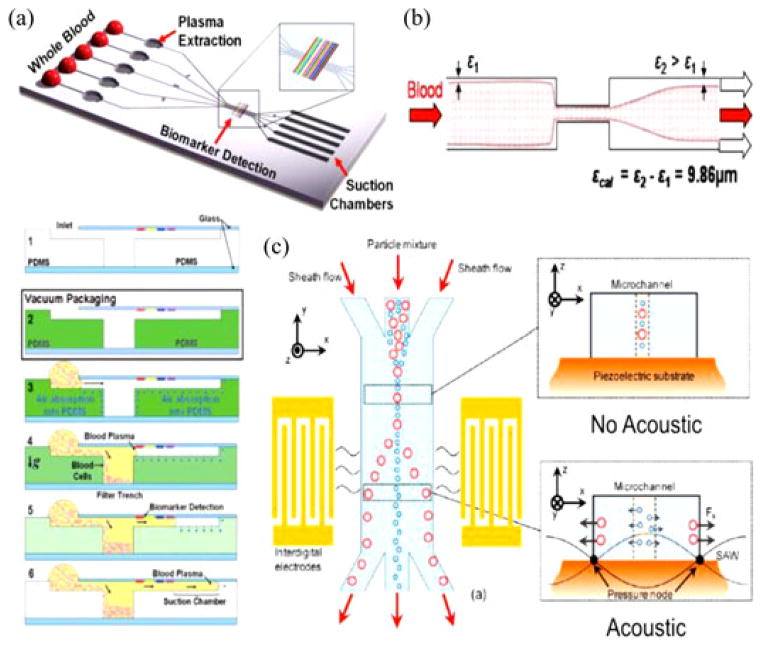
The presence of bacteria cells in crude blood samples offers direct confirmation toward the presence of pathogenic material. Recently, a microfluidic device developed by Ai et al. utilized surface acoustic waves (SAWs) to separate particles and cells of varying properties [18]. Acoustic radiation force, highly dependent on a component’s mechanical characteristics, would thereby separate unidentical cells toward different nodes and ultimately different outlets as seen in Fig. 2. Using this microfluidic device allows the separation of bacteria and normal blood cells, providing specific organisms for efficient downstream analyses.
Fig. 2.
(a) Schematic diagram for SIMBAS. Cross-sectional views describing the principle of microfluidic trench-based filtration. The presence of degas-driven flow removes the need for an external pumping system. Reprinted from [19] permission from the Royal Society of Chemistry (RSC). (b) Restriction effect of blood separation. (a) Schematic view of the blood flow in microfluidic channel. (b) A 1:20 diluted sample injected at 100 μL/min with an 18-μm difference after restriction enables the device to separate. Reprinted from [28] with permission from Springer. (c) Standing SAWs enable effective separation of bacteria from blood. Reprinted from [18] with permission from ACS.
In certain cases, the preservation of whole blood enables diagnosis of unique diseases. Specific pathogens, such as plasmodium, parasites (malaria), and HIV virus, replicate in blood cells and have higher counts than in plasma. For serum analysis, once blood cells are filtrated, pathogens in the serum will be lysed and the DNA/RNA/proteins will be stored.
To endeavor autonomous sample preparation, our laboratory have developed SIMBAS (Self-powered Integrated Microfluidic Blood Analysis Systems) as shown in Fig. 2 [19]. To effectively harvest the pathogenic information, we successfully separated a large portion of blood cells and plasma through a sedimentation-based sample fractionation system. Well-optimized trenches separate blood cells (red blood cells and white blood cells) effectively allowing for the extraction of human genes and proteins (i.e., hemoglobin). The downstream plasma after an array of trenches contains smaller and lighter cells such as bacterial, plasmodium, and viral cells, which can be delivered to a lysis module for further pathogenic module separation. An advantage of SIMBAS is that it is self-powered by a prevacuumed polymer, polydimethylsiloxane (PDMS). This becomes a simple solution for powerless microfluidic operations that can be optimized for various field applications like a POC device.
The need for continuous flow separation methods has also led to the emergence of several innovative methods of mechanical separation, most notably in the field of microfiltration. Selective segregation based upon particle size differences between red blood cells (2 μm) and white blood cells (5 μm) allows for varying degrees of extraction before complications in critical pore size, blood passing capability, and trapping or filtering efficiency. Recently however, research by Ji et al. has led to filtrate designs appropriately named U-Filter and Snake, demonstrating an efficient cross-flow design while allowing for high blood passing capability [20]. Microfilter devices, operating entirely on capillary action and transverse-flow, also play a role in a passive on-chip method for blood plasma separation [21]. Transverse or cross-flow filtration produces high separation efficiencies by reducing the accumulation of clogging elements on the filter face. Filtration is thus a highly efficient, but short-lived preparation method. Problems with clogging, preferential low flow rate, and typical blood dilution requirements require the development of innovative designs to properly fulfill POC practicality.
As opposed to mechanical separation, Haeberle et al. implemented a decanting chamber on a rotating lab-disk to extract plasma by means of centrifugation [22]. While such a device requires a rotating motor, further research has led to designs inducing the centrifugal force by flow through curved channels. Sollier et al. developed a spiral design allowing for injection in the center, where the centrifugal force induces flow in the curved channels to obtain continuous fragmentation and flow particulates and plasma into an end outer and inner channel, respectively [23]. Unfortunately, while the centrifugal force makes particles with a higher density than plasma travel toward the outer channel wall, the presence of a Dean drag force from high flow rates causes particles to circulate and mix. Recent development of a 180° bend device is suggested to allow opposing Dean forces to contribute in separation, prevailing over the centrifugal effect [24], [25]. Sample preparation through use of centrifugation supports a high flow rate, simple fabrication, no clogging design that through further development in coupling with hydrodynamic principles shows great promise.
Hydrodynamic effects, rather than particle retention or the use of external centrifugal forces, exploit a natural phenomenon in blood flow found in networks of branched blood capillaries through the use of channel bifurcations. Blood flows in microcirculation are characterized by a narrow cell-free layer located nearby channel walls. Several devices have been noted to use wide and shallow channels in a rectangular cross section to create an inverted T-bifurcation with one inlet, the feed channel, and two outlets, a large drain channel with a smaller perpendicular plasma collection duct [26], [27]. A similar device proposed by Faivre et al. utilizes the cross section of a microfluidic channel to modify the spatial distribution of cells downstream of a narrowed channel and increase the cell-free layer adjacent to the boundary [28] as shown in Fig. 2. According to the hydrodynamic effect, cells are drawn into the higher flow rate drainage vessel from the asymmetric distribution of shear forces on the surface of cell, allowing plasma to then enter the two outer outlets. Yang et al. experimented in increasing the total plasma volume by placing five parallel plasma channels within the device instead of a single bifurcating region [29]. Higher volumes of plasma were thereby extracted allowing for greater concentration of desired biomolecules for downstream analysis. The hydrodynamic effect ultimately implements a robust and efficient blood plasma separation method utilizing a high flow rate. Unfortunately, a low extraction yield due to a limited number of bifurcating channels gives room for further design improvement.
Cell lysis is another desired but an optional component of the sample preparation in fulfilling iMDx. Any pathogenic cells, bacteria or viruses in plasma can undergo lysis—breaking down biological containment and enabling downstream molecule-based detection. Lysis modules can also be tuned for the selective disruption of mammalian cells when whole blood is contaminated with bacteria or other pathogen. Selective lysis allows for the widest flexibility of the sample output for downstream assay (lysate of select pathogens).
1) Electrochemical Lysis
Electrochemical lysis allows one to selectively lyse the cells in the blood sample. Since there are no chemical reagents involved, electrochemical lysis would not interfere with downstream assay. Low power will also be possible since electrical fields can be highly concentrated in microscale geometries [30]. Research with alkali solutions has also allowed researchers to create an on-chip cell lysis based on local hydroxide electro-generation [31], [32]. Hydroxide ions, generated electrochemically within the device by a palladium electrode, irreversibly porate the membrane of a cell and lead to cellular lysis.
2) Chemical Lysis
Chemical disruption is a common and theoretically simple method of cellular lysis used in a wide range of experiments. At its core, the lysis revolves around the use of detergents or alkali materials to chemically deteriorate/solubilize the proteins and lipids present within the membrane of targeted cells. Recent discoveries have led to the integration of chemical lysis in bioanalytical assays on a microfluidic chip. Schilling et al. developed a relatively simple diffusion-based microfluidic system that allows the continuous lysis of bacterial cells in addition to the fractionation and detection of a large intracellular enzyme [33]. A cell suspension and a chemical lytic agent enter through separate inlets into the lysis channel, where the two liquids mix only through lateral diffusion. In a very similar integration, Irimia et al. use a microfabricated device for the controlled mixing of a picoliter cell suspension and lysis solution [34]. Virtual walls formed by pockets of air within the fluids were allowed to expand and detract through electrically-driven heaters, pressurizing the liquids and forcing them to move from channel to channel in intended directions. Drawing the air out of the capillary allowed a mechanically gentle nature of lysis, mixing the cell solution and chemical lysate together. Another device developed by Sethu et al. notably achieves complete lysis of erythrocytes and approximately total recovery of leukocytes by exposing cells to an isotonic buffer for 40 s [35]. Methods using the device for purposes of a massively parallel lytic experiment can reportedly process several milliliters of whole blood in less than 15 min. While the device has focused specifically on erythrocytes, adaption of the platform mechanism may allow it to become incorporated on a fully integrated device for future sample preparation. Chemical methods are particularly attractive because extensive experience and well-established protocols for large samples are available. Unlike other lysis methods though, a separate fluid or lysate is typically needed to treat the experimental cell suspension.
3) Acoustic Lysis
A relatively uncommon, acoustic lysis involves the use of ultrasonic waves to generate localized areas of high pressure and create cavitation. Cavitation, referring to the formation and instant collapse of cavities (bubbles or nuclei) within a liquid, in turn destroys nearby cells and performs lysis. To do this, an equipment such as a horn, bell, or other specialized tool is necessary to create the proper acoustic waves. Taylor et al. discovered a microfluidic model that rapidly captures bacterial spores on a filter, washes the spores with water, and then disrupts the spores in the presence of glass beads by applying ultrasonic energy through a thin-film flexible interface [36]. By placing the ultrasound horn tip in contact with a flexible interface, pulses of positive pressure, temperature increase, and streaming flow acted together to get the beads moving in violent motion and produce a sufficient force to achieve cell disruptions. With relatively high lysing speeds, ultrasound lysing destroys the integrity of the cell wall and provides an extract of intracellular components (typically DNA) in an efficient and uninhibited manner. This method however has the disadvantages of a nonuniform energy distribution, large volume from the ultrasonic transducer, and high-power consumption from the general ultrasonic operation. Costly and difficult to reproduce, acoustic lysis would have to undergo several price and equipment changes before becoming practical for iMDx.
