Abstract
The production and dissemination of spores by members of the fungal kingdom is a major reason for the success of this eukaryotic lineage in colonizing most terrestrial ecosystems. Ballistospores are a type of spore produced by basidiomycete fungi, such as the mushrooms and plant pathogenic rusts. These spores are forcefully discharged through a unique liquid-drop fusion mechanism, enabling the aerosolization of these particles that can contribute to plant disease and human allergies. The genes responsible for this process are unknown due to technical challenges in studying many of the fungi that produce ballistospores. Here, we applied newly-developed techniques in a forward genetic screen to identify genes required for ballistospore formation or function in a tractable red yeast, a species of Sporobolomyces. One strain bearing a mutation in the PHS1 gene was identified as a mirror mutant. PHS1 encodes 3-hydroxyacyl-CoA dehydratase required for the third step in very long chain fatty acid biosynthesis. The Sporobolomyces PHS1 gene complements the essential functions of a S. cerevisiae phs1 mutant. The Sporobolomyces phs1 mutant strain has less dehydratase activity and a reduction in very long chain fatty acids compared to wild type. The mutant strain also exhibits sensitivity to cell wall stress agents and loss of shooting due to a delay in ballistospore formation, indicating that the role of Phs1 in spore dissemination may be primarily in cellular integrity.
Introduction
Life on land without the convenience of an aqueous environment presents many challenges to organisms, one of which is how to spread from one location to another to find new nutrient sources or mating partners. The three macroscopic kingdoms of eukaryotes have solved this problem in different ways. For instance, animals are able to move physically. Plants disperse by pollen and seed, leading to the elaborate methods present today for pollination or encasing seed within fruits to attract animals. The third group, the fungi, often use spores for dispersal in the air, requiring physical force to launch the spores away from the colony or fruiting body.
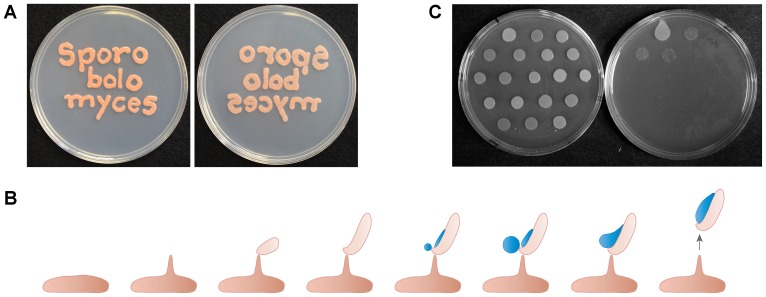
Fungi have evolved mechanism to disperse spores on independent occasions. These mechanisms include firing the ascospores from fruiting structures in Ascomycetes, the water canon of Pilobolus spp., water splash dispersal in the Nidulariaceae (bird's nest fungi) and a springboard in Sphaerobolus spp. [1]. One spore type that is unique to the basidiomycetes is the ballistospore (Fig. 1). Ballistospores can be produced either asexually (ballistoconidia) or sexually (basidiospores). The process has attracted interest for more than a century, with ballistospore-forming yeasts termed “mirror” yeasts, taking this name by being able to produce a mirror image of the culture from one Petri dish on an uninoculated second dish (Fig. 1A). For instance, noted mycologist A.H.R. Buller (1874–1944) dedicated a considerable part of his career to research on ballistospore formation in the red yeasts [2], and the characteristic liquid that forms on the ballistospore is termed “Buller's drop” as a consequence. More recent analyses, including high-speed video photography, reveals that spore release involves a fusion event between Buller's and a second drop of liquid that forms on the side of the ballistospore [3], [4] (Fig. 1B).
Figure 1. The “mirror” capability of ballistospore-forming yeast species.
(A) The plate on the left was inoculated with Sporobolomyces sp. wild type strain IAM 13481, and inverted onto an uninoculated plate on the right. (B) Diagram of the steps in the formation of a ballistospore from a yeast cell, based on [2], [10]. (C) The 18 mirror mutants isolated in this study growing on the top plate (left) and their replication to a bottom plate (right). The wild type strain is in the top left hand corner of the bottom plate.
Ballistospores are prevalent particles in the air during certain times of the year when their associated species are sporulating, and when these spores form the majority of bioaerosols [5], [6]. Two important influences that ballistospores have on humans are their roles in the life cycles of the rust fungi and as airborne particles that cause allergies in people. First, the rusts are obligate plant parasites, i.e. they cannot be cultured in vitro. They cause disease in thousands of plant species, the most economically serious being rusts of grain crops. Second, basidiomycete spores are a common cause of allergic asthma. Rates of affected individuals vary depending on country, environment and season. The limited access to antigens hinder detection of reactivity to allergens in patients, leading to underestimates in incidence. Nevertheless, metadata analyses of clinical studies indicate that 3 to 35% of asthma patients react to basidiomycetes [7]–[9]. The sources of these allergens are ballistospores fired from fungi, and that circulated in the air until inhaled by susceptible people.
The process of ballistospore biosynthesis requires intricate cell morphogenesis and developmental timing, with highly polarized growth, partitioning of the spore cell into different areas, and asymmetrical release of sugars to trigger the formation of Buller's and the adaxial drops of liquid (Fig. 1B). Another striking feature of ballistospores is the force generated for propulsion. This is of relevance because spores fired from mushrooms must avoid impact on the adjacent side of the gill. In contrast, spore release from red yeasts is optimized for maximum dispersion in air currents [10]. The main forces acting against the spores are drag, with additional input in repulsion and attraction from electrostatic forces [11]–[13].
Despite the importance of ballistospores for agriculture and human health, and the fascinating underlying cell biology and biophysics, no gene is known that affects ballistospore production or release. Two challenges in understanding ballistospores are that (a) rust fungi cannot be cultured away from a host plant and (b) the fruiting bodies of mushrooms contain two nuclei, limiting the employment of mutant screens for gene discovery since most mutations are likely to be recessive loss-of-function when compared to the wild type allele in the other nucleus. In contrast, the red yeasts are haploid, can be cultured easily in vitro, and generate ballistospores. We and others have recently developed transformation protocols for red yeasts, including a species of ballistospore-shooting Sporobolomyces [14]–[17], providing the means to dissect the gene functions in this group of organisms. In this study, we apply these tools towards understanding the genetic requirements of ballistospores formation and release. We report a mutant screen to identify strains with impaired shooting, and the identification and characterization of the first gene required for ballistospore production, demonstrating the requirement of very long chain fatty acids (VLCFAs) in this process.
Materials and Methods
Strains and transformation
The Sporobolomyces sp. ura5 uracil auxotroph AIS2 was transformed using Agrobacterium-mediated delivery of T-DNA carrying the wild type URA5 gene [14]. Strains were maintained on yeast extract peptone dextrose medium (YPD). Impaired ballistospore production or release was assessed by inoculating transformants on YPD medium in 10 mm deep Petri dishes, and inverting these over an uninoculated YPD plate. The wild type strain produces a mirror image on the uninoculated plate, whereas mirror mutants do not. The URA5 marker was recycled by selection for mutations within it by plating strain GI209 on medium containing 1 g/L 5-fluoroorotic acid. The S. cerevisiae phs1::KanMX/PHS1 heterozygote “magic marker” strain (Thermo Scientific Open Biosystems, Waltham, MA [18]) was transformed with plasmids as described below. Transformed strains were sporulated in 1% potassium acetate 0.005% zinc acetate, and plated onto magic medium to select for haploid strains carrying the phs1::KanMX allele [19]. Cafenstrole (analytical standard, Sigma-Aldrich, St. Louis, MO) was dissolved in DMSO and added to plates in one set of experiments. The fungal strains and their genotypes are listed in table 1.
