SUMMARY
The extent to which bacterial ribosomes and the significantly larger eukaryotic ribosomes share the same mechanisms of ribosomal elongation is unknown. Here, we present sub-nanometer resolution cryo-electron microscopy maps of the mammalian 80S ribosome in the post-translocational state and in complex with the eukaryotic eEF1A•Val-tRNA•GMPPNP ternary complex, revealing significant differences in the elongation mechanism between bacteria and mammals. Surprisingly, and in contrast to bacterial ribosomes, a rotation of the small subunit around its long axis and orthogonal to the well-known intersubunit rotation distinguishes the post-translocational state from the classical pre-translocational state ribosome. We term this motion “subunit rolling”. Correspondingly, a mammalian decoding complex visualized in sub-states before and after codon recognition reveals structural distinctions from the bacterial system. These findings suggest how codon recognition leads to GTPase activation in the mammalian system and demonstrate that in mammalia subunit rolling occurs during tRNA selection.
INTRODUCTION
Genetic information within messenger RNA (mRNA) is translated into protein during the elongation phase of translation (Voorhees and Ramakrishnan, 2013). Ribosomes decode one codon of the mRNA sequence per elongation cycle by using transfer RNA (tRNA) substrates and append the encoded amino acid to the nascent peptide. An elongation cycle can be subdivided into three steps: (i) delivery of aminoacyl-tRNA (aa-tRNA), which involves decoding and accommodation, (ii) peptide-bond formation, and (iii) tRNA translocation. Peptide-bond formation is catalyzed by the ribosome’s peptidyl transferase center and is a fast and spontaneous step. However, during decoding and translocation the ribosome has to overcome large activation energy barriers. Accordingly, the pre-translocational (PRE) and the post-translocational (POST) states of the ribosome are relatively stable (Schilling-Bartetzko et al., 1992). These activation energy barriers are reduced by translational GTPase elongation factors, which are responsible for speed and accuracy of protein synthesis.
The bacterial elongation cycle and its sub-steps have been extensively studied over the past decades using a battery of functional, genetic and structural methods (Frank and Spahn, 2006; Voorhees and Ramakrishnan, 2013). An elongation cycle starts when an aa-tRNA is delivered to the POST state ribosome carrying a peptidyl-tRNA in the P-site and a deacylated tRNA in the E-site. A-site occupation is a complex, multistep process (Schmeing et al., 2009; Schuette et al., 2009; Voorhees and Ramakrishnan, 2013). In the initial decoding step the aa-tRNA•EF-Tu•GTP ternary complex binds to the ribosome. Recognition of the cognate codon leads to a complex rearrangement of the tRNA. In the resulting A/T-state the tRNA can bind to the A-site of the ribosomal 30S subunit and remains simultaneously bound to EF-Tu, which in turn interacts with the ribosome’s factor binding site. The conformational rearrangements of the ternary complex, combined with changes in the ribosome, transmit decoding signals from cognate codon-anticodon interactions to EF-Tu stimulating GTP hydrolysis and subsequent dissociation of EF-Tu•GDP. Decoding is completed when the body of the tRNA swings into the ribosome through the accommodation corridor and establishes interactions with the A-site of the 50S subunit during accommodation (Sanbonmatsu et al., 2005). After peptide-bond formation the resulting PRE complex contains a peptidyl-tRNA in the A-site and a deacylated tRNA in the P-site. In order to reset the ribosome for the next round of elongation, the EF-G-dependent translocation reaction moves the tRNA2•mRNA complex by one codon through the ribosome, establishing the POST state(Voorhees and Ramakrishnan, 2013).
While the structure and function of the ribosome is substantially conserved across the domains of life, comparatively little is known about the detailed mechanism of eukaryotic translation. The eukaryotic (80S) ribosome is significantly larger and more complex than its bacterial counterpart (Anger et al., 2013; Ben-Shem et al., 2011; Klinge et al., 2011; Spahn et al., 2004a) and mechanistic investigations of the translation mechanisms in higher organisms are lacking. Substantial disparities in the translation mechanism between eukaryotic and prokaryotic systems are implied by the elaborated initiation, termination and recycling mechanisms, requiring numerous additional factors in eukaryotes (Melnikov et al., 2012). In contrast, the elongation phase is thought to be highly conserved across domains of life because core elements of the ribosome, including the substrate binding sites and the general elongation factors, are largely conserved. Conversely, the existence of ribosome-targeting antibiotics that display domain specificity (Wilson, 2009) and evidence of essential, domain-specific translation factors (Andersen et al., 2006), indicates that aspects of the translation mechanism must differ. A first analysis of the mammalian PRE complex revealed distinctions of the mammalian 80S ribosome with respect to the dynamic behaviour of the complex and the exact nature of the tRNA binding sites (Budkevich et al., 2011). Here, we explore the mammalian elongation cycle through structural investigations of the mammalian 80S ribosome decoding complexes and the POST state from rabbit liver using cryo-electron microscopy (cryo-EM). Strikingly, substantial differences to analogous bacterial complexes can be observed, revealing that the bacterial and the mammalian elongation cycle diverge more than previously thought.
RESULTS
CryoEM maps for mammalian POST, PRE and decoding complexes
In order to provide the structural foundation for the A-site occupation/decoding step in the mammalian system we used cryo-EM to analyze ribosomal decoding and POST complexes. Both specimens were prepared in vitro from mammalian components (Budkevich et al., 2011; Budkevich et al., 2008). To yield the decoding complex the ternary complex Val-tRNA•eEF1A•GMPPNP was stalled by the non-hydrolyzable GTP analogue. For the POST complex, PRE ribosomes were translocated by eEF2 to move N-acylated and deacylated tRNAPhe from A- and P- sites to P- and E- sites, respectively (Experimental Procedures).
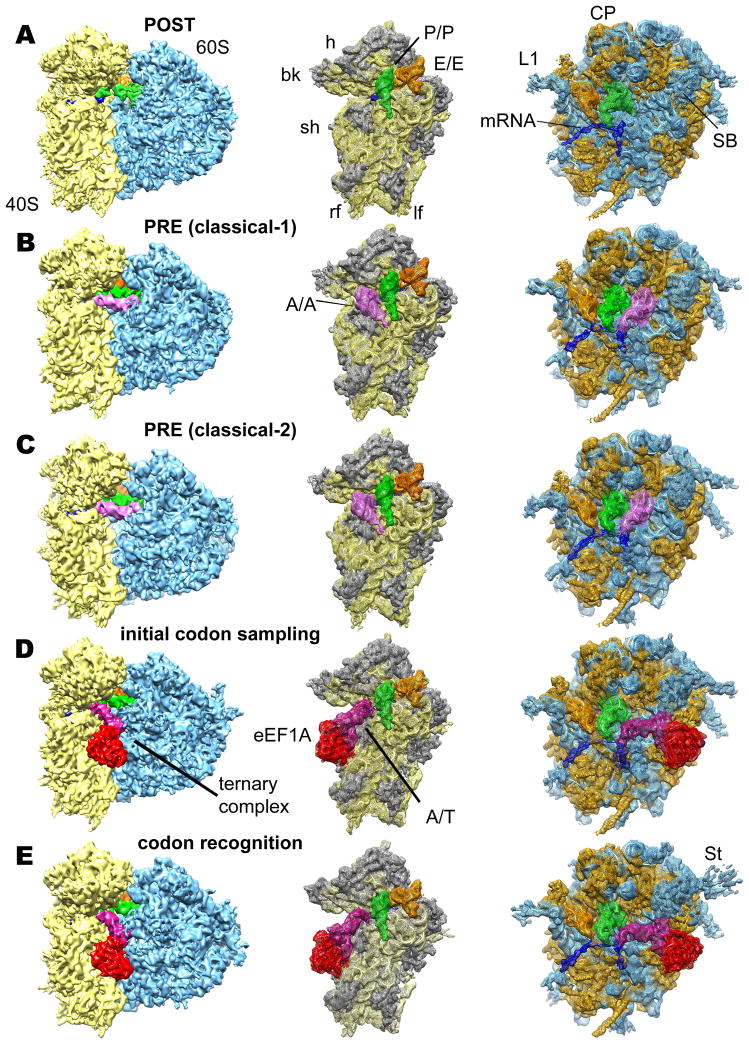
The resulting complexes were analyzed by multiparticle cryo-EM (Loerke et al., 2010). The compositional heterogeneity of the POST specimen was larger than expected from the biochemical data and the subpopulation corresponding to the desired 80S POST complex consisted of only about 35% of ribosomal complexes (Fig. S1A and Supplemental Experimental Procedure). This major subpopulation (236,113 particle images) representing a POST complex with two tRNAs in classical P- and E -sites (Fig. 1A) was further refined to a resolution of 6.9 Å (Fig. S1B).
