Abstract
Bacterial symbionts of insects have received increasing attention due to their prominent role in nutrient acquisition and defense. In social bees, symbiotic bacteria can maintain colony homeostasis and fitness, and the loss or alteration of the bacterial community may be associated with the ongoing bee decline observed worldwide. However, analyses of microbiota associated with bees have been largely confined to the social honeybees (Apis mellifera) and bumblebees (Bombus spec.), revealing – among other taxa – host-specific lactic acid bacteria (LAB, genus Lactobacillus) that are not found in solitary bees. Here, we characterized the microbiota of three Australian stingless bee species (Apidae: Meliponini) of two phylogenetically distant genera (Tetragonula and Austroplebeia). Besides common plant bacteria, we find LAB in all three species, showing that LAB are shared by honeybees, bumblebees and stingless bees across geographical regions. However, while LAB of the honeybee-associated Firm4–5 clusters were present in Tetragonula, they were lacking in Austroplebeia. Instead, we found a novel clade of likely host-specific LAB in all three Australian stingless bee species which forms a sister clade to a large cluster of Halictidae-associated lactobacilli. Our findings indicate both a phylogenetic and geographical signal of host-specific LAB in stingless bees and highlight stingless bees as an interesting group to investigate the evolutionary history of the bee-LAB association.
Introduction
Mutualistic interactions are widespread in the animal and plant kingdoms and have left their footprints in the evolutionary history of many organisms. One of the most common groups of mutualists associated with multicellular organisms are bacteria, which are particularly prevalent across insects [1]. Mutualistic bacteria can provide a range of ecological benefits to their insect hosts, including nutritional upgrading of deficient diets, degradation of dietary polymers, and defense against antagonists (reviewed by [2]).
Bees represent an ecologically and economically important group of insects due to their functional role as pollinators in most ecosystems. Lately, they have declined in both abundance and species richness [3]–[6], with negative consequences for the quality and stability of pollination services to wild plants and agricultural crops [7]–[12]. Among bees, the (highly) social species play a particularly important role as pollinators due to the sheer numbers of foragers from single colonies, the diversity of species in some ecosystems (particularly in the tropics: [13]), the individual flower constancy (e.g., in bumblebees: [14],[15],[16]), the early onset of foraging (e.g., bumblebees: [17]) and year-long foraging in many species.
In this context, the microbial community associated with social bees has received considerable attention, and previous studies found a consistent core microbiota across honeybees and bumblebees [18]–[20]. While some of these symbiotic microbes have been hypothesized to aid in nutrient acquisition [21], others play an important role for the social immunity of bee colonies [21]–[23]. This function may be particularly relevant, as the diversity of immune-related genes is strongly reduced in honeybees (and likely other eusocial bee species) compared to other insects [24]. In the bumblebee Bombus terrestris, gamma- and betaproteobacterial symbionts convey resistance against an intestinal parasitic protozoan (Crithidia bombi) that negatively impacts the bees' fecundity [25]. Within honeybee nests, several Bacillus strains, actinomycetes, as well as some fungal associates have been isolated from pollen, honey and nest building material and are thought to protect bee colonies and/or enhance their growth [26]–[28]. Moreover, lactic acid bacteria (LAB), primarily belonging to the genera Lactobacillus and Bifidobacterium, have been described for several bee species, including honeybees (Apis mellifera) [29]–[32], bumblebees [32],[33], stingless bees [29] and several solitary bee species (e.g., Xylocopa, halictids, [19],[34]). LAB have been suggested to contribute to pollen fermentation within nests [30] and are known to also protect honeybees and bumblebees against pathogens [29],[35].
The microbiota of all social and solitary bees analyzed so far includes widespread LAB that are not host-specific. These LAB are closely related to flower-inhabiting, fructophilic lactobacilli or other lactobacilli found in the environment, which are likely obtained by bees when foraging at flowering plants [34]. In contrast, highly host-specific and diverged strains (i.e. strains of the Firm3, Firm4 and, to a lesser extent, Firm5 cluster) were reported for social bees, but not any other social insects [19],[32]. Based on this finding, McFrederick et al. [32] suggested that host-specificity of LAB is rare in Hymenoptera and may be maintained in social bees by spreading the symbionts among nestmates and transmitting them from one generation to the next via workers during colony fissioning.