Table 1. Strains used in this study.
| Name | Genotype* | Background | Reference |
| Sporobolomyces sp. | |||
| IAM 13481 | Wild type | [41] | |
| AIS2 | ura5 | IAM 13481 | [14] |
| GI209 | phs1::URA5 ura5 | AIS2 | This study |
| AIS11 | phs1::ura5 ura5 | GI209 | This study |
| AIS13 | URA5 phs1::ura5 ura5 | AIS11 | This study |
| AIS15 | PHS1-URA5 phs1::ura5 ura5 | AIS11 | This study |
| Saccharomyces cerevisiae | |||
| BY4741 | his3 leu2 met15 ura3 | [42] | |
| YSC 4034-97/041153 | MATa/α ura3 leu2 his3 lys2/LYS2 met15/MET15 can1::LEU2-MFA1pr-HIS3/CAN1 phs1::KanMX/PHS1 | RESGEN KanMx YKO heterozygous knock out strain + pXP346 | [19], [43] |
| AIY6 | PHS1/phs1 + pTH19 | YSC 4034-97/041153 | This study |
| AIY9 | PHS1 + pTH19 | BY4741 | This study |
| AIY5 | PHS1/phs1 + pTH19-PHS1 | YSC 4034-97/041153 | This study |
| AIY8 | phs1::KanMX + pTH19-PHS1 lys2 | AIY5 | This study |
| R1158 | URA3::CMV-tTA | BY4741 | [26] |
| TH-3237 | pTetO7-PHS1 | R1158 | [28] |
*Unless specified, the genotype of the background strain is also present in the strain, and is not re-written.
DNA manipulation
Inverse PCR was used to amplify flanks of the T-DNA in Sporobolomyces as described previously [14]. The amplicons were sequenced and compared to the Sporobolomyces genome database (http://genome.jgi-psf.org/Sporo1/Sporo1.home.html) by BLASTn.
The ends of the PHS1 gene were defined by 5′ and 3′ rapid amplification of cDNA ends (RACE) using the GeneRacer kit (Invitrogen, Life Technologies, Grand Island, NY) with gene specific primers (RA004 5′-ATGTGCGACATCATGAAGGC-3′ and RA009 5′-AGACCAGGCGGTAACCATC-3′ for 5′ RACE, and RA010 (5′-ATGTCTGCCGCTTCGCGC-3′) and ALID1374 5′-CTGCGTCGTCTTCCCTCAGGTCCCTC-3′ for 3′ RACE). The RNAs used for RACE were purified with TRIzol reagent (Invitrogen) from overnight cultures grown in yeast nitrogen base +2% dextrose. The full-length PHS1 cDNA was amplified with primers RA010 (5′-ATGTCTGCCGCTTCGCGC-3′) and RA011 (5′-TCAGTGAGACGAAGTGATC-3′), and cloned into plasmid pCR2.1 (Invitrogen) to produce plasmid pRA1. The cDNA was subcloned with EcoRI into pTH19 [20], a 2 µ circle plasmid for replication in S. cerevisiae and that expressed the insert from the ADH1 promoter. For complementation of the phs1 mutation, a wild type copy of the gene was amplified with primers ALID1288 (5′-CCGCTCGAGCTGTCCTCAGAATGGTCTCG-3′) and ALID1289 (5′-CCGCTCGAGACATTTGACAGGCTTGGTCG-3′), digested with XhoI, and cloned into the XhoI site of plasmid pAIS4 [14]. The plasmid was electroporated into A. tumefaciens strain EHA105, and the PHS1-URA5 T-DNA or the empty URA5 T-DNA from pAIS4 trans-conjugated into Sporobolomyces strain AIS11.
Lipid characterization
Sphingolipid metabolism and labeling experiments in Sporobolomyces strains and S. cerevisiae followed those as reported previously [21]. The elongation assay was performed as reported [22], [23].
Stress sensitivity tests
For sensitivity tests, mirror mutants were serially diluted 10-fold in liquid YPD and spotted onto solid YPD agar containing caspofungin, congo red, fluconazole, hydrogen peroxide, sodium chloride, or sodium dodecyl sulfate. Non-shooting mutants were also exposed to UV irradiation and to 37°C for 7 h. Experiments were replicated three times.
Results
Identification of mirror mutants by T-DNA insertional mutagenesis
To identify factors that influence that ability of fungi to produce or shoot ballistospores, a forward genetic screen was used in a species of Sporobolomyces (strain IAM 13481). Approximately 5,000 T-DNA insertional mutants were generated using Agrobacterium-mediated transformation. The transformants were patched on agar medium to form colonies, and inverted over uninoculated plates of media. 18 mirror mutants were identified with loss or reduction of the ability to transfer to the bottom plate (Fig. 1C).
Examination of the colonies of the 18 strains by microscopy revealed that the most common reason (17 cases) for the inability to produce the mirror image was the loss or large reduction in the ability to produce the ballistospore. One strain, GI277, still produced ample ballistospores, but was unable to release them (Fig. S1).
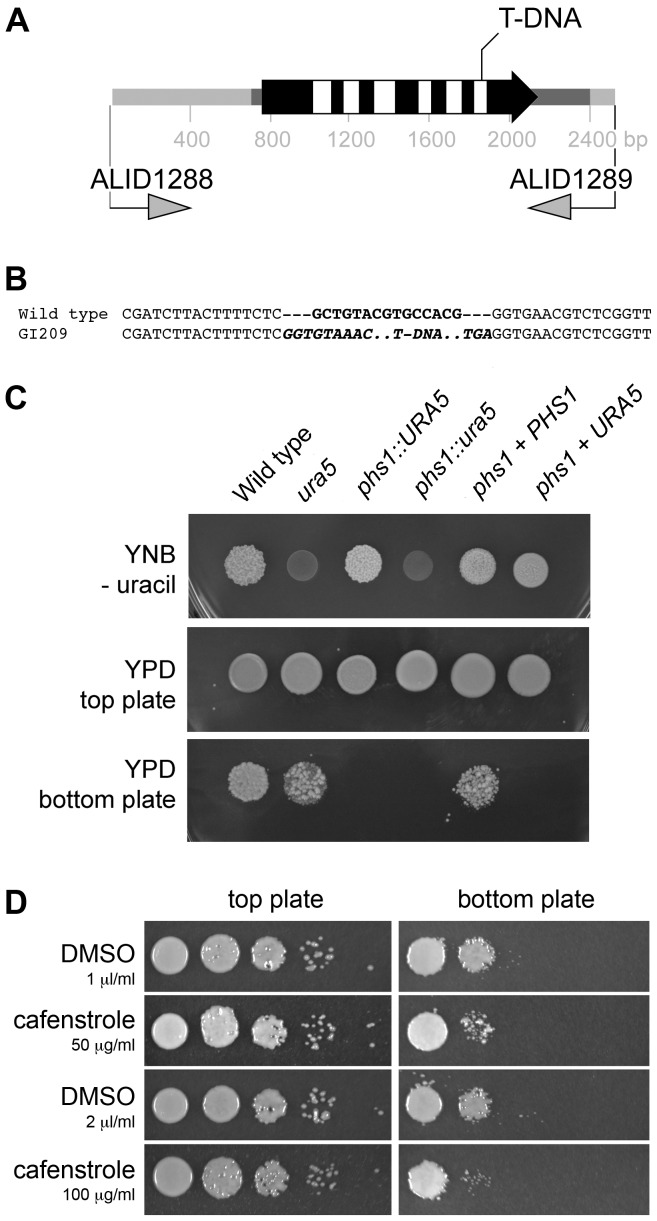
Strain GI209, with reduced spore production and with a single T-DNA insertion in its genome as based on Southern blot analysis, was chosen for further study. The junctions of its T-DNA insertion were obtained by inverse PCR and compared by BLASTn to the genome sequence database of Sporobolomyces. The T-DNA inserted close to a gene annotated as a PHS1 homolog, that encodes 3-hydroxyacyl-CoA dehydratase for the third step in very long chain fatty acid biosynthesis (Fig. 2).