Figure 1. Reconstruction of eukaryotic 80S POST (A), PRE (classical-1) (B), PRE (classical-2) (C), codon sampling (D) and codon recognition / GTPase activation (E) complexes from rabbit liver.
Left panels: Overall view of the cryo-EM reconstructions, displaying the 60S subunit (blue), 40S subunit (yellow), A-tRNA (pink), P-tRNA (green), E-site tRNA (orange), A/T-tRNA (dark violet), eEF1A (red) and mRNA (blue). Middle and right panels: Individual mesh representation of the subunit maps with docked models, showing the ribosomal ligands relative to the 40S subunit (ribosomal RNA yellow and S proteins gray) and the 60S subunit (rRNA blue and L proteins orange), respectively. Landmarks are denoted for 40S: beak (bk), left foot (lf), right foot (rf), head (h), shoulder (sh) and 60S: central protuberance (CP), L1 stalk (L1), stalk base (SB) and stalk (St).
See also
Figure S1. Heterogeneity in the 80S ribosome POST complexes from rabbit liverand resolution curves.
Table S1. Summary of the modelled proteins in the rabbit ribosome.
The classical PRE state is the final product of the ribosomal A-site occupation/decoding step. We made use of the presence of significantly populated classical PRE states within the POST dataset (Fig. S1A) to furthermore obtain cryo-EM maps of the 80S ribosome in classical-1 (66,618 particle images) and classical-2 (73,951 particle images) PRE states (Fig. 1B,C) at 7.6 Å and 7.5 Å resolution, respectively (Fig. S1B). Both maps exhibited density for tRNAs in classical A-, P- and E-sites. They agree well with our previously reported structures (Budkevich et al., 2011) but are significantly improved in terms of map quality and resolution. Moreover, the PRE and the POST maps were obtained from the same sample, thus facilitating a direct comparison of both states. For the ribosomal decoding complex, our multiparticle approach resulted in two maps (Fig. 1D,E) in subtly different conformations (see below) with resolutions of 8.7 Å and 8.9 Å, respectively (Fig. S1B).
In total, we obtained five cryo-EM maps delineating the decoding process from the POST state via two decoding states to the classical-1 and -2 PRE states (Fig. 1). All maps show typical features expected at sub-nanometer resolution such as major and minor grooves of RNA double helices, α-helices and extended protein tails. To interpret our maps in molecular terms, we created a cryo-EM based homology model of the mammalian 80S ribosome. As secondary structure maps for rRNAs from rabbit are not available, and rabbit and human ribosomes are very similar (Budkevich et al., 2011; Spahn et al., 2004b), we chose the human ribosome as target for our modelling. The high sequence conservation (see Supplemental Experimental Procedures) also suggests that a human based model is a valid approximation for the presented cryo-EM maps. Importantly, our model will aid subsequent studies in the human system. Recent X-ray structures of the ribosomal 40S and 60S subunits from Tetrahymena thermophila (Klinge et al., 2011; Rabl et al., 2011) and the yeast 80S ribosome (Ben-Shem et al., 2011) allow homology modelling not only for the evolutionary conserved inner core of the ribosome but also for many of the eukaryotic-specific components (Fig. 1A, Table S1). Recently, a model for the human 80S ribosome in the rotated state became available based on a cryo-EM map at 5.4 Å resolution (Anger et al., 2013). The model presented here is in good overall agreement with this model regardless of our use of an alternative sequence for the ribosomal RNAs.
The path of the mRNA in eukaryotic ribosomes and codon-anticodon interactions with P- and E-site tRNAs
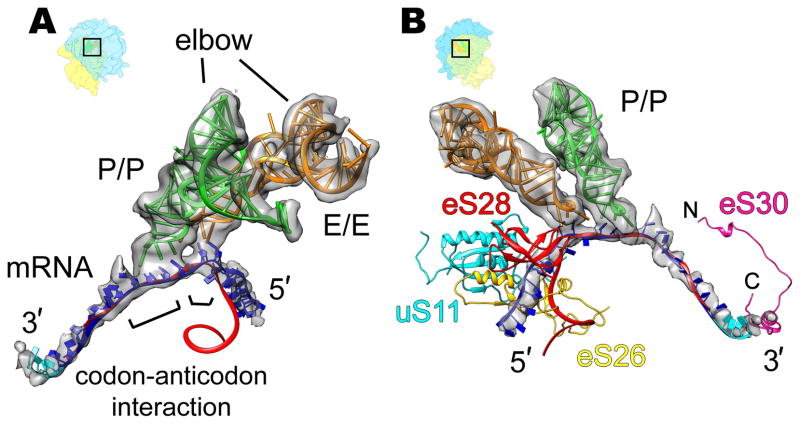
The presented cryo-EM map of the mammalian POST complex allows a direct visualization of an mRNA fragment with a length of approximately 34 nucleotides (Fig. 1A and 2A, B). The modelled mRNA ranges from position −15 on the 5′-side to position +19 on the 3′-side. This agrees well with the part of the mRNA that is shielded by the ribosome according to mRNA protection studies (Steitz, 1969) and recent ribosome profiling data (Ingolia et al., 2011). As in bacterial ribosomes (Jenner et al., 2010), the mRNA enters the mRNA groove of the 40S subunit from the solvent side through the mRNA entry tunnel, wraps around the neck of the 40S subunit, where it interacts with the tRNAs, and leaves the 40S through the mRNA exit tunnel. On the 5′-side the overall path of the mRNA through the mRNA exit tunnel deviates significantly from the bacterial system (Jenner et al., 2010), whereas on the 3′-side, the path of the mRNA through the mRNA entry tunnel is remarkably similar (Fig. 2B). However, there is a kink in the mRNA at the solvent side of the entry tunnel and about 4 nucleotides of mRNA (+16 to +19), which are not visible in the bacterial system, lead upwards towards the 40S head and 18S rRNA helix 16 (h16).
Figure 2. Eukaryotic-specific features of the mRNA path visible from intersubunit space (A) and solvent side (B) of 40S subunit.