While the microbial community of honeybees has received considerable attention, much less is known about the microbiota associated with stingless bees (Apidae: Meliponini) (but see [29]). Stingless bees represent the sister group of honeybees within the Apidae family, but in contrast to honeybees, they are restricted to tropical and subtropical regions, where they have achieved an impressive diversity with approximately 500 species described to date [36]. In a comparative study of the microbiota associated with several honeybee and stingless bee species, Vasquez et al. [29] found Firm4 and Firm5-associated LAB in South American, African and South East Asian stingless bee species, whereas LAB associated with Australian stingless bee species have not yet been investigated.
Here, we characterized the microbial community of three sympatric Australian stingless bee species (Austroplebeia australis, Tetragonula carbonaria, and Tetragonula hockingsii), with particular focus on the bee-associated LAB. According to the hypothesis that host-specific LAB are maintained by obligate colony fissioning (see McFrederick et al. (2013), we assume that LAB associated with the Firm3-5 clusters occur across all Meliponini. While the home ranges of the three species overlap broadly [37],[38], and they are similar in size and color (worker body size: [37],[38]), the two genera fall within different phylogenetic clades that diverged approximately 60 Mya ago [39]. The genus Austroplebeia (comprising five species) is endemic to Australia and Papua New Guinea and genetically more closely related to stingless bee lineages from the ancient African clade, whereas the closest relatives of the genus Tetragonula (comprising seven species in Australia) are found across Southeast Asia [39]. They also largely differ in their cuticular surface profiles [40] as well as their resource intake and nesting behavior, with only Tetragonula collecting [41] and incorporating substantial amounts of plant resins in their nest structures ([42]; Leonhardt SD, Drescher N, Wallace H, unpublished data). These differences in chemical ecology may result in a different nest and body environment for associated microbes.
Methods
Ethics statement
The stingless bee species investigated in this study are commonly found and not protected in Australia. As all bees were collected on private property, collecting permits were not required.
Sampling of bees
Bee specimens for genetic analyses were collected from colonies located at the Glenmount Research Station in Buderim (South East Queensland, Australia) in March 2011 and 2012. All colonies had access to the same resource environment and faced the same ecological conditions, with a mixed rainforest and eucalypt forest as well as gardens included in their foraging range (approximately 500 m radius of the hive).
Specimens were collected from five colonies of Tetragonula carbonaria, four colonies of Austroplebeia australis and one colony of Tetragonula hockingsii, a species closely related to T. carbonaria [43], by placing a clean clear plastic bag over the hive entrance, thereby catching foragers leaving the nest. To kill the bees, the plastic bag was placed in a freezer for approximately 10 minutes. Following a close inspection of their bodies to exclude contamination with plant or hive material (e.g., pollen or resin), bees were then stored in 70% ethanol for molecular analysis.
Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) and data analysis
DNA was extracted from six individual worker bees of all ten colonies sampled, respectively, using the MasterPure DNA Purification Kit (Epicentre Technologies) according to the manufacturer's instructions. For each colony, a pooled DNA sample was sent to an external service provider (Molecular Research LP, MR DNA, Shallowater, TX, USA) for bTEFAP with 16S rRNA primers Gray28F (5′-GAGTTTGATCNTGGCTCA-3′) and Gray519R (5′-GTNTTACNGCGGCKGCTG -3′) [44],[45]. A sequencing library was generated through one-step PCR with 30 cycles, using a mixture of HotStar and HotStar HiFidelity Taq polymerases (Qiagen). Sequencing extended from Gray28F, using a Roche 454 FLX instrument with Titanium reagents and procedures described at Molecular Research LP (http://www.mrdnalab.com/). Quality control and analysis of 454 reads was done in QIIME [46]. Low-quality ends of the sequences were trimmed with a sliding window size of 50 and an average quality cut-off of 25. Subsequently, all low quality reads (quality cut-off = 25) and sequences <200 bp were removed. High-quality reads were clustered into operational taxonomic units (OTUs) using a multiple OTU picking strategy with cdhit [47] and uclust [48], with 97% similarity cut-offs, respectively. For each OTU, the longest sequence was chosen as representative sequence (Data S1). Within the set of representative sequences, chimeras were identified using UCHIME (uchime_denovo) [49] and removed from further analysis. RDP classifier [50] and BLASTn against the NCBI database were used for taxonomy assignment. An OTU table was generated describing the occurrence of bacterial phylotypes within the samples (Table S1). OTUs were combined on the order level to display relative abundances.
