Abstract
Background
Selenium (Se) is an important nutrient that carries out many biological processes including maintaining optimal immune function. Here, inorganic selenite (Se(IV)) was evaluated for its pathogen resistance and potential-associated factors in Caenorhabditis elegans. The immune effects of Se(IV) were investigated by examining the responses of C. elegans to Pseudomonas aerugonisa PA14 strain.
Principal Findings
Se(IV)-treated C. elegans showed increased survival under PA14 infection compared with untreated controls. The significant pathogen resistance of Se(IV) on C. elegans might not be attributed to the effects of Se(IV) on PA14 as Se(IV) showed no effect on bacterial quorum-sensing and virulence factors of PA14. This study showed that Se(IV) enhanced the expression of a gene pivotal for the innate immunity in C. elegans. The study found that the pathogen-resistant phenotypes contributed by Se(IV) was absent from the skn-1 mutant worms. Moreover, Se(IV) influenced the subcellular distribution of SKN-1/Nrf in C. elegans upon PA14 infection. Furthermore, Se(IV) increased mRNA levels of SKN-1 target genes (gst-4 and gcs-1).
Conclusions
This study found evidence of Se(IV) protecting C. elegans against P. aeruginosa PA14 infection by exerting effects on the innate immunity of C. elegans that is likely mediated via regulation of a SKN-1-dependent signaling pathway.
Introduction
Dietary selenium (Se), an essential trace mineral, is involved in several key metabolic activities via selenoproteins that are essential for human health to protect against oxidative damage and to maintain optimal immune function [1]. Thus, Se plays an important role in many crucial cellular processes in nearly all tissues and cell types including those involved in innate and adaptive immune responses [2], [3], [4].
Dietary Se and selenoproteins are not only important for initiating or enhancing immunity, but also involved in immuno-regulation, which is crucial for preventing excessive responses that may lead to autoimmunity or chronic inflammation [5], [6]. Se deficiency exacerbates flu symptoms and alters cytokine responses in mice fed a Se-deficient diet compared with those on an adequate Se diet [7]. Dietary Se supplementation has been shown to attenuate the lipopolysaccharide-induced expression of pro-inflammatory agents, cyclooxygenase-2 and tumor necrosis factor-alpha in macrophages [8]. Se deficiency also causes hyper-oxidant production in T cells, which results in suppression of T cell proliferation [9]. In human immunodeficiency virus (HIV)-infected children, low levels of plasma Se were significantly and independently related to mortality and faster disease progression [10]. Se deficiency is also linked to the occurrence, virulence, or disease progression of some viral infections, such as hepatitis virus (B or C) [11], [12]. These findings demonstrate an emerging and important role for Se in immune system function and this essentiality of Se has also been summarized by another report [13]. However, whether the mechanisms involved in immune systems are regulated by Se remained to be further elucidated.
The ubiquitous bacterium, Pseudomonas aeruginosa, is an opportunistic human pathogen commonly causing nosocomial contamination in medical care facilities [14], [15], [16]. P. aeruginosa may also cause serious infection in immunocompromised, HIV, and cancer patients, resulting in morbidity and mortality [16], [17]. Pathogenesis of P. aerugonisa is regulated by secreted virulence factors which include secondary metabolites (e.g., pyocyanin and hydrogen cyanide) and bacterial enzymes such as elastase and alkaline protease [18], [19], [20]. P. aerugonisa also adopts the biofilm mode of growth to make the bacteria recalcitrant to antibiotic treatments and to increase pathogenesis [21], [22]. Moreover, to facilitate the establishment of infection, P. aerugonisa produces both cell-associated biofilm and extracellular virulence factors regulated by two well-defined quorum-sensing systems, namely lasIR system and rhlIR system, which are cell-to-cell communication systems of the bacteria [23], [24].
P. aeruginosa causes lethal infection not only to human, but also to the nematode C. elegans, a powerful model organism for studying developmental biology and host-pathogen interactions [18], [25], [26]. C. elegans can be infected with numerous human pathogens and has a high degree of conservation in innate immune signaling pathways of mammals; hence, it has been widely employed to examine virulence factors, host innate defense mechanism, and drug discovery [18], [27], [28]. P. aeruginosa strain PA14 is a clinical isolate that can infect C. elegans; and screening assays of this pathosystem have been established to identify novel virulence factors and innate immune mechanisms [18], [29].
In view of the effects of Se on immune systems, it was hypothesized that pretreatment with Se(IV) can improve the survival of P. aeruginosa strain PA14-infected worms. Herein, the protective potential of Se(IV) on C. elegans under P. aeruginosa infection was investigated. In addition, the effects of Se(IV) on the secreted virulence factors, biofilm formation, and quorum sensing of P. aeruginosa were also examined Moreover, immune response gene regulation that may be responsible for the Se(IV)-induced protection of C. elegans against PA14 infection was explored. Finally, the possible regulatory mechanism in Se(IV)-elicited immune response in C. elegans was investigated.
Materials and Methods
C. elegans and bacterial strains
Inorganic selenite (Na2SeO3, Se(IV)) was purchased from Sigma-Aldrich (Poole, Dorset, UK). Strains used in this study were Bristol N2 (wild-type); EU1, skn-1 (zu67); LD1, IdIs7 [skn-1B/C::GFP; pRF4(rol-6(su1006))] [30]; SAL105, pha-1 (e2123); and denEx2[lys-7::GFP + pha-1(+)] [31]. All C. elegans strains used in this work were provided by the Caenorhabditis Genetics Center. C. elegans was normally maintained and assayed (unless otherwise stated) at 20°C on nematode growth medium (NGM) agar plates using E. coli OP50 bacteria as food source. Pseudomonas aeruginosa strain PA14 (referred hereafter as PA14) was grown with aeration at 37°C overnight in King's B broth (KB broth) [32].
C. elegans slow-killing assay
For PA14 infection, the C. elegans slow-killing assay was adapted from the previous studies [18], [29]. Briefly, synchronized L1 larvae (wild-type or mutants) were pretreated with Se(IV) of various concentrations in liquid S-basal medium containing E. coli OP50 bacteria at 109 cells/ml for 72 h at 20°C. Subsequently, adult C. elegans was subjected to the slow-killing assay using the modified nematode growth medium (0.35% peptone instead of 0.25%, NG medium) [33]. Treatments were conducted by diluting the overnight PA14 culture with NG medium to OD600 0.1 as the assay medium containing Se(IV) of various concentrations. The Se(IV)-containing media were then aliquoted 500 µl/ well in a 24 well plate. About 20 worms were transferred to each well and incubated at 20°C; and the survival of the infected C. elegans was scored at 1-day intervals. At least three biological and three technical replicates were performed for each experiment.
Total protease activity assay
Protease activity was examined by measuring the skimmed milk hydrolysis efficacy of the secreted protease in the bacterial culture [34]. PA14 was cultured in KB broth without or with Se (IV) (0.01 µM) overnight at 37°C. A 100-µl aliquot of PA14 KB culture with or without Se(IV) was added to 900 µl of skimmed milk (0.5% (w/v)) in Tris HCl buffer (50 mM, pH 8). Total protease activity was determined by measuring the absorbance at OD600 nm at 24-h incubation. The protease activity was expressed as OD 600 per µg of protein [34]. At least three biological and three technical replicates were performed for each experiment.