(A) Interaction of the P- and E-site tRNAs of the 80S POST complex (transparent gray) with mRNA. (B) Comparison of eukaryotic (transparent gray) and prokaryotic (red ribbon, Jenner et al., 2010, PDB ID 3I8G) mRNA paths. The model for the additional four nucleotides on the 3′-end of the eukaryotic mRNA is shown in cyan. Densities for the ligands were segmented from the cryo-EM map presented in Figure 1A. The models for tRNAs, mRNA and ribosomal proteins from the homology model of the human 80S ribosome presented in this paper. Ribosome orientations are indicated by orientation aids.
See also
Figure S2. Eukaryotic-specific contacts between P-tRNA and large ribosomal proteins.
In the P-site the three-nucleotide helix formed by codon-anticodon interaction can be directly observed (Fig. 2A). Interestingly, we also observe a shorter interaction interface between the mRNA codon and the tRNA anticodon in the E-site, indicating one or at most two base pairing interactions (Fig. 2A). This is consistent with bacterial X-ray structures where Watson-Crick base pairing of the E-site tRNA anticodon with the first nucleotide of the E-site codon was described (Jenner et al., 2010). Thus, codon-anticodon interaction at the E-site may be a feature of the mammalian POST complex, but weakened with respect to the P-site.
Interactions of tRNA and mRNA with ribosomal proteins specific to eukaryotes
The domain-specific differences in the path through the mRNA exit tunnel on the mRNA’s 5′-side can be explained by the presence of the eukaryotic-specific ribosomal proteins eS26 and eS28 (we use the new system for naming ribosomal proteins according to Ban et al., 2014). The N- and C-terminal parts of eS26 block the bacterial path of the mRNA and at the same time shield the 3′-end of 18S rRNA preventing formation of Shine-Dalgarno-like interactions. Moreover, eS26, eS28 and also uS11 (rpS14) appear to interact with the eukaryotic mRNA and thus line out an alternative path of the mRNA exit (Fig. 2B).
At the solvent side of the mRNA entry tunnel, a eukaryotic-specific contact with the 3′-side of the mRNA may also take place, facilitated by the C-terminal part of protein eS30. Interestingly, eS30 wraps around the shoulder, and reaches into the decoding center (Rabl et al., 2011), where its N-terminus has been proposed to interact with the A-site tRNA in the mammalian PRE complex (Budkevich et al., 2011). Thus, eS30 could provide a structural link between the outer and inner ends of the mRNA entry tunnel. Hence, our structure suggests a functional role for the eukaryotic ribosomal proteins uS11, eS26, eS28 and eS30 in escorting the mRNA through the 40S subunit. Furthermore, eukaryotic-specific contacts also contribute to the tRNA binding sites (Fig. S2).
40S subunit rolling: a novel mode of intersubunit rearrangement
The presented POST complex contains classically configured tRNAs in the P/P- and E/E-sites (Fig. 1A). It is therefore expected to correspond, in terms of overall ribosome conformation, to the classical-1 PRE state that also carries classically configured tRNAs (Fig. 1B). Unexpectedly, the subunit arrangement of the POST state differs markedly from that of the classical-1 PRE state (Fig. 3A, Movie S1). The underlying conformational change, which we term “subunit rolling”, can be described as a ~6° rotation of the 40S subunit toward the L1 stalk around the long axis of the small subunit. The axis co-localizes approximately with the upper part of h44 of 18S rRNA and is roughly orthogonal to the well-known intersubunit rotation (Fig. S3A).
Figure 3. 40S subunit rolling.
Comparison of the 40S subunit positions in classical-1 state (orange) to the POST state (yellow) represented by (A) cryo-EM maps and (B) ribbons. Comparisons are based on a common 60S alignment. Arrows indicate the direction of movement during transition between the two different states. The distance changes in the 40S subunit positions resulting from the rigid body transformation are color-coded in Å units. Landmarks for 40S aredenoted: head (h), body (b) and beak (bk).
See also
Figure S3. 40S rolling and its effect on integrity of ribosomal intersubunit bridges and tRNA positions.
Movie S1. Effects of 40S rolling on the intersubunit space.
Interestingly, subunit rolling shapes the openings to the intersubunit space and causes reciprocal opening and closing of the A- and E-site regions (Movie S1). The distance between the 40S and 60S subunits on the A-site side of the 80S ribosome decreases by about 13–15 Å during subunit rolling from the POST state to the classical-1 PRE state. As a consequence, the A-site region is more widely open in the POST state. The opposite is observed for the E-site, which is narrower in the POST state than in the classical-1 PRE state. However, due to the smaller distance from the rotation axis, the underlying movements are only in the range of 6–7 Å at the E-site. Subunit rolling also affects the interactions between the 40S and the 60S subunit, i.e. the inter-subunit bridges (Fig. S3D). Most of the conserved bridges are present in the current POST 80S state with the exception of B6 and B7 (Fig. S3D, in green), which are, however, found in the mammalian classical-1 PRE state. Subunit rolling provides an explanation for this observation as it results in movement on the order of ~5–7 Å in the lower part of 40S (Fig. 3B). Moreover, subunit rolling is expected to affect tRNA positions. Indeed, the elbow region of the P/P-tRNA in the PRE state is shifted by ~6 Å toward the E-site in comparison with the mammalian POST state (and also the bacterial PRE state; Fig. S3E & F). Thus, some differences in tRNA positioning exist for the mammalian 80S ribosome between the classical PRE and POST states, with either a deacylated or peptidyl-tRNA being present in the P-site, respectively.
Cryo-EM analysis of the mammalian decoding complex
The observed difference in the subunit arrangement of the 80S ribosome in POST and classical PRE states has consequences for the mechanism of tRNA selection. It implies that subunit rolling has to occur when the POST complex is converted into the PRE complex. To explore mammalian A-site occupation, we analyzed a mammalian decoding complex. Unexpectedly, we were able to observe two subpopulations of the 80S•Val-tRNA•eEF1A•GMPPNP complex (Fig. 1D, E). Both cryo-EM maps display clear density for the ternary complex (Fig. 4A, B) but are distinguished by more subtle changes in position, conformation and interaction patterns between the ternary complex and the 80S ribosome (Figs. 4, 5, 6, S4–5).
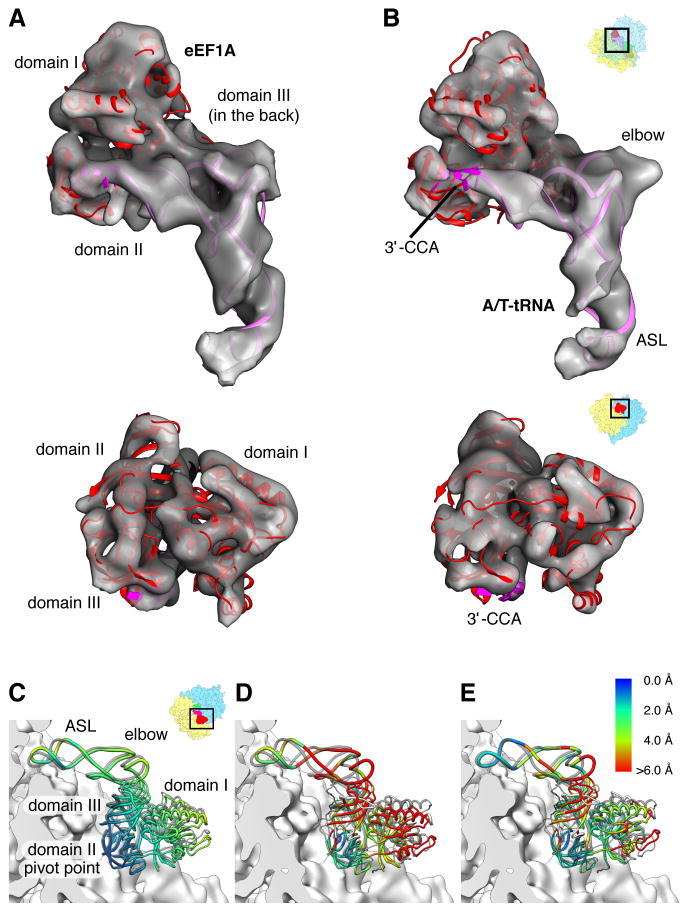
Figure 4. Conformation of the mammalian ternary complex and differences from the bacterial counterpart.
