Abstract
The microRNAs (miRNAs) are small size non-coding RNAs that regulate expression of target mRNAs at post-transcriptional level. miRNAs differentially expressed under pathological conditions may help identifying mechanisms underlying the disease and may represent biomarkers with prognostic value. However, this kind of studies are difficult in the brain because of the cellular heterogeneity of the tissue and of the limited access to fresh tissue. Here, we focused on a pathology affecting specific cells in a subpopulation of epileptic brains (hippocampal granule cells), an approach that bypasses the above problems. All patients underwent surgery for intractable temporal lobe epilepsy and had hippocampal sclerosis associated with no granule cell pathology in half of the cases and with type-2 granule cell pathology (granule cell layer dispersion or bilamination) in the other half. The expression of more than 1000 miRNAs was examined in the laser-microdissected dentate granule cell layer. Twelve miRNAs were differentially expressed in the two groups. One of these, miR487a, was confirmed to be expressed at highly differential levels in an extended cohort of patients, using RT-qPCR. Bioinformatics searches and RT-qPCR verification identified ANTXR1 as a possible target of miR487a. ANTXR1 may be directly implicated in granule cell dispersion because it is an adhesion molecule that favors cell spreading. Thus, miR487a could be the first identified element of a miRNA signature that may be useful for prognostic evaluation of post-surgical epilepsy and may drive mechanistic studies leading to the identification of therapeutic targets.
Introduction
The microRNAs (miRNAs) are small size endogenous non-coding RNAs that regulate the expression of target mRNAs at post-transcriptional level [1]. To date, more than 1000 human miRNAs have been identified, about 50% of which are expressed in the brain. miRNAs have been demonstrated to be involved in several brain functions, many of which may be implicated in epilepsy and epileptogenesis, like cell death, neurogenesis, synaptic plasticity [2],[3]. Indeed, silencing miR-134 using a specific antagomir exerted prolonged seizure-suppressant and neuroprotective actions in a murine model [4]. Thus, understanding which specific miRNAs are differentially expressed in epilepsy may help to identify the mechanisms underlying the disease. Moreover, differentially expressed miRNAs may represent biomarkers that identify specific subpopulations of epileptic patients, holding a prognostic value [5].
Microarray platforms allow screening and identifying miRNAs differentially expressed under pathological conditions. Experimental studies have profiled miRNA expression in animal models of epilepsy [6],[7],[8],[9] and profiling studies have been also recently published using hippocampi resected from temporal lobe epilepsy (TLE) patients [10],[11]. However, some outstanding obstacles make difficult the interpretation of data from microarray analysis of human brain samples. First, in most studies tissue is derived from autopsies or, potentially even worse, pathological tissue is from surgery samples and control tissue from autopsies. Post-mortem modifications are very likely to dramatically alter the molecular composition of the tissue, making the results questionable. Second, each brain area has a specific and complex cellular composition that changes (often markedly) in the course of diseases. Again, this makes interpretation of molecular data very difficult, because analysis of heterogeneous tissue homogenates does not allow identification of the cells where changes occur and because up-regulation of a molecule in one cell population may be obscured by down-regulation in another cell population.
One approach to overcome these problems is focusing on a well-defined cell population. For example, we focused here on a TLE-associated pathology of the granule cells of the hippocampus. Drug-resistant TLE is the most common type of epilepsy requiring surgical treatment, with a favorable postsurgical outcome in 60-70% of the patients. Based on the underlying etiology, TLE subtypes with different surgical prognosis have been described. Neuropathological classifications of epileptogenic lesions, including focal cortical dysplasias (FCD) [12], hippocampal sclerosis (HS) [13] and granule cell pathology (GCP) [14], define histopathological features and subtypes, allowing attempts to correlate clinical and pathological findings. Correlations with molecular markers, however, are still unavailable.
All patients included in this study underwent surgery for pharmacoresistant TLE and had HS type 1 [13]. All were similar for age, gender, clinical features of the disease. The most relevant difference was that half of the patients had no granule cell pathology (no GCP), whereas the other half had granule cell dispersion or bilamination (GCP type 2) [14], i.e. the single differential pathological feature was in a specific, isolable cell population. Therefore, the granule cell layer was laser-microdissected from all samples, total RNA was extracted from dissected tissues and the miRNAome profile was obtained using a miRNA microarray.