Phylogenetic analysis
For phylogenetic analysis, all OTUs with Lactobacillus sp. as the first BLASTn hit were selected (20 OTUs). The representative sequences for these OTUs were trimmed to 350 bp in order to remove potential low-quality ends that were not detected by the preceding quality-trimming steps (see above). The trimmed reads were combined with the Lactobacillus sequences used in McFrederick et al. [32] as well as the Meliponini-associated lactobacilli reported by Vasquez et al. [29]. The resulting 656 sequences were aligned to the SILVA SSU database [51] using the SINA aligner [52] (Data S2). An approximately-maximum-likelihood tree was reconstructed with FastTree 2.1 using the GTR model [53]. Local support values were estimated with the Shimodaira-Hasegawa test based on 1,000 resamples without reoptimizing the branch lengths for the resampled alignments [53].
Results
Bacterial community composition
Using bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP), we characterized the microbial communities associated with worker bees from four colonies of A. australis, five colonies of T. carbonaria, and one colony of T. hockingsii. In total, 139,771 reads were obtained (mean ± standard error = 13,977±2,184 per sample), 126,919 of which passed quality filtering and chimera screening (mean ± standard error = 12,692±1,843 per sample). Rarefaction analyses indicate that the microbiota associated with the individual colonies was exhaustively sampled, with the possible exception of the T. carbonaria colony H89 (Fig. S1). Based on 97% similarity clustering with cdhit [47] and uclust [48], the sequences were grouped into 241 OTUs (Table S1 and Data S1).
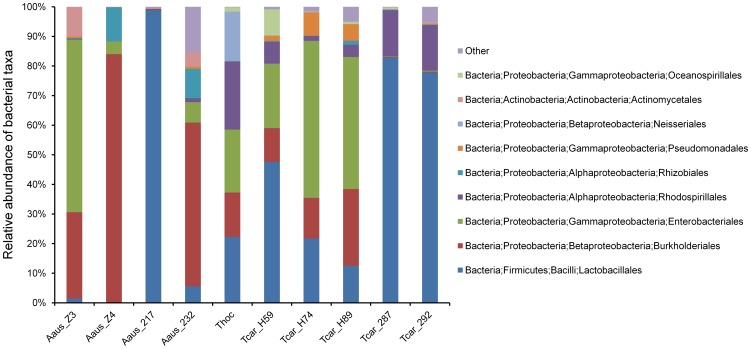
Lactobacillales (Firmicutes) as well as Beta-, Gamma-, and Alpha-Proteobacteria were the dominant taxa across colonies, but their relative abundance varied considerably within and between species (Fig. 1). One OTU associated with the genus Ralstonia (Beta-Proteobacteria, Burkholderiales; 99% similarity to Ralstonia pickettii) was consistently present across all colonies of the three bee species, two OTUs related to the genus Pantoea (Gamma-Proteobacteria, Enterobacteriales) were detected in six of the ten colonies across the three species, and one OTU associated with the family Acetobacteriaceae (Alpha-Proteobacteria, Rhodospirillales) occurred in all six Tetragonula colonies, but not in A. australis (Table S1). With the exception of the A. australis colony Z4, Lactobacillales were present in all colonies, with relative abundances ranging from 1.4 to 98.9% of the total microbiota.
Figure 1. Bacterial community associated with three species of Australian stingless bees, as revealed by 16S tag-encoded FLX amplicon pyrosequencing.
Different numbers denote different bee colonies. Aaus = Austroplebeia australis, Tcar = Tetragonula carbonaria, Thoc = Tetragonula hockingsii.