Microtiter plate biofilm formation assay
Assay of the biofilm-forming ability of the bacteria was adapted from previous studies [35], [36]. To assay biofilm formation, overnight KB broth cultures without or with Se (IV) (0.01 µM) were diluted to 1∶100 into fresh KB broth containing 0.01 µM Se(IV) or distilled water as control; and 100 µl of freshly inoculated culture was then dispensed into tissue culture-treated, 96-well polystyrene microtiter plates (Costar 3599, Corning Inc., NY, USA). The plates were incubated at 37°C for 24 h with well-controlled humidity without agitation. Biofilm formation was observed by staining with 0.1% (w/v) crystal violet at room temperature for 15 min. Subsequently, the wells were washed with distilled water to remove cells and residual dye. Crystal violet from the biofilms was eluted by absolute ethanol and then the solubilized dye was measured at OD590 nm. At least three biological and three technical replicates were performed for each experiment.
Quantitative real-time RT-PCR analysis of C. elegans and P. aeruginosa PA14
For C. elegans preparation, wild-type C. elegans was treated similar to that described in previous sections except that the nematodes were collected on the first day of PA14 infection. For bacteria preparation, PA14 was cultured in KB broth without or with Se(IV) (0.01 µM) overnight at 37°C. Subsequently, the overnight PA14 cultures were centrifuged for 10 min at 1,500×g to collect the pellet. Total RNA from C. elegans or PA14 pellet was isolated using TRIzol according to manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized using Super-Script III First-strand synthesis super-Mix for qRT-PCR according to manufacturer's instructions (Invitrogen). The qRT-PCR reaction was performed on a Step One real-time cycler (Applied Biosystems, Carlsbad, CA, USA) using a SYBR Green qPCR kit (Affymetrix, Inc., Cleveland, Ohio, USA). mRNA levels were normalized to that of ACT-1 for C. elegans and 16S rRNA for PA14, respectively. Primer sequences used for qRT-PCR are listed in Table S1. The fold change was normalized to that observed in untreated C. elegans samples or untreated PA14 sample. At least three biological and three technical replicates were performed for each experiment.
Fluorescence analysis of transgenic SAL105 (lys-7::GFP) C. elegans
Synchronized L1-stage transgenic SAL105 (lys-7::GFP) was cultured as described in previous sections. SAL105 cultures without or with Se(IV) (0.01 µM) were fed with OP50 or PA14, respectively, at 20°C for 24 h. The expressions of LYS-7 were directly measured by observing the reporter green fluorescent protein (GFP) fluorescence. Fluorescence in 20 nematodes randomly selected from each set of treatments was analyzed using microscopy [37]. At least three biological and three technical replicates were performed for each experiment.
SKN-1 localization assays
Synchronized L1-stage transgenic LD1 (SKN-1::GFP) [30] was incubated in liquid S-basal containing E. coli OP50 bacteria at 109 cells/mL without or with Se (IV) (0.01 µM) for 72 h at 20°C. Subsequently, LD1 was fed with OP50 or PA14 for 5 h [38], [39]. Subsequent to this treatment, 20 nematodes randomly selected from each set of treatments were mounted onto microscope slides coated with 2% agarose, anaesthetized with 2 mM sodium azide, and capped with coverslips. The subcellular SKN-1 distribution was analyzed by fluorescence microscopy [37]. Expression patterns of LD1 worms were classified into three categories (low, medium, and high) with respect to major localization of the SKN-1::GFP fusion protein in intestinal cells. “Low” refers to animals in which SKN-1::GFP was detected in less than 5 intestinal nuclei; “medium,” SKN-1::GFP detected in 5–15 intestinal nuclei; and “high,” SKN-1::GFP present in more than 15 intestinal nuclei [39]. At least three biological and three technical replicates were performed for each experiment.
Data analysis
Statistical analysis of data was performed using SPSS, version 17.0 (SPSS Inc, Chicago, IL, USA). Results are expressed as the mean ± standard error of mean (SEM). The statistical significance of differences between the experimental groups was analyzed by one-way ANOVA and LSD post hoc test. Statistical differences were indicated at p<0.05 (*), p<0.01 (**) or p<0.001 (***) (see figures).
Results
Se(IV) protects wild-type C. elegans against P. aeruginosa PA14 infection
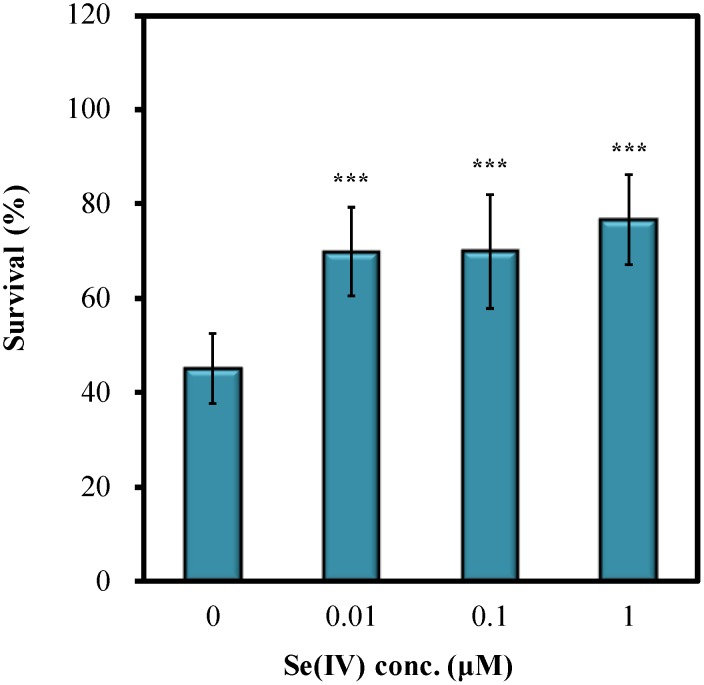
Previous research indicated that trace amount of Se(IV) exerts beneficial effects on development, reproduction, cholinergic signaling, neuroprotection, and oxidative stress defense in C. elegans [37], [40], [41]. In the present study, Se(IV)'s protective action against pathogen infection in C. elegans was further explored. To evaluate the effects of Se(IV), P. aerugonisa strain PA14 was used as the target pathogen. Wild-type N2 synchronized L1 larvae were pretreated with various concentrations of Se(IV) for 72 h at 20°C followed by PA14 infections for 72 h. Figure 1 showed that Se(IV) significantly enhanced the survival of the wild-type N2 nematodes upon PA14 infection compared with that of the control (0 µM Se(IV)). Only about 45% of PA14-infected worms survived in the control group (0 µM), whereas Se(IV)-treated nematodes showed 25% to 30% higher survival than the control group (Fig. 1). Taken together, the results indicated that Se(IV) protects wild-type C. elegans against P. aeruginosa PA14 infection.
Figure 1. Effects of Se(IV) on survival of wild-type C. elegans N2 under P. aeruginosa PA14 infection.
Synchronized wild-type L1 larvae were pretreated with Se(IV) of various concentrations (0, 0.01, 0.1, and 1 µM) for 72 h at 20°C. Subsequently, adult worms were prepared for PA14 infection. Survival of the PA14-infected worm population was scored at 1-day intervals. Graph shows the survival of C. elegans with or without Se(IV) treatment on the 3rd day of infection as the mean ± standard error of mean (SEM). Experiments were independently performed at least three times and approximated 60 worms for each treatment were scored in each experiment. Differences compared with the control (0 µM) were considered statistically significant at p<0.001 (***) by one-way ANOVA and LSD post hoc test.