Figure 2. Loss of PHS1 impairs the ability to produce ballistospores.
(A) The structure of the PHS1 gene, and the T-DNA insertion position within it in strain GI209. Exons are in black and introns in white, with the 5′ and 3′ untranslated regions in darker grey. The two primers, ALID1288 and ALI1289, were used to amplify the wild type copy for gene complementation. (B) DNA sequence alignments of the wild type or T-DNA insertional mutant strain. The 15-bp black region in the wild type is replaced with the T-DNA molecule, whose left and right border sequences are in bold italics. (C) The loss of PHS1 can be complemented by reintroducing a wild type copy of the PHS1 gene. Yeast nitrogen base (YNB) without uracil is the selection medium for transformation. Ballistospores were fired from the top plate onto the bottom plate, and colonies allowed to grow. (D) An inhibitor of VLCFA synthesis, cafenstrole, reduces ballistospore production. Wild type Sporobolomyces were 10-fold serially diluted, and grown on YPD supplemented with cafenstrole or the solvent DMSO for two days (top plate) inverted over the bottom plates. The bottom plates were cultured two additional days.
A phs1 mutant has reduced ballistospore production
The structure of the PHS1 gene in Sporobolomyces was ambiguous in the genome sequencing database. Based on BLAST comparisons, we predicted the gene extended beyond the annotated version, and that the T-DNA insertion would lie within the final intron of the gene to generate an allele with residual function. The 5′ and 3′ ends of the PHS1 transcript were determined by rapid amplification of cDNA ends, allowing us to annotate this gene correctly and confirm the hypothesis that the T-DNA insertion in strain GI209 is in the final intron (Fig. 2A, B). A 15-bp deletion following the T-DNA insertion event also occurred (Fig. 2B). The strain GI209, ura5 phs1::URA5, is indicated in the text and in the figures as Sporobolomyces mutant phs1, with the exception of figure 2B where it is indicated as phs1::URA5. The PHS1 gene sequence is deposited in GenBank as accession JQ068150.
To show that the phenotype of strain GI209 is due to mutation of PHS1, functional complementation was carried out. Due to the lack of gene markers in Sporobolomyces, the complementation reported here is based on a uracil marker recycling system. An uracil auxotroph, AIS11, was isolated from strain GI209, by selection on YNB +5-FOA, and transformed by A. tumefaciens either with the URA3 or URA5 gene of Sporobolomyces. The URA5 gene complemented the mutation, indicating that AIS11 is ura5 phs1::ura5. This strain is indicated in figure 2B as phs1::ura5. One stable ura5 phs1::ura5 URA5 transformant, AIS13, was used as a control for further experiments; this strain is indicated in the text and in the figures as phs1 + URA5. For complementation, the wild type copy of the PHS1 gene was amplified from Sporobolomyces, cloned in the plasmid pAIS4 containing the Sporobolomyces URA5 gene, and introduced by A. tumefaciens into the uracil auxotroph AIS11. One stable ura5 phs1::ura5 PHS1 URA5 transformant, AIS15, was selected; this strain is indicated in the text and in the figures as phs1 + PHS1. In strain AIS15, the wild type gene complemented the mirror mutant phenotype, providing evidence that the mutation in PHS1 causes the loss of ballistospore production (Fig. 2C).
There are several possible consequences to transcript production of having the T-DNA insertion in the final intron of PHS1 in strain GI209. The 3′ end for the transcript produced by this mutant allele was sought by RACE and amplicons cloned and sequenced. This revealed at least one scenario is that the original 5′ splice site still functions, but a new 3′ splice site within the T-DNA sequence is used. Further, a second and novel intron within the T-DNA is also spliced. This information is illustrated as Fig. S2.
The chloroacetamides are a class of chemicals used in agriculture for weed control, and that act by inhibition of VLCFA synthesis [24], [25]. Cafenstrole was tested on the wild type Sporobolomyces strain. At concentrations that allowed equal growth compared to the solvent control, the ability to mirror to the bottom plate was reduced almost an order of magnitude (Fig. 2D). This result provides additional evidence that VLFCA synthesis is important for the proper formation of ballistospores.
The Sporobolomyces PHS1 gene complements the lethality of deleting PHS1 in Saccharomyces cerevisiae
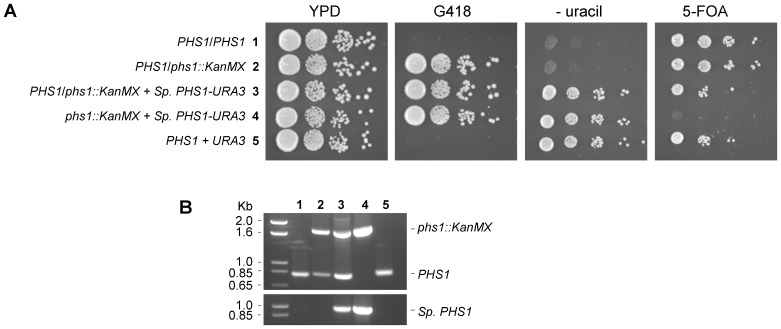
Comparison of the Sporobolomyces Phs1 amino acid sequence with its characterized homologs that are available in GenBank suggests that the protein would function in VLCFA biosynthesis. For instance, alignment of the homolog with the Phs1 protein sequence of S. cerevisiae reveals that all amino acid residues known to be important for function are conserved in the Sporobolomyces protein [[26], [27] Fig. S3]. To infer similar biochemical function, we tested if the Sporobolomyces gene could complement the loss of the PHS1 gene from S. cerevisiae, where it is essential for viability. The Sporobolomyces cDNA was cloned into a yeast expression plasmid, and the plasmid introduced into a phs1/PHS1 heterozygote strain of S. cerevisiae. As a control, the empty plasmid was also transformed into this phs1/PHS1 heterozygote strain. After meiotic sporulation and selection for the haploids [19], the strains transformed with the Sporobolomyces PHS1 gene yet carrying a deletion of the S. cerevisiae homolog were able to grow, whereas no progeny were obtained for the strain carrying the empty plasmid (Fig. 3A). This was further supported by the observation that plasmids could be “cured” from strains by selection on medium containing 5-FOA that is toxic to strains carrying a functional URA3 gene, except in the case of the phs1::KanMX haploid strain (Fig. 3A). The absence or presence of the PHS1 genes in the S. cerevisiae strains was confirmed by PCR analysis (Fig. 3B). These suggest that the Sporobolomyces PHS1 gene has the same functions as the S. cerevisiae homolog.