Materials and Methods
Patients
This study was approved by the Ethics Committee of Bologna (full name: Comitato Etico Indipendente dell'Azienda USL della Città di Bologna). A comprehensive written informed consent (also approved by the Ethics Committee of Bologna) was signed for the surgical treatment that produced the tissue samples, the related diagnostic procedures and the research use. All information regarding the human material used in this study was managed using anonymous numerical codes and samples were handled in compliance with the Helsinki declaration (http://www.wma.net/en/30publications/10policies/b3/).
Fourteen drug-resistant TLE patients candidate to epilepsy surgery were collected at the Epilepsy Surgery Center of the IRCCS Institute of Neurological Sciences of Bologna. All patients underwent detailed epileptological evaluation and wakefulness/sleep EEG. All patients also underwent continuous (24 hours) long-term video-EEG monitoring for seizure recording. Analysis of ictal clinical and EEG semiology and electroclinical correlations aimed to identify the epileptogenic area were performed.
Three Tesla MRI, and brain CT scan when necessary, were carried out. Electroclinical, neuroimaging, and neuropsychological data were discussed by the Epilepsy Surgery Team (epileptologists, neuroradiologists, neuropsychologists, neurosurgeons) to establish the site of the epileptogenic area and the surgical strategy.
Surgery
All patients underwent tailored temporal lobe resection to remove the epileptogenic area, according to the data obtained during pre-surgical investigation. Essentially, surgery consisted of removing the temporal pole, the anterior neocortical lateral cortex, the uncus–entorhinal area and the hippocampus and parahippocampal gyrus. The main surgical specimens (hippocampus and/or temporal pole) were removed “en bloc” and spatially oriented to allow a proper histopathological examination.
Histology and microdissection
Specimens were formalin fixed and paraffin embedded. They were de-waxed using Bio-Clear (Bio-Optica, Milan, Italy), washed in ethanol and stained with hematoxylin and eosin for histological diagnosis. Neuropathological evaluation was performed using the most recent classifications of HS, GCP and FCD [12],[13],[14], applying the recommended histochemical and immunohistochemical stains. Specimens either displayed no GCP or GCP type 2. Four different types of neuropathological features can define GCP type 2: 1) dispersion: rows of granule cells spread into the molecular layer and the distance between granule cells is increased; 2) ectopic granule cells: single ectopic granule cells are dispersed into the molecular layer; 3) clusters: ectopic granule cells form clusters within the molecular layer; 4) bilaminar: two granule cell layers, separated by a cell-free gap [14]. While patterns of granule cell loss (thinning and/or cell free gaps, GCP 1) occur isolated, patterns of architectural abnormalities (GCP 2) can came along with cell loss. Therefore, only sections in which no cell loss was detected (based on NeuN staining) where included in analysis.
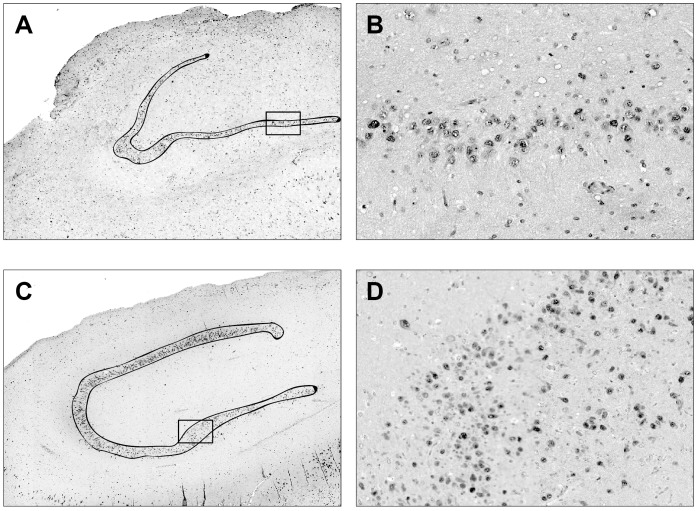
Ten-micron-thick sections were cut using a microtome and the dentate granule layer of the dentate gyrus was laser-dissected ( Fig. 1 ) using the SL microcut microtest dissector (Nikon, Tokyo, Japan). Microdissected cells were captured in microcut transfer film (Nikon). Granule cells were collected in this manner from at least 3–4 slices per patient, in order to obtain an adequate quantity of RNA. Material from all sections of the same patient was pooled together, and total RNA extracted using an RNA purification kit (RecoverAll Total Nucleic Acid Isolation Kit, Ambion Life Technologies, CA, USA). Approximately 1.5 µg total RNA were obtained from each patient. Since miRNAs are more stable than mRNAs, they can be used for microarray analysis from formalin-fixed paraffin-embedded tissues [15]. We performed quality control checks on microarray hybridizations using the Agilent quality control (QC) tool in the Feature Extraction software. All samples passed the QC check.