Overall fitting of crystallographic models of Aeropyrum pernix aEF1A (Kobayashi et al., 2012) (red ribbon, PDB ID 3VMF) and T.thermophilus A/T-tRNA (pink ribbon, modified from PDB IDs 2XQD and 1TTT for ASL and tRNA body, respectively) to the ternary complex cryo-EM maps (transparent gray) of (A) codon sampling state and (B) codon recognition / GTPase activation state. Densities for the ternary complex are extracted from the cryo-EM maps presented in Figure 1D and E. (C) Superposition of the codon sampling state (transparent gray) and codon recognition / GTPase activation state ternary complex molecular models on the 40S subunit surface. The alignment was based on the 40S subunit densities. (D–E) Comparisons of the mammalian (D) codon sampling state and (E) codon recognition / GTPase recognition state (E) ternary complexes with the bacterial ternary complex stalled by GTP analog (Voorhees et al, Science 2010) (PDB ID 2XQD; transparent gray). The alignment was based on conserved parts of the 18S/16S rRNA. Ribosome orientations are indicated by orientation aids. (C–E) The distances between positions of the ternary complexes are color coded (capped at 6 Å). We note that these distances essentially reflect rigid body transformations of eEF1A/EF-Tu and ASL or body of tRNA.
See also
Figure S4. Protein sequence alignment of EF1A homologs from different species.
Movie S2. Codon recognition induces conformational changes in the A/T-tRNA, leading to a seesaw-like motion of the eEF1A on the 40S surface.
Fig. 5. Interactions of the ternary complexes with the 80S ribosome.
(A) Contacts of eEF1A with the shoulder of the 40S ribosomal subunit. Contacts of (B) eEF1A and (C) A/T-tRNA inside of the ternary complexes with SRL of 28S rRNA. (D) Contact of the eEF1A domain III with the C-terminus of uS12. Left column represents the codon sampling state, right column represents the codon recognition / GTPase activation state. Ribosome orientations are indicated by orientation aids. The UniProt numbering which includes the leading methionine, was used, resulting in a +1sequence shift, such that e.g. eukaryotic His94 is His95 according to our numbering.
See also
Figure S5. Re-organisation of the mammalian ternary complex to adapt to a new ribosomal environment.
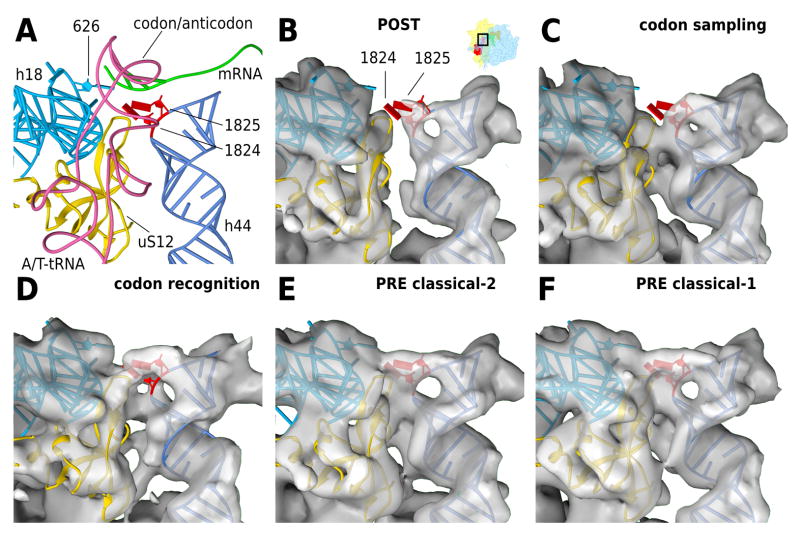
Figure 6. Codon recognition and 40S domain closure at the decoding center.
(A) Models for the codon recognition / GTPase activation state complex. The decoding center is shown from the 60S side, with h44 in light blue, h18 containing the 626 (530, E.colinumbering ) loop in dark blue, A/T-tRNA in pink, mRNA in green and uS12 in gold. Bases 1824 and 1825 (1492 and 1493, E.coli numbering) of h44 are highlighted in red and are shown in the flipped out positions modelled to the codon recognition state. Panels B–F show density maps for the (B) POST state, (C) the codon sampling state, (D) the codon recognition / GTPase activation state, (E) the PRE classical-2 and (F) the PRE classical-1 state. The models are kept fixed in the recognition state in all panels for better comparison. Electron density maps are aligned to h44 and 40S platform. For clarity, the density around h44 and uS12 has been segmented to show only the surface-most layer.
See also
Movie S3. 40S domain closure in the decoding center and shoulder domain.
In terms of subunit configuration, both substates of the 80S•Val-tRNA•eEF1A•GMPPNP complex are similar to the POST complex. When the larger substate of the decoding complex (79,705 particle images) and the smaller one (52,686 particle images) are compared to the POST complex only small rotations of ~ 0.5° and ~ 1°, respectively, between the 40S subunits were found. In contrast, comparing the two subpopulations of the decoding complex with the classical-1 PRE complex reveals respective rotations of ~ 5.6° and ~ 5.0°. Thus, subunit rolling occurs after the decoding step during accommodation of the tRNA from the A/T- to the A/A-state. By contrast, structural investigations in bacteria did not show evidence for such a major subunit rearrangement (Schmeing et al., 2009; Schuette et al., 2009; Voorhees et al., 2010).
Conformational heterogeneity of the mammalian decoding complex
For a molecular interpretation of the mammalian decoding complexes we used the X-ray structure of aEF1A from the archaeal EF1A/RF1 complex (Kobayashi et al, 2012), which we could fit well as a rigid body into the cryo-EM maps of both subpopulations (Fig. 4A, B). Archaeal EF1A and human eEF1A are closely related by sequence (Fig. S4A). Remarkably, density for the only major insertion helix 5 (aa 214–225) (human numbering), which is present in eEF1A but not in aEF1A, is observed in our cryo-EM map and creates a significant mismatch between model and map at the C-terminal part of the G-domain of eEF1A before the junction to domain II (Fig. S4B). To account for the bend between the anticodon-stem loop (ASL) and the D-stem, the structure of the tRNA was flexibly docked (Fig. 4A, B).