Figure 3. Sporobolomyces PHS1 complements loss-of-function of the S. cerevisiae phs1 homolog.
Five strains (numbered 1–5) were cultured on different media (A) and the presence or absence of the native PHS1 or the Sporobolomyces PHS1 gene in S. cerevisiae tested by PCR (B). (A) Ten fold serial dilutions of yeast strains. YPD allows equal growth of the strains. G418 selects for strains carrying a phs1::KanMX allele. The YNB – uracil selects for strains carrying a plasmid with the URA3 selectable marker. 5-FOA is used to counter-select or “cure” plasmids containing the URA3 marker from a ura3 mutant strain background. Strain 1 is a diploid with both copies of PHS1. Strain 2 is a heterozygote strain with one wild type and one mutated copy of the PHS1 gene. Strain 3 is strain 2 expressing the Sporobolomyces PHS1 gene from a 2 µ plasmid. Strain 4 is a haploid progeny from meiosis of strain 3. Strain 5 is a control haploid strain with the 2 µ plasmid. (B) The top panel shows PCR of the wild type allele of S. cerevisiae PHS1 (near the 0.85 kb ladder marker) or the replacement allele phs1::KanMX (near to 1.6 kb ladder marker). The bottom panel is amplification of the Sporobolomyces PHS1 gene from the same strains.
Mutation of PHS1 in Sporobolomyces sp. impairs 3-hydroxyacyl-CoA dehydratase activity and alters the lipid profiles of the cells
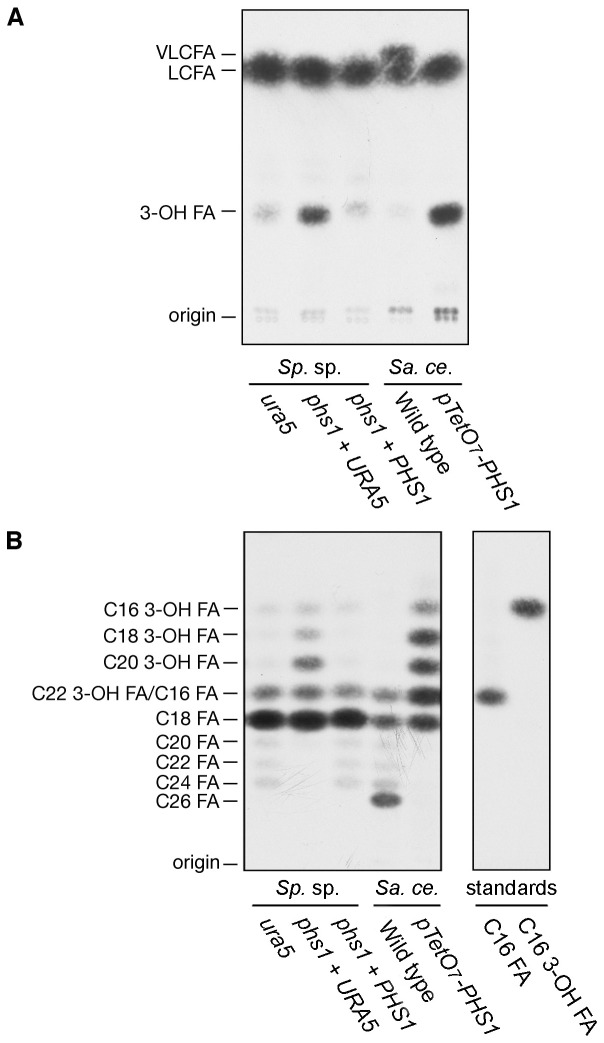
The second evidence for Sporobolomyces Phs1 being a 3-hydroxyacyl-CoA dehydratase is altered enzymatic activity in the T-DNA phs1 mutant GI209. Membranes were extracted from Sporobolomyces strains and S. cerevisiae control strains. The two S. cerevisiae strains, R1158 (wild type) and TH-3237 (pTetO7-PHS1), were used as controls since their activities and lipid profiles have been characterized previously. TH-3237 cells carry the PHS1 gene under control of the TetO7 promoter. Doxycyclin can shut off expression [26], [28]. The extracts were incubated with [14C]3-hydroxypalmitoyl-CoA, then lipids processed and separated by normal phase thin layer chomatography. The radioactivities associated with the reaction product 2,3-trans hexadecenoic acid were compared between strains. In mutants phs1 and phs1+ URA5, the T-DNA insertion caused a reduction in the 3-hydroxyacyl-CoA dehydratase activity to about half compared to the wild type cells of IAM 13481 and AIS2 (ura5), or the complemented control strain AIS15 (phs1 + PHS1). Interestingly, the reduction in 2,3-trans hexadecenoic acid levels in the Sporobolomyces mutant was equivalent to that seen in the PHS1-repressed strain of S. cerevisiae (Fig. 4A). Furthermore, the radioactivity associated with the reaction product 2,3-trans hexadecenoic acid was significantly higher in strains bearing a functional PHS1 (strains AIS2 and complemented phs1 + PHS1) compared to the phs1 mutant (Fig. 4B).
Figure 4. The phs1 mutation in Sporobolomyces causes reduction in 3-hydroxyacyl-CoA dehydratase activity.
(A) Sporobolomyces (Sp. sp.) wild type (IAM 13481), mutant phs1 (GI209), ura5 auxotroph (AIS2), uracil prototroph phs1+ URA5 (AIS13), and complementing strain phs1 + PHS1 (AIS15) cells were grown in YPD medium at 25°C. As controls, S. cerevisiae (Sa. ce.) wild type (R1158) and pTetO7-PHS1 (TH-3237) were grown in YPD medium containing 10 µg/ml doxycyclin for 6.5 h at 30°C. Total membranes (4 µg) prepared from the strains were incubated for 15 min at 37°C [14C]3-hydroxypalmitoyl-CoA (3.6 µM; 10 nCi/µl). After termination of the reactions, lipids were saponified, acidified, extracted, and separated by normal phase TLC. (B) The radioactivities associated with the reaction product 2,3-trans hexadecenoic acid were quantified using a bioimaging analyzer BAS-2500. Values represent the mean ± S.D. from three independent experiments. Statistically significant differences are indicated (**p<0.01; t-test).