Figure 1. Laser microdissection of the human granule cell layer.
Neu-N stained hippocampal sections prepared form a patient without granule cell pathology (A) and a patient with granule cell dispersion, i.e. type 2 granule cell pathology (C). The dissection line is in black. (B) and (D) are higher magnifications of the boxes in (A) and (C), respectively.
Microarray
Total RNA was used for microarray analysis (Human microRNA Microarray V3, #G4470C, Agilent Technologies, Santa Clara, CA, USA). This chip consists of 60-mer DNA probes and allows simultaneous analysis of almost 1200 human miRNA obtained from the Sanger miR-BASE database (Release 10.1). We employed approximately 100 ng total RNA per sample in each experiment. RNA labeling and hybridization were performed in accordance to manufacturer's indications. Agilent scanner and the Feature Extraction 10.5 software (Agilent Technologies) were used to obtain the microarray raw-data.
Microarray results were analyzed using the GeneSpring GX 12 software (Agilent Technologies). Data transformation was applied to set all the negative raw values at 1.0, followed by Quantile normalization and log2 transformation. Filters on gene expression were used to keep only the miRNAs detected in at least one sample (n = 536). The number of expressed miRNAs was 493 in the no GCP group, 464 in the GCP 2 group. Differentially expressed miRNAs were identified by comparing GCP type 2 vs. no GCP samples. A 2 fold-change filter (n = 141) and the unpaired t-test were applied (p<0.05; False Discovery Rate-FDR = 7%). Differentially expressed genes were employed in Cluster Analysis, using the Pearson correlation as a measure of similarity. For Cluster image generation, an additional step of normalization on gene median across all samples was added.
miRNA qRT-PCR
Quantitative real-time PCR (qRT-PCR) analysis of hsa-miR-338-3p, 219-5p and 487a was performed using a TaqMan miRNA assay kit (Applied Biosystems) according to the manufacturer's instructions. Samples were run in triplicate at 95°C for 15 s and 60°C for 1 min using a CFX96 Touch Real-Time PCR Detection System (Applied Biosystems). Analysis was performed by the comparative threshold cycle (CT) method. rRNA U48 was used as reference gene. The relative amount of each miRNA in epileptic samples was calculated using the equation RQ = 2−CT, where CT = (CT miRNA − CT U6 RNA). Similar results were obtained using rRNA U6 as reference gene.
Bioinformatics
Target prediction was performed by comparative analysis of several databases, using an open-source database [16].
ANTXR1 qRT-PCR
mRNA levels of antrax receptor 1 (ANTXR1) (assay ID: Hs01120394) and neuronal enolase 2 (ENO 2) (assay ID: Hs01102367), were determined using TaqMan Real-Time PCR, according to manufacturer's instructions (Applied Biosystems). Ten ng of total RNA were retro-transcribed using iScript Reverse Transcription Supermix (BIO-RAD). cDNA templates were amplified with TaqMan PreAmp Master Mix (Applied Biosystems), using pooled assay mix for ANTXR1 and ENO 2 and then assayed for gene expression as described previously. Each sample was analyzed in triplicate, in two independent experiments. The level of each mRNA was measured using Ct (threshold cycle) and the amount of target was calculated as described above for miRNAs. Gene expression levels were normalized using ENO 2 expression, as reported previously for this particular tissue [17].
ANTXR1 immunohistochemistry
ANTXR1 immunostaining was performed by an automatic and clinically validated instrument based on Ventana Benchmark Ultra systems from Roche Tissue Diagnostics. This immunohistochemistry technique takes advantage of a new enhanced sensitivity biotin-free multimer technology system, based on direct linkers between peroxidase and secondary antibodies (ultraView Universal DAB Detection Kit, Ventana Medical System). The protocol provided for the automatic CC1 Ventana pre-treatment (Cell Conditioning Solution, Ventana) for 52 min, then the incubation for 2 hours with the anti-ANTXR1 antibody by titration (rabbit polyclonal, by ThermoFisher Scientific; 1: 100). Staining was visualized with the UltraView DAB procedure by Benchmark Ultra System. Sections were then counterstained with haematoxylin. Negative controls were treated identically except that the primary antibody was omitted. Sections of metastatic breast cancer were used as positive controls [18],[19]. Evaluation of data was performed by two expert neuropathologists (GM and BP) under double-blind conditions.