Se(IV) does not affect quorum-sensing and virulence factors of PA14
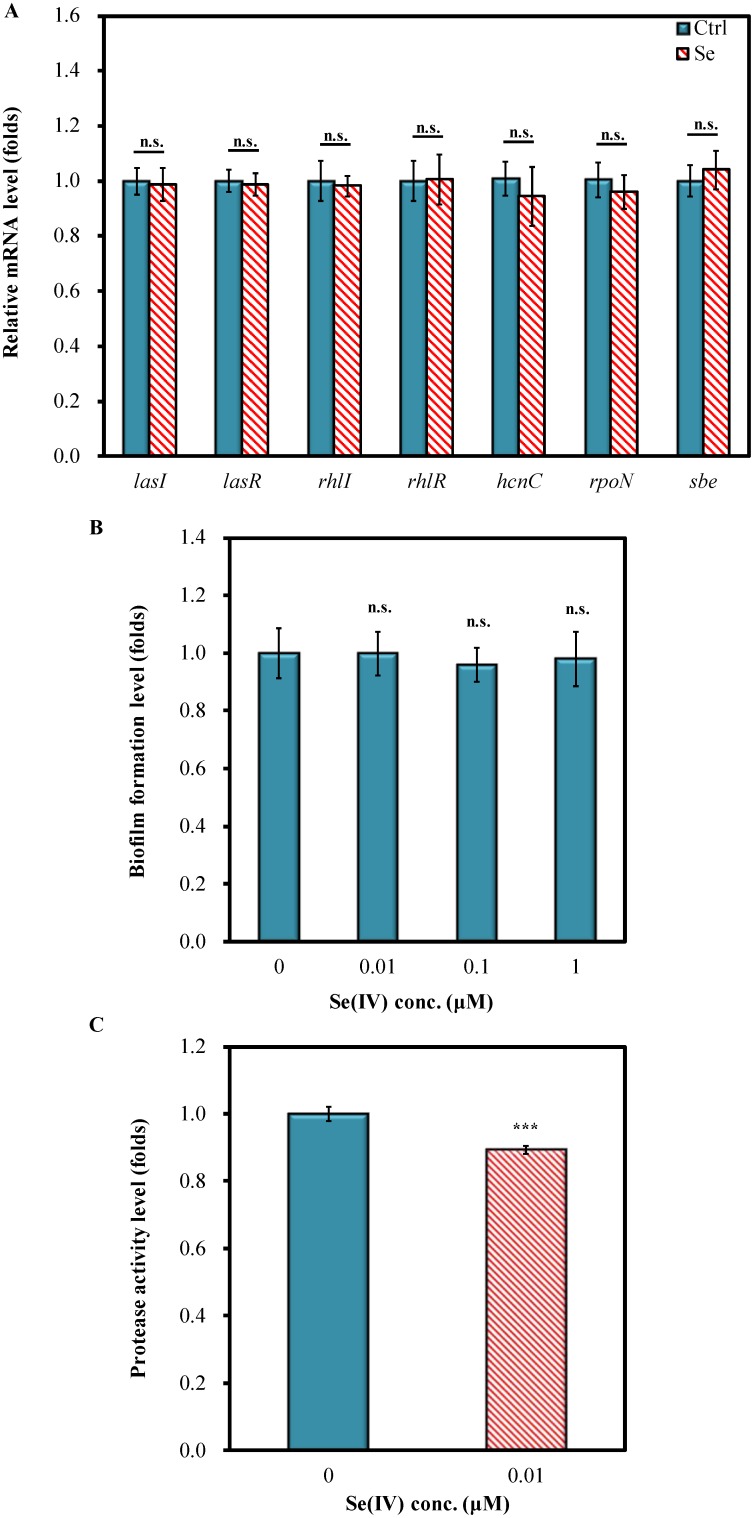
A possible mechanism that contributes to decrease the death of wild-type N2 nematodes following PA14 exposure is the inhibition of virulence factors by Se(IV) treatment. Therefore, the effects of Se(IV) on the expression of quorum-sensing genes and virulence factor genes of PA14 were examined. In all concentrations tested (0.01–1 µM), no adverse effect on PA14 growth was observed (data not shown). qRT-PCR analysis showed that 0.01 µM Se(IV) did not affect the mRNA levels of quorum-sensing genes (lasI, lasR, rhlI, and rhlR) (Fig. 2A). Moreover, the mRNA levels of several virulence factor genes, including hcnC, rpoN, and sbe, showed no significant changes under treatment with 0.01 µM Se(IV) in the culture medium (Fig. 2A). These results suggested that Se(IV) might not affect the pathogenesis-related genes of P. aeruginosa PA14.
Figure 2. Effects of Se(IV) on quorum-sensing and virulence factors of PA14.
(A). Gene expression of quorum-sensing genes and virulence factor genes of PA14 without or with Se(IV) (0.01 µM). mRNA levels were determined by quantitative real-time PCR (qRT-PCR). All measurements were normalized to mRNA levels of 16S rRNA for PA14, and fold change of each gene was normalized to that observed in control (0 µM) samples. (B). Influence of Se(IV) treatment on PA14 biofilm formation. Biofilm formation was stained with 0.1% (w/v) crystal violet and determined by measuring the absorbance at OD590. Fold change of each Se(IV) treatment was normalized to that observed in control (0 µM) samples. (C). Effect of Se(IV) on total protease activity of PA14. The enzyme activity was determined by measuring the absorbance at OD600 at 24-h incubation. Fold change of Se(IV) treatment was normalized to that observed in control (0 µM) samples. Results are presented as mean ± standard error of mean (SEM). Experiments were independently performed at least three times. Differences compared with the control (0 µM) were considered statistically significant at p<0.05 (*), p<0.001 (***) by one-way ANOVA and LSD post hoc test. n.s., no significance.
In addition, biofilm formation and secretory virulence factors such as protease in the PA14 culture grown in King's B broth supplemented with Se(IV) were examined. The effect of Se(IV) on biofilm formation was studied by crystal violet staining. The results showed that Se(IV) did not inhibit the biofilm formation of PA14 (Fig. 2B). Moreover, the protease activity of PA14 was determined by analyzing the ability of culture supernatants to lyse skimmed milk powder via measurement of absorbance at OD600 nm. Se(IV) treatment slightly decreased the total protease activity of pathogen-secreted enzyme by about 10% (Fig. 2C). Taken together, the results showed that Se(IV) does not protect wild-type C. elegans against P. aeruginosa PA14 pathogenicity by inhibiting virulence factors and reducing quorum-sensing signals of PA14.