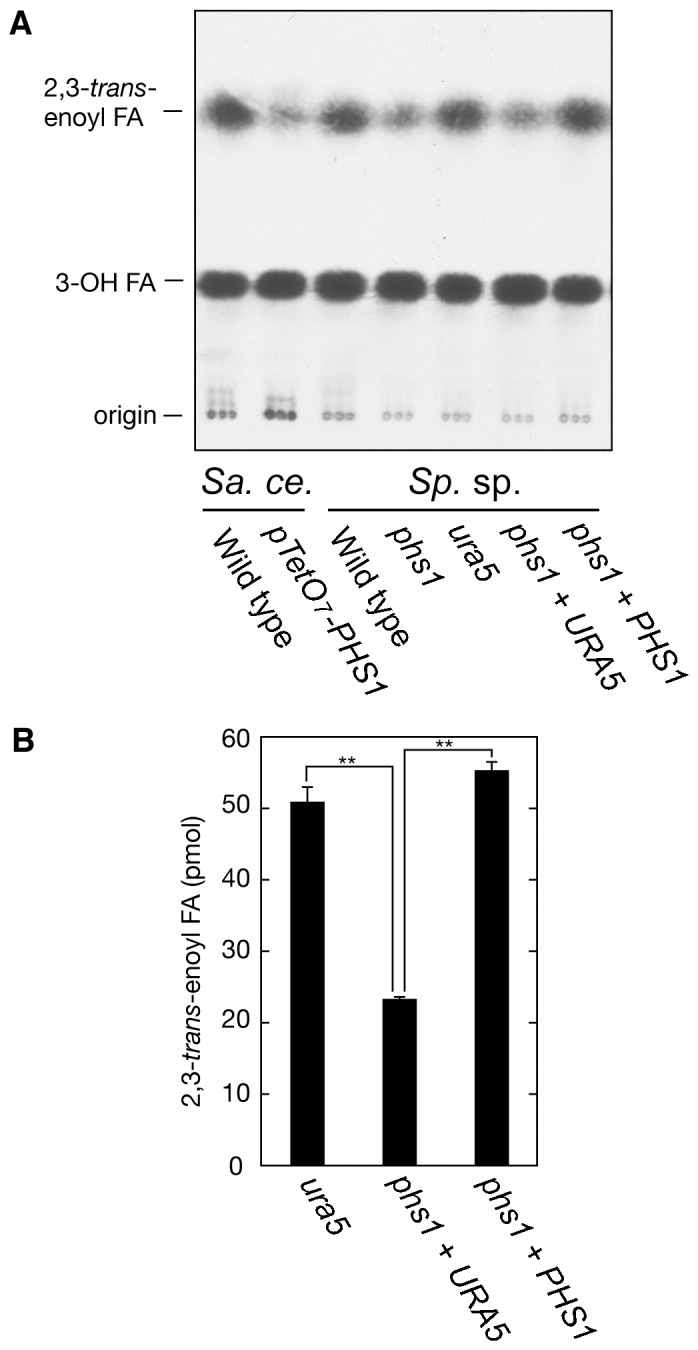
A fatty acid elongation assay was used with the substrates palmitoyl-CoA and malonyl-CoA in the presence of NADPH. In wild type S. cerevisiae R1158, C16-CoA was elongated to C26-CoA (Fig. 5) as previously reported. In the engineered S. cerevisiae strain TH-3237, pTetO7-PHS1, a differential spot corresponding to 3-hydroxy acyl-CoAs (3-OH FA) was detected (Fig. 5A). Separation of the reaction products by reverse phase TLC revealed that the chain-lengths of the accumulated 3-hydroxyacyl-CoAs were a mixture of C16, C18, C20, and C22 (Fig. 5B). In strains of Sporobolomyces having a wild type copy of the PHS1 gene, C16-CoA was mainly elongated to C18-CoA and slightly to C24-CoA, similarly to the wild type of S. cerevisiae R1158 with the exception of C26-CoA. In Sporobolomyces phs1 + URA5, 3-hydroxyacyl-CoAs accumulated like those in the pTetO7-PHS1 S. cerevisiae cells (Fig. 5A and B), further confirming the role of the Sporobolomyces PHS1 gene in the elongation of VLCFAs.
Figure 5. The phs1 mutation causes reduction in elongation of C16:0-CoA.
(A) Sporobolomyces (Sp. sp.) ura5 auxotroph (AIS2), uracil prototroph phs1+ URA5 (AIS13) and complementing strain phs1 + PHS1 (AIS15) cells were grown in YPD medium at 25°C. S. cerevisiae (Sa. ce.) wild type (R1158) and pTetO7-PHS1 (TH-3237) strains were grown in YPD medium containing 10 µg/ml doxycyclin for 6.5 h at 30°C. Total membranes (20 µg) prepared from the strains were incubated with C16:0-CoA (20 µM) and 100 µM (0.075 µCi) [14C]malonyl-CoA in the presence of 1 mM NADPH for 1 h at 37°C. (A) After termination of the reactions, lipids were saponified, acidified, extracted, and separated by normal-phase TLC, followed by detection by autoradiography. (B) After termination of the reactions, lipids were saponified, acidified, and extracted. The resulting fatty acids were then converted to fatty acid methyl ester, extracted, separated by reverse-phase TLC, and detected by autoradiography. [3H]Palmitic acid (C16 FA) and [14C]3-hydroxypalmitic acid (C16 3-OH FA) were used as standards.
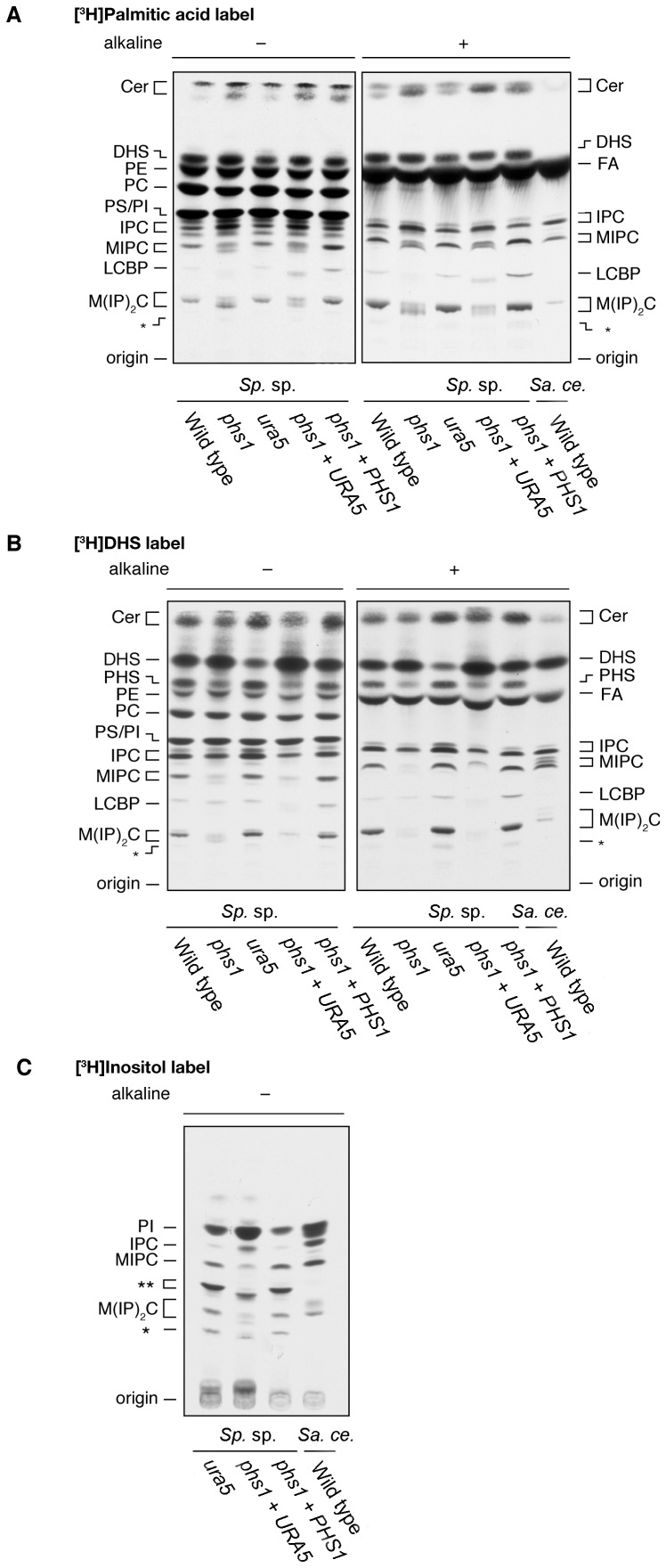
Eukaryotes synthesize a suite of VLCFAs or derived molecules. The fungal strains were cultured in [3H]palmitic acid, [3H]dihydrosphingosine, or [3H]inositol, the lipids extracted and resolved by normal phase TLC. Sporobolomyces likely contains the same sphingolipids as S. cerevisiae, that is IPC, MIPC, and M(IP)2C (Fig. 6). We speculate that the fatty acid portion of sphingolipids in Sporobolomyces may be C18 or C24, but not C26. The T-DNA insertion into the phs1 gene caused reductions in MIPC and M(IP)2C but an increase in IPC. Synthesis of glycerophospholipids was not affected. MIPC and M(IP)2C in the phs1 mutants migrated slower than those in the wild type strain on TLC plates. It is possible that MIPC and M(IP)2C in the phs1 mutants contain an abnormal hydroxy group or a shorter fatty acid compared to S. cerevisiae. The asterisks in Fig. 6 indicate unidentified lipids. One could be a M(IP)2C derivative, such as M(IP)2C containing an additional hydroxy group or sugar.