4) Mechanical Lysis
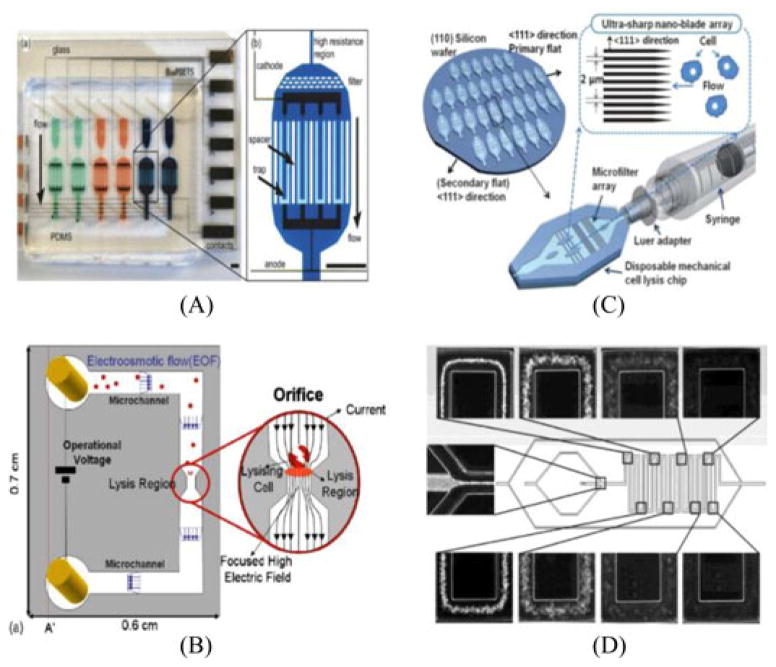
Biological cells are not rigid particles. This philosophy is the key to mechanical lysis. Mechanical lysis occurs when the cell membrane (and/or cell wall) is sheared, punctured, pressured, or mechanically disrupted in some way to release the contents of its cytoplasm. Di Carlo et al. discovered a micromechanical filter where cells, forced through at a certain flow rate, would come into contact with a series of nanoscaled barbs (nanoknives) [37]. Cells passing through the filter would first be punctured by the nanoknives, and then proceed by forming and disassociating multiple protrusions of the cell body into the gaps between the knives. Improvements to this design were developed by Yun et al. by fabricating handheld ultrasharp nanoblade arrays which would lyse cells both at a faster speed and minimization of the lysate dead volume [38]. An alternative design was discovered by Kim et al. that uses pure mechanical stress with a deflected PDMS membrane between two microchannels [39]. As the pressure from the controlled channel begins, compressive stress is directly applied to the cell through the PDMS. Increasing the pressure slowly brings all cells into direct contact with the deflected membrane, forcing cells to begin to burst and release their intracellular content. Unfortunately, construction of a microfluidic device incorporating the aspects needed for mechanical lysis (nanoknives, PDMS, and beads) may become complex and costly. To improve, research must be made in finding simpler and more effective methods for not only lysing, but also obtaining the biomolecules necessary for downstream analysis.
5) Thermal Lysis
One of the more firmly established techniques for DNA analysis, thermal lysis has made a strong presence in numerous research groups. Heat produced from an external generator is applied to targeted cells, and as a result, poration occurs within the cellular membrane. Given enough energy, the formation of pores becomes severe and irreversible, thereby leading to lysis and a release of intracellular components. Waters et al. created a microfluidic device incorporating thermal cell lysis, multiplex PCR amplification, and electrophoresis of the amplified products [40]. In order to perform the lysis, however, the entire microchip was thermally cycled in a commercial thermal cycler. More recently, Privorotskaya et al. reported a rapid thermal lysis of cells through the use of silicon–diamond microcantilever heaters [41]. These microcantilever heaters, fabricated from selectively doped single crystal silicon, provide a uniform distribution of local resistive heating and cell immobilization on the cantilever surface. After electrically heating the cantilevers, all the cells compromised, with no clear intact membrane observed. DNA molecules were seen to still reside on the cantilever surface during the lysis process, allowing for potential downstream analysis. Thermal lysis offers a reliable and straightforward method to the extraction of both bulk and single-cell DNA. Unlike other lysis methods, the clean application of heat requires only a simple device and avoids all the complications of lysate purification, high-voltage safety, or complex device manipulation. However, despite the numerous advantages, necessity for an external power source adds both to the cost and storage of thermal experimentation. High temperatures also denature a majority of non-DNA biomolecules within the cell, and the violent bursting of the cell membrane can potentially create a lysate in need of purification.
6) Electrical Lysis
Due to its speed and reagentless feature, electrical lysis has become a growing field in bioanalytical research. The science behind electrical disruption revolves around the fact that the bilayer structure of a cell membrane is dielectric. When a cell becomes exposed to an external field, a trans-membrane potential is produced and the attraction of opposite charges between the inner and outer membrane generates the compression pressure. The resulting cell membrane becomes thinner and increasingly permeable, and if the electrical field strength surpasses a critical value, an irreversible breakdown of the cell membrane will cause the cell to spill its intracellular content. Integrating several steps required for bioanalytical experimentation, McClain et al. created a device that incorporated cell handling, rapid cell lysis, electrophoretic separation, and detection of fluorescent cytosolic dyes [42]. Loaded cells were then hydrodynamically transported to a region on a microfluidic device where they were focused and rapidly lysed using an electric field. The voltage applied to lyse the cells was supplied by a function generator coupled with an ac amplifier (with dc offsets) in a desire to avoid electrolysis. Although the device offers a relatively fast and efficient method for full biomolecular analysis, after 45 min of continuous use, the separation performance on the microfluidic device began to degrade. Contrary to the use of an ac current, Wang et al. developed a microfluidic flow-through device with a specialized geometric modification for high-throughput electrical lysis based completely on the continuous dc voltage [43]. Although a dc field simplifies the need of using electrical pulses, it generates bubbles due to the electrolysis of water and Joule heating associated with the high field strength needed. By geometrically modifying the size of a microchannel, the overall voltage needed to lyse the cells dramatically decreases. In a very similar discovery, Lee and Cho added on to the idea of a geometric modification by creating a continuous and low-voltage cell lysis device in which the width and length of a channel constrict to generate focused high electric field strength for cell lysis as shown in Fig. 3 [44]. More recently, Bao et al. created a simple microfluidic device that physically traps and then performs electrical lysis on E. coli cells [45]. The device integrates the capture of cells using a microscale bead array that acts as a microscale matrix filter. The microfluidic bead array then provides an ideal, reusable platform for the incorporation of applied periodic electrical pulses in the channel. Electrical lysis stands as an advantageous method due to both its rapid lysis and selectivity toward the cell membrane, leaving organelle membranes and a majority of other intracellular biomolecules undamaged [46]. Many biochemical reactions occur on time scales of seconds or less, and as a result, the microsecond speed of electrical disruption becomes vital in the analysis of cellular properties such as enzymatic activity. However, due to the need for an external electrical voltage, cost and complexity of the procedure may substantially increase. Device fabrication and control may also be relatively difficult, and electrodes may have a very limited lifespan and restrict further experimentation. While electrical lysis is more complicated and costly than methods such as thermal, its speed and efficiency may be a beneficial tradeoff for certain experiments.
Fig. 3.
(A) Integrated microfluidic cell culture and lysis on a chip. Electrodes are on either side of the trapping region, which is preceded by a high resistance, or pinched, section. Scale bars are 1.3 mm. Reprinted from [32] with permission of RSC. (B) The schematic view of the electrical lysis device including the orifice. The electric field was highly focused through geometrical constriction of the fluidic channel. The concentrated electric field effectively lysed cells in a brief period of time. Reprinted from [40] with permission of Elsevier. (C) On-chip development of ultrasharp nanoblade arrays improving previous nanoknives mechanical lysis experimentation. Reprinted from [38] with permission of RSC. (D) On-chip chemical lysis using continuous flow for rapid erythrocyte lysis. Lysis efficiency correlated with the distance traveled in the microfluidic system. Reprinted from [35] with permission of ACS.
Another critical part to sample preparation is the mixing of reagents with collected samples as a precursor for satisfying requirements of any downstream assay. So far, many microfluidic systems have utilized valve-based systems to route the sample to many reaction chambers [47]. The Ismagilov laboratory at Caltech has introduced an innovative technique, so-called, “SlipChip” to perform multiplex microfluidic reaction by mixing without any pumps or valves [48]. No bonding during fabrication of devices and no active/passive valves for operation are very attractive features as a POC diagnostic platform. Similar techniques were introduced based on the preloaded plate-based method [49], [50]. Many other sample preparation methods exist such as a preconcentration module for concentrating extremely low analytes [51] and embedded actuation system (e.g. electrochemical pump [52].
III. Signal Amplification and Transduction
In order to perform reliable signal transduction, biospecimens and their related signals must be amplified immediately after sample preparation. Signal enhancement of desired biomolecules ranging from oligonucleotides to proteins will greatly enhance laboratory or physician disease detection through the observation of key biomarkers. In the case of infectious diseases, one can aim at amplifying strain specific DNAs or mRNAs for verifying the existence of foreign entities such as virus (HIV) and bacteria [Malaria, Tuberculosis (TB)]. In addition to infectious diseases, diseases caused by genetic anomalies such as cancer can be diagnosed by detecting certain abnormalities in specific gene expressions. A common example arises in lung cancer, where patients are known to express three times the amount of the hTERT gene as fragments found as circulating DNA in blood [53]. Similarly, lung cancer patients on average are found to express 2.3 times more exosomal miRNA than a typical control group [54]. As depicted in Fig. 4, the timely progression of several key experiments are coming up as stepping-stones for future avenues of integrating signal amplification and transductions on a self-reliable iMDX device.
Fig. 4.
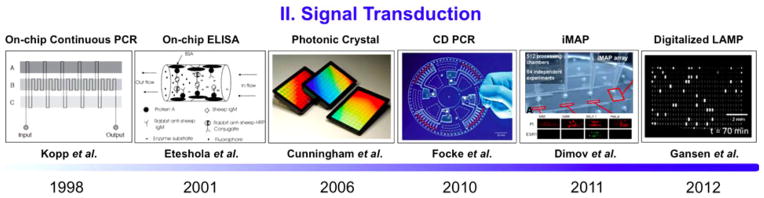
Historical advancement of on-chip amplification for signal transduction: spatiotemporal variation in temperature allows for quick DNA amplification (1998) [55], on-chip integration of ELISA for protein detection (2001) [81], photonic crystal structures enabling label-free and ultrasensitive protein detection, (2006) [87], CD platform for on-chip PCR to detect Methicillin-resistant Staphylococcus aureus (2010) [61] integrated microfluidic array for simultaneous gene and protein expression monitoring from a few cells (2011) [97], and digital improvement of bench top LAMP into on-chip integration (2012) [68]. Reproduced from [55], [61], [68], [81], [87], and [97] with permission.