Se(IV) enhances immune response gene expression in C. elegans under PA14 infection
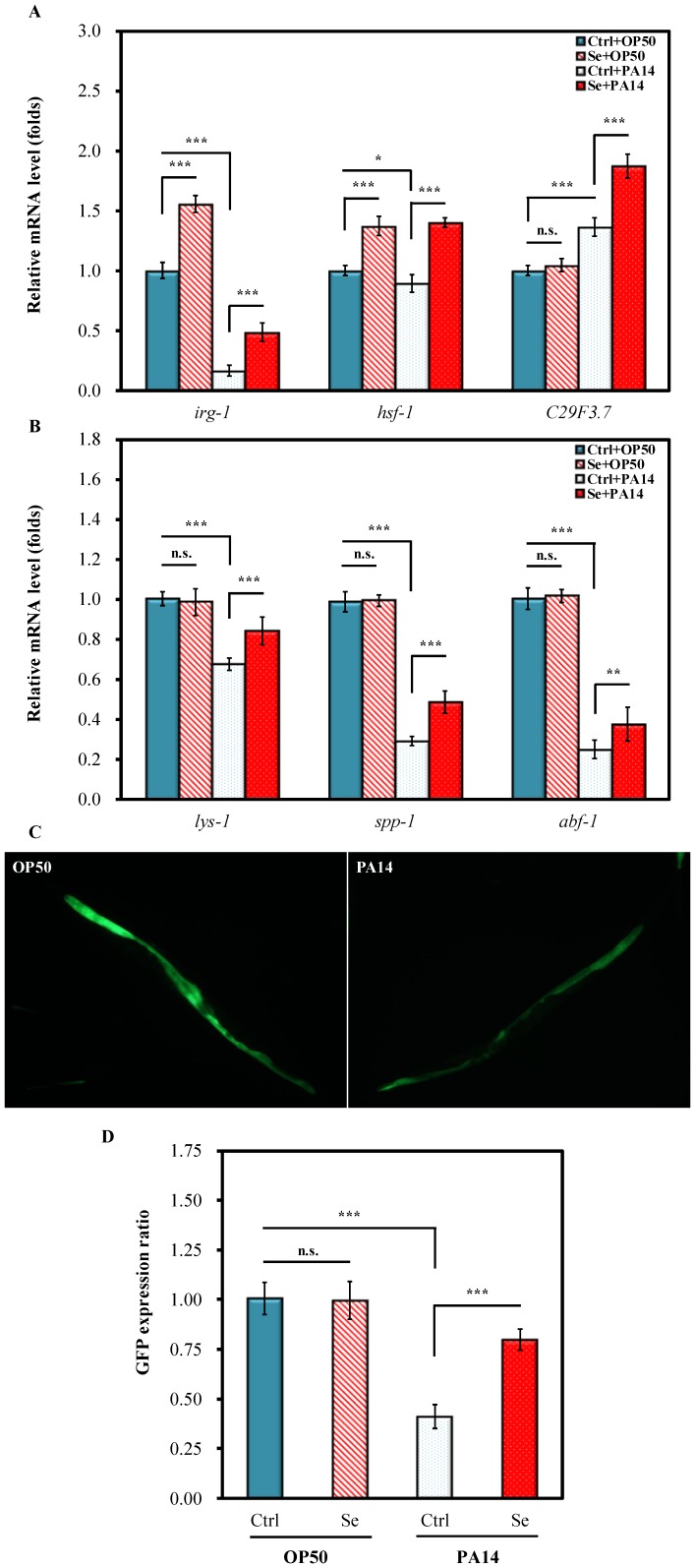
Whether immune response gene regulation may be responsible for the Se(IV)-induced protection of C. elegans against PA14 pathogenicity was examined. Three early response genes: irg-1 (infection response gene 1), hsf-1 (heat shock factor 1), and C29F3.7 (CUB-like domain) [42], [43] and three late response genes: lys-1 (lysozyme-like protein), spp-1 (saponin-like protein), and abf-1 (antibacterial protein) [44], [45] were selected. The mRNA levels in PA14-infected and uninfected worms with Se(IV) treatment were investigated. E. coli OP50, the standard laboratory food for C. elegans, served as the uninfected control to investigate the effects of Se(IV) under normal diet. Under normal diet, the mRNA levels of all tested immune-related genes, except irg-1 and hsf-1, were not significantly altered by Se(IV) compared with those of the control fed with OP50 (Ctrl + OP50) (Fig. 3A and 3B). In addition, after 24-h PA14 infection, the mRNA levels of all tested immune genes, except C29F3.7 (Ctrl + PA14), were significantly suppressed by 20% to 80% compared with that of the uninfected group (Ctrl + OP50) (Fig. 3A and 3B). Noticeably, Se(IV) treatment led to more significant activation of all six immune genes in PA14-infected worms (Se + PA14) than in the uninfected control (Ctrl + PA14) (Fig. 3A and 3B). Taken together, the results showed that Se(IV) enhanced immunity in C. elegans.
Figure 3. Effects of Se(IV) on immune response genes in C. elegans.
Synchronized L1 wild-type (N2) or transgenic strain (SAL105) larvae were pretreated without (0 µM) or with Se(IV) (0.01 µM) for 72 h at 20°C. Subsequently, Se(IV)-pretreated and control (0 µM) adult worms were divided into two aliquots and fed with OP50 or PA14 for 24 h at 20°C. The relative gene expressions were then determined by quantitative real-time PCR (qRT-PCR) or by GFP quantification. (A). Relative mRNA levels of the early immune response genes in wild-type C. elegans. (B). Relative mRNA levels of the late immune response genes in wild-type C. elegans. Ctrl + OP50: worm raised with OP50 for 72 h and then exposed to OP50 for another 24 h without Se(IV) exposure; Se + OP50: worm raised with OP50 for 72 h and then exposed to OP50 for another 24 h with 0.01 µM Se(IV) exposure; Ctrl + PA14: worm raised with OP50 for 72 h and then infected with PA14 for another 24 h without Se(IV) exposure; Se + PA14: worm raised with OP50 for 72 h and then infected with PA14 for another 24 h with 0.01 µM Se(IV) exposure. All measurements were normalized to mRNA levels of ACT-1 for C. elegans, and fold change of each gene was normalized to that observed in “Ctrl + OP50” samples. (C). (Left) Representative image of transgenic SLA105 (lys-7::GFP) cultured in normal conditions with OP50 as food source; (Right) Representative image of transgenic SLA105 (lys-7::GFP) infected with PA14 for 24 h without Se(IV) treatment. Worms infected with PA14 for 24 h and treated with 0.01 µM Se(IV) showed increased GFP fluorescence intensity similar to that presented in (left). (D) Quantifications of GFP expression of lys-7 gene in each treatment condition normalized to that of the control (Ctrl fed with OP50). Results are presented as mean ± standard error of mean (SEM). Experiments were independently performed at least three times and approximated 40 worms for each treatment were scored in each experiment. Differences compared with the control (0 µM) were considered statistically significant at p<0.05 (*), p<0.01 (**), and p<0.001 (***) by one-way ANOVA and LSD post hoc test. n.s., no significance.
To further provide evidences for the beneficial effects of Se(IV) on immune systems, transgenic strain SAL105 (lys-7::GFP) was employed to observe the effect of PA14 infection and Se(IV) on lys-7 expression. It has been reported that PA14 suppresses C elegans immunity by repressing the expression of the lysozyme-like LYS-7 [45]. The effect of Swietenia macrophylla on the expression of lys-7 was previously described [46]. The current results showed that PA14 infection diminished the overall GFP fluorescence intensity in C. elegans, indicating decreased expression of lys-7 compared with that in the uninfected C. elegans (Fig. 3C). Furthermore, Se(IV) treatment enhanced the GFP fluorescence intensity to a level comparable to that in uninfected C. elegans.
The GFP fluorescence intensity in each treatment group was further quantified. Quantitated data showed that lys-7 expression was not affected by Se(IV) under normal diet (E. coli OP50) but Se(IV) prevented the decrease of lys-7 gene expression during PA14 infection (Fig. 3D). This evidence was in agreement with above qRT-PCR data. Overall, the results suggested that Se(IV) enhanced immunity in C. elegans via activation of the immune genes under PA14 infection.