Figure 6. The phs1 mutation affects sphingolipid synthesis.
Sporobolomyces (Sp. sp.) wild type (IAM 13481), mutant phs1 (GI209), ura5 auxotroph (AIS2), uracil prototroph phs1+ URA5 (AIS13), and complementing strain phs1 + PHS1 (AIS15) cells and S. cerevisiae (Sa. ce.) wild type (R1158) were grown in YPD medium medium (A and B) or in SC medium lacking inositol (C) at 25°C. Cells were labeled with [3H]palmitic acid (A), [3H]dihydrosphingosine (B), or [3H]inositol (C) for 2 h. Lipids were extracted, treated with nothing or alkaline solution, and separated by normal phase TLC, followed by detection by autoradiography. Abbreviations are CER, ceramide; DHS, dihydrosphingosine; FA, fatty acid; PE, phosphatidylethanolamine; PHS, phytosphingosine; PC, phosphatidylcholine; PS, phosphatidylserine; PI, phosphatidylinositol; IPC, inositolphosphorylceramide; MIPC, mannosylinositol phosphorylceramide; LCBP, long chain base phospholipid; M(IP)2C, mannosyldiinositol phosphorylceramide. The asterisks indicate unidentified lipids.
Additional phenotypes are associated with mutation of PHS1 and other mirror mutants
VLCFAs are associated with a number of cellular processes. We expected that additional phenotypes should be observed for the phs1 T-DNA insertion mutant, and compared its growth under different conditions, along with the other 17 mirror mutants for comparison. Of the 18 strains examined, 14 also showed impaired growth under one or more stress conditions (Fig. S1B). In the case of the phs1 mutant, it showed strong sensitivity to agents like the Fks1 inhibitor caspofungin, congo red, sodium chloride and the detergent sodium dodecyl sulfate (Fig. 7). Caspofungin and congo red act by inhibiting the enzyme (1, 3)-β-D-glucan synthase to disturb the integrity of the fungal cell wall; sodium dodecyl sulfate affects membrane stability and also, indirectly, cell wall construction; and NaCl interferes with the normal osmolarity of the cells, thus damaging cells with cell wall defects. The re-introduction of the wild type copy of the PHS1 gene into the mutant phs1 restored the tolerance to the stress compounds back to the wild type level, indicating additional roles of the Sporobolomyces PHS1 gene in cell wall function and maintenance. This information suggests that some genes, such as PHS1, required for ballistospore production or release from the parent yeast cell also have other roles in the cell.
Figure 7. Stress phenotypes of the phs1 mutant of Sporobolomyces suggest that defects in other cellular processes can impair the formation of spores.
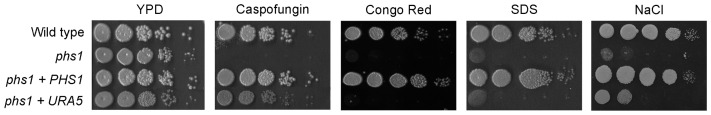
10-fold serial dilutions of overnight cultures of Sporobolomyces wild type (IAM 13481), mutant phs1 (GI209), complementing strain phs1 + PHS1 (AIS15) and uracil prototroph phs1+ URA5 (AIS13) were plated onto YPD media without or with stress agents, and cultured 3–4 days. Stress agents were caspofungin (50 µg/ml), congo red (0.4%), sodium dodecyl sulfate (SDS, 0.015%) and sodium chloride (NaCl, 1 M).
Discussion
Ballistospores are a mobilized spore type found in one lineage of the fungi, the Basidiomycota. Although these spores play important roles in life cycles of these fungi, no gene involved in their biosynthesis or release has been characterized. In part this is due to the technical challenges in working with rust fungi or mushrooms. As a first step towards identifying factors that are required for ballistospore formation or release, we performed a genetic screen on a collection of mutants. The screen is far from saturated (5,000 transformants; the Sporobolomyces sp. IAM 13481 genome project estimates 5,500 genes). 18 transformants (3.6%) were isolated as impaired in efficient spore shooting, being unable to make a mirror image of the colonies onto an adjacent Petri dish. Characterization of other phenotypes in these mutants suggests that many are affected in cell wall, membrane or polarity in generating the ballistospores, because the strains are also sensitive to agents that cause such stress.
We characterized one Sporobolomyces strain bearing a mutation in the PHS1 gene in detail: this gene encodes the 3-hydroxyacyl-CoA dehydratase enzyme required for VLCFA biosynthesis. The VLCFAs have chain lengths of more than 20 carbons. These molecules are synthesized by a four-step process of sequential elongation, that add two carbons to the chain for each cycle. The pathway for VLCFA synthesis is conserved in eukaryotes and it plays essential roles in physiology related to lipid membranes [23]. The importance of this pathway is illustrated by the factor that loss of VLFCA synthesis is lethal in fungi like S. cerevisiae or S. pombe [29]. PHS1 first emerged in studies in S. cerevisiae, where genetic screens revealed that it was essential for cell viability [30]–[32]. At about the same time, mutant screens in the plant Arabidopsis thaliana identified a gene required for cell differentiation and proliferation. Mutants form callus tissue instead of leaves and stems, and the phenotype is exasperated by addition of cytokinin plant hormones. The PASTICCINO2 (or PEPINO) gene was subsequently cloned, and is able to complement the S. cerevisiae phs1 mutation [33]–[36]. Key studies elucidating the function of Phs1 were conducted in S. cerevisiae, showing that the protein catalyzes the third step in biosynthesis of very long chain fatty acids [26], [37]. Human and plant homologs have similar activities [28], [38]. Recently, a human inbred family has been identified with childhood myopathogies attributable to a mutation in one of the four human homologs of PHS1, HACD1 [39]. The identity of this human gene was in part guided by a dog centronuclear myopathy model, which is linked to a transposon insertion in the homolog of this gene [40].