Phylogenetic affiliation of lactic acid bacteria (LAB)
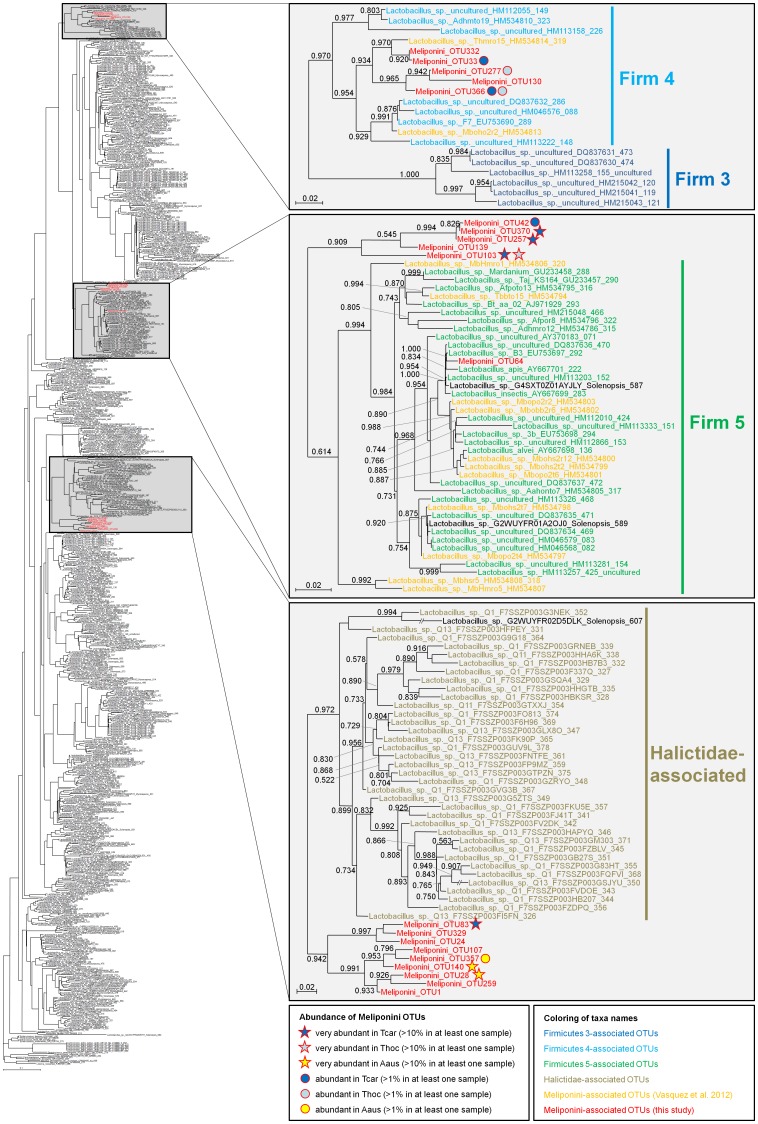
Among the 241 Meliponini-associated OTUs reported in this study, 20 were identified by RDP classification and BLASTn searches as members of the genus Lactobacillus. Phylogenetic analyses including 620 additional lactobacilli and outgroup sequences revealed the placement of the OTUs in four clusters (Fig. 2). (i) Five OTUs exclusively found in the two Tetragonula species were most closely related to a sequence obtained previously from a South East Asian stingless bee, Trigona sp. [29], and grouped within the bee-associated Firm4 cluster. (ii) A single sequence from the T. carbonaria colony 292 (OTU64) fell within the Firm5 cluster comprising honeybee- and stingless bee-associated LAB. (iii) Five OTUs only found in the two Tetragonula species formed the sister clade to the Firm5 cluster. And (iv) a monophyletic group of nine OTUs that were present in all three stingless bee species investigated in this study formed the sister clade to a large cluster of Halictidae-associated LAB.
Figure 2. Phylogenetic affiliation of lactic acid bacteria associated with Australian stingless bees. Numbers at the tree nodes represent local support values based on the approximately maximum likelihood analysis performed in FastTree 2.1.
Sequences obtained in the present study are highlighted in red font, stingless bee-associated sequences reported by Vasquez et al. (2012) are given in yellow font. Abundance of OTUs in the three investigated Australian stingless bee species is indicated by circles and asterisks, respectively, behind the OTU names. Aaus = Austroplebeia australis, Tcar = Tetragonula carbonaria, Thoc = Tetragonula hockingsii.
Discussion
While the microbial community of honeybees has been thoroughly investigated, the microbiota associated with stingless bees (Apidae: Meliponini) has only been addressed in a single comparative study with honeybees conducted by Vasquez et al. [29], which excluded Australian stingless bee taxa. Here we analyzed the microbiota associated with three Australian stingless bee species from two distinct phylogenetic lineages, in order to investigate the occurrence of host-specific LAB across stingless bees.
We found Lactobacillales (Firmicutes) as well as Beta-, Gamma-, and Alpha-Proteobacteria as the dominant bacterial taxa in all three stingless bee species. Samples from all ten colonies contained bacteria related to the genus Ralstonia (Burkholderiales), which are known as common pathogens [54],[55], but also as laboratory contaminants. Six of the ten colonies (including all species) additionally harbored bacteria associated with the genus Pantoea (Enterobacteriales, Gamma-Proteobacteria), which is a genus commonly found on plant roots, leaves [56] and flowers [57]. It has also been found in the hive environment and intestines of honeybees [58] [59]. Likewise, the family Acetobacteriaceae (Alpha-Proteobacteria, Rhodospirillales) that occurred in all six Tetragonula colonies (but not in A. australis) is often found in floral nectar and in the environment of bees (reviewed by [60]). Acetobacteriaceae belong to the acetic acid bacteria (AABs) and are known to break down carbohydrates in an acidic environment. They have recently been found to regulate the immune system homeostasis of Drosophila [61] and were suggested to be secondary symbionts across many insects [60]. They were also detected in the gut of a solitary bee species, Osmia bicornis [62]. In agreement with Koch et al. [20], we did not find the honeybee symbionts Gilliamella apicola and Snodgrassella alvi. The latter was implicated in the protection of bumblebees against Crithidia bombi [25].