SKN-1 is essential for Se(IV)-induced protection of C. elegans against PA14 infection
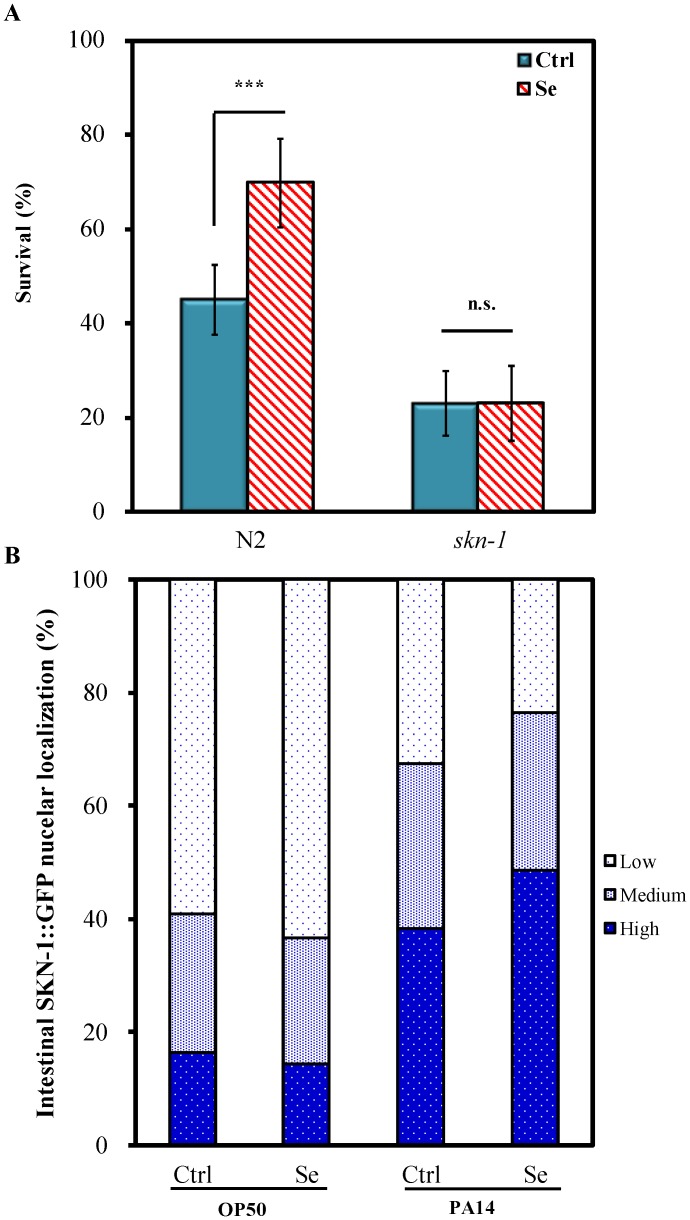
In C. elegans, SKN-1/ Nrf transcription factor plays an important role not only in oxidative and xenobiotic stress responses [47], [48] but also in innate immunity [38], [39]. To investigate whether Se(IV)-enhanced C. elegans against P. aeruginosa PA14 pathogenicity was modulated by SKN-1, skn-1 (zu67) mutant in response to Se(IV) was examined. Unlike the wild-type N2 C. elegans (Fig. 1), skn-1 mutant did not show significantly increased survival after 0.01 µM Se(IV) treatment for 3 days at 20°C followed by PA14 infection compared with no treatment (Fig. 4A), suggesting that Se(IV) may provide PA14 pathogen resistance in C. elegans via SKN-1.
Figure 4. Influences of Se(IV) on skn-1 mutant and subcellular SKN-1 localization under PA14 infection.
Synchronized wild-type N2, skn-1 mutant, or transgenic LD1 strain (SKN-1::GFP) L1 larvae were pretreated without (0 µM) or with Se(IV) (0.01 µM) for 72 h at 20°C. Subsequently, adult worms were prepared for PA14 infection. (A). Survival of the PA14-infected worm population was scored at 1-day intervals. Graph shows the survival of C. elegans without or with Se(IV) treatment on the 3rd day of infection as mean ± standard error of mean (SEM). Experiments were independently performed at least three times and approximated 60 worms for each treatment were scored in each experiment. Differences compared with the control (0 µM) were considered statistically significant at p<0.001 (***) by one-way ANOVA and LSD post hoc test. n.s., no significance. (B). Expression patterns of LD1 worms were classified into three categories (low, medium, and high) with respect to major localization of the SKN-1::GFP fusion protein in intestinal cells. “Low” refers to animals in which SKN-1::GFP was detected in less than 5 intestinal nuclei; “medium,” SKN-1::GFP detected in 5–15 intestinal nuclei; “high,” SKN-1::GFP present in more than 15 intestinal nuclei [39]. Experiments were independently performed at least three times and approximated 40 worms for each treatment were scored in each experiment.
SKN-1 is a transcription factor playing multiple essential roles; and localization of SKN-1 in nuclei is an essential prerequisite for activating transcription of target genes, such as gst-4 and gcs-1 [30], [47], [48], [49]. Therefore, to further explore the role of SKN-1 in regulating Se(IV)-enhanced PA14 pathogen resistance, the nuclear translocation of SKN-1 was examined using the transgenic strain LD1 (SKN-1B/C::GFP). Transgenic LD1 strain was cultured to adulthood without or with Se (IV) (0.01 µM) as described above. The adult worms were then fed with E. coli OP50 for 5 h as uninfected controls or infected with PA14, and the patterns of SKN-1 nuclear localization in intestinal cells were scored using the fluorescence microscope. The results showed no significant difference in SKN-1 nuclear localization between untreated worms and Se(IV)-treated worms under OP50 diet (Fig. 4B). Without Se(IV) treatment, a massive accumulation of SKN-1::GFP could be observed in intestinal nuclei of PA14-infected worms compared with those fed with nonpathogenic OP50 (Fig. 4B), indicating that PA14 triggered SKN-1 nuclear localization in intestinal cells of C. elegans. Moreover, the results also showed increase in SKN-1 nuclear localization in intestinal cells of the Se(IV)-treated group compared with the untreated group under PA14 infection (Fig. 4B). Taken together, the results showed that the PA14 pathogen resistance in Se(IV)-treated C. elegans could be attributed to Se(IV)-triggered SKN-1 nuclei translocation in intestinal cells.
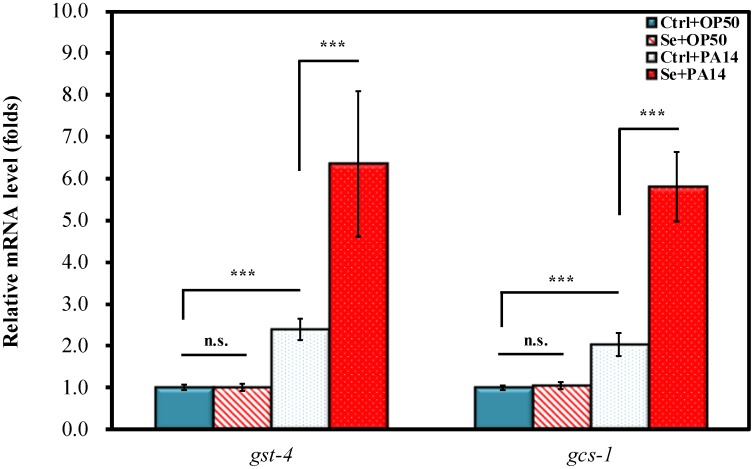
Se(IV) enhances expressions of glutathione-S-transferase (GST-4) and gamma-glutamine cysteine synthetase (GCS-1) in C. elegans under PA14 infection
To elucidate whether the increase in PA14 pathogen resistance described above was due to Se(IV) regulating SKN-1-dependent gene expressions, the mRNA levels of glutathione-S-transferase (GST-4) and gamma-glutamine cysteine synthetase (GCS-1) in response to Se(IV) treatment and PA14 infection were examined. Under normal E. coli OP50 diet, Se(IV) did not significantly affect the mRNA levels of both gst-4 and gcs-1 (Ctrl + OP50 vs. Se + OP50) (Fig. 5). Upon PA14 infection, the mRNA levels of gst-4 and gcs-1 were significantly elevated compared with those in uninfected C. elegans on OP50 diet (Ctrl + OP50 vs. Ctrl + PA14, p<0.001) (Fig. 5). Moreover, the results showed that Se(IV) treatment caused up-regulation of gst-4 and gcs-1 gene expression under PA14 infection (Ctrl + PA14 vs. Se + PA14, p<0.001) (Fig. 5). Taken together, the results indicate that Se(IV) triggered increased expression of SKN-1 downstream target genes such as gst-4 and gcs-1 under PA14 pathogen infection.