The isolation of the Sporobolomyces allele of phs1 in this study was fortuitous since the gene is essential in other fungi. The T-DNA insertion is within the final intron, past the essential amino acid residues required for function. Characterization of the transcript produced in the strain revealed a chimeric form that utilized information within the T-DNA region, including for splicing novel introns. We speculate that this aberrant protein maintains some residual hydroxyacyl-CoA dehydratase activity. The mechanism by which reduction of Phs1 function impairs ballistospore formation is currently unknown, but likely is by alteration of cell polarity, membrane properties, or vesicle transport. Overall, the identification of the VLCFA pathway as a requirement for ballistospore formation adds another striking property to the growing list of important biological processes – Phs1 is essential in yeasts, regulates differentiation and division in plants, and impacts muscle function in mammals – controlled by these molecules.
This study demonstrates that the genes for ballistospores no longer need to remain unknown. Future developments in molecular tools in Sporobolomyces sp. or other ballistospore-forming yeasts, and the identification of genes affected in other mutants, will reveal insights into this unique cell biology process. Of particular interest are mutants that can produce ballistospores, but are unable to fire them. The identification of the genes required for ballistospore formation may also have practical applications. For instance, chemical reagents that inhibit VLCFA biosynthesis or the development of drugs targeting genes involved in sporulation could be used in control of rust fungi on infected crops or their alternative host plants, or to reduce allergenic spores to improve the indoor environmental quality in office buildings, schools, and other non-industrial buildings.
Supporting Information
Summary of spore production and the sensitivity tests to cell wall and plasma membrane stress-generating compounds of non-shooting mutants of Sporobolomyces . (A) Images of the surface of colonies of the wild type or mirror mutant strains. The photographs were modified to provide contrast such that each dark dot represents a ballistospore. (B) Ten-fold serial dilutions of yeast cells were spotted onto YPD without any supplements or containing caspofungin (50 µg/ml), congo red (0.4%), fluconazole (FLC, 32 and 64 µg/ml), hydrogen peroxide (H2O2, 2 mM), sodium chloride (NaCl, 1 M), sodium dodecyl sulfate (SDS, 0.015%). The strains were also exposed to UV irradiation (120 J/m2) and to 37°C for 7 h.
(TIF)
Comparison of 3′ ends of the PHS1 alleles from wild type and strain GI209. Coding nucleotides are in upper case, with the difference in amino acid sequence between the predicted proteins encoded by the two alleles in green. Introns are in grey lower case, or orange for the two new intron sequences in strain GI209. Blue sequence represents 3′ untranslated regions (one intron in GI209 falls in this region). The underlined region is the T-DNA sequence.
(TIF)
Alignment of Sporobolomyces Phs1 predicted amino acid sequence with characterized homologs. The homologs are from S. cerevisiae (PHS1), human (HACD1) and A. thaliana (PASTICCINO2). Conserved sites with characterized functions are marked with green highlighted asterisks. Grey m letters above the black font indicate transmembrane residues. Amino acid residues in blue indicate cytoplasmic and in orange indicates ER lumen localization.
(TIF)
Acknowledgments
We thank Steven Henning and Christopher Eddleman for technical help.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The PHS1 gene sequence (genomic and mRNA) were deposited to GenBank and are available under accession JQ068150.
Funding Statement
This research was supported by a Grant-in-Aid for Scientific Research (B) (23370057) from the Japan Society for the Promotion of Science (JSPS), the University of Missouri Research Board, National Science Foundation (grant MCB-0920581) and National Institutes of Health (NIAID grant R21 AI094364). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ingold CT (1971) Fungal spores: their liberation and dispersal. Oxford: Oxford University Press. [Google Scholar]
- 2.Buller AHR (1933) Researches on Fungi, Vol. V. London: Longmans, Green & Co. [Google Scholar]
- 3. Pringle A, Patek SN, Fischer M, Stolze J, Money NP (2005) The captured launch of a ballistospore. Mycologia 97: 866–871. [DOI] [PubMed] [Google Scholar]
- 4. Noblin X, Yang S, Dumais J (2009) Surface tension propulsion of fungal spores. J Exp Biol 212: 2835–2843. [DOI] [PubMed] [Google Scholar]
- 5. Pashley CH, Fairs A, Free RC, Wardlaw AJ (2012) DNA analysis of outdoor air reveals a high degree of fungal diversity, temporal variability, and genera not seen by spore morphology. Fungal Biol 116: 214–224. [DOI] [PubMed] [Google Scholar]
- 6. Fröhlich-Nowoisky J, Pickersgill DA, Després VR, Pöschl U (2009) High diversity of fungi in air particulate matter. Proc Natl Acad Sci USA 106: 12814–12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Simon-Nobbe B, Denk U, Pöll V, Rid R, Breitenbach M (2008) The spectrum of fungal allergy. Int Arch Allergy Immunol 145: 58–86. [DOI] [PubMed] [Google Scholar]
- 8. Horner WE, Helbling A, Lehrer SB (1998) Basidiomycete allergens. Allergy 53: 1114–1121. [DOI] [PubMed] [Google Scholar]
- 9. Horner WE, Helbling A, Salvaggio JE, Lehrer SB (1995) Fungal allergens. Clin Microbiol Rev 8: 161–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stolze-Rybczynski JL, Cui Y, Stevens MHH, Davis DJ, Fischer MWF, et al. (2009) Adaptation of the spore discharge mechanism in the basidiomycota. PLoS ONE 4: e4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buller AHR (1909) Researches on Fungi, Vol. 1. London: Longmans. [Google Scholar]
- 12. Gregory PH (1957) Electrostatic charges on spores of fungi. Nature 130: 330. [Google Scholar]
- 13. Webster J, Proctor MCF, Davey RA (1988) Measurement of the electrical charge on some basidiospores and an assessment of two possible mechanisms of ballistospore propulsion. T Brit Mycol Soc 91: 193–203. [Google Scholar]
- 14. Ianiri G, Wright SAI, Castoria R, Idnurm A (2011) Development of resources for the analysis of gene function in Pucciniomycotina red yeasts. Fungal Genet Biol 48: 685–695. [DOI] [PubMed] [Google Scholar]
- 15. Abbott EP, Ianiri G, Castoria R, Idnurm A (2013) Overcoming recalcitrant transformation and gene manipulation in Pucciniomycotina yeasts. Appl Microbiol Biotechnol 97: 283–295. [DOI] [PubMed] [Google Scholar]
- 16. Liu Y, Koh CMJ, Sun L, Hlaing MM, Du M, et al. (2013) Characterization of glyceraldehyde-3-phosphate dehydrogenase gene RtGPD1 and development of genetic transformation method by dominant selection in oleaginous yeast Rhodosporidium toruloides . Appl Microbiol Biotechnol 97: 719–729. [DOI] [PubMed] [Google Scholar]
- 17. Lin X, Wang Y, Zhang S, Zhu Z, Zhou YJ, et al. (2014) Functional integration of multiple genes into the genome of the oleaginous yeast Rhodosporidium toruloides . FEMS Yeast Res 14: 547–555. [DOI] [PubMed] [Google Scholar]