Lactic acid bacteria (LAB) were present in nine out of ten colonies, but only the two Tetragonula species contained LAB that are closely related to other stingless bees and honeybee associated LAB, i.e. belong to the host-specific Firm4 and Firm5 clusters. Interestingly, neither our study nor the study of Vasquez et al. [29] found LAB of the Firm3 cluster in stingless bees. The Firm3 cluster is the most derived cluster of LAB associated with bees and may be honey- and bumblebee-specific, while Firm4–5 LAB are shared by honeybees, bumblebees and stingless bees across geographical regions. However, the absence of LAB of the Firm4–5 cluster in A. australis indicates that they are not present in all corbiculate bee species that propagate through colony fissioning as suggested by McFederick et al. [32].
In addition to the Firm4–5 clusters, we identified a novel cluster of LAB that is closely related to Halictidae-associated LAB in both A. australis and the two Tetragonula species. Despite its phylogenetic affiliation with the Lactobacillus buchneri group, this monophyletic group (comprising LAB associated with Halictidae and Meliponini) may represent a novel host-specific clade of bee-associated LAB.
Considering the occurrence of host-specific LAB (particularly Firm4–5) across corbiculate bees, their presence in stingless bees appears to be the ancestral state, with A. australis having secondarily lost the Firm4–5 cluster. This agrees with earlier studies detecting LAB of the Firm4–5 cluster in other stingless bees of the genera Trigona, Melipona, and Meliponula [29]. Given the close phylogenetic affiliation of Austroplebeia with the African genus Lisotrigona [39], it will be interesting to characterize the microbial community of additional species in these two genera, in order to find out how widespread the loss of Firm4–5 LAB is across stingless bees. Furthermore, investigating the distribution of LAB of the Halictidae-Meliponini cluster identified in this study across social and non-social bees may yield novel insights into the occurrence of host-specific LAB in bees. The widespread occurrence and potential host specificity of LAB in stingless bees suggests an important function in the protection against pathogens [29],[35] or in pollen fermentation within nests [30], as has been demonstrated for honeybees.
Our analysis of the microbiota associated with three Australian stingless bee species shows that the LAB community associated with stingless bees resembles that associated with honeybees, but lacks LAB of the highly host-specific Firm3 cluster and instead comprises an additional clade of likely host-specific LAB that form a sister clade to a large cluster of Halictidae-associated lactobacilli. This finding suggests that LAB are of similar ecological importance to stingless bees as they are to other corbiculate bees, but that their composition depends on the phylogenetic background and geographic region of their hosts. Therefore, stingless bees represent interesting organisms for understanding the evolutionary history of the bee-LAB association.
Supporting Information
Rarefaction analysis with the sequencing data for 10 colonies belonging to three different species of Australian stingless bees. Different numbers denote different bee colonies. Aaus = Austroplebeia australis, Tcar = Tetragonula carbonaria, Thoc = Tetragonula hockingsii.
(TIF)
Abundance of 241 operational taxonomic units (OTUs) across ten colonies of three different Australian stingless bee species (Aaus = Austroplebeia australis , Tcar = Tetragonula carbonaria , Thoc = Tetragonula hockingsii ). Taxonomic assignment was done with the RDP classifier based on the representative sequences for the OTUs. OTUs associated with the genus Lactobacillus (BLASTn results) are highlighted in bold print.
(XLSX)
Representative sequences for 241 OTUs identified across ten colonies of three different Australian stingless bee species. Each sequence is identified by the OTU number, the species and colony it was found in (Aaus = Austroplebeia australis, Tcar = Tetragonula carbonaria, Thoc = Tetragonula hockingsii), and a unique sequence identifier. If an OTU was detected in multiple colonies, only one colony/species is indicated.
(FASTA)
Alignment of 656 lactobacilli and outgroup sequences used for the phylogenetic analyses of Meliponini-associated lactic acid bacteria. The sequences obtained in this study were combined with the Lactobacillus sequences used in McFrederick et al. [32] as well as the Meliponini-associated lactobacilli reported by Vasquez et al. [29], and the resulting sequence set was aligned to the SILVA SSU database [51] using the SINA aligner [52].
(FASTA)
Acknowledgments
We thank Alexander Keller for his helpful comments on an earlier version of this manuscript.