Figure 5. Effects of Se(IV) on expression of SKN-1 target genes upon PA14 infection in C. elegans.
Synchronized L1 wild-type larvae were pretreated without (0 µM) or with Se(IV) (0.01 µM) for 72 h at 20°C. Subsequently, Se(IV)-pretreated and control adult worms were divided into two aliquots and fed with OP50 or PA14 for 24 h at 20°C. Subsequently, the mRNA levels of GST-4 and GCS-1 were determined by qRT-PCR. All measurements were normalized to mRNA levels of ACT-1 for C. elegans, and fold change of each gene was normalized to that observed in “Ctrl + OP50” samples. Results are presented as mean ± standard error of mean (SEM). Experiments were independently performed at least three times. Differences compared with “Ctrl + OP50” were considered statistically significant at p<0.001 (***) by one-way ANOVA and LSD post hoc test. n.s., no significance.
Discussion
Previous studies reported that trace amount of Se(IV) exerts multiple beneficial effects on development, reproduction, cholinergic signaling, neuroprotection, and oxidative stress resistance in C. elegans [37], [40], [41]. In the present study, the protective potential of Se(IV) on C. elegans against P. aeruginosa infection was further investigated. The C. elegans/P. aeruginosa slow-killing infection model was employed to test the ability of Se(IV) to induce immune responses in C. elegans. In addition, antibacterial potential of Se(IV) against the pathogenic P. aeruginosa strain PA14 was examined. The results showed that trace amount of Se(IV) protects wild-type N2 C. elegans against P. aeruginosa PA14 infection (Fig. 1), suggesting that supplementation of Se(IV) may enhance immune responses to C. elegans. Dietary Se has been indicated as an essential micronutrient for optimal immune responses; however, the mechanisms accounting for such requirement are not fully understood [50], [51]. A number of studies have investigated the effects of Se-deficiency and Se-supplementation on immune responses, demonstrating an elevation of both cell-mediated and humoral immune responses by increasing Se intake levels [6], [51]. Antiviral immune responses have been generally shown to be affected by host Se status. However, the effect of Se on the immune responses to non-viral pathogens is more complicated [13], [51]. Currently available data suggested that host resistance of Se to pathogen infections varies with the microorganism involved [13], [51]. Thus, Fig. 1 provided further evidence showing that supplementation of Se(IV) enhances immune responses against P. aeruginosa PA14 infection, suggesting the importance of Se intake to immune system of organisms.
The increased survival of PA14-infected C. elegans under Se(IV) treatment (Fig. 1) may result from enhanced host immune defense systems or weakened pathogen activities and viability. Recent studies from water extract of TWE made from the brown seaweed (Ascophyllum nodosum) and water extract of red seaweed Chondrus crispus (CCWE) have been shown to enhance host C. elegans immunity by suppressing the mRNA levels of quorum sensing and the virulence factors of PA14 [44], [52]. The antioxidant curcumin was shown to enhance C. elegans survival by limiting P. aeruginosa pathogenicity through the repression of several quorum-sensing genes [53]. Moreover, anti-quorum-sensing activity provided by extractions from plants or marine sponge-associated bacteria (e.g., Syzygium aromaticum, Terminalia chebula Retz., and Haliclona spp.) was shown to improve protections of C. elegans against pathogen infections [54], [55], [56]. The effects of Se(IV) on both C. elegans and PA14 were examined in the present study. Se(IV) did not affect the growth of PA14 (data not shown). qRT-PCR analysis showed that 0.01 µM Se(IV) did not affect the mRNA levels of the tested quorum-sensing genes (lasI, lasR, rhlI, and rhlR) and virulence factor genes (hcnC, rpoN, and sbe) of PA14 (Fig. 2A). Moreover, PA14 grown with Se(IV) (0.01 µM) supplementation showed that Se(IV) did not inhibit the biofilm formation of PA14 (Fig. 2B), whereas total protease activity was slightly decreased by Se(IV) treatment (Fig. 2C). Therefore, Se(IV) did not protect wild-type C. elegans against P. aeruginosa PA14 pathogenicity by inhibiting virulence factors and reducing quorum-sensing signals of PA14.
Whether the increased survival of PA14-infected C. elegans by Se(IV) treatment (Fig. 1) was attributable to the enhancement of immune systems of C. elegans was examined. Repression of host defense genes is often associated with suppression of host defense pathways by the pathogen. P. aeruginosa infection is responsible for the impairment of host defense through the down-regulation of host's antimicrobial factors [45], [57]. The present findings showed that after PA14 infection, the mRNA levels of selected immune-related genes (namely, irg-1, hsf-1, lys-1, spp-1, and abf-1) (Ctrl + PA14) were significantly suppressed 20% to 80% compared with that of the uninfected group (Ctrl + OP50) (Fig. 3A and 3B) without Se(IV) treatment. The PA14-suppressed mRNA levels were attenuated when PA14-infected C. elegans were treated with 0.01 µM Se(IV) (Fig. 3A and 3B), suggesting that Se(IV) enhanced immunity in C. elegans by enhancing the immune responses of C. elegans. These similar effects were observed in water extracts from the brown seaweed (Ascophyllum nodosum) and red seaweed Chondrus crispus [44], [52]. PA14 infection was further shown to suppress the overall GFP fluorescence intensity in transgenic C. elegans carrying lys-7::GFP compared with that of uninfected C. elegans, whereas Se(IV) treatment enhanced the GFP fluorescence intensity to a level comparable to that of uninfected C. elegans (Fig. 3C and 3D). This similar effect was observed in Swietenia macrophylla, in which the plant extract restored the initially repressed lys-7 expression in PA14-infected C. elegans [46], [52]. Taken together, the results showed that Se(IV) might enhance immunity in C. elegans via activation of immune genes under PA14 infection.
In C. elegans, several conserved signal transduction pathways including the mitogen-activated protein kinase (MAPK) pathways, insulin/IGF-like signaling (IIS), and TGF-β pathways are involved in the immune responses [58],[59]. Both p38 MAPK and IIS pathways regulate SKN-1 that masters both oxidative and xenobiotic stress responses in C. elegans [47], [48], though the role of SKN-1 in the regulation of pathogen response is not well understood. SKN-1 has recently been shown as a prerequisite for C. elegans pathogen resistance, suggesting SKN-1 as a regulator of the innate immunity [39]. Therefore, to gain a mechanistic view of Se(IV) regulating immune responses in C. elegans, whether Se(IV) is linked to SKN-1 activity in immune responses was examined. Figure 4A showed that lack of SKN-1 resulted in sensitivity to PA14 infection, a finding consistent with previous studies [38], [39]. In contrast to the observation in wild-type C. elegans, the enhanced survival against PA14 infection by Se(IV) was not observed in skn-1 deletion mutant (Fig. 4A), suggesting the essential role of SKN-1 in Se(IV)-enhanced PA14 pathogen resistance in C. elegans.