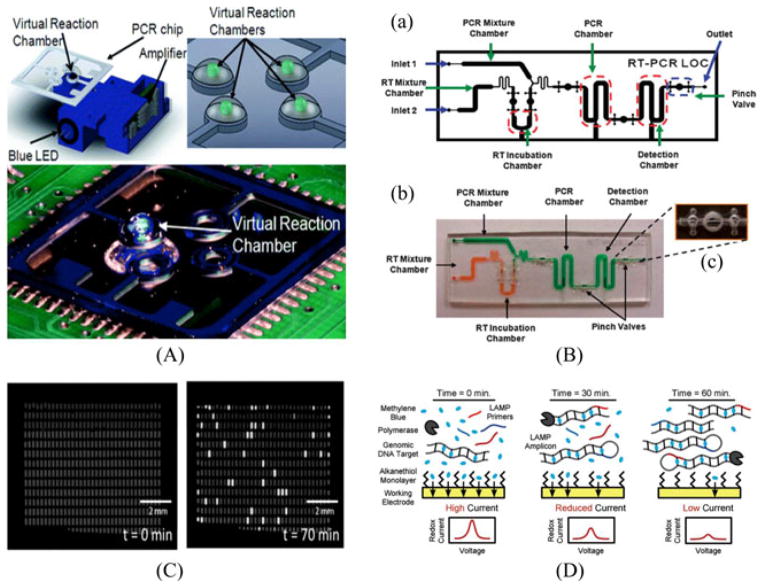
All genetic material, including DNA, RNA, and miRNA, can be specifically sequence-based amplified through a common laboratory practice known as polymerase chain reaction (PCR). Initial attempts for on-chip integration of the PCR were made in 1998. A report published in Science demonstrated the use of a micromachined chemical amplifier to perform the PCR in a continuous flow at a high speed [55]. The research group converted standard thermocycling into a spatiotemporal flow pattern, having liquid flow through two defined heat blocks for annealing and denaturation of nucleic acids, with an entire amplification achieved in approximately 10 min. In the same year, Burns et al. demonstrated the use of an integrated nanoliter DNA analysis device with microheaters, thermistors, and detectors in a microfluidic platform [56]. Many small but noticeable variations have additionally tried to accomplish the PCR on a small scale as shown in Fig. 5 [57]. Kim et al. show the successful PCR on chip in nanodroplets dispersed in an oil phase based on the application of low-power (~30 mW) laser radiation as an optical heating source [58]. For RNA species, the reverse transcription step was additionally incorporated into a single chip for POC clinical diagnostics [59]. An additional DNA extraction step was added to a microchip along with the real-time PCR for rapid pathogen detection [60]. Various other integrated PCRs on-chip have been reported over the two last decades [61] and their review can be found in Lab Chip Focus [62]. Although the design and applications of many on-chip PCRs vary widely, utilizing microfabricated devices holds a clear advantage in demonstrating enhanced analytical performance, superior component miniaturization, enabling high-throughput, and improved automation.
Fig. 5.
(A) Drawing of the on-chip PCR system with the optical configuration underneath. The integrated heater enables the light to interact with the sample placed above the heater. (Top right) A figure of four reactions at the glass on the top of the heaters. (Bottom) Photograph of the total system for PCR amplification. Reprinted from [57] with permission of RSC. (B) On-chip integration of reverse-transcription PCR for detection of pathogenic RNA species. Reprinted from [59] with permission of RSC. (C) Digitalized quantification of lambda DNA concentration by LAMP. An array of 535 wells is shown before (left) and after (right) incubation at 65 °C for 70 min. By counting the number of chambers with florescence, one can conjecture Reprinted from [68] with permission from RSC. (D) Microfluidic electrochemical detection via LAMP to detect pathogenic DNA for POC purposes. Reprinted from [69] with permission of Wiley.
Unfortunately, the nature of the PCR thermocycling makes it a challenge to integrate onto an existing microfluidic system. Advancements by groups such as Shah et al. have worked on noncontact heating through microwave energy to simplify and improve amplification setups [63]. Still, the PCR unavoidably necessitates a cycling of temperatures, adding significant power consumption, complexity, and cost to the whole process. In order to compensate the thermocycling steps of the PCR, isothermal amplification methods have recently received much attention due to its simplicity in maintaining the same temperature during the entire amplification. One isothermal amplification method known as LAMP (loop mediated isothermal amplification) incorporates a versatility that allows it to function on many different microfluidic POC systems. LAMP is a single-tube amplification process in which the DNA of interest is amplified using a set of primers at a constant temperature of 65–70 °C [64]. LAMP also has the ability to amplify miRNA species, which are 25 bases long on average. This amplification scheme is especially amenable to adaptation onto a portable iMDx setup. Li et al. demonstrated ultrasensitive amplification of Let-7a with selectivity by amplifying similar miRNAs to Let-7a [65], [66]. Since Let-7a is closely related to human cancer and plays a key role in suppressing tumors, isothermal amplification methods widen the horizon for possible cancer diagnostics. Our lab has begun to optimize the integration of LAMP-based assays on-chip using a self-powered degassing method for automated molecular detection. Current assays allow for visualization with the naked eye through proper illumination facilitated by fluorescent excitation for quantitative analysis [67]. For further quantitative analysis of existing pathogenic DNA in a sample, Gansen et al. developed a self-digitized LAMP using a microfluidic chamber array. Fig. 5 demonstrated an accurate quantification of relative and absolute DNA concentrations with sample volumes of less than 2 μl [68]. Increasing motivation toward on-chip integration of LAMP has led Hsieh et al. to develop a microfluidic electrochemical quantitative LAMP (MEQ-LAMP) allowing for the rapid, sensitive, and quantitative detection of designated DNA [69]. As shown in Fig. 5, MEQ-LAMP reaction achieves real-time quantitative detection at a high specificity through the employment of six different primers. Overall platform effectiveness has led to the detection of at least 16 copies of genomic DNA of food-borne illnesses in less than an hour, establishing a powerful position in utilizing LAMP for potential iMDx.
Other isothermal amplification techniques have also been developed more recently, and researchers are now trying to incorporate these techniques into a given POC platform. Transcription mediated amplification [70], nucleic acid sequence-based amplification (NASBA) [71], single primer isothermal amplification [72], rolling circle amplification [73], helicase-dependent amplification [74], strand displacement amplification [75], recombinase polymerase amplification [76], and smart amplification process version 2 [77] are the best-known isothermal amplification methods for readily integrated devices using a microfluidic system. The detailed description of these amplification method’s descriptions can be found in [78]–[80].
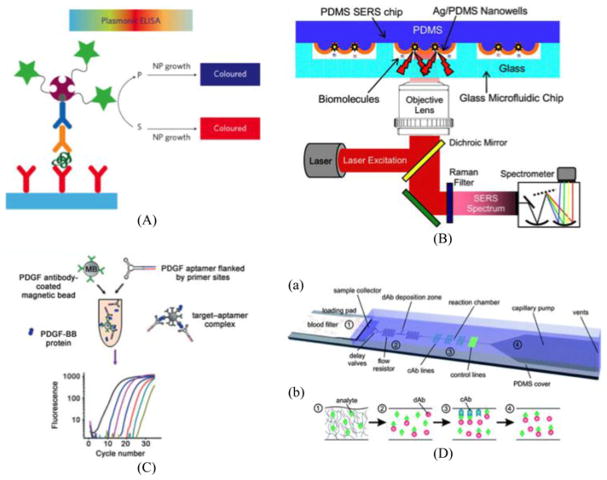
While oligonucleotides play a key role in disease detection, biomarkers, indicators of biological states or conditions, are majorly protein-based. Unfortunately, unlike their nucleotide-based counterparts, proteins cannot be amplified directly by well-known polymerases. Conventional methods for signal amplification have therefore utilized indirect signal transduction such as ELISA (Enzyme Linked Immunosorbent Assay). These immunoassays use an enzymatic substrate to transduce a signal that is proportional to the original quantity of the antigen in the sample. Quantification of these assays then measures the absorbance for the intensity of a given solution color through a plate reader. The ease-of-concept and efficiency of ELISA has made it a gold standard for protein-based amplification; however, complexity in terms of protocol makes it hard to implement in resource limited settings. To solve this problem, many variations on signal transduction have been discovered for POC applications. Lateral flow immunoassay is a variation from ELISA that allows users to visibly distinguish end results with their naked eye. A classic example of a lateral flow immunoassay is thepregnancy kit. The pregnancy kit demonstrates a compact, cost-efficient naked eye detection method for a specific hormone found in the urine of pregnant women (see Fig. 6). Human chorionic gonadotropin (hCG) is targeted for determining pregnancy because of a doubling rate of 72 h after conception, allowing for simple detection through a clean lateral flow immunoassay.
Fig. 6.
(A) A schematic of plasmonic ELISA. Color change by nanoparticle formation enables naked eye detection of pathogenic biomarkers such as p24. Reprinted from [83] with permission from the Nature Publishing Group (NPG). (B) Aptamer-based protein detection scheme aided by micromagnetic beads and microfluidics. Reprinted from [90] with permission from Wiley. (C) A diagram of an integrated microfluidic chip using a Raman imaging system for label-free bimolecular detection. Reprinted from [85] with permission from AIP. (D) Capillary-driven microfluidics for one-step POC immunodiagnostics using antibodies (dAbs) and capture antibodies (cAbs). Reprinted from [82] with permission from RSC.
Unfortunately, most biomarkers targeting infectious and/or genetic diseases are low in abundance, poor in stability, and sometimes go through significant interference incomplex biosamples such as blood. In order to efficiently generalize the use of point-of-care diagnostics, it is necessary to demonstrate ultrasensitive detection schemes through a reliable method of amplification. Initial efforts toward on-chip integration of ELISA was initially developed and characterized by Eteshola et al. The group demonstrated the first preliminary evaluation of a heterogeneous sandwich enzyme-linked immunoassay through an innovative method design in a PDMS microfluidic device [81]. In another effort to integrate reliable protein detection, Gervais et al. incorporated microfluidic functional elements, reagents, and analyte molecules collectively into a one-step immunoassay using a capillary-driven microfluidic platform [82]. Addition of only a small sample volume is noted to trigger a large cascade of events leading from sample preparation to signal transduction through a fluorescence microscope. Amplification and the ability to transduce signals from biomolecules of low quantity are critical to the generalized function of a disease diagnostic device. For signal amplification applications in biospecimens, the Stevens laboratory used nanoparticle growth to obtain significant color change from a blue to red solution in the presence or absence of target molecule p24 for naked-eye detection of HIV (see Fig. 6) [83]. Their data revealed extreme sensitivity in a range as low as 1 × 10−18 g/ml, using clinical trials to confirm the validity against HIV patients. The sera of 30 donors was assayed with plasmonic ELISA with their assay determining HIV infection even with less than 50 copies of viral load from ten patients. In a similar design, our laboratory has developed a plasmonic method to distinguish the presence and absence of amyloid beta through nanoparticle aggregation, enabling the naked eye to detect Alzheimer’s disease diagnostic [84]. However, due to many of these techniques being based on a 96-well plate technology, they are not readily available for sample-to-answer iMDx.
A promising extension of plasmonic application on signal transduction is Raman Scattering or SERS (Surface Enhanced Raman Spectroscopy). As shown in Fig. 6, Liu and Lee utilized a nanowell array fabricated by soft-lithography achieving label-free detection of biomolecules with gigantic signal enhancement of 107 [85]. Park et al. demonstrated a microfluidic and nanofluidic interface to enhance the signal from dengue virus by using SERS [51]. Many SERS-based detection schemes are attractive because of their extreme signal amplification capability; however, this methodology is not readily available for standalone diagnostics since it requires a complicated external signal reader. In addition to SERS, another variation of plasmonic detection includes photonic crystals designed to reflect selected bands of wavelengths when illuminated. The field of biophotonbic crystals offers an alternate label-free and ultrasensitive biochemical or cell-based assay that has been researched extensively by Cunningham et al. [86], [87].