A comparison between both subpopulations of the mammalian 80S•Val-tRNA•eEF1A•GMPPNP complex reveals a rotational movement of eEF1A (and tRNA) around a hinge at the interaction between the two evolutionary conserved 258GIGTV and 289VKS/312VKN loops (human numbering) of eEF1A domain II with the shoulder region of the 40S subunit (Fig. 5A, Movie S2). It closes an apparent gap between the G-domain (domain I) of eEF1A and the sarcin-ricin loop (SRL, H95 of 28S rRNA) so that interactions between both elements can be observed only in the smaller subpopulation (Fig. 5B, right panel). With 12 nucleotides, the SRL is the longest universally conserved sequence of rRNAs and an essential part of the factor binding site. Because interactions between the SRL and the switch regions of EF-Tu have been assigned a prominent role in the molecular mechanism of GTPase activation in the bacterial system (Voorhees and Ramakrishnan, 2013), we propose that the smaller subpopulation of the 80S•Val-tRNA•eEF1A•GMPPNP complex is at least close to the GTPase activation state.
This interpretation is corroborated by the appearance of the ribosomal decoding center in our cryo-EM maps (Fig. 6). For the smaller subpopulation of the decoding complex (Fig. 6D) as well as for the classical-2 and -1 PRE complexes (Fig. 6E and 6F, respectively) the respective cryo-EM densities are compatible with a fully flipped-out conformation of the bases A1824 and A1825 of 18S rRNA at the top of h44 (A1492/A1493 in E. coli) allowing minor groove interactions with the codon-anticodon helix of the A/T- or A-tRNA. These minor-groove interactions are at the heart of the ribosomal decoding mechanism and ensure fidelity of tRNA selection (Voorhees and Ramakrishnan, 2013) (Fig. 6A). Because codon-recognition is a prerequisite for GTPase activation, the SRL interactions and the state of the decoding center both suggest that the smaller subpopulation of our mammalian 80S•Val-tRNA•eEF1A•GMPPNP complex represents the codon-recognition / GTPase activation state of the mammalian system, and is functionally equivalent to the bacterial structure of the GDPCP stalled 70S•tRNA•EF-Tu complex (Voorhees et al., 2010).
In contrast, the POST complex (Fig. 6B) and the larger substate of the decoding complex (Fig. 6C) do not display density for stably flipped-out 18S rRNA bases A1824/A1825 indicating a more dynamic state of the top of h44. Therefore, and in line with the observation that eEF1A does not yet interact with the SRL (Fig. 5B, left panel), we suggest that the larger substate of the mammalian decoding complex represents an initial codon sampling state. Here, codon-anticodon interaction occurs but is not yet stabilized. So far such a state of the ribosomal decoding complex has not been directly visualized.
Concomitant with the occupation of the 40S decoding center by an ASL and the stabilization of the codon-anticodon duplex by the flipped-out 18S rRNA bases A1824/A1825, a progressive tightening of the interaction surface between the top of 18S rRNA h44 and the shoulder region (h18 and uS12) can be observed (Fig. 6A). These more local changes appear embedded in an overall tightening of the 40S subunit around the ASL, which involves a subtle movement of the shoulder region, in particular h16, and the 40S head (Movie S3). These changes are reminiscent of the bacterial domain closure movement of the 30S subunit (Ogle et al., 2002). However, the mammalian conformational change is not completed at the decoding step and still progresses from the two decoding subpopulations to the classical PRE states (Fig. 6, Movie S3).
Differences in conformation and interaction pattern of the ternary complexes with the ribosome
When the X-ray structures of the 70S•tRNA•EF-Tu complexes stalled by kirromycin (Schmeing et al., 2009) or GDPCP (Voorhees et al., 2010) or the cryo-EM map of the 70S•tRNA•EF-Tu•GDP•kirromycin complex (Schuette et al., 2009) are compared to the present 80S•Val-tRNA•eEF1A•GMPPNP cryo-EM maps by aligning the large ribosomal subunits, the bacterial 30S subunit is found to be rotated by ~2° and in-between the positions of the mammalian 40S subunit of the 80S•Val-tRNA•eEF1A•GMPPNP and PRE complexes, respectively. This is smaller than the full subunit rolling between the mammalian POST and PRE complexes but leads to a more open factor binding site of the mammalian ribosome and significant differences in the relative positions of ribosomal elements that interact with the ternary complex. The difference in position of the tip of the functionally important SRL, for example, is in the range of ~5 Å when the 80S and 70S complexes are compared in a common alignment of the small ribosomal subunits. From geometric consideration some corresponding adjustment in the ternary complex can be expected.
Indeed, when the ternary complexes are compared from the perspective of the 40S subunit, differences in the positioning can be detected between the bacterial codon-recognition complex and the mammalian codon sampling state (Fig. 4D), or codon recognition states (Fig. 4E), respectively. Positional differences between bacterial and eukaryotic complexes manifest mainly at the tRNA elbow, whereas the positions and interactions of the two ends of the A/T-tRNA, i.e. the ASL and the 3′-CCA-end are similar (Fig. 4D, E). The interaction of the 3′-CCA-end with the 40S subunit occurs in the context of an interaction with domain II of eEF1A with the shoulder region of the 40S subunit. Consistent with their functional importance for decoding (Voorhees and Ramakrishnan, 2013), these interactions appear highly conserved from bacteria to mammals. Thus, similar to the changes between the codon sampling and codon recognition states of the 80S•Val-tRNA•eEF1A•GMPPNP complex, the interaction of domain II with the shoulder of the 40S subunit serves as anchor point (Fig. 5A).
However, the presented mammalian model of the ternary complex shows a subtle change in the relative orientations of eEF1A and tRNA. Compared to the ribosome-bound Thermus thermophilus ternary complex (Voorhees et al., 2010) the T-arm is moved away from the core of the protein by 4–5 Å (Fig. S5A). In light of the limited resolution of our study this could be regarded as not meaningful. However, a movement in this range can be predicted from a structural comparison of EF-Tu (Voorhees et al., 2010) and aEF1A (Kobayashi et al., 2012). Due to sequence variation the interaction surface of domain III of aEF1A is grown out and would clash with the bacterial tRNA (Fig. S5A). Tentative interactions of the mammalian ternary complex are formed by the loops around His349/Pro350 (human numbering) and Asp428/Met429 (human numbering) with the tRNA around positions 52 and 63/64, respectively (Fig. S5A). Thus, our structural findings are corroborated by evolutionary changes of EF1A, which apparently adapt the ternary complex to a changed ribosomal environment.
Moreover, the different position of the ternary complex in the two states of the mammalian decoding complex and in the bacterial complex leads to a differential interaction pattern with the ribosome. For the mammalian codon sampling state there is a contact between between uS12 of the 40S shoulder and domain III of eEF1A nearby Pro409 (human numbering) (Fig. 5D) facilitated by a eukaryotic-specific rearrangement of the C-terminal tail of uS12 (rpS23). In the mammalian codon recognition state, this specific contact and also the nearby evolutionary conserved contact between nucleotide 68 of the A/T-tRNA and protein uS12 is less obvious than for the mammalian codon sampling state (Fig. 5D, right panel) and can be observed only at lower contour level.
Most interestingly, the changed position of the elbow of the eukaryotic A/T-tRNA is stabilized by specific interactions with the 60S subunit (Fig. 5C). In the mammalian codon sampling state the T-loop (near position 62/63) and D-loop (near position 17) appear to interact with the prominent SRL and helix H89, respectively (Fig. 5C, left panel). Direct interaction of the A/T-tRNA with 28S rRNA (outside of the stalk base) constitutes a surprising difference between bacterial and eukaryotic systems. These unique eukaryotic-specific contacts could stabilize binding of the A/T-tRNA and make the reconstruction of the initial codon sampling state possible. For the mammalian codon recognition / GTPase activation state there is a complex interaction pattern between the SRL and eEF1A (Fig. 5B, right panel). It is not entirely clear whether the contact between the apical loop of the SRL and the ternary complex directly involves the T-loop of the A/T-tRNA as well or occurs indirectly via the eEF1A region around Gln431, which is adjacent to the A/T-tRNA. In addition, the cryo-EM density shows several contact regions between the SRL and eEF1A. One may involve the P-loop of eEF1A around Asp17 and/or His95 from the switch II region (Fig.5B, right panel, d) and a second one Arg69 of the switch I region (Fig. 5B, right panel, c). Interestingly, also eukaryotic specific elements participate in the interaction network, i.e. the helical insertion of eEF1A near Gly121 and Gly131, respectively (Fig. 5B. right panel, e and f).