To further validate that SKN-1 is necessary for the pathogen resistance induced by Se(IV), the effect of Se(IV) on accumulation of SKN-1 in intestinal nuclei was examined (Fig. 4B). Exposure to P. aeruginosa leads to SKN-1 accumulation in intestinal nuclei; and transcriptional activation of SKN-1 target genes, gcs-1 and gst-4, has been described [38], [39]. Without Se(IV) treatment, SKN-1 accumulation in intestinal nuclei could be observed in PA14-infected worms compared with those on nonpathogenic OP50 diet (Fig. 4B); and Se(IV) led to further increase in SKN-1 nuclear localization in intestinal cells (Fig. 4B). These observations suggested that PA14 pathogen resistance induced by Se(IV) was due to triggered SKN-1 nuclei translocation in intestinal cells of C. elegans. Further evidence showed that Se(IV) significantly up-regulated the SKN-1 target genes: gst-4 and gcs-1 gene expression under PA14 infection (Fig. 5), suggesting that Se(IV) triggered increased expression of SKN-1 downstream target genes such as gst-4 and gcs-1 under PA14 pathogen infection. Taken together, the results showed that Se(IV) protects C. elegans from P. aeruginosa infection via SKN-1.
There has been increasing evidence suggesting that selenoproteins play important roles in regulating inflammation and immunity, providing important insight into mechanisms by which Se influences inflammation and immunity [13]. In contrast to results observed in other animals, the only selenoprotein in C. elegans TRXR-1, an ortholog of the human thioredoxin reductase-1, was shown not to directly protect C. elegans from oxidative stress [60], [61]. This implies a unique role of C. elegans selenoprotein. A recent study indicated that TRXR-1 is involved in Se(IV) regulating oxidative stress resistance in C. elegans [37]. The role of TRXR-1 in Se(IV)-mediated immune responses in C. elegans requires further elucidation.
In conclusion, the present study using the model animal C. elegans obtained evidence supporting a beneficial effect of Se(IV) in host immune regulation. The findings revealed Se(IV) protecting C. elegans against P. aeruginosa PA14 infection by exerting effects on the innate immunity of C. elegans but having no direct effects on bacterial quorum-sensing and virulence factors. Se(IV) was also found to enhance the expression of a gene pivotal for the innate immunity in C. elegans. Finally, mechanistic study indicated that the protective effects of Se(IV) is likely mediated via regulation of a SKN-1/Nrf-dependent signaling pathway by inducing the expression of the target genes (gst-4 and gcs-1), thereby enhancing immune resistance on C. elegans against P. aeruginosa PA14 infection. These findings advance the understanding of the regulatory mechanism of Se in immune systems of intact organisms.
Supporting Information
Sequences of primers used for real time PCR.
(DOC)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was financially supported by grant (NSC 101-2313-B-002-041-MY3) from the Ministry of Science and Technology of Taiwan (http://www.most.gov.tw/) to VHCL. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Brown KM, Arthur JR (2001) Selenium, selenoproteins and human health: a review. Public Health Nutr 4: 593–599. [DOI] [PubMed] [Google Scholar]
- 2. Bainbridge DR (1976) Use of (75Se)L-Selenomethionine as a label for lymphoid cells. Immunology 30: 135–144. [PMC free article] [PubMed] [Google Scholar]
- 3. Behne D, Wolters W (1983) Distribution of selenium and glutathione peroxidase in the rat. J Nutr 113: 456–461. [DOI] [PubMed] [Google Scholar]
- 4. Gromer S, Eubel JK, Lee BL, Jacob J (2005) Human selenoproteins at a glance. Cell Mol Life Sci 62: 2414–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bellinger FP, Raman AV, Reeves MA, Berry MJ (2009) Regulation and function of selenoproteins in human disease. Biochem J 422: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spallholz JE, Boylan LM, Larsen HS (1990) Advances in understanding selenium's role in the immune system. Ann N Y Acad Sci 587: 123–139. [DOI] [PubMed] [Google Scholar]
- 7. Beck MA, Levander OA, Handy J (2003) Selenium deficiency and viral infection. J Nutr 133: 1463S–1467S. [DOI] [PubMed] [Google Scholar]
- 8. Vunta H, Belda BJ, Arner RJ, Channa Reddy C, Vanden Heuvel JP, et al. (2008) Selenium attenuates pro-inflammatory gene expression in macrophages. Mol Nutr Food Res 52: 1316–1323. [DOI] [PubMed] [Google Scholar]
- 9. Shrimali RK, Irons RD, Carlson BA, Sano Y, Gladyshev VN, et al. (2008) Selenoproteins mediate T cell immunity through an antioxidant mechanism. J Biol Chem 283: 20181–20185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campa A, Shor-Posner G, Indacochea F, Zhang G, Lai H, et al. (1999) Mortality risk in selenium-deficient HIV-positive children. J Acquir Immune Defic Syndr Hum Retrovirol 20: 508–513. [DOI] [PubMed] [Google Scholar]
- 11. Yu MW, Horng IS, Hsu KH, Chiang YC, Liaw YF, et al. (1999) Plasma selenium levels and risk of hepatocellular carcinoma among men with chronic hepatitis virus infection. Am J Epidemiol 150: 367–374. [DOI] [PubMed] [Google Scholar]
- 12. Yu SY, Zhu YJ, Li WG (1997) Protective role of selenium against hepatitis B virus and primary liver cancer in Qidong. Biol Trace Elem Res 56: 117–124. [DOI] [PubMed] [Google Scholar]
- 13. Huang Z, Rose AH, Hoffmann PR (2012) The role of selenium in inflammation and immunity: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal 16: 705–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Richards MJ, Edwards JR, Culver DH, Gaynes RP (1999) Nosocomial infections in medical intensive care units in the United States. National Nosocomial Infections Surveillance System. Crit Care Med 27: 887–892. [DOI] [PubMed] [Google Scholar]
- 15. Govan JR, Deretic V (1996) Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia . Microbiol Rev 60: 539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vandeputte OM, Kiendrebeogo M, Rajaonson S, Diallo B, Mol A, et al. (2010) Identification of catechin as one of the flavonoids from Combretum albiflorum bark extract that reduces the production of quorum-sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Appl Environ Microbiol 76: 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner VE, Filiatrault MJ, Picardo KF, Iglewski BH (2008) Pseudomonas aeruginosa Virulence and Pathogenesis Issues. In: Cornelis P, editor. Pseudomonas: Genomics and Molecular Biology. 1 ed. Norfolk, UK: Caister Academic Press. pp. 129–158.