Statistical analysis
For qRT-PCR data, comparisons between experimental groups were performed by using the Mann–Whitney U test. Differences between groups were considered significant when P < 0.05.
Results
Patients
Tissues from patients indicated in numbers in Table 1 were employed for microarray analysis. Neuropathological examination evidenced that these patients had HS type 1 [13], which was associated with no granule cell pathology (no GCP) in 5 patients and with granule cell pathology (GCP type 2) in the other 5 [14]: GCP consisted of granule cell dispersion in 4 cases and bilaminar granule cell layer in one. The no GCP group was composed of 3 males and 2 females, with mean age at surgery of 44 (33–60), mean years after epilepsy diagnosis of 24 (7–38) and approximately 10 seizures per month before surgery (2 to >30); the GCP group was composed of 5 females, with mean age at surgery of 33 (31–37), mean years after epilepsy diagnosis of 23 (10–35) and approximately 10 seizures per month before surgery (3 to 15). An epileptogenic insult could be identified in only one of the no GCP cases (febrile convulsions), whereas all GCP cases had a history of febrile convulsions, one also a possible brain trauma ( Table 1 ).
Table 1. Patients included in the study.
| Patient number | Gender | Age at surgery | Epileptogenic insult | Years after diagnosis | Seizures per month | Drug therapy (current) | Pathology MTS [29] | Pathology MTS [30] | Pathology GCP [14] | Outcome [31] |
| 01 | M | 60 | none | 38 | >30 | VPA, CBZ, TGB | Grade IV | MTS 1B | no GCP | Ia |
| 02 | M | 44 | none | 12 | 5–9 | TPM, LVT | Grade III | MTS 1A | no GCP | Ia |
| 03 | M | 36 | none | 7 | 8–10 | LVT, PB, CLB | Grade III | MTS 1A | no GCP | Ia |
| 04 | F | 47 | none | 33 | 2–3 | PB, CBZ | Grade III | MTS 1A | no GCP | Ic |
| 05 | F | 33 | febrile convulsions | 30 | 5–10 | TPM, CBZ, VPA, PB | Grade III | MTS 1A | no GCP | Ia |
| I | M | 55 | none | 51 | 10–15 | OXC, LTG, CLB | Grade III | MTS 1A | no GCP | Ia |
| II | F | 31 | none | 16 | 3–4 | CBZ, ZNS | Grade III | MTS 1A | no GCP | Ia |
| 06 | F | 31 | febrile convulsions | 10 | 8–12 | PB, TPM | Grade III | MTS 1A | GCP 2 | IIa |
| 07 | F | 33 | febrile convulsions | 24 | 3–4 | LTG, LVT, PB | Grade III | MTS 1A | GCP 2 | Ia |
| 08 | F | 32 | febrile convulsions | 27 | 4–10 | CBZ | Grade III | MTS 1A | GCP 2 | Ia |
| 09 | F | 32 | febrile conv. (trauma?) | 20 | 9–10 | OXC, LVT | Grade IV | MTS 1B | GCP 2 | Ia |
| 10 | F | 37 | febrile convulsions | 35 | 12–15 | CBZ, TPM | Grade IV | MTS 1B | GCP 2 | Ia |
| III | F | 41 | febrile convulsions | 38 | 3–4 | CBZ, PGB | Grade III | MTS 1A | GCP 2 | Ia |
| IV | M | 36 | febrile convulsions | 22 | 5–6 | TPM, LVT | Grade III | MTS 1A | GCP 2 | Ia |
CBZ, carbamazepine; CLB, clobazam; LTG, lamotrigine; LVT, levetiracetam; OXC, oxcarbazepine; PB, pentobarbital; TGB, tiagabine; TPM, topiramate; VPA, valproic acid.