Contributions
Conceived and designed the experiments: SDL MK. Performed the experiments: SDL MK. Analyzed the data: MK. Contributed to the writing of the manuscript: SDL MK.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The representative sequences for all OTUs identified in this study as well as the OTU table and an alignment of the sequences with other Lactobacillus sequences are included as supplementary online material.
Funding Statement
The authors gratefully acknowledge funding from the German Science Foundation (LE2750/1-1 to SDL, and KA2846/2-1 to MK; www.dfg.de) and the Max Planck Society (MK; www.mpg.de). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Buchner P (1965) Endosymbiosis of animals with plant microorganisms. New York: Interscience Publishers.
- 2. Feldhaar H (2011) Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol Entomol 36: 533–543. [Google Scholar]
- 3.Banaszak J (1995) Changes in fauna of wild bees in Europe. Bydgoszcz, Poland: Pedagogical University.
- 4. Biesmeijer JC, Roberts SPM, Reemer M, Ohlemuller R, Edwards M, et al. (2006) Parallel declines in pollinators and insect-pollinated plants in Britain and the Netherlands. Science 313: 351–354. [DOI] [PubMed] [Google Scholar]
- 5. Winfree R, Aguilar R, Vazquez DP, LeBuhn G, Aizen MA (2009) A meta-analysis of bees' responses to anthropogenic disturbance. Ecology 90: 2068–2076. [DOI] [PubMed] [Google Scholar]
- 6. Potts SG, Biesmeijer JC, Kremen C, Neumann P, Schweiger O, et al. (2010) Global pollinator declines: Trends, impacts and drivers. Trends Ecol Evol 25: 345–353. [DOI] [PubMed] [Google Scholar]
- 7. Kremen C, Williams NM, Aizen MA, Gemmill-Herren B, LeBuhn G, et al. (2007) Pollination and other ecosystem services produced by mobile organisms: a conceptual framework for the effects of land-use change. Ecol Lett 10: 299–314. [DOI] [PubMed] [Google Scholar]
- 8. Kremen C, Williams NM, Thorp RW (2002) Crop pollination from native bees at risk from agricultural intensification. Proc Natl Acad Sci USA 99: 16812–16816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garibaldi LA, Steffan-Dewenter I, Kremen C, Morales JM, Bommarco R, et al.. (2011) Stability of pollination services decreases with isolation from natural areas despite honey bee visits. Ecol Lett doi: 10.1111/j.1461-0248.2011.01669.x. [DOI] [PubMed]
- 10. Steffan-Dewenter I, Potts SG, Packer L (2005) Pollinator diversity and crop pollination services are at risk. Trends Ecol Evol 20: 651–652. [DOI] [PubMed] [Google Scholar]
- 11. Klein AM, Vaissiere BE, Cane JH, Steffan-Dewenter I, Cunningham SA, et al. (2007) Importance of pollinators in changing landscapes for world crops. Proc R Soc Lond B 274: 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein AM, Vaissiere BE, Cane JH, Cunningham S, Kremen C, et al.. (2008) The role of pollinators for global crop production. Ecological Society of America Annual Meeting Abstracts.
- 13. Heithaus ER (1979) Community structure of neotropical flower visiting bees and wasps: diversity and phenology. Ecology 60: 190–202. [Google Scholar]
- 14. Heinrich B (1976) The foraging specializations of individual bumblebees. Ecol Monogr 46: 105–128. [Google Scholar]
- 15. Heinrich B, Mudge PR, Deringis PG (1977) Laboratory analysis of flower constancy in foraging bumblebees: Bombus ternarius and B. terricola . Behav Ecol Sociobiol 2: 247–265. [Google Scholar]
- 16. Kleijn D, Raemakers I (2008) A retrospective analysis of pollen host plant use by stable and declining bumble bee species. Ecology 89: 1811–1823. [DOI] [PubMed] [Google Scholar]
- 17.Goulson D (2003) Bumblebees: their behaviour and ecology. New York, USA: Oxford University Press.