- 18. Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM (1999) Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci U S A 96: 2408–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, et al. (2001) Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol 183: 6454–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith RS, Iglewski BH (2003) P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol 6: 56–60. [DOI] [PubMed] [Google Scholar]
- 21. Gander S (1996) Bacterial biofilms: resistance to antimicrobial agents. J Antimicrob Chemother 37: 1047–1050. [DOI] [PubMed] [Google Scholar]
- 22. Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jorgensen A, et al. (2003) Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48: 1511–1524. [DOI] [PubMed] [Google Scholar]
- 23. Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, et al. (2012) The multiple signaling systems regulating virulence in Pseudomonas aeruginosa . Microbiol Mol Biol Rev 76: 46–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Juhas M, Eberl L, Tummler B (2005) Quorum sensing: the power of cooperation in the world of Pseudomonas . Environ Microbiol 7: 459–471. [DOI] [PubMed] [Google Scholar]
- 25. Silverman GA, Luke CJ, Bhatia SR, Long OS, Vetica AC, et al. (2009) Modeling molecular and cellular aspects of human disease using the nematode Caenorhabditis elegans . Pediatr Res 65: 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tan MW, Shapira M (2011) Genetic and molecular analysis of nematode-microbe interactions. Cell Microbiol 13: 497–507. [DOI] [PubMed] [Google Scholar]
- 27. Aballay A, Ausubel FM (2002) Caenorhabditis elegans as a host for the study of host-pathogen interactions. Curr Opin Microbiol 5: 97–101. [DOI] [PubMed] [Google Scholar]
- 28. Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, et al. (2002) A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297: 623–626. [DOI] [PubMed] [Google Scholar]
- 29. Tan MW, Mahajan-Miklos S, Ausubel FM (1999) Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A 96: 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. An JH, Blackwell TK (2003) SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev 17: 1882–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA (2007) Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol 27: 5544–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44: 301–307. [PubMed] [Google Scholar]
- 33.Sulston J, Hodgkin J (1988) Methods. In: Wood WB, editor. The Nematode Caenorhabditis elegans. New York: Cold Spring Harbor Laboratory Press. pp. 587–606.
- 34. Dow JM, Clarke BR, Milligan DE, Tang JL, Daniels MJ (1990) Extracellular proteases from Xanthomonas campestris pv. campestris, the black rot pathogen. Appl Environ Microbiol 56: 2994–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Müsken M, Di Fiore S, Romling U, Haussler S (2010) A 96-well-plate-based optical method for the quantitative and qualitative evaluation of Pseudomonas aeruginosa biofilm formation and its application to susceptibility testing. Nat Protoc 5: 1460–1469. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole GA (2011) Microtiter dish biofilm formation assay. J Vis Exp: e2437. [DOI] [PMC free article] [PubMed]
- 37. Li WH, Shi YC, Chang CH, Huang CW, Hsiu-Chuan Liao V (2014) Selenite protects Caenorhabditis elegans from oxidative stress via DAF-16 and TRXR-1. Mol Nutr Food Res 58: 863–874. [DOI] [PubMed] [Google Scholar]
- 38. Hoeven R, McCallum KC, Cruz MR, Garsin DA (2011) Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans . PLoS Pathog 7: e1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Papp D, Csermely P, Soti C (2012) A role for SKN-1/Nrf in pathogen resistance and immunosenescence in Caenorhabditis elegans . PLoS Pathog 8: e1002673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li WH, Shi YC, Tseng IL, Liao VH (2013) Protective efficacy of selenite against lead-induced neurotoxicity in Caenorhabditis elegans . PLoS One 8: e62387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li WH, Hsu FL, Liu JT, Liao VH (2011) The ameliorative and toxic effects of selenite on Caenorhabditis elegans . Food Chem Toxicol 49: 812–819. [DOI] [PubMed] [Google Scholar]
- 42. Estes KA, Dunbar TL, Powell JR, Ausubel FM, Troemel ER (2009) bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans . Proc Natl Acad Sci U S A 107: 2153–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, et al. (2006) p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans . PLoS Genet 2: e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kandasamy S, Khan W, Evans F, Critchley AT, Prithiviraj B (2012) Tasco(R): a product of Ascophyllum nodosum enhances immune response of Caenorhabditis elegans against Pseudomonas aeruginosa infection. Mar Drugs 10: 84–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Evans EA, Kawli T, Tan MW (2008) Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans . PLoS Pathog 4: e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dharmalingam K, Tan BK, Mahmud MZ, Sedek SA, Majid MI, et al. (2012) Swietenia macrophylla extract promotes the ability of Caenorhabditis elegans to survive Pseudomonas aeruginosa infection. J Ethnopharmacol 139: 657–663. [DOI] [PubMed] [Google Scholar]
- 47. Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, et al. (2008) Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans . Cell 132: 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, et al. (2005) The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev 19: 2278–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE (2008) Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans . Biochem J 409: 205–213. [DOI] [PubMed] [Google Scholar]
- 50. Arthur JR, McKenzie RC, Beckett GJ (2003) Selenium in the immune system. J Nutr 133: 1457S–1459S. [DOI] [PubMed] [Google Scholar]
- 51. Hoffmann PR, Berry MJ (2008) The influence of selenium on immune responses. Mol Nutr Food Res 52: 1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu J, Hafting J, Critchley AT, Banskota AH, Prithiviraj B (2013) Components of the cultivated red seaweed Chondrus crispus enhance the immune response of Caenorhabditis elegans to Pseudomonas aeruginosa through the pmk-1, daf-2/daf-16, and skn-1 pathways. Appl Environ Microbiol 79: 7343–7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rudrappa T, Bais HP (2008) Curcumin, a known phenolic from Curcuma longa, attenuates the virulence of Pseudomonas aeruginosa PAO1 in whole plant and animal pathogenicity models. J Agric Food Chem 56: 1955–1962. [DOI] [PubMed] [Google Scholar]
- 54. Husain FM, Ahmad I, Asif M, Tahseen Q (2013) Influence of clove oil on certain quorum-sensing-regulated functions and biofilm of Pseudomonas aeruginosa and Aeromonas hydrophila . J Biosci 38: 835–844. [DOI] [PubMed] [Google Scholar]
- 55. Durai S, Vigneshwari L, Balamurugan K (2013) Caenorhabditis elegans-based in vivo screening of bioactives from marine sponge-associated bacteria against Vibrio alginolyticus . J Appl Microbiol 115: 1329–1342. [DOI] [PubMed] [Google Scholar]
- 56. Sarabhai S, Sharma P, Capalash N (2013) Ellagic acid derivatives from Terminalia chebula Retz. downregulate the expression of quorum sensing genes to attenuate Pseudomonas aeruginosa PAO1 virulence. PLoS One 8: e53441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, et al. (2002) Inducible antibacterial defense system in C. elegans . Curr Biol 12: 1209–1214. [DOI] [PubMed] [Google Scholar]
- 58.Ewbank JJ (2006) Signaling in the immune response. WormBook: 1–12. [DOI] [PMC free article] [PubMed]
- 59. Gravato-Nobre MJ, Hodgkin J (2005) Caenorhabditis elegans as a model for innate immunity to pathogens. Cell Microbiol 7: 741–751. [DOI] [PubMed] [Google Scholar]
- 60. Li W, Bandyopadhyay J, Hwaang HS, Park BJ, Cho JH, et al. (2012) Two thioredoxin reductases, trxr-1 and trxr-2, have differential physiological roles in Caenorhabditis elegans . Mol Cells 34: 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stenvall J, Fierro-Gonzalez JC, Swoboda P, Saamarthy K, Cheng Q, et al. (2011) Selenoprotein TRXR-1 and GSR-1 are essential for removal of old cuticle during molting in Caenorhabditis elegans . Proc Natl Acad Sci U S A 108: 1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of primers used for real time PCR.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.