miRNA microarray
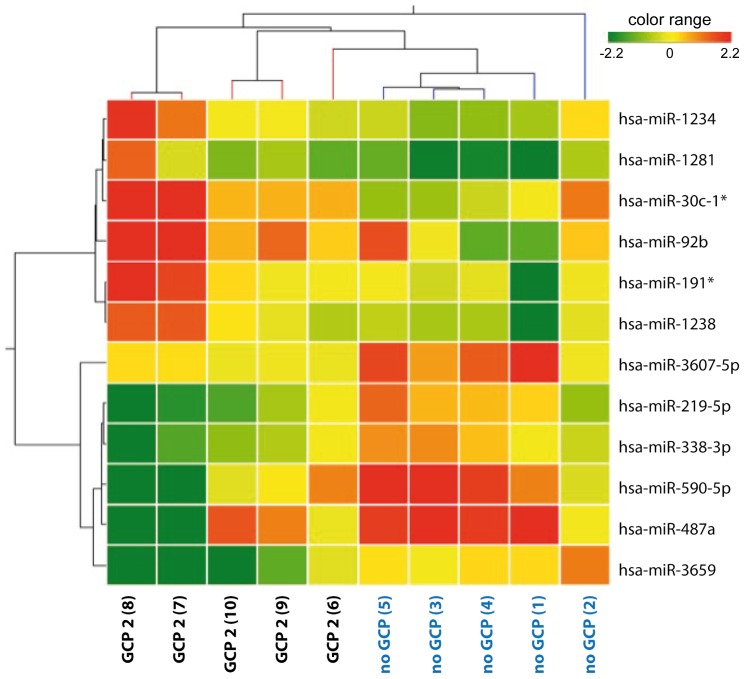
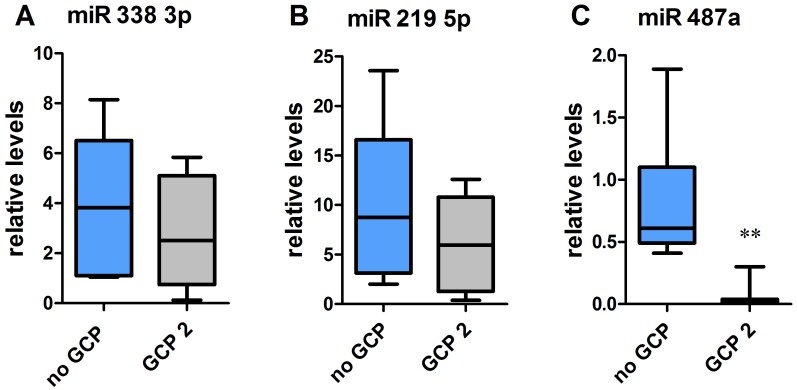
Twelve miRNAs were differentially expressed in patients without GCP compared with patients with type 2 GCP ( Fig. 2 ). Of these, 6 had relatively higher expression in tissue from patients without GCP and 6 were higher in those with GCP 2 ( Fig. 2 ). Differential expression of a subset of 3 miRNAs (namely miR-338-3p, miR-219-5p and miR-487a) was validated in an extended cohort of patients (the ten original patients plus another 2 per group, indicated in roman numbers in Table 1 ) using qRT-PCR. Expression levels of all these miRNAs were apparently different in the two groups, confirming microarray findings, but data were dispersed for miR-338-3p and miR-219-5p, not reaching significance level ( Fig. 3A and 3B ). In contrast, miR-487a expression was confirmed to be highly significantly reduced in GCP 2 ( Fig. 3C ).
Figure 2. miRNAs differentially expressed in patients without granule cell pathology (no GCP) or with type 2 GCP (GCP 2).
Heat-map representation of the average expression of the 12 differentially expressed miRNAs in no GCP and GCP 2 from ten different tissues. The colors of the genes represented on the heat map correspond to the expression values normalized on miRNA mean expression across all samples: green indicates down-regulated; red indicates up-regulated in the tissue. Patients are identified by numbers (in parenthesis) that correspond to those reported in the Table 1.
Figure 3. Relative expression of miR-338-3p (A), miR-219-5p (B) and miR-487a (C), evaluated by qRT-PCR in patients without granule cell pathology (no GCP, blue bars) or with type-2 GCP (black bars).
Seven patients per group. **P<0.01 Mann-Whitney U test.