- 18. Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, et al. (2007) A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318: 283–287. [DOI] [PubMed] [Google Scholar]
- 19. Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, et al. (2010) A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol 20: 619–628. [DOI] [PubMed] [Google Scholar]
- 20. Koch H, Abrol DP, Li J, Schmid-Hempel P (2013) Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol Ecol 22: 2028–2044. [DOI] [PubMed] [Google Scholar]
- 21. Engel P, Martinson VG, Morana NA (2012) Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci USA 109: 11002–11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anderson KE, Sheehan TH, Eckholm BJ, Mott BM, DeGrandi-Hoffmann G (2011) An emerging paradigm of colony health: microbial balance of the honey bee and hive (Apis mellifera). Insect Soc 58: 431–444. [Google Scholar]
- 23. Cremer S, Sixt M (2009) Analogies in the evolution of individual and social immunity. Philos Trans R Soc B-Biol Sci 364: 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Evans JD, Aronstein K, Chen YP, Hetru C, Imler JL, et al. (2006) Immune pathways and defence mechanisms in honeybees Apis mellifera . Insect Mol Biol 15: 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koch H, Schmid-Hempel P (2011) Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci USA 108: 19288–19292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gilliam M (1997) Identification and roles of non-pathogenic microflora associated with honey bees. Microbiol Lett 155: 1–10. [Google Scholar]
- 27. Gilliam M, Taber S, Lorenz BJ, Prest DB (1988) Factors affecting development of chalkbrood disease in colonies of honeybees, Apis mellifera, fed pollen contaminated with Ascosphaera apis . J Invertebr Pathol 52: 314–325. [Google Scholar]
- 28. Promnuan Y, Kudo T, Chantawannakul P (2009) Actinomycetes isolated from beehives in Thailand. World J Microbiol Biotechnol 25: 1685–1689. [Google Scholar]
- 29. Vasquez A, Forsgren E, Fries I, Paxton RJ, Flaberg E, et al. (2012) Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PLoS One 7: e33188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vasquez A, Olofsson TC (2009) The lactic acid bacteria involved in the production of bee pollen and bee bread. J Apicult Res 48: 189–195. [Google Scholar]
- 31. Olofsson TC, Vasquez A (2008) Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera . Curr Microbiol 57: 356–363. [DOI] [PubMed] [Google Scholar]
- 32.McFrederick SQ, Cannone JJ, Gutell RR, Kellner K, Plowes RM, et al.. (2013) Host specificity between Hymenoptera and lactobacilli is the exception rather than the rule. Appl Environ Microbiol 03681–12. [DOI] [PMC free article] [PubMed]
- 33. Olofsson TC, Vasquez A (2009) Phylogenetic comparison of bacteria isolated from the honey stomachs of honeybees Apis mellifera and bumblebees Bombus spp. J Apicult Res 48: 233–237. [Google Scholar]
- 34. McFrederick SQ, Wcislo WT, Taylor DR, Ishak HD, Dowd SE, et al. (2012) Environment or kin: whence do bees obtain acidophilic bacteria? Mol Ecol Notes 21: 1754–1768. [DOI] [PubMed] [Google Scholar]
- 35. Forsgren E, Olofsson TC, Vasquez A, Fries I (2010) Novel lactic acid bacteria inhibiting Paenibacillus larvae in honeybee larvae. Apidologie 41: 99–108. [Google Scholar]
- 36.Michener CD (2007) The bees of the world. Baltimore & London: John Hopkins University Press. 953 p. [Google Scholar]
- 37.Dollin A, Walker K, Heard T (2009) “Trigona carbonaria” sugarbag bee (Tetragonula carbonaria). PaDIL - http://wwwpadilgovau: 2012.
- 38.Walker K (2010) “Austroplebeia australis” sugarbag bee (Austroplebeia australis). PaDIL – Available: http://wwwpadilgovau 2012.
- 39. Rasmussen C, Cameron SA (2010) Global stingless bee phylogeny supports ancient divergence, vicariance, and long distance dispersal. Biol J Linn Soc 99: 206–232. [Google Scholar]
- 40. Leonhardt SD, Wallace HM, Schmitt T (2011) The cuticular profiles of Australian stingless bees are shaped by resin of the eucalypt tree Corymbia torelliana . Austral Ecol 36: 537–543. [Google Scholar]
- 41.Leonhardt SD, Heard TA, Wallace HM (2013) Differences in the resource intake of two sympatric Australian stingless bee species. Apidologie, in press. DOI: 10.1007/s13592-013-0266-x.