Research in the field of micro- and nanocantilevers has allowed label-free detection of a variety of different biomolecules, on chip. The Craighead laboratory established the world record of measuring the weight of a single virus particle by using a laser actuated cantilever device [88], later on incorporating this ultrasensitive cantilever in a microfluidic platform [89]. Arntz et al. developed a functional label-free protein detection device using an array of microfabricated cantilevers functionalized with covalently anchored antibodies [90]. Biomarker proteins were detected via sensitive measurements of surface stress placed by antigen–antibody molecular recognitions. Sensitivity for myoglobin detection, the biomolecule of choice in the experiment, was found to be as low as 20 μg/ml. Use of micro-cantilevers has been repeated and slightly varied by labs such as Wee et al. [91] and branched into areas of DNA detection as well [92]. Label-free detection also includes fabrications of silicon nanowire devices. Found to be effectively used for a wide variety of biomolecular applications, the general idea revolves around coating the wires with desired receptors and then measuring the conductance through the wires. Several groups have done extensive research into both the fabrication and biomolecular detection capabilities of silicon nanowires, showing promising results for the application to be incorporated for on-chip diagnostic purposes [93]–[95].
Currently, antibodies serve as standard reagents that can bind various disease targets with high affinity and specificity. Antibodies have been extensively utilized for many applications in medical and life sciences research, but have shortcomings in their general use as disease capture reagents for high-throughput capture and detection. For the long-term storage, antibodies also show vulnerability against heat and are susceptible to degradation over time. In contrast to antibodies, target-specific ap-tamers can easily be generated regardless of immunogenicity or target toxicity. Once a specific aptamer has been selected and sequenced, unlimited amounts of the exact same aptamer can be synthesized with little effort and investment [96]. Avoiding the cost of reagent maintenance, storage, and transportation, aptamers are the perfect candidate as an essential recognition component of diagnostic devices [97]. In an attempt to utilize its growing capabilities, Csordas et al. demonstrated indirect amplification of proteins in serum through micromagnetic aptamers PCR technology [98]. After tagging aptamers with micromagnetic beads, the group selected target-aptamer complexes and selectively amplified them with the real-time PCR. Protein signal transduction was ultimately made possible by aptamer amplification. Collectively, aptamers characteristics and current research looking into further applications put them as a strong candidate for integration into an iMDx platform.
Ever since a family of aptamers has been approved by U.S. Food and Drug Administration (FDA) as a therapeutic reagent (Macugen) for macular degeneration by targeting Vascular Endothelial Growth Factor [99], clinical use of aptamers has now become a hot topic [100]. However, one major hurdle to overcome is its low-throughput and labor-intensive selection process, so-called Systematic Evolution of Ligands by Exponential Enrichment). Many microfluidic platforms [15], [101]–[104] have been introduced in order to reduce this burdensome work-flow, but still more efficient and automated platforms are required for clinical use of the aptamers.
Finally, the combination of gene expression and protein immunoassay was developed by Dimov et al. from a few mammalian cells enabling sample prepartion of the input cells with effective perturbation of the cell and simultaneous real-time optical analysis for gene and protein assay [105].
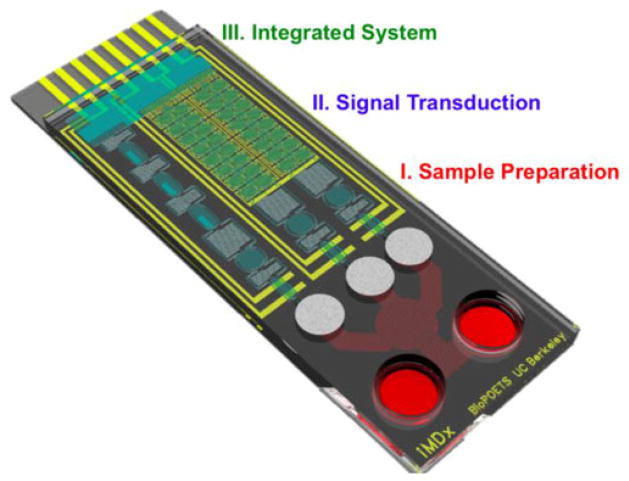
IV. Integrated System
The last part of this review is the integration of modules introduced previously and their possible clinical use in proposed devices with patient samples. Starting with the development of the first pregnancy kit [106], integration has grown as a field of functionality and innovation where numerous modules and inputs/outputs are elegantly arranged for complete optimization [107]. Several attempts for a fully functional integrated platform have already been made (Fig. 7), and reviews such as Soper et al., have already gone over different methods of biosensor integration in field such as cancer diagnostics and general POC devices [108]. General consensus remains that in order to implement highly multiplexed but low-complexity iMDx, the final product should be capable to integrate the technologies required for portable analysis of clinical samples. To add to this discussion, we will also include a review of paper microfluidic systems, a relatively new field that incorporates all necessary modules in one paper platform.
Fig. 7.
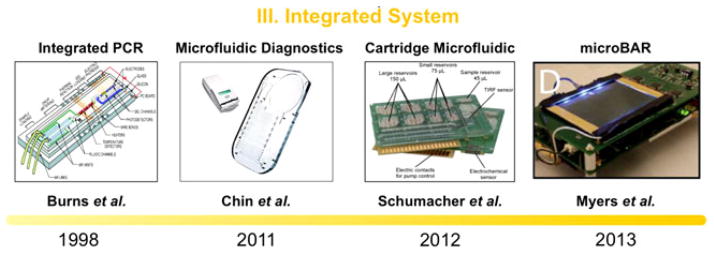
Timeline progression of on-chip system integration: initial attempt in microfabricating on-chip PCR along with several sample detection methods on a single device (1998) [56], fully integrated system brought to developing countries for the detection of infectious disease by incorporating a sample preparation and POC ELISA like assay for amplification (2011) [110], highly-integrated LOC system for multiparameter analysis in a cartridge microfluidic platform (2012) [128], and MicroBAR system to reliably confirm the existence of pathogenic DNA via LAMP (2013) [67]. Parallel development of integrated paper microfluidic including various aspects needed from a fully functional iMDx platforms. Reproduced from [56], [67], [110], and [128] with permission.
Fabrication of a complete sample-in answer-out diagnostic platform slowly came to fruition when Easley et al. connected injected whole blood with endpoint genetic analysis on a single device [109]. The device claims to perform DNA purification and PCR-based amplification all on-chip, connecting modules of sample preparation and amplification together. Injections of nanoliter aliquots of amplicon enabled the device to also perform electrophoretic separation, complementing the first two modules with a form of signal transduction. While the device does contain certain practical drawbacks, it helped pave the way as a sample for a fully integrated POC system.
A model example for a fully integrated diagnostic microfluidic platform nicknamed the mChip (mobile microfluidic chip for immunoassay on protein markers) was developed by the Sia laboratory to be applied for medical testing in areas of Rwanda, India, and Tanzania [110]. The mChip contains the complete integration of multiple microfluidic technologies for manufacturing, fluid handling, and signal transduction based with a miniaturized ELISA protocol. Sample preparation starts by retrieving only several microliters of blood, which can be obtained on site from a needle prick. Reagents are then delivered to the sample through a bubble-based method, where metered plugs of reagents are introduced sequentially into a tubing separated by air spaces, to deliver antibodies, washing solutions, and signal development solutions. Fig. 8 shows signal amplification by on-chip ELISA, followed shortly after by transduction both performed using minimal instrumentation. A device as simple as a cellular phone is then used to detect any signals and create a following prognosis on the presence of HIV or syphilis. Everything in the chip is self-driven with no moving parts, electricity, or external instrumentation needed. Using a reproducible and low-cost fabrication method, the mChip shows promise in clinical settings and is one of the few fully integrated autonomous diagnostic platforms to treat samples from retrieval to transduction. Fluctuations in temperature are recorded to likely affect the performance of the mChip, and while such obstacles are important to overcome to create a fully applicable iMDx device, the mChip is a first step to future diagnostic research.
Fig. 8.
(a) Schematic of an early-integrated PCR on chip integrating microfluidic channels, microheaters, temperature sensors, and detectors. Reprinted from [56] with permission from AAAS. (b) An integrated device for POC assays. The red O-rings were used to interface with external pumping systems. Reprinted from [107] with permission from NPG. (c) An example of system integration and pictures of integrated microfluidic device. The data represent fluid handling of a POC ELISA-like assay. This device is tested in the developing countries targeting Syphilis and HIV. Reprinted from [110] NPG with permission. (d) The design of the MicroBAR system and operation. (a) A disposable microfluidic cartridge is loaded with sample fluid (e.g., blood) via SIMBAS degas-driven flow. (b)–(c) The instrument features blue excitation LEDs on the side of the chip. An ITO substrate provides uniform heating across the reaction chambers of the chip. Below this substrate is a green plastic emission filter followed by a neoprene foam barrier with holes. The microfluidic chip sticks to the surface of the ITO. Reprinted from [67] with permission from PLOS.
Greatly inspired by the simplicity behind pregnancy diagnostics, developments in paper-based iMDx have flourished in recent times by showing the ability to integrate several functional modules into a basic, low-cost paper device (Fig. 9). In an effort to create an innovative, pump-free, and extremely low-cost integrated microfluidic platform, the Whitesides Group at Harvard University helped establish what is now known as the rapidly emerging field of paper microfluidics [111]. Working on the principle of passive capillary-driven flow through patterned paper channels, paper microfluidics has been considered as an easy-to-prototype, inexpensive, lightweight, and biocompatible design offering a useful new method for the fabrication of microfluidic devices. Due to the rapid popularization of the field, many research groups have designed a variety of new fabrication methods starting from baseline photolithography methods [112]–[114] to inkjet etching [115], [116], inkjet printing [117], [118], wax printing [119]–[121], and laser treatment [122]. Methods of simply cutting designs have also led Fenton et al. to a cutting technique where paper patterns are first cut precisely by a computer-controlled plotter and then sandwiched between vinyl or polyester plastic sheets to protect assay surfaces [123]. With fabrication methods moving toward simple and low-cost processes, even the type of paper and the following preparation steps were brought into account. Entire fabrications taking less than 10 min have been recorded, using materials such as an omniphobic RF paper prepared from the reaction of hydrophilic cellulose paper with fluoroalkyl trichlorosilanes in the gas phase [124].
Fig. 9.
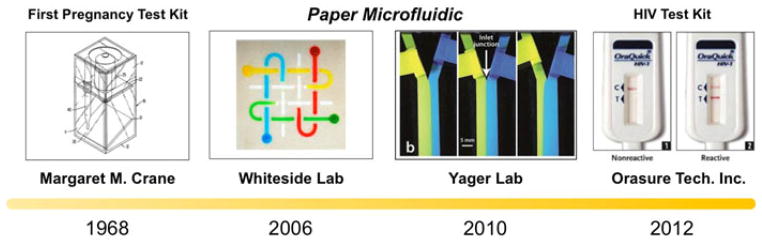
Past progression of paper microfluidic into integrated diagnostic platform: Primary take-home biomolecular testing kit used to detect hCG in a single comprehensive platform (1968) [106], Whiteside lab popularized the concept of using paper as a comprehensive microfluidic device (2006) A figure from [113]. (2010). Low-cost, pumpless T-sensor and H-filter in paper networks (2010) [128]. The first in-home test kit for HIV by swapping gums [135]. Reproduced from [98], [113], [128], and [125] with permission.