Target identification and validation for miR-487a
Comparative analysis of several databases [16] indicated at least 10 highly likely targets for miR-487a, namely FAM126A, ANTXR1, NUDCD1, AP1S3, AP3D1, AFTPH, KIAA1217, ZNF57 PAG1 and PTGER3. All these mRNAs are expressed in the brain (www.genecards.org). A subset of these are associated with vesicle trafficking (AP1S3, AP3D1, AFTPH), others code for receptors (PTGER3), intracellular signaling (FAM126A) or transcription factors (ZNF57; www.genecards.org). More interestingly, two of these mRNAs (PAG1 and ANTXR1) may be associated with cell adhesion (www.genecards.org). In particular, ANTXR1 (also known as tumor endothelial marker 8, TEM8) is expressed in the mouse dentate gyrus granule cell layer (Allen atlas; http://mouse.brain-map.org/gene/show/45380) and has been reported to promote cell spreading in human tumor tissues [20],[21]: therefore, it was hypothesized that reduced expression of miR487a will increase ANTXR1 levels, leading to granule cell spreading (i.e. dispersion or bilamination, i.e. GCP 2).
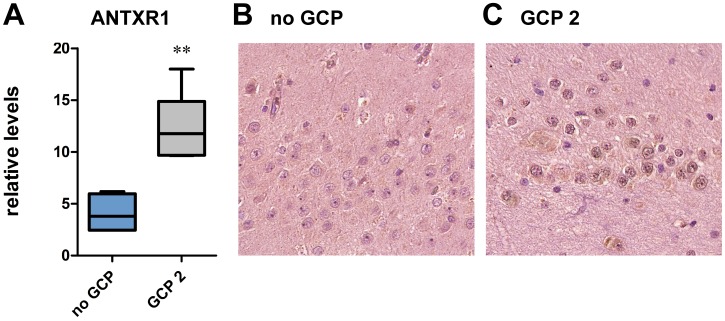
Evidence in support of this hypothesis has been pursued by analyzing ANTXR1 mRNA and protein levels in the same samples employed for validation of miR-487a. As predicted, using qRT-PCR ANTXR1 mRNA levels were increased in the GCP 2 group ( Fig. 4A ). Moreover, a clear increase of ANTXR1 immunoreactivity was observed in the granule cell layer of patients with type-2 GCP, as compared with those with no GCP ( Fig. 4B–C ).
Figure 4. Relative expression of ANTXR1 (A), evaluated by qRT-PCR, in patients without granule cell pathology (no GCP, blue bars) or with type-2 GCP (black bars).
Seven patients per group. **P<0.01 Mann-Whitney U test. Representative granule cell layer hippocampal sections from patients without granule cell pathology (B) or with type-2 GCP (C) exhibiting DAB-labeled ANTXR1-like immunoreactivity (LI). Omitting the primary antibody to estimate nonspecific signal yielded completely negative labeling (data not shown). Note a widespread increase in ANTXR1-LI in granule cells from patients with type-2 GCP (C).
Discussion
The main findings of this study were: (1) the identification of 12 miRNAs differentially expressed in the hippocampal granule cell layer of patients with hippocampal sclerosis associated with GCP 2 as compared with patients with no GCP; (2) the RT-qPCR confirmation of one of these, miR-487a, in an extended cohort of patients; (3) the identification of ANTXR1 as a possible target of miR-487a.
An important issue in evaluation of this data is the possible influence of medical treatments on miRNA expression. Indeed, there is evidence that antiepileptic drugs can interfere with miRNA expression: it has been reported that valproate can modulate miR-24, miR-34a, and miR-128 [22] and that phenobarbital can down-regulate miR-122 [23]. Although two patients in the no-GCP group were treated with valproate and five patients (three in the no-GCP group and two in the GCP 2 group) were treated with phenobarbital, none of the above miRNAs was found to be differentially expressed. More in general, a systematic bias due to pharmacological treatments seems unlikely, because all patients in both groups were using many drugs in combination. Therefore, although we cannot rule out an influence of the antiepileptic treatment on miRNAs expression, it seems more likely that the changes we observed are due to the pathology.
The prognosis of patients undergoing epilepsy surgery has been hypothesized to depend on the absence or presence of GCP but, thus far, results have been inconsistent. While some studies reported that GCP does not affect post-surgical outcome [24],[25], others suggested association between GCP and a favorable prognosis [14],[26]. Identification of molecular biomarkers that parallel and/or integrate the pathology findings would provide a valuable prognostic element, and miR-487a may represent a first component of this molecular signature that will allow better patient stratification. In the cohort of patients we analyzed in this study, however, no clear distinction of the outcome was observed in the early timeframe of post-surgical follow-up. Therefore, extension of the follow-up will be needed to verify this possibility.