- 42. Milborrow BV, Kennedy JM, Dollin A (1987) Composition of wax made by the Australian stingless bee Trigona australis . Austr J Biol Sci 40: 15–25. [Google Scholar]
- 43. Rasmussen C, Cameron SA (2007) A molecular phylogeny of the Old World stingless bees (Hymenoptera: Apidae: Meliponini) and the non-monophyly of the large genus Trigona . Syst Entomol 32: 26–39. [Google Scholar]
- 44.Sun Y, Wolcott RD, Dowd SEIE (2011) Tag-encoded FLX amplicon pyrosequencing for the elucidation of microbial and functional gene diversity in any environment. In: Kwon YM, Ricke SC, editors. High-Throughput Next Generation Sequencing: Methods and Application. New York, USA: Springer. pp.129–141. [DOI] [PubMed]
- 45. Ishak HD, Plowes R, Sen R, Kellner K, Meyer E, et al. (2011) Bacterial diversity in Solenopsis invicta and Solenopsis geminata ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb Ecol 61: 821–831. [DOI] [PubMed] [Google Scholar]
- 46. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- 48. Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- 49. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, et al. (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pruesse E, Peplies J, Gloeckner FO (2012) SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Price MN, Dehal PS, Arkin AP (2010) FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Genin S, Boucher C (2004) Lessons learned from the genome analysis of Ralstonia solanacearum . Annu Rev Phytopathol 42: 107–134. [DOI] [PubMed] [Google Scholar]
- 55. Ryan MP, Adley CC (2014) Ralstonia spp.: emerging global opportunistic pathogens. Eur J Clin Microbiol 33: 291–304. [DOI] [PubMed] [Google Scholar]
- 56. Mergaert J, Verdonck L, Kesters K, Swings J, Boeufgras J-M, et al. (1984) Numerical taxonomy of Erwinia species using API systems. J Gen Microbiol 130: 1893–1910. [Google Scholar]
- 57. Junker RR, Loewel C, Gross R, Dötterl S, Keller A, et al. (2011) Composition of epiphytic bacterial communities differs on petals and leaves. Plant Biol 13: 918–924. [DOI] [PubMed] [Google Scholar]
- 58. Loncaric I, Heigl H, Licek E, Moosbeckhofer R, Busse HJ, et al. (2009) Typing of Pantoea agglomerans isolated from colonies of honey bees (Apis mellifera) and culturability of selected strains from honey. Apidologie 40: 40–54. [Google Scholar]
- 59. Lyapunov YE, Kuzyaev RZ, Khismatullin RG, Bezgodova OA (2008) Intestinal enterobacteria of the hibernating Apis mellifera mellifera L. bees. Microbiology 77: 373–379. [PubMed] [Google Scholar]
- 60. Crotti E, Rizzi A, Chouaia B, Ricci I, Favia G, et al. (2010) Acetic acid bacteria, newly emerging symbionts of insects. Appl Environ Microbiol 76: 6963–6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ryu J-H, Kim S-H, Lee H-Y, Bai JY, Nam Y-D, et al. (2008) Innate immune homeostasis by the homeobox gene Caudal and commensal-gut mutualism in Drosophila . Science 319: 777–782. [DOI] [PubMed] [Google Scholar]
- 62. Keller A, Grimmer G, Steffan-Dewenter I (2013) Diverse microbiota identified in whole intact nest chambers of the red mason bee Osmia bicornis (Linnaeus 1758). PLoS One 8: e78296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rarefaction analysis with the sequencing data for 10 colonies belonging to three different species of Australian stingless bees. Different numbers denote different bee colonies. Aaus = Austroplebeia australis, Tcar = Tetragonula carbonaria, Thoc = Tetragonula hockingsii.
(TIF)
Abundance of 241 operational taxonomic units (OTUs) across ten colonies of three different Australian stingless bee species (Aaus = Austroplebeia australis , Tcar = Tetragonula carbonaria , Thoc = Tetragonula hockingsii ). Taxonomic assignment was done with the RDP classifier based on the representative sequences for the OTUs. OTUs associated with the genus Lactobacillus (BLASTn results) are highlighted in bold print.
(XLSX)
Representative sequences for 241 OTUs identified across ten colonies of three different Australian stingless bee species. Each sequence is identified by the OTU number, the species and colony it was found in (Aaus = Austroplebeia australis, Tcar = Tetragonula carbonaria, Thoc = Tetragonula hockingsii), and a unique sequence identifier. If an OTU was detected in multiple colonies, only one colony/species is indicated.
(FASTA)
Alignment of 656 lactobacilli and outgroup sequences used for the phylogenetic analyses of Meliponini-associated lactic acid bacteria. The sequences obtained in this study were combined with the Lactobacillus sequences used in McFrederick et al. [32] as well as the Meliponini-associated lactobacilli reported by Vasquez et al. [29], and the resulting sequence set was aligned to the SILVA SSU database [51] using the SINA aligner [52].
(FASTA)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The representative sequences for all OTUs identified in this study as well as the OTU table and an alignment of the sequences with other Lactobacillus sequences are included as supplementary online material.