In addition to fabrication processes, structure developments have widened the areas of applications for paper platforms. Martinez et al. started off by fabricating 3-D fluidic networks in paper simply by patterning and stacking multiple layers of network designed papers [113] as shown in Fig. 9. The push of paper microfluidics into 3-D expanded the capabilities of the field to incorporate multiplexing tests and multiple functional zones. Key diagnostic amplification methods such as ELISA have also been adapted to fit paper-based platforms, as described by Cheng et al. [125]. Paper-based ELISA, also known as P-ELISA, shows promise in terms of convenience, low cost, and ease-of-use due to its compatibility with paper platforms. Unfortunately, P-ELISA is also less sensitive than conventional ELISA on the order of a magnitude, while also suffering from concerns over contamination and exposed environment conditions when working in tests zones open to the atmosphere. Research from Martinez et al. utilizes the development of 3-D microfluidic paper-based platforms that can be programmed, after fabrication, by the user to generate multiple patterns of flow [126]. These devices are controlled by pressing one-time “on” buttons that closes a small space between two vertically aligned channels and allow fluid to transfer from one channel to the other. The basis of these devices give the end-user freedom in controlling time-sensitive fluid movements between a wide variety of samples, opening up the possibility to choose which biomarkers to test for at which times. Finally, incorporation of electrodes and circuits onto paper platforms is a budding field that has garnered some recent research but has yet to be fully established [127]. In addtion to the White-side lab, the Yager group complimented the advancement of paper microfluidics through the implementation of their own devices [128].
Along with the integration of the functional modules introduced previously, a portable detection system plays as a final component for full iMDx performance in resource-limited settings. Because a variety of signal transduction methods currently exist, corresponding detection modules should accompany methods used to provide direct interpretation of results. In light of this, Schumacher et al. fabricated a device incorporating several diagnostic steps typically seen in laboratories. After necessary sample preparation and amplification, the device performs a diagnostic assay through a self-performing assay protocol, data acquisition, and data analysis in just 15 min [129]. Our laboratory has developed a portable detection system, microfluidic bioamplification reader (MicroBAR) in order to interpret the final stages of iMDx performance throughout a variety of resource-limited areas. This portable instrument, as shown in Fig. 8, uses microfluidic cartridges to perform a rapid genotyping assay on bacterial or viral pathogens in blood, saliva, or other bodily fluids. Powered by a rechargeable lithium polymer battery, it features a touchscreen interface, cell phone and GPS modules, USB interface, and SD card reader for storing assay results for direct interpretation from the genotyping assay. LAMP technology introduced previously was used to detect SNPs of pathogenic DNA for TB [67].
The field of paper microfluidics has garnered much applause recently due to its innovative approach in solving complex fabrication processes and restrictive costs. Valuable advantages of paper microfluidics include: 1) utilization of a common, flexible, and extremely cheap cellulosic material, 2) capillary wicking of solutions that provides a means of transporting fluids without the assistance of an external source, 3) biocompatibility allowing the storage and immobilization of a variety of reagents, and 4) fabrication processes that range between a large variety of equipment, speed, precision, and costs. These components, in addition to new discoveries into structure and fluid flow designs have put paper microfluidics on the forefront of potential integrated medical diagnostic devices in limited-resource settings. Several drawbacks however have been labeled in a detailed review by Li et al. in the use of paper-based platforms [130]. Solutions reaching the detection zone at the end of the device typically retain only 50% of the total initial volume, creating significant concerns with sample retention when the sample quantity is small or retrieval of a large sample is either costly or impractical. Inefficient sample retention presents a problem in low-resource settings where the abundance is a luxury. Certain hydrophobic agents, such as wax, used by previously discussed paper fabrication methods may not be strong enough to withstand samples of low surface tension. This is because hydrophobic materials such as wax that are used to detail patterns on paper do not specifically block the paper’s pores, allowing for low surface tension samples to bypass the barriers and render the device useless. A low limit of detection of paper-based microfluidic platforms for samples of very low concentration presents a third critical drawback. Especially in areas where even slight water contamination or presence of toxin in blood may be critical to health diagnostics, paper microfluidics may be too ill-equipped to sufficiently detect and offer sound diagnosis to the medical staff.
V. Prospectives
Although tremendous advancements have been made in microfluidic diagnostic platforms, the majority of microfluidic works introduced previously is a “proof of principle” or “ validation of single functionality.” To calibrate the readiness of a device for clinical use, the Technology Readiness Level (TRL) matrices can be used as an effective protocol [131]. TRL is a measure to assess the readiness of evolving technologies including materials, components, and devices proposed by the United States Department of Defense. According to its criteria, the reviewed works above are in the range of TRL3 or TRL4, categorized as “Analytic Proof of Concept” or “Standalone component validated,” respectively. In order to integrate each functional module into a unit, the demonstration of a complete iMDx prototype at the Alpha level (TRL5, Integrated Component Validated) is required according to recent DARPA’s RFA (Request for Application, DARPA-11-38 and DARPA-11-39). The demonstrated prototype should be consistent with the requirements for FDA clearance and Clinical Laboratory Improvement Amendment. An Alpha level (TRL 5) iMDx is expected to be self-calibrating, carrying all necessary reagents to test samples along with negative controls. Other requirements to achieve a TRL5 iMDx include an ability to dispose and fabricate at low costs (e.g., injection molding).
As an additional guideline to formulate a sturdy iMDx, we would like to suggest a 3S criteria—speed, sensitivity and selectivity. Successful integration in compliance with the 3S criteria will help commercialize the given product. As an example, the first in-home FDA-approved HIV test, OraQuick, was released in 2012. According to our 3S criteria, the device complies with 1) speed—the result can be obtained in 20 min, 2) sensitivity—antibodies generated by the human immune system three months after suspected incident can even be obtained by gum swab, 3) selectivity—diagnosing HIV infection with more than 99% accuracy. Furthermore, clinical validation regarding the performance of the device can be found in [132]–[135].
A successful iMDx will comply with the 3S criteria suggested above for POC diagnostics in resource-limited settings, showing the necessary level of independence to be fully functional as a standalone platform as shown in Fig. 10. By incorporating the methods of sample preparation, amplification, and signal transduction indicated in this review, future research into the development of an innovative microfluidic platform will undoubtedly offer a simple and efficient alternative to current medical equipment. Furthermore, development of a low-cost, portable, sensitive, and fully integrated iMDx will revolutionize medical diagnostics in numerous settings across the world and the field of diagnostic devices.
Fig. 10.
An example of a schematic drawing of iMDx. A fully functional POC device should include: 1) methods of biospecimen preparation, 2) signal amplification and transduction, and 3) a cost-efficient and easy to fabricate method of integration and application to clinical settings.
Acknowledgments
This work was supported by the US National Institutes of Health under Award U54CA151459 (Center for Cancer Nanotechnology Excellence and Translation).
Contributor Information
Seung-min Park, Email: sp293@berkeley.edu, Department of Bioengineering, and the Berkeley Sensor and Actuator Center, UC Berkeley, University of California, Berkeley, Berkeley, CA 94720 USA, and also with the Department of Radiology, School of Medicine, Stanford University, Stanford, CA 94305 USA.
Andrew F. Sabour, Email: afsabour@berkeley.edu, Department of Bioengineering, University of California, Berkeley, Berkeley, CA 94720 USA
Jun Ho Son, Email: jhson78@berkeley.edu, Department of Bioengineering, and the Berkeley Sensor and Actuator Center, University of California, Berkeley, Berkeley, CA 94720 USA.
Sang Hun Lee, Email: sanghun.lee@berkeley.edu, Department of Bioengineering, and the Berkeley Sensor and Actuator Center, University of California, Berkeley, Berkeley, CA 94720 USA.
Luke P. Lee, Email: lplee@berkeley.edu, Department of Bioengineering, and the Berkeley Sensor and Actuator Center, University of California, Berkeley, Berkeley, CA 94720 USA
References
- 1.Fiorini GS, Chiu DT. Disposable microfluidic devices: Fabrication, function, and application. Biotechniques. 2005 Mar;38:429–446. doi: 10.2144/05383RV02. [DOI] [PubMed] [Google Scholar]
- 2.Vo-Dinh T, Cullum B. Biosensors and biochips: Advances in biological and medical diagnostics. Fresenius J Anal Chem. 2000 Mar-Apr;366:540–551. doi: 10.1007/s002160051549. [DOI] [PubMed] [Google Scholar]
- 3.Bashir R. BioMEMS: State-of-the-art in detection, opportunities and prospects. Adv Drug Del Rev. 2004 Sep 22;56:1565–1586. doi: 10.1016/j.addr.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Westh P, Ramlov H. Trehalose accumulation in the tardigrade Adorybiotus coronifer during anhydrobiosis. J Exp Zoology. 1991 Jun;258:303–311. [Google Scholar]