The conformation of the switch I region differs between the bacterial and the eukaryotic/archaeal systems. We note a helical insertion next to the switch I region that is specific for eukaryotes and archae. When the G-domains of yeast eEF1A in complex with the catalytic C-terminus of its nucleotide exchange factor eEF1Bα (Andersen et al., 2000) and aEF1A•GTP (Kobayashi et al., 2012) are compared, this helical insertion is found to be rotated by ~90°, suggesting that it is part of an extended switch I sub-domain (Fig. S5B). Interestingly, density for this expanded sequence suggests an interaction with h14 of 18S rRNA in the mammalian codon sampling state (Fig. 5A, left panel, d). In bacteria, h14 has been proposed as a contact site for the switch I region of the translational GTPases EF-G (Connell et al., 2007), EF-Tu (Schuette et al., 2009; Villa et al., 2009) and EF4 (Connell et al., 2008). In X-ray maps of ribosome-bound EF-Tu, this interaction has not been observed directly, but the importance of the h8/h14 region for decoding has been demonstrated (Fagan et al., 2013).
DISCUSSION
Conformational modes of the mammalian 80S ribosome
The ribosome is a dynamic macromolecular machine, in which the elongation steps of translation are facilitated by large-scale conformational changes of the ribosome and its ligands. As outlined by the metastable energy landscape view (Munro et al., 2009), the ribosome is rather a stochastic Brownian machine than a mechanical one. The spontaneous nature of conformational modes causes intrinsic conformational heterogeneity, even for compositionally and functionally defined ribosomal complexes. Thus, rather than displaying a single, unique structure, dynamic heterogeneous ensembles are exhibited that reflect an exchange between distinct, functionally relevant structural configurations. A prominent example is the PRE state of the bacterial 70S ribosome (for review see (Munro et al., 2009)), where spontaneous inter-subunit rotation and the fluctuation between classical and hybrid tRNA configurations have been well documented. Bacterial and eukaryotic ribosomes differ in their preferential PRE state: the bacterial 70S ribosome is found predominantly in the classical non-rotated conformation. In contrast, vacant yeast 80S ribosomes (Ben-Shem et al., 2011; Spahn et al., 2004a) or the mammalian 80S PRE complex have been found to prefer the rotated intersubunit state, unless an excess of deacylated tRNA or the antibiotic cycloheximide shifts the landscape towards the classical, non-rotated state (Budkevich et al., 2011). It follows that functional differences between the translational apparatus from different kingdoms or species are not necessarily caused by gross differences in binding sites or conformation. More subtle changes to the energy landscape may lead to differences in the distributions of states.
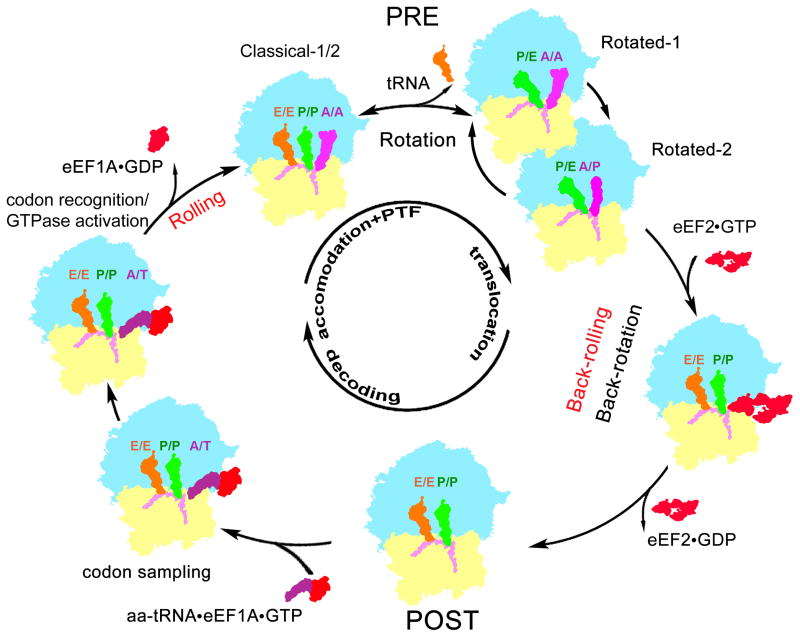
Here we have determined the cryo-EM map of the mammalian POST state (Fig. 1A). Whereas the mammalian PRE complex co-exists in at least four structurally distinct and interchangeable substates (Budkevich et al., 2011), intrinsic large-scale conformational changes such as the intersubunit rotation within the POST complex have not been found. Thus, the POST state may be distinguished by a deep minimum of energy/enthalpy in the energy landscape of the elongating ribosome to compensate for the relatively low entropy. Surprisingly, the POST state deviates with respect to overall conformation not only from the rotated PRE states but also from the classical PRE states, because a novel conformational mode, termed subunit rolling, exists for mammalian 80S ribosomes (Fig. 3, S3 and Movie S1). Rolling converts the POST to the classical PRE state subunit configurations and occurs largely during the accommodation step of A-site occupation (Fig. 7). Back-rolling occurs in the opposite direction during translocation from the PRE to the POST state; however, as translocation involves large ratchet-like intersubunit rearrangements including the rotation of 40S subunit relative to 60S subunit and swivel-like rotation of the small subunit head, back-rolling may happen not as a stand-alone step, but in combination with intersubunit rotation/back-rotation movements (Fig. 7).
Figure 7. The mammalian elongation circle features unique motions of the 40S subunit.
Cartoon representation of the eukaryotic elongation circle highlighting the individual subunit motions necessary to convert one functional intermediate state to the next. Movements of the small ribosomal subunit shared with the prokaryotic system are noted in black, while eukaryotic specific intersubunit movements, i.e. rolling and back-rolling, are noted in red.
It follows that the conformational landscape of the mammalian 80S ribosome is more complex than the prokaryotic one with three overall types of subunit configuration: the classical PRE, the rotated PRE and the POST conformations. The underlying basis for this new conformational degree of freedom may relate to the presence of additional, dynamic intersubunit bridges in the eukaryotic system (Fig. S3D). Moreover, bound ligands can influence the preferred conformation and can lead to additional intermediate conformational states. As a consequence of subunit rolling, the distinction between PRE and POST states is much more pronounced in the mammalian system, which also has implications for the exact nature of classical tRNA binding sites (Fig. S2).
The mammalian decoding complex
We have shown here that the mammalian decoding complex with the ternary complex being trapped in the A/T-state by GMPPNP also exhibits subtle intrinsic structural dynamics as it coexists in (at least) two substates. Because of the local configuration of the ribosomal decoding center (Fig. 6) and the differential interaction pattern of eEF1A with the SRL (Fig. 5B and C), we interpret them as states representing codon sampling and codon recognition /GTPase activation, respectively. Both substates are present essentially in a non-rotated, non-rolled POST-like configuration. As the A-site is more open than in the bacterial 70S complex, the binding site for the ternary complex is altered between the bacterial and eukaryotic domains of life. Indeed, we can observe conserved as well as divergent features in the conformation/structure of the ternary complex itself and in the interaction patterns of ribosomal ligands with the ribosome to account for this difference.