MiR-487a has been reported to be down-regulated in Alzheimer disease [27] and up-regulated in schizophrenia [28]. Could it play a role in GCP? All miRNAs can have hundreds of targets, but target prediction based bioinformatics approaches is difficult for many reasons, most of all because of imperfect complementarity. However, comparative analysis of several databases [16] indicates at least 10 highly likely targets for miR-487a. ANTXR1 emerged as the most interesting, because it has been reported to promote cell spreading: ANTXR1 is a transmembrane protein that functions as an adhesion molecule, coupling binding of an immobilized extracellular ligand and cell spreading through association to the actin cytoskeleton [20]. Thus, reduced expression of miR-487a could increase ANTXR1 mRNA and protein levels and thereby favor granule cell dispersion. Here, we have provided circumstantial evidence that this could indeed be the case. Further studies in vitro and in animal models are currently ongoing to directly demonstrate this hypothesis.
In conclusion, miR-487a may be the first identified element of a miRNA signature that may be useful for a prognostic evaluation of post-surgical epilepsy and form the basis for mechanistic studies that may lead to the identification of new therapeutic targets, like ANTXR1.
Acknowledgments
The authors are grateful to Dr. Pitt Niehusmann (University of Bonn) for helpful discussions; to Dr. Fulvio Chiesa (Roche, Italy) for technical advice; to Cristina Zampini, Patrizia Raisi, Maura Masiero, Maria Novi, Rosaria Morelli, Anna Cherubino and Elisa Zaffoni for technical support.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This work has been supported by grants from the European Community [FP7-PEOPLE-2011-IAPP project 285827 (EPIXCHANGE) and FP7-HEALTH project 602102 (EPITARGET), to MS], and from the Ri.MED foundation (to PC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 2. Im HI, Kenny PJ (2012) MicroRNAs in neuronal function and dysfunction. Trends Neurosci 35: 325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McNeill E, Van Vactor D (2012) MicroRNAs shape the neuronal landscape. Neuron 75: 363–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jimenez-Mateos EM1, Engel T, Merino-Serrais P, McKiernan RC, Tanaka K, et al. (2012) Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat Med 18: 1087–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Henshall DC (2014) MicroRNA and epilepsy: profiling, functions and potential clinical applications. Curr Opin Neurol 27: 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Song YJ, Tian XB, Zhang S, Zhang YX, Li X, et al. (2011) Temporal lobe epilepsy induces differential expression of hippocampal miRNAs including let-7e and miR-23a/b. Brain Res 1387: 134–140. [DOI] [PubMed] [Google Scholar]
- 7. Hu K, Xie YY, Zhang C, Ouyang DS, Long HY, et al. (2012) MicroRNA expression profile of the hippocampus in a rat model of temporal lobe epilepsy and miR-34a-targeted neuroprotection against hippocampal neurone cell apoptosis poststatus epilepticus. BMC Neurosci 13: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8. Bot AM, Debski KJ, Lukasiuk K (2013) Alterations in miRNA levels in the dentate gyrus in epileptic rats. PLoS ONE 8: e76051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gorter JA, Iyer A, White I, Colzi A, van Vliet EA, et al. (2013) Hippocampal subregion-specific microRNA expression during epileptogenesis in experimental temporal lobe epilepsy. Neurobiol Dis 62: 508–520. [DOI] [PubMed] [Google Scholar]
- 10. Kan AA, van Erp S, Derijck AA, de Wit M, Hessel EV, et al. (2012) Genome-wide microRNA profiling of human temporal lobe epilepsy identifies modulators of the immune response. Cell Mol Life Sci 69: 3127–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McKiernan RC, Jimenez-Mateos EM, Bray I, Engel T, Brennan GP, et al. (2012) Reduced mature microRNA levels in association with dicer loss in human temporal lobe epilepsy with hippocampal sclerosis. PLoS ONE 7: e35921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blümcke I, Thom M, Aronica E, Armstrong DD, Vinters HV, et al. (2011) The clinicopathologic spectrum of focal cortical dysplasias: a consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia 52: 158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blümcke I, Thom M, Aronica E, Armstrong DD, Bartolomei F, et al. (2013) International consensus classification of hippocampal sclerosis in temporal lobe epilepsy: a task force report from ILAE Commission on Diagnostic Methods. Epilepsia 54: 1315–1329. [DOI] [PubMed] [Google Scholar]