- 5.Welnicz W, Grohme MA, Kaczmarek L, Schill RO, Frohme M. Anhydrobiosis in tardigrades—The last decade. J Insect Physiol. 2011 May;57:577–583. doi: 10.1016/j.jinsphys.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 6.Fridley GE, Le HQ, Fu E, Yager P. Controlled release of dry reagents in porous media for tunable temporal and spatial distribution upon rehydration. Lab Chip. 2012 Nov 7;12:4321–4327. doi: 10.1039/c2lc40785j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevens DY, Petri CR, Osborn JL, Spicar-Mihalic P, McKenzie KG, Yager P. Enabling a microfluidic immunoassay for the developing world by integration of on-card dry reagent storage. Lab Chip. 2008;8:2038–2045. doi: 10.1039/b811158h. [DOI] [PubMed] [Google Scholar]
- 8.Garcia E, Kirkham JR, Hatch AV, Hawkins KR, Yager P. Controlled microfluidic reconstitution of functional protein from an anhydrous storage depot. Lab Chip. 2004;4:78–82. doi: 10.1039/b308914b. [DOI] [PubMed] [Google Scholar]
- 9.Lins RD, Pereira CS, Hunenberger PH. Trehalose-protein interaction in aqueous solution. Proteins-Structure Function Bioinformatics. 2004 Apr 1;55:177–186. doi: 10.1002/prot.10632. [DOI] [PubMed] [Google Scholar]
- 10.Carpenter JF, Crowe JH. Modes of stabilization of a protein by organic solutes during desiccation. Cryobiology. 1988 Oct;25:459–470. [Google Scholar]
- 11.Lee SB, Clabaugh KC, Silva B, Odigie KO, Coble MD, Loreille O, Scheible M, Fourney RM, Stevens J, Carmody GR, Parsons TJ, Pozder A, Eisenberg AJ, Budowle B, Ahmad T, Miller RW, Crouse CA. Assessing a novel room temperature DNA storage medium for forensic biological samples. Forensic Sci Int Genet. 2012 Jan;6:31–40. doi: 10.1016/j.fsigen.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Allen-Hall A, McNevin D. Human tissue preservation for disaster victim identification (DVI) in tropical climates. Forensic Sci Int Genet. 2012 Sep;6:653–657. doi: 10.1016/j.fsigen.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Stevens DS, Crudder CH, Domingo GJ. Post-extraction stabilization of HIV viral RNA for quantitative molecular tests. J Virol Methods. 2012 Jun;182:104–110. doi: 10.1016/j.jviromet.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez GE, Mondala TS, Head SR. Assessing a novel room-temperature RNA storage medium for compatibility in microarray gene expression analysis. Biotechniques. 2009 Aug;47:667–670. doi: 10.2144/000113209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park SM, Ahn JY, Jo M, Lee DK, Lis JT, Craighead HG, Kim S. Selection and elution of aptamers using nanoporous sol-gel arrays with integrated microheaters. Lab Chip. 2009 May 7;9:1206–1212. doi: 10.1039/b814993c. [DOI] [PubMed] [Google Scholar]
- 16.Wang W. Lyophilization and development of solid protein pharmaceuticals. Int J Pharmaceutics. 2000 Aug 10;203:1–60. doi: 10.1016/s0378-5173(00)00423-3. [DOI] [PubMed] [Google Scholar]
- 17.Chen CJ, Han DD, Cai CF, Tang X. An overview of liposome lyophilization and its future potential. J Controlled Release. 2010 Mar 19;142:299–311. doi: 10.1016/j.jconrel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 18.Ai Y, Sanders CK, Marrone BL. Separation of Escherichia coli bacteria from peripheral blood mononuclear cells using standing surface acoustic waves. Anal Chem. 2013 Oct 1;85:9126–9134. doi: 10.1021/ac4017715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimov IK, Basabe-Desmonts L, Garcia-Cordero JL, Ross BM, Park Y, Ricco AJ, Lee LP. Stand-alone self-powered integrated microfluidic blood analysis system (SIMBAS) Lab Chip. 2011 Mar 7;11:845–850. doi: 10.1039/c0lc00403k. [DOI] [PubMed] [Google Scholar]
- 20.Ji HM, Samper V, Chen Y, Heng CK, Lim TM, Yobas L. Silicon-based microfilters for whole blood cell separation. Biomed Microdevices. 2008 Apr;10:251–257. doi: 10.1007/s10544-007-9131-x. [DOI] [PubMed] [Google Scholar]
- 21.Crowley TA, Pizziconi V. Isolation of plasma from whole blood using planar microfilters for lab-on-a-chip applications. Lab Chip. 2005;5:922–929. doi: 10.1039/b502930a. [DOI] [PubMed] [Google Scholar]
- 22.Haeberle S, Brenner T, Zengerle R, Ducree J. Centrifugal extraction of plasma from whole blood on a rotating disk. Lab Chip. 2006 Jun;6:776–781. doi: 10.1039/b604145k. [DOI] [PubMed] [Google Scholar]
- 23.Sollier E, Rostaing H, Pouteau P, Fouillet Y, Achard JL. Passive microfluidic devices for plasma extraction from whole human blood. Sens Actuators B-Chem. 2009 Sep 7;141:617–624. doi: 10.1109/IEMBS.2009.5333314. [DOI] [PubMed] [Google Scholar]
- 24.Di Carlo D, Irimia D, Tompkins RG, Toner M. Continuous inertial focusing, ordering, and separation of particles in microchannels. Proc Nat Acad Sci USA. 2007 Nov 27;104:18892–18897. doi: 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Carlo D, Edd JF, Irimia D, Tompkins RG, Toner M. Equilibrium separation and filtration of particles using differential inertial focusing. Anal Chem. 2008 Mar 15;80:2204–2211. doi: 10.1021/ac702283m. [DOI] [PubMed] [Google Scholar]
- 26.Jaggi RD, Sandoz R, Effenhauser CS. Microfluidic depletion of red blood cells from whole blood in high-aspect-ratio microchannels. Microfluidics Nanofluidics. 2007 Feb;3:47–53. [Google Scholar]
- 27.Sollier E, Cubizolles M, Fouillet Y, Achard JL. Fast and continuous plasma extraction from whole human blood based on expanding cell-free layer devices. Biomed Microdevices. 2010 Jun;12:485–497. doi: 10.1007/s10544-010-9405-6. [DOI] [PubMed] [Google Scholar]
- 28.Faivre M, Abkarian M, Bickraj K, Stone HA. Geometrical focusing of cells in a microfluidic device: An approach to separate blood plasma. Biorheology. 2006;43:147–159. [PubMed] [Google Scholar]
- 29.Yang S, Undar A, Zahn JD. A microfluidic device for continuous, real time blood plasma separation. Lab Chip. 2006;6:871–880. doi: 10.1039/b516401j. [DOI] [PubMed] [Google Scholar]
- 30.Lee HJ, Kim JH, Lim HK, Cho EC, Huh N, Ko C, Park JC, Choi JW, Lee SS. Electrochemical cell lysis device for DNA extraction. Lab Chip. 2010 Mar 7;10:626–633. doi: 10.1039/b916606h. [DOI] [PubMed] [Google Scholar]
- 31.Di Carlo D, Ionescu-Zanetti C, Zhang Y, Hung P, Lee LP. On-chip cell lysis by local hydroxide generation. Lab Chip. 2005 Feb;5:171–178. doi: 10.1039/b413139h. [DOI] [PubMed] [Google Scholar]
- 32.Nevill JT, Cooper R, Dueck M, Breslauer DN, Lee LP. Integrated microfluidic cell culture and lysis on a chip. Lab Chip. 2007 Dec;7:1689–1695. doi: 10.1039/b711874k. [DOI] [PubMed] [Google Scholar]
- 33.Schilling EA, Kamholz AE, Yager P. Cell lysis and protein extraction in a microfluidic device with detection by a fluorogenic enzyme assay. Anal Chem. 2002 Apr 15;74:1798–1804. doi: 10.1021/ac015640e. [DOI] [PubMed] [Google Scholar]
- 34.Irimia D, Tompkins RG, Toner M. Single-cell chemical lysis in picoliter-scale closed volumes using a microfabricated device. Anal Chem. 2004 Oct 15;76:6137–6143. doi: 10.1021/ac0497508. [DOI] [PubMed] [Google Scholar]
- 35.Sethu P, Anahtar M, Moldawer LL, Tompkins RG, Toner M. Continuous flow microfluidic device for rapid erythrocyte lysis. Anal Chem. 2004 Nov 1;76:6247–6253. doi: 10.1021/ac049429p. [DOI] [PubMed] [Google Scholar]
- 36.Taylor MT, Belgrader P, Furman BJ, Pourahmadi F, Kovacs GTA, Northrup MA. Lysing bacterial spores by sonication through a flexible interface in a microfluidic system. Anal Chem. 2001 Feb 1;73:492–496. doi: 10.1021/ac000779v. [DOI] [PubMed] [Google Scholar]
- 37.Di Carlo D, Jeong KH, Lee LP. Reagentless mechanical cell lysis by nanoscale barbs in microchannels for sample preparation. Lab Chip. 2003;3:287–291. doi: 10.1039/b305162e. [DOI] [PubMed] [Google Scholar]
- 38.Yun SS, Yoon SY, Song MK, Im SH, Kim S, Lee JH, Yang S. Handheld mechanical cell lysis chip with ultra-sharp silicon nano-blade arrays for rapid intracellular protein extraction. Lab Chip. 2010 Jun 7;10:1442–1446. doi: 10.1039/b925244d. [DOI] [PubMed] [Google Scholar]
- 39.Kim YC, Kang JH, Park SJ, Yoon ES, Park JK. Microfluidic biomechanical device for compressive cell stimulation and lysis. Sens Actuators B-Chem. 2007 Dec 12;128:108–116. [Google Scholar]
- 40.Waters LC, Jacobson SC, Kroutchinina N, Khandurina J, Foote RS, Ramsey JM. Microchip device for cell lysis, multiplex PCR amplification, and electrophoretic sizing. Anal Chem. 1998 Jan 1;70:158–162. doi: 10.1021/ac970642d. [DOI] [PubMed] [Google Scholar]
- 41.Privorotskaya N, Liu YS, Lee JC, Zeng HJ, Carlisle JA, Radadia A, Millet L, Bashir R, King WP. Rapid thermal lysis of cells using silicon-diamond microcantilever heaters. Lab Chip. 2010;10:1135–1141. doi: 10.1039/b923791g. [DOI] [PubMed] [Google Scholar]
- 42.McClain MA, Culbertson CT, Jacobson SC, Allbritton NL, Sims CE, Ramsey JM. Microfluidic devices for the high-throughput chemical analysis of cells. Anal Chem. 2003 Nov 1;75:5646–5655. doi: 10.1021/ac0346510. [DOI] [PubMed] [Google Scholar]
- 43.Wang HY, Bhunia AK, Lu C. A microfluidic flow-through device for high throughput electrical lysis of bacterial cells based on continuous dc voltage. Biosens Bioelectron. 2006 Dec 15;22:582–588. doi: 10.1016/j.bios.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 44.Lee DW, Cho YH. A continuous electrical cell lysis device using a low dc voltage for a cell transport and rupture. Sens Actuators B-Chem. 2007 Jun 10;124:84–89. [Google Scholar]
- 45.Bao N, Lu C. A microfluidic device for physical trapping and electrical lysis of bacterial cells. Appl Phys Lett. 2008 May 26;92:214103. [Google Scholar]
- 46.Lu H, Schmidt MA, Jensen KF. A microfluidic electroporation device for cell lysis. Lab Chip. 2005;5:23–29. doi: 10.1039/b406205a. [DOI] [PubMed] [Google Scholar]
- 47.Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002 Oct 18;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 48.Du W, Li L, Nichols KP, Ismagilov RF. SlipChip. Lab Chip. 2009 Aug 21;9:2286–2292. doi: 10.1039/b908978k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moerman R, Knoll J, Apetrei C, van den Doel LR, van Dedem GW. Quantitative analysis in nanoliter wells by prefilling of wells using electrospray deposition followed by sample introduction with a coverslip method. Anal Chem. 2005 Jan 1;77:225–231. doi: 10.1021/ac0400515. [DOI] [PubMed] [Google Scholar]