The interactions of the shoulder region of the 40S subunit with two evolutionary conserved loops of domain II of eEF1A and the 3′-CCA-end of the aa-tRNA acts as an evolutionarily preserved anchor (Figs. 4D&E, 5A). On the other hand, local changes in the ribosome exist in the elbow position of tRNA and the exact location of eEF1A (Fig. S5A). For example, the divergent structure of the C-terminal end of uS12 (rpS23) is seen to interact with eEF1A predominantly in the mammalian codon sampling state (Fig. 5D). Also, an insertion sequence in eEF1A domain III dictates a change in relative orientation to the T-loop/stem of the aa-tRNA (Fig. S5A). Apparently, the ternary complex evolved to interact with the altered configuration of factor binding sites between bacteria and mammals. Most unexpected is the observed interaction of the A/T-tRNA elbow with the apical loop of the highly conserved SRL and helix H89 in the mammalian complex (Fig. 5C). In the bacterial system the tRNA elbow interacts only with the mobile stalk base region of 23S rRNA. This indicates a more rigid and tighter binding state for the tRNA elbow in the mammalian system.
The mammalian decoding pathway
Selection of the cognate tRNA has to proceed with optimal speed and accuracy. This is achieved by a complex, multistep pathway involving an initial selection step and a kinetic proofreading step (Rodnina and Wintermeyer, 2001; Geggier et al., 2010). Crucial for tRNA selection is the codon recognition step in the decoding center, and in particular the stabilization of codon-anticodon interaction by A-minor interactions with A1492/A1493 of 16S rRNA (A1824/A1825 in human) in the flipped-out conformation (Ogle et al., 2002). Thereby, enforcing Watson-Crick geometry of the codon-anticodon duplex provides discrimination energy between the cognate and near-cognate interactions. Conformational changes in the ribosomes decoding center in turn provide the signal for activation of GTP hydrolysis by EF-Tu/eEF1A about 80 Å away. For the bacterial system, cryo-EM (Schuette et al., 2009; Villa et al., 2009) and finally X-ray crystallography (Schmeing et al., 2009; Voorhees et al., 2010) have revealed a series of coupled conformational changes that start at the decoding center and are transferred via domain closure of the 30S subunit and a distortion of the tRNA to EF-Tu, inducing the activated conformation. Less clear is the preceeding step, because direct structural information about the codon-sampling state is lacking.
For the mammalian sytem, however, we have now obtained the cryo-EM map of a substate of the GMPPNP-stalled mammalian decoding complex that shows all hallmark features of a codon sampling state (Figs. 4–5). The ASL of the ternary complex is located in the decoding center allowing the formation of the codon-anticodon duplex. However, the appearance of the ribosomal decoding center indicates that the stabilizing A-minor interactions with A1824/A1825 of 18S rRNA do not yet occur, in contrast to the codon recognition state (Fig. 6C, D). Interestingly, the conformation of the A/T-tRNA is in a bent conformation also for the codon sampling state, likely facilitating efficient monitoring of the codon. In the mammalian system, codon recognition is apparently not essential for keeping the tRNA in the distorted conformation.
Comparison of both subpopulations of the GMPPNP-stalled decoding complex suggests a model for the mammalian decoding pathway with similarities, but also surprising distinctions to the generally accepted model for the bacterial system (Voorhees and Ramakrishnan, 2013). As in the bacterial system, stabilization of the codon-anticodon duplex by A-minor interactions with A1824/A1825 of 18S rRNA and long-range signalling of this event to the GTPase center of eEF1A are key for decoding. However, in eukaryotes codon recognition results in a subtle movement of the ASL deeper into the decoding cleft (Movie S2). With the interaction between the two evolutionary conserved 258GIGTV and 289VKS/312VKN loops of eEF1A domain II and the shoulder region of the 40S subunit serving as an anchor, pulling on the ASL leads in first approximation to a rotation of the ternary complex as a whole. Due to a lever arm effect, movements are largest at the elbow region of the tRNA and the G-domain of eEF1A. We do not exclude the presence of concomitant conformational changes in the ternary complex such as a smaller rotation of the G-domain of the factor relative to domains II and III that has been observed in the bacterial system (Voorhees et al., 2010). As a result of the rotational movement of the ternary complex, the G-domain of eEF1A becomes positioned at the SRL by a seesaw-like mechanism. The SRL has been suggested to play a prominent role by facilitating a rearrangement of a catalytically important His84 (E. coli nomenclature) within the switch II regions of translational GTPase factors (Connell et al., 2007; Schuette et al., 2009; Voorhees et al., 2010). Thus, the codon-recognition-dependent movement of eEF1A may lead to further conformational changes within the G-nucleotide binding site resulting in the GTPase activated state.
Conclusions
We have presented here cryo-EM maps of the mammalian 80S ribosome POST and decoding complexes at sub-nanometer resolution. The maps reveal mechanistic differences between the mammalian and the bacterial elongation cycle. Most surprisingly, a unique rolling of the ribosomal 40S subunit by ~6° around the upper region of the h44 occurs during the transition from the POST to the PRE state. Accordingly, there must be changes during the tRNA selection step and indeed we can describe some prominent differences in the conformation of the ribosome-bound aa-tRNA•eEF1A•GMPPNP ternary complex as well as its interaction pattern with the ribosome. Evolutionary changes in key steps of elongation demonstrate that in structural terms the bacterial and the mammalian elongation cycle are less similar than previously thought. Further studies will be needed to understand whether and how these structural changes translate into changes of mechanisms or functions in order to achieve optimal translation or translational control in the eukaryotic environment.
EXPERIMENTAL PROCEDURES
Sample Preparation
Re-associated 80S ribosomes from rabbit liver, free of endogenous tRNAs and mRNAs, were prepared according to (Bommer et al., 1997). The POST 80S complex bearing deacylated tRNAPhe in the E site and in the P-site was prepared by addition of elongation factor 2 (eEF2) in the presence of 200 μM GTP to PRE complex (0.8 μM) programmed with MFK-mRNA (Budkevich et al., 2008). The occupancy of N-acylated was approximately 0.66 per 80S ribosome and more than 90% of the bound tRNA was reactive with puromycin, indicating a nearly quantitative translocation and P-site location. In order to prepare the decoding complex, 80S ribosomes programmed with MFV-mRNA and occupied by Ac[3H]Phe-tRNAPhe were incubated with pre-formed ternary complex eEF1A•GMPPNP•[14C]Val-tRNAVal in the presence of 400 μM GMPPNP (Budkevich et al., 2008).
Electron Microscopy and Image Processing
The 80S complexes were diluted to a final concentration of 30 nM and flash-frozen in liquid ethane and images were recorded under low-dose conditions using an FEI Tecnai G2 Polara operating at 300 kV and a nominal magnification of 39,000. The resulting micrographs were digitized on a drum-scanner (Heidelberg) with a pixel size of 1.26 Å on the object scale. Multiparticle refinement using SPIDER (Frank et al., 1996) was carried out as described previously (Budkevich et al., 2011; Loerke et al., 2010; Penczek et al., 2006) in order to overcome sample heterogeneity caused by substochiometric binding of the ligands and inhomogeneity in the preparation of the mammalian 80S ribosome. Final reconstructions were calculated with SPARX (Hohn et al., 2007). The final resolution for all maps was estimated using the 0.5 cutoff criteria from the Fourier shell correlation curves. For further details see Supplemental Experimental Procedures.