- 14. Blümcke I, Kistner I, Clusmann H, Schramm J, Becker AJ, et al. (2009) Towards a clinico-pathological classification of granule cell dispersion in human mesial temporal lobe epilepsies. Acta Neuropathol 117: 535–544. [DOI] [PubMed] [Google Scholar]
- 15. Peiró-Chova L1, Peña-Chilet M, López-Guerrero JA, García-Giménez JL, Alonso-Yuste E, et al. (2013) High stability of microRNAs in tissue samples of compromised quality. Virchows Arch 463: 765–774. [DOI] [PubMed] [Google Scholar]
- 16. Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk - database: prediction of possible miRNA binding sites by "walking" the genes of 3 genomes. J Biomed Inform 44: 839–847. [DOI] [PubMed] [Google Scholar]
- 17. Maurer-Morelli CV, de Vasconcellos JF, Reis-Pinto FC, Rocha Cde S, Domingues RR, et al. (2012) A comparison between different reference genes for expression studies in human hippocampal tissue. J Neurosci Methods 208: 44–47. [DOI] [PubMed] [Google Scholar]
- 18. Chen D, Bhat-Nakshatri P, Goswami C, Badve S, Nakshatri H (2013) ANTXR1, a stem cell-enriched functional biomarker, connects collagen signaling to cancer stem-like cells and metastasis in breast cancer. Cancer Res 73: 5821–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gutwein LG, Al-Quran SZ, Fernando S, Fletcher BS, Copeland EM, Grobmyer SR (2011) Tumor endothelial marker 8 expression in triple-negative breast cancer. Anticancer Res 31: 3417–3422. [PubMed] [Google Scholar]
- 20. Werner E, Kowalczyk AP, Faundez V (2006) Anthrax toxin receptor 1/tumor endothelium marker 8 mediates cell spreading by coupling extracellular ligands to the actin cytoskeleton. J Biol Chem 281: 23227–23236. [DOI] [PubMed] [Google Scholar]
- 21. Gu J, Faundez V, Werner E (2010) Endosomal recycling regulates anthrax toxin receptor 1/tumor endothelial marker 8-dependent cell spreading. Exp Cell Res 316: 1946–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou R, Yuan P, Wang Y, Hunsberger JG, Elkahloun A, et al. (2009) Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology 34: 1395–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shizu R1, Shindo S, Yoshida T, Numazawa S (2012) MicroRNA-122 down-regulation is involved in phenobarbital-mediated activation of the constitutive androstane receptor. PLoS ONE 7: e41291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thom M, Liagkouras I, Elliot KJ, Martinian L, Harkness W, et al. (2010) Reliability of patterns of hippocampal sclerosis as predictors of postsurgical outcome. Epilepsia 51: 1801–1808. [DOI] [PubMed] [Google Scholar]
- 25. da Costa Neves RS, Jardim AP, Caboclo LO, Lancellotti C, Marinho TF, et al. (2013) Granule cell dispersion is not a predictor of surgical outcome in temporal lobe epilepsy with mesial temporal sclerosis. Clin Neuropathol 32: 24–30. [DOI] [PubMed] [Google Scholar]
- 26. Marucci G, Rubboli G, Giulioni M (2010) Role of dentate gyrus alterations in mesial temporal sclerosis. Clin Neuropathol 29: 32–35. [DOI] [PubMed] [Google Scholar]
- 27. Wang WX, Huang Q, Hu Y, Stromberg AJ, Nelson PT (2011) Patterns of microRNA expression in normal and early Alzheimer's disease human temporal cortex: white matter versus gray matter. Acta Neuropathol 121: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beveridge NJ, Cairns MJ (2012) MicroRNA dysregulation in schizophrenia. Neurobiol Dis 46: 263–271. [DOI] [PubMed] [Google Scholar]
- 29. Wyler AR, Dohan FC Jr, Schweitzer JB, Berry AD III (1992) A grading system for mesial temporal pathology (hippocampal sclerosis) from anterior temporal lobectomy. J Epil 5: 220–225. [Google Scholar]
- 30. Blümcke I, Pauli E, Clusmann H, Schramm J, Becker A, et al. (2007) A new clinico-pathological classification system for mesial temporal sclerosis. Acta Neuropathol 113: 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engel J Jr, Van Ness P, Rasmussen T, Ojemann L (1993) Outcome with respect to epileptic seizures. In Engel J Jr editor.Surgical treatment of the epilepsies, 2nd edNew YorkRaven Press 609–621. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.