- 50.Zhou X, Lau L, Lam WW, Au SW, Zheng B. Nanoliter dispensing method by degassed poly(dimethylsiloxane) microchannels and its application in protein crystallization. Anal Chem. 2007 Jul 1;79:4924–4930. doi: 10.1021/ac070306p. [DOI] [PubMed] [Google Scholar]
- 51.Park SM, Huh YS, Craighead HG, Erickson D. A method for nanofluidic device prototyping using elastomeric collapse. Proc Nat Acad Sci USA. 2009 Sep 15;106:15549–15554. doi: 10.1073/pnas.0904004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park SM, Lee KH, Craighead HG. On-chip coupling of electrochemical pumps and an SU-8 tip for electrospray ionization mass spectrometry. Biomed Microdevices. 2008 Dec;10:891–897. doi: 10.1007/s10544-008-9203-6. [DOI] [PubMed] [Google Scholar]
- 53.Sozzi G, Conte D, Leon M, Ciricione R, Roz L, Ratcliffe C, Roz E, Cirenei N, Bellomi M, Pelosi G, Pierotti MA, Pastorino U. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol. 2003 Nov 1;21:3902–3908. doi: 10.1200/JCO.2003.02.006. [DOI] [PubMed] [Google Scholar]
- 54.Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: A diagnostic marker for lung cancer. Clin Lung Cancer. 2009 Jan;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 55.Kopp MU, de Mello AJ, Manz A. Chemical amplification: Continuous-flow PCR on a chip. Science. 1998 May 15;280:1046–1048. doi: 10.1126/science.280.5366.1046. [DOI] [PubMed] [Google Scholar]
- 56.Burns MA, Johnson BN, Brahmasandra SN, Handique K, Webster JR, Krishnan M, et al. An integrated nanoliter DNA analysis device. Science. 1998 Oct 16;282:484–487. doi: 10.1126/science.282.5388.484. [DOI] [PubMed] [Google Scholar]
- 57.Neuzil P, Novak L, Pipper J, Lee S, Ng LFP, Zhang CY. Rapid detection of viral RNA by a pocket-size real-time PCR system. Lab Chip. 2010;10:2632–2634. doi: 10.1039/c004921b. [DOI] [PubMed] [Google Scholar]
- 58.Kim H, Dixit S, Green CJ, Faris GW. Nanodroplet real-time PCR system with laser assisted heating. Opt Exp. 2009 Jan 5;17:218–227. doi: 10.1364/oe.17.000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee SH, Kim SW, Kang JY, Ahn CH. A polymer lab-on-a-chip for reverse transcription (RT)-PCR based point-of-care clinical diagnostics. Lab Chip. 2008;8:2121–2127. doi: 10.1039/b811131f. [DOI] [PubMed] [Google Scholar]
- 60.Lee JG, Cheong KH, Huh N, Kim S, Choi JW, Ko C. Microchip-based one step DNA extraction and real-time PCR in one chamber for rapid pathogen identification. Lab Chip. 2006;6:886–895. doi: 10.1039/b515876a. [DOI] [PubMed] [Google Scholar]
- 61.Focke M, Stumpf F, Faltin B, Reith P, Bamarni D, Wadle S, Muller C, Reinecke H, Schrenzel J, Francois P, Mark D, Roth G, Zengerle R, von Stetten F. Microstructuring of polymer films for sensitive genotyping by real-time PCR on a centrifugal microfluidic platform. Lab Chip. 2010 Oct 7;10:2519–2526. doi: 10.1039/c004954a. [DOI] [PubMed] [Google Scholar]
- 62.de Mello AJ. DNA amplification: Does ‘small’ really mean ‘ efficient’? Lab Chip. 2001;1:24N–29N. [Google Scholar]
- 63.Shah JJ, Jon G, Michael G. Microwave-induced adjustable nonlinear temperature gradients in microfluidic devices. J Micromech Microeng. 2010;20:105025. [Google Scholar]
- 64.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000 Jun 15;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li CP, Li ZP, Jia HX, Yan JL. One-step ultrasensitive detection of microRNAs with loop-mediated isothermal amplification (LAMP) Chem Commun. 2011;47:2595–2597. doi: 10.1039/c0cc03957h. [DOI] [PubMed] [Google Scholar]
- 66.Jia HX, Li ZP, Liu CH, Cheng YQ. Ultrasensitive detection of microRNAs by exponential isothermal amplification. Angewandte Chemie-Int Ed. 2010;49:5498–5501. doi: 10.1002/anie.201001375. [DOI] [PubMed] [Google Scholar]
- 67.Myers FB, Henrikson RH, Bone J, Lee LP. A handheld point-of-care genomic diagnostic system. PLoS One. 2013;8:e70266. doi: 10.1371/journal.pone.0070266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gansen A, Herrick AM, Dimov IK, Lee LP, Chiu DT. Digital LAMP in a sample self-digitization (SD) chip. Lab Chip. 2012;12:2247–2254. doi: 10.1039/c2lc21247a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hsieh K, Patterson AS, Ferguson BS, Plaxco KW, Soh HT. Rapid, sensitive, and quantitative detection of pathogenic DNA at the point of care through microfluidic electrochemical quantitative loop-mediated isothermal amplification. Angew Chem Int Ed Engl. 2012 May 14;51:4896–4900. doi: 10.1002/anie.201109115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramalingam N, San TC, Kai TJ, Mak MYM, Gong HQ. Microfluidic devices harboring unsealed reactors for real-time isothermal helicase-dependent amplification. Microfluidics Nanofluidics. 2009 Nov;7:325–336. doi: 10.1007/s10404-008-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dimov IK, Garcia-Cordero JL, O’Grady J, Poulsen CR, Viguier C, Kent L, Daly P, Lincoln B, Maher M, O’Kennedy R, Smith TJ, Ricco AJ, Lee LP. Integrated microfluidic tmRNA purification and real-time NASBA device for molecular diagnostics. Lab Chip. 2008 Dec;8:2071–2078. doi: 10.1039/b812515e. [DOI] [PubMed] [Google Scholar]
- 72.Kurn N, Chen PC, Heath JD, Kopf-Sill A, Stephens KM, Wang SL. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clinical Chem. 2005 Oct;51:1973–1981. doi: 10.1373/clinchem.2005.053694. [DOI] [PubMed] [Google Scholar]
- 73.Konry T, Smolina I, Yarmush JM, Irimia D, Yarmush ML. Ultrasensitive detection of low-abundance surface-marker protein using isothermal rolling circle amplification in a microfluidic nanoliter platform. Small. 2011 Feb 7;7:395–400. doi: 10.1002/smll.201001620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mahalanabis M, Do J, Muayad HAL, Zhang JY, Klapperich CM. An integrated disposable device for DNA extraction and helicase dependent amplification. Biomed Microdevices. 2010 Apr;12:353–359. doi: 10.1007/s10544-009-9391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He JL, Wu ZS, Zhou H, Wang HQ, Jiang JH, Shen GL, Yu RQ. Fluorescence aptameric sensor for strand displacement amplification detection of cocaine. Anal Chem. 2010 Feb 15;82:1358–1364. doi: 10.1021/ac902416u. [DOI] [PubMed] [Google Scholar]
- 76.Lutz S, Weber P, Focke M, Faltin B, Hoffmann J, Muller C, Mark D, Roth G, Munday P, Armes N, Piepenburg O, Zengerle R, von Stetten F. Microfluidic lab-on-a-foil for nucleic acid analysis based on isothermal recombinase polymerase amplification (RPA) Lab Chip. 2010 Apr 7;10:887–893. doi: 10.1039/b921140c. [DOI] [PubMed] [Google Scholar]
- 77.Mitani Y, Lezhava A, Kawai Y, Kikuchi T, Oguchi-Katayama A, Kogo Y, Itoh M, Miyagi T, Takakura H, Hoshi K, Kato C, Arakawa T, Shibata K, Fukui K, Masui R, Kuramitsu S, Kiyotani K, Chalk A, Tsunekawa K, Murakami M, Kamataki T, Oka T, Shimada H, Cizdziel PE, Hayashizaki Y. Rapid SNP diagnostics using asymmetric isothermal amplification and a new mismatch-suppression technology. Nature Methods. 2007 Mar;4:257–262. doi: 10.1038/nmeth1007. [DOI] [PubMed] [Google Scholar]
- 78.Gill P, Ghaemi A. Nucleic acid isothermal amplification technologies: A review. Nucleosides Nucleotides Nucleic Acids. 2008 Mar;27:224–243. doi: 10.1080/15257770701845204. [DOI] [PubMed] [Google Scholar]
- 79.Asiello PJ, Baeumner AJ. Miniaturized isothermal nucleic acid amplification, a review. Lab Chip. 2011 Apr 21;11:1420–1430. doi: 10.1039/c0lc00666a. [DOI] [PubMed] [Google Scholar]
- 80.Craw P, Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: A critical review. Lab Chip. 2012;12:2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 81.Eteshola E, Leckband D. Development and characterization of an ELISA assay in PDMS microfluidic channels. Sens Actuators B: Chem. 2001 Jan 25;72:129–133. [Google Scholar]
- 82.Gervais L, Delamarche E. Toward one-step point-of-care immunodiagnostics using capillary-driven microfluidics and PDMS substrates. Lab Chip. 2009 Dec 7;9:3330–3337. doi: 10.1039/b906523g. [DOI] [PubMed] [Google Scholar]
- 83.de la Rica R, Stevens MM. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat Nanotechnol. 2012 Dec;7:821–824. doi: 10.1038/nnano.2012.186. [DOI] [PubMed] [Google Scholar]
- 84.Choi I, Lee LP. Rapid detection of abeta aggregation and inhibition by dual functions of gold nanoplasmic particles: Catalytic activator and optical reporter. ACS Nano. 2013 Jul 23;7:6268–6277. doi: 10.1021/nn402310c. [DOI] [PubMed] [Google Scholar]
- 85.Liu GL, Lee LP. Nanowell surface enhanced Raman scattering arrays fabricated by soft-lithography for label-free biomolecular detections in integrated microfluidics. Appl Phys Lett. 2005;87:074101. [Google Scholar]
- 86.Block ID, Chan LL, Cunningham BT. Photonic crystal optical biosensor incorporating structured low-index porous dielectric. Proc IEEE Sens. 2005:742–745. [Google Scholar]
- 87.Block ID, Chan LL, Cunningham BT. Photonic crystal optical biosensor incorporating structured low-index porous dielectric. Sens Actuators B: Chem. 2006 Dec 14;120:187–193. [Google Scholar]
- 88.Ilic B, Yang Y, Craighead HG. Virus detection using nano-electromechanical devices. Appl Phys Lett. 2004 Sep 27;85:2604–2606. [Google Scholar]
- 89.Aubin KL, Huang J, Park SM, Yang Y, Kondratovich M, Craighead HG, Ilic BR. Microfluidic encapsulated nanoelectromechanical resonators. J Vacuum Sci Technol B. 2007 Jul-Aug;25:1171–1174. [Google Scholar]
- 90.Arntz Y, Seelig JD, Lang HP, Zhang J, Hunziker P, Ramseyer JP, Meyer E, Hegner M, Gerber C. Label-free protein assay based on a nanomechanical cantilever array. Nanotechnology. 2003 Jan;14:86–90. [Google Scholar]
- 91.Wee KW, Kang GY, Park J, Kang JY, Yoon DS, Park JH, Kim TS. Novel electrical detection of label-free disease marker proteins using piezoresistive self-sensing micro-cantilevers. Biosens Bioelectron. 2005 Apr 15;20:1932–1938. doi: 10.1016/j.bios.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 92.McKendry R, Zhang JY, Arntz Y, Strunz T, Hegner M, Lang HP, Baller MK, Certa U, Meyer E, Guntherodt HJ, Gerber C. Multiple label-free biodetection and quantitative DNA-binding assays on a nanomechanical cantilever array. Proc Nat Acad Sci USA. 2002 Jul 23;99:9783–9788. doi: 10.1073/pnas.152330199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang WU, Chen C, Lin KH, Fang Y, Lieber CM. Label-free detection of small-molecule-protein interactions by using nanowire nanosensors. Proc Nat Acad Sci USA. 2005 Mar 1;102:3208–3212. doi: 10.1073/pnas.0406368102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hahm J-I, Lieber CM. Direct ultrasensitive electrical detection of DNA and DNA sequence variations using nanowire nanosensors. Nano Lett. 2004;4:51–54. [Google Scholar]