Modelling
A secondary structure map for rabbit 80S ribosomes is not available, so the human ribosome as target for the modelling was chosen. Initial structural models of the human ribosomal RNAs were built following a homology modelling approach (Tung and Sanbonmatsu, 2004). For homology modelling of the ribosomal proteins we used prime (from the Schrodinger suite) (Jacobson et al., 2002; Jacobson et al., 2004). Initial structural models of the 80S ribosome were fit to the cryo-EM map of the 80S POST complex using MDfit (Ratje et al., 2010) which allows flexible docking while maintaining stereochemistry present in the initial model. Simulations were performed at structure-based potential temperature of 20. EMweight was adjusted to the number of atoms in the model.
Supplementary Material
Highlights.
The structure of mammalian POST translocationcomplex at sub -nanometer resolution
Structuresof two sub-states of themammalian d ecoding complex
Subunit rolling: a eukaryotic-specific conformational change in the ribosome
A seesaw-like model for GTPase activation during mammalian tRNA selection
Acknowledgments
The present work was supported by grants from the Deutsche Forschungsgemeinschaft DFG (SFB 740 to C.M.T.S., P.W.H. and T.M.; 436 UKR 113/64/1-1 to T.V.B. and HI 1502 to P.W.H.), HSFP and Senatsverwaltung für Wissenschaft, Forschung und Kultur Berlin (UltraStructureNetwork, Anwenderzentrum). K.Y.S. was supported by HFSP, NIH Grant R01-GM072686 and Los Alamos Institutional Computing. We acknowledge the use of computational resources supplied by the North-German Supercomputing Alliance (HLRN; project beb00001 to C.M.T.S. and bec00085 to P.W.H.).
Footnotes
Accession numbers
The electron density maps of the mammalian POST, PRE (classical-1/2) and decoding (codon sampling and codon recognition/ GTPase activation) complexes have been deposited in the 3D-EM data base (European Molecular Biology Laboratory-European Bioinformatics Institute, Cambridge, UK) with the accession codes EMD-2620, EMD-2621, EMD-2622, EMD-2623 and EMD-2624, respectively. The human models for 40S and 60S subunits have been deposited in the Protein Data Bank database with PDB IDs 4cxc (40S), 4cxd (60S ribosomal proteins), 4cxe (60S rRNA), 4cxb (ligands). The models for the ternary complexes have been deposited with PDB IDs 4cxg (initial codon sampling state) and 4cxh (codon recognition state).
Author contribution
T.V.B. prepared the 80S-POST and decoding complexes. T.V.B. and J.G. and T.M. collected the cryo-EM data. T.V.B., J.G., J.L. and C.M.T.S. carried out the image processing. E.B., J.L., D.J.F.R., J.I., P.W.H., K.Y.S. and C.M.T.S. carried out the modelling of the human 80S ribosomes. T.V.B., E.B., J.L., K.H.N. and C.M.T.S. discussed the results and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen CB, Becker T, Blau M, Anand M, Halic M, Balar B, Mielke T, Boesen T, Pedersen JS, Spahn CM, et al. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;443:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- Andersen GR, Pedersen L, Valente L, Chatterjee I, Kinzy TG, Kjeldgaard M, Nyborg J. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Bα. Molecular Cell. 2000;6:1261–1266. doi: 10.1016/s1097-2765(00)00122-2. [DOI] [PubMed] [Google Scholar]
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, et al. A new system for naming ribosomal proteins. Current opinion in structural biology. 2014 doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Bommer U, Burkhardt N, Junemann R, Spahn CMT, Triana-Alonso F, Nierhaus KH. Ribosomes and polysomes. In: Graham J, Rickwood D, editors. Subcellular Fractionation: A practical Approach. Washington, DC: IRL Press; 1997. pp. 271–301. [Google Scholar]
- Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CM. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Molecular cell. 2011;44:214–224. doi: 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budkevich TV, El’skaya AV, Nierhaus KH. Features of 80S mammalian ribosome and its subunits. Nucleic Acids Res. 2008;36:4736–4744. doi: 10.1093/nar/gkn424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell SR, Takemoto C, Wilson DN, Wang H, Murayama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007;25:751–764. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- Connell SR, Topf M, Qin Y, Wilson DN, Mielke T, Fucini P, Nierhaus KH, Spahn CM. A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat Struct Mol Biol. 2008;15:910–915. doi: 10.1038/nsmb.1469. [DOI] [PubMed] [Google Scholar]
- Fagan CE, Dunkle JA, Maehigashi T, Dang MN, Devaraj A, Miles SJ, Qin D, Fredrick K, Dunham CM. Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state. Proc Natl Acad Sci U S A. 2013;110:9716–9721. doi: 10.1073/pnas.1301585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. Journal of Structural Biology. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Frank J, Spahn CM. The ribosome and the mechanism of protein synthesis. Rep Prog Phys. 2006;69:1383–1417. [Google Scholar]
- Geggier P, Dave R, Feldman MB, Terry DS, Altman RB, Munro JB, Blanchard SC. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J Mol Biol. 2010;399:576–595. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ. SPARX, a new environment for Cryo-EM image processing. Journal of structural biology. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MP, Friesner RA, Xiang Z, Honig B. On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol. 2002;320:597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, Friesner RA. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Saito K, Ishitani R, Ito K, Nureki O. Structural basis for translation termination by archaeal RF1 and GTP-bound EF1α complex. Nucleic acids research. 2012;40:9319–9328. doi: 10.1093/nar/gks660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerke J, Giesebrecht J, Spahn CM. Multiparticle cryo-EM of ribosomes. Methods Enzymol. 2010;483:161–177. doi: 10.1016/S0076-6879(10)83008-3. [DOI] [PubMed] [Google Scholar]
- Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 2012;19:560–567. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC. Navigating the ribosome’s metastable energy landscape. Trends Biochem Sci. 2009;34:390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Penczek PA, Yang C, Frank J, Spahn CM. Estimation of variance in single-particle reconstruction using the bootstrap technique. J Struct Biol. 2006;154:168–183. doi: 10.1016/j.jsb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annual review of biochemistry. 2001;70:415–435. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- Sanbonmatsu KY, Joseph S, Tung CS. Simulating movement of tRNA into the ribosome during decoding. Proc Natl Acad Sci U S A. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling-Bartetzko S, Bartetzko A, Nierhaus KH. Kinetic and thermodynamic parameters for tRNA binding to the ribosome and for the translocation reaction. The Journal of biological chemistry. 1992;267:4703–4712. [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVt, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuette JC, Murphy FVt, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. The EMBO journal. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004a;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004b;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz JA. Nucleotide sequences of the ribosomal binding sites of bacteriophage R17 RNA. Cold Spring Harb Symp Quant Biol. 1969;34:621–630. doi: 10.1101/sqb.1969.034.01.072. [DOI] [PubMed] [Google Scholar]
- Tung CS, Sanbonmatsu KY. Atomic model of the Thermus thermophilus 70S ribosome developed in silico. Biophys J. 2004;87:2714–2722. doi: 10.1529/biophysj.104.040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Ramakrishnan V. Structural basis of the translational elongation cycle. Annual review of biochemistry. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.