- 18. Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, et al. (2004) A robust toolkit for functional profiling of the yeast genome. Mol Cell 16: 487–496. [DOI] [PubMed] [Google Scholar]
- 20. Harashima T, Heitman J (2005) Gα subunit Gpa2 recruits kelch repeat subunits that inhibit receptor-G protein coupling during cAMP-induced dimorphic transitions in Saccharomyces cerevisiae . Mol Biol Cell 16: 4557–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nakahara K, Ohkuni A, Kitamura T, Abe K, Naganuma T, et al. (2012) The Sjögren-Larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Mol Cell 46: 461–471. [DOI] [PubMed] [Google Scholar]
- 22. Ohno Y, Suto S, Yamanaka M, Mizutani Y, Mitsutake S, et al. (2010) ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc Natl Acad Sci USA 107: 18439–18444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kihara A (2012) Very long-chain fatty acids: elongation, physiology and related disorders. J Biochem 152: 387–395. [DOI] [PubMed] [Google Scholar]
- 24. Böger P, Matthes B, Schmalfuβ J (2000) Towards the primary target of chloroacetamides-new findings pave the way. Pest Manag Sci 56: 497–508. [Google Scholar]
- 25. Takahashi H, Ohki A, Kanzaki M, Tanaka A, Sato Y, et al. (2001) Very-long-chain fatty acid biosynthesis is inhibited by cafenstrole, N,N-diethyl-3-mesitylsulfonyl-1H-1,2,4-triazole-1-carboxamide and its analogs. Z Naturforsch C 56: 781–786. [DOI] [PubMed] [Google Scholar]
- 26. Kihara A, Sakuraba H, Ikeda M, Denpoh A, Igarashi Y (2008) Membrane topology and essential amino acid residues of Phs1, a 3-hydroxyacyl-CoA dehydratase involved in very long-chain fatty acid elongation. J Biol Chem 283: 11199–11209. [DOI] [PubMed] [Google Scholar]
- 27. Yazawa T, Naganuma T, Yamagata M, Kihara A (2013) Identification of residues important for the catalysis, structure maintenance, and substrate specificity of yeast 3-hydroxyacyl-CoA dehydratase Phs1. FEBS Lett 587: 804–809. [DOI] [PubMed] [Google Scholar]
- 28. Ikeda M, Kanao Y, Yamanaka M, Sakuraba H, Mizutani Y, et al. (2008) Characterization of four mammalian 3-hydroxyacyl-CoA dehydratases involved in very long-chain fatty acid synthesis. FEBS Lett 582: 2435–2440. [DOI] [PubMed] [Google Scholar]
- 29. Kim D-U, Hayles J, Kim D, Wood V, Park H-O, et al. (2010) Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe . Nat Biotechnol 28: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miosga T, Boles E, Schaaff-Gerstenschläger I, Schmitt S, Zimmermann FK (1994) Sequence and function analysis of a 9.74 kb fragment of Saccharomyces cerevisiae chromosome X including the BCK1 gene. Yeast 10: 1481–1488. [DOI] [PubMed] [Google Scholar]
- 31. Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, et al. (2005) Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123: 507–519. [DOI] [PubMed] [Google Scholar]
- 32. Yu L, Peña Castillo L, Mnaimneh S, Hughes TR, Brown GW (2006) A survey of essential gene function in the yeast cell division cycle. Mol Biol Cell 17: 4736–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Faure J-D, Vittorioso P, Santoni V, Fraisier V, Prinsen E, et al. (1998) The PASTICCINO genes of Arabidopsis thaliana are involved in the control of cell division and differentiation. Development 125: 909–918. [DOI] [PubMed] [Google Scholar]
- 34. Bellec Y, Harrar Y, Butaeye C, Darnet S, Bellini C, et al. (2002) Pasticcino2 is a protein tyrosine phosphatase-like involved in cell proliferation and differentiation in Arabidopsis . Plant J 32: 713–722. [DOI] [PubMed] [Google Scholar]
- 35. Haberer G, Erschadi S, Torres-Ruiz RA (2002) The Arabidopsis gene PEPINO/PASTICCINO2 is required for proliferation control of meristematic and non-meristematic cells and encodes a putative anti-phosphatase. Dev Genes Evol 212: 542–550. [DOI] [PubMed] [Google Scholar]
- 36. Nobusawa T, Okushima Y, Nagata N, Kojima M, Sakakibara H, et al. (2013) Synthesis of very-long-chain fatty acids in the epidermis controls plant organ growth by restricting cell proliferation. PLoS Biol 11: e1001531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Denic V, Weissman JS (2007) A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 130: 663–677. [DOI] [PubMed] [Google Scholar]
- 38. Bach L, Michaelson LV, Haslam R, Bellec Y, Gissot L, et al. (2008) The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc Natl Acad Sci USA 105: 14727–14731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Muhammad E, Reish O, Ohno Y, Scheetz T, Deluca A, et al. (2013) Congenital myopathy is caused by mutation of HACD1 . Hum Mol Genet 22: 5229–5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pelé M, Tiret L, Kessler J-L, Blot S, Panthier J-J (2005) SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum Mol Genet 14: 1417–1427. [DOI] [PubMed] [Google Scholar]
- 41. Yamazaki M, Komagata K (1983) An electrophoretic comparison of enzymes of ballistosporogenous yeasts. J Gen Appl Microbiol 29: 115–143. [Google Scholar]
- 42. Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, et al. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- 43. Tong AHY, Evangelista M, Parsons AB, Xu H, Bader GD, et al. (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of spore production and the sensitivity tests to cell wall and plasma membrane stress-generating compounds of non-shooting mutants of Sporobolomyces . (A) Images of the surface of colonies of the wild type or mirror mutant strains. The photographs were modified to provide contrast such that each dark dot represents a ballistospore. (B) Ten-fold serial dilutions of yeast cells were spotted onto YPD without any supplements or containing caspofungin (50 µg/ml), congo red (0.4%), fluconazole (FLC, 32 and 64 µg/ml), hydrogen peroxide (H2O2, 2 mM), sodium chloride (NaCl, 1 M), sodium dodecyl sulfate (SDS, 0.015%). The strains were also exposed to UV irradiation (120 J/m2) and to 37°C for 7 h.
(TIF)
Comparison of 3′ ends of the PHS1 alleles from wild type and strain GI209. Coding nucleotides are in upper case, with the difference in amino acid sequence between the predicted proteins encoded by the two alleles in green. Introns are in grey lower case, or orange for the two new intron sequences in strain GI209. Blue sequence represents 3′ untranslated regions (one intron in GI209 falls in this region). The underlined region is the T-DNA sequence.
(TIF)
Alignment of Sporobolomyces Phs1 predicted amino acid sequence with characterized homologs. The homologs are from S. cerevisiae (PHS1), human (HACD1) and A. thaliana (PASTICCINO2). Conserved sites with characterized functions are marked with green highlighted asterisks. Grey m letters above the black font indicate transmembrane residues. Amino acid residues in blue indicate cytoplasmic and in orange indicates ER lumen localization.
(TIF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The PHS1 gene sequence (genomic and mRNA) were deposited to GenBank and are available under accession JQ068150.