- 95.Patolsky F, Zheng G, Lieber CM. Fabrication of silicon nanowire devices for ultrasensitive, label-free, real-time detection of biological and chemical species. Nat Protoc. 2006;1:1711–1724. doi: 10.1038/nprot.2006.227. [DOI] [PubMed] [Google Scholar]
- 96.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990 Aug 30;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 97.Cass AEG, Zhang YY. Nucleic acid aptamers: Ideal reagents for point-of-care diagnostics? Faraday Discussions. 2011;149:49–61. doi: 10.1039/c005487a. [DOI] [PubMed] [Google Scholar]
- 98.Csordas A, Gerdon AE, Adams JD, Qian J, Oh SS, Xiao Y, Soh HT. Detection of proteins in serum by micromagnetic aptamer PCR (MAP) technology. Angew Chem Int Ed Engl. 2010;49:355–358. doi: 10.1002/anie.200904846. [DOI] [PubMed] [Google Scholar]
- 99.Lee JH, Canny MD, De Erkenez A, Krilleke D, Ng YS, Shima DT, Pardi A, Jucker F. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF(165) Proc Nat Acad Sci USA. 2005 Dec 27;102:18902–18907. doi: 10.1073/pnas.0509069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ni X, Castanares M, Mukherjee A, Lupold SE. Nucleic acid aptamers: Clinical applications and promising new horizons. Current Medicinal Chem. 2011 Sep;18:4206–4214. doi: 10.2174/092986711797189600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Meyer S, Maufort JP, Nie J, Stewart R, McIntosh BE, Conti LR, Ahmad KM, Soh HT, Thomson JA. Development of an efficient targeted cell-SELEX procedure for DNA aptamer reagents. PLoS One. 2013;8:e71798. doi: 10.1371/journal.pone.0071798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ahmad KM, Oh SS, Kim S, McClellen FM, Xiao Y, Soh HT. Probing the limits of aptamer affinity with a microfluidic SELEX platform. PloS One. 2011 Nov 14;6:e27051. doi: 10.1371/journal.pone.0027051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cho M, Xiao Y, Nie J, Stewart R, Csordas AT, Oh SS, Thomson JA, Soh HT. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. Proc Nat Acad Sci USA. 2010 Aug 31;107:15373–15378. doi: 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Latulippe DR, Szeto K, Ozer A, Duarte FM, Kelly CV, Pagano JM, White BS, Shalloway D, Lis JT, Craighead HG. Multiplexed microcolumn-based process for efficient selection of RNA aptamers. Anal Chem. 2013 Mar 19;85:3417–3424. doi: 10.1021/ac400105e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dimov IK, Kijanka G, Park Y, Ducree J, Kang T, Lee LP. Integrated microfluidic array plate (iMAP) for cellular and molecular analysis. Lab Chip. 2011 Aug 21;11:2701–2710. doi: 10.1039/c1lc20105k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Crane M. Diagnostic test device. 3 579 306. US Patent. 1969
- 107.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Microfluidic diagnostic technologies for global public health. Nature. 2006 Jul 27;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 108.Soper SA, Brown K, Ellington A, Frazier B, Garcia-Manero G, Gau V, Gutman SI, Hayes DF, Korte B, Landers JL, Larson D, Ligler F, Majumdar A, Mascini M, Nolte D, Rosenzweig Z, Wang J, Wilson D. Point-of-care biosensor systems for cancer diagnostics/prognostics. Biosens Bioelectron. 2006 Apr 15;21:1932–1942. doi: 10.1016/j.bios.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 109.Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, Hughes MA, Hewlett EL, Merkel TJ, Ferrance JP, Landers JP. A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proc Nat Acad Sci USA. 2006 Dec 19;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, Wang J, Moore H, Rouse R, Umviligihozo G, Karita E, Mwambarangwe L, Braunstein SL, van de Wijgert J, Sahabo R, Justman J, El-Sadr EW, Sia SK. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat Med. 2011 Aug;17:1015–1019. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 111.Mao XL, Huang TJ. Microfluidic diagnostics for the developing world. Lab Chip. 2012;12:1412–1416. doi: 10.1039/c2lc90022j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Martinez AW, Phillips ST, Butte MJ, Whitesides GM. Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew Chem Int Ed Engl. 2007;46:1318–1320. doi: 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Martinez AW, Phillips ST, Whitesides GM. Three-dimensional microfluidic devices fabricated in layered paper and tape. Proc Nat Acad Sci USA. 2008 Dec 16;105:19606–19611. doi: 10.1073/pnas.0810903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Klasner SA, Price AK, Hoeman KW, Wilson RS, Bell KJ, Culbertson CT. Paper-based microfluidic devices for analysis of clinically relevant analytes present in urine and saliva. Anal Bioanal Chem. 2010 Jul;397:1821–1829. doi: 10.1007/s00216-010-3718-4. [DOI] [PubMed] [Google Scholar]
- 115.Abe K, Suzuki K, Citterio D. Inkjet-printed microfluidic multi-analyte chemical sensing paper. Anal Chem. 2008 Sep 15;80:6928–6934. doi: 10.1021/ac800604v. [DOI] [PubMed] [Google Scholar]
- 116.Abe K, Kotera K, Suzuki K, Citterio D. Inkjet-printed paperfluidic immuno-chemical sensing device. Anal Bioanal Chem. 2010 Sep;398:885–893. doi: 10.1007/s00216-010-4011-2. [DOI] [PubMed] [Google Scholar]
- 117.Delaney JL, Hogan CF, Tian JF, Shen W. Electrogenerated chemiluminescence detection in paper-based microfluidic sensors. Anal Chem. 2011 Feb 15;83:1300–1306. doi: 10.1021/ac102392t. [DOI] [PubMed] [Google Scholar]
- 118.Li X, Tian JF, Garnier G, Shen W. Fabrication of paper-based microfluidic sensors by printing. Colloids Surfaces B-Biointerfaces. 2010 Apr 1;76:564–570. doi: 10.1016/j.colsurfb.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 119.Lu Y, Shi WW, Jiang L, Qin JH, Lin BC. Rapid prototyping of paper-based microfluidics with wax for low-cost, portable bioassay. Electrophoresis. 2009 May;30:1497–1500. doi: 10.1002/elps.200800563. [DOI] [PubMed] [Google Scholar]
- 120.Carrilho E, Martinez AW, Whitesides GM. Understanding wax printing: A simple micropatterning process for paper-based microfluidics. Anal Chem. 2009 Aug 15;81:7091–7095. doi: 10.1021/ac901071p. [DOI] [PubMed] [Google Scholar]
- 121.Leung V, Shehata AAM, Filipe CDM, Pelton R. Streaming potential sensing in paper-based microfluidic channels. Colloids Surfaces a-Physicochemical Eng Aspects. 2010 Jul 20;364:16–18. [Google Scholar]
- 122.Chitnis G, Ding Z, Chang CL, Savran CA, Ziaie B. Laser-treated hydrophobic paper: An inexpensive microfluidic platform. Lab Chip. 2011 Mar 21;11:1161–1165. doi: 10.1039/c0lc00512f. [DOI] [PubMed] [Google Scholar]
- 123.Fenton EM, Mascarenas MR, Lopez GP, Sibbett SS. Multiplex lateral-flow test strips fabricated by two-dimensional shaping. ACS Appl Mater Interfaces. 2009 Jan;1:124–129. doi: 10.1021/am800043z. [DOI] [PubMed] [Google Scholar]
- 124.Glavan AC, Martinez RV, Maxwell EJ, Subramaniam AB, Nunes RMD, Soh S, Whitesides GM. Rapid fabrication of pressure-driven open-channel microfluidic devices in omniphobic R-F paper. Lab Chip. 2013;13:2922–2930. doi: 10.1039/c3lc50371b. [DOI] [PubMed] [Google Scholar]
- 125.Cheng CM, Martinez AW, Gong JL, Mace CR, Phillips ST, Carrilho E, Mirica KA, Whitesides GM. Paper-based ELISA. Angewandte Chemie-Int Ed. 2010;49:4771–4774. doi: 10.1002/anie.201001005. [DOI] [PubMed] [Google Scholar]
- 126.Martinez AW, Phillips ST, Nie ZH, Cheng CM, Carrilho E, Wiley BJ, Whitesides GM. Programmable diagnostic devices made from paper and tape. Lab Chip. 2010;10:2499–2504. doi: 10.1039/c0lc00021c. [DOI] [PubMed] [Google Scholar]
- 127.Maxwell EJ, Mazzeo AD, Whitesides GM. Paper-based electroanalytical devices for accessible diagnostic testing. Mater Res Soc Bulletin. 2013 Apr;38:309–314. [Google Scholar]
- 128.Osborn JL, Lutz B, Fu E, Kauffman P, Stevens DY, Yager P. Microfluidics without pumps: Reinventing the T-sensor and H-filter in paper networks. Lab Chip. 2010 Oct 21;10:2659–2665. doi: 10.1039/c004821f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schumacher S, Nestler J, Otto T, Wegener M, Ehrentreich-Forster E, Michel D, Wunderlich K, Palzer S, Sohn K, Weber A, Burgard M, Grzesiak A, Teichert A, Brandenburg A, Koger B, Albers J, Nebling E, Bier FF. Highly-integrated lab-on-chip system for point-of-care multiparameter analysis. Lab Chip. 2012 Feb 7;12:464–473. doi: 10.1039/c1lc20693a. [DOI] [PubMed] [Google Scholar]
- 130.Li X, Ballerini DR, Shen W. A perspective on paper-based microfluidics: Current status and future trends. Biomicrofluidics. 2012 Mar;6:011301-1–011301-13. doi: 10.1063/1.3687398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Technology readiness assessment (TRA) guidance. U.S. Dept. Defense; Washington, DC, USA: 2011. [Google Scholar]
- 132.O’Connell RJ, Merritt TM, Malia JA, VanCott TC, Dolan MJ, Zahwa H, Bradley WP, Branson BM, Michael NL, De Witt CC. Performance of the OraQuick rapid antibody test for diagnosis of human immunodeficiency virus type 1 infection in patients with various levels of exposure to highly active antiretroviral therapy. J Clin Microbiol. 2003 May;41:2153–2155. doi: 10.1128/JCM.41.5.2153-2155.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cha YJ, Park Q, Kang ES, Yoo BC, Park KU, Kim JW, Hwang YS, Kim MH. Performance evaluation of the OraQuick hepatitis C virus rapid antibody test. Ann Lab Med. 2013 May;33:184–189. doi: 10.3343/alm.2013.33.3.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Granade TC, Parekh BS, Phillips SK, McDougal JS. Performance of the OraQuick and Hema-Strip rapid HIV antibody detection assays by non-laboratorians. J Clin Virol. 2004 Jul;30:229–232. doi: 10.1016/j.jcv.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 135.Holguin A, Gutierrez M, Portocarrero N, Rivas P, Baquero M. Performance of OraQuick advance rapid HIV-1/2 antibody test for detection of antibodies in oral fluid and serum/plasma in HIV-1+ subjects carrying different HIV-1 subtypes and recombinant variants. J Clin Virol. 2009 Jun;45:150–152. doi: 10.1016/j.jcv.2009.04.002. [DOI] [PubMed] [Google Scholar]