Abstract
Bean yellow mosaic virus (BYMV), genus Potyvirus, has an extensive natural host range encompassing both dicots and monocots. Its phylogenetic groups were considered to consist of an ancestral generalist group and six specialist groups derived from this generalist group during plant domestication. Recombination was suggested to be playing a role in BYMV's evolution towards host specialization. However, in subsequent phylogenetic analysis of whole genomes, group names based on the original hosts of isolates within each of them were no longer supported. Also, nine groups were found and designated I-IX. Recombination analysis was conducted on the complete coding regions of 33 BYMV genomes and two genomes of the related Clover yellow vein virus (CYVV). This analysis found evidence for 12 firm recombination events within BYMV phylogenetic groups I–VI, but none within groups VII–IX or CYVV. The greatest numbers of recombination events within a sequence (two or three each) occurred in four groups, three which formerly constituted the single ancestral generalist group (I, II and IV), and group VI. The individual sequences in groups III and V had one event each. These findings with whole genomes are consistent with recombination being associated with expanding host ranges, and call into question the proposed role of recombination in the evolution of BYMV, where it was previously suggested to play a role in host specialization. Instead, they (i) indicate that recombination explains the very broad natural host ranges of the three BYMV groups which infect both monocots and dicots (I, II, IV), and (ii) suggest that the three groups with narrow natural host ranges (III, V, VI) which also showed recombination now have the potential to reduce host specificity and broaden their natural host ranges.
Introduction
Bean yellow mosaic virus (BYMV), genus Potyvirus, occurs worldwide, and has an extensive natural host range that encompasses domesticated and wild plants species, including both monocots and dicots. It causes serious diseases in a wide range of crops [1]–[4], e.g recent studies found that late infection with BYMV causes black pod syndrome (BPS) in Lupinus angustifolius (narrow-leafed lupin) and substantial yield losses [5], [6]. BYMV is transmitted non-persistently by many different aphid species [1], [7]. It consists of an RNA single stranded plus sense genome of about 10 kb. Its genome comprises two open reading frames (ORFs). There is one large polyprotein which is processed into ten proteins (biological characteristics linked to each in parentheses): P1 (symptomatology); HC-Pro (aphid transmission, systemic movement, suppression of gene silencing, self-interaction); P3 (plant pathogenicity); 6K1; CI (cell to cell movement); 6K2 (membrane attachment); VPg (genome replication); Nia-Pro (protein-protein interaction, cellular localization); Nib (RNA-dependent RNA polymerase, involved in replication); CP (aphid transmission, virus assembly, movement) [8], [9]. The second ORF, called PIPO, is embedded within P3, is around 180 nucleotides in length and translated in the +2 reading frame relative to the polyprotein [10]. PIPO has been linked to virulence determinacy in potyvirus resistant plants of Pisum sativum (pea) and long distance virus movement [11], [12].
Wylie et al. [13] analyzed the coat protein (CP) gene sequences of 64 BYMV isolates and based the names of the phylogenetic groups found on the types of original plant hosts that the isolates within each group came from. They proposed that these groups consisted of an ancestral generalist group with a wide natural host range and six specialist groups with narrow natural host ranges derived from the generalist group. They suggested that host specialization of BYMV had arisen within isolated crop domestication centers in different parts of the world [14], [15]. When Kehoe et al. [16] analyzed 40 whole BYMV genomes, they found nine phylogenetic groups which they named I–IX. The former ancestral group (called the general group) was split into three separate groups (I, II and IV). The genera the original isolation host species came from within each group were: Lupinus, Vicia (Fabaceae), Freesia (Iridaceae) and Diurus (Orchidaceae) in group I; Lupinus and Diurus in group II; Gladiolus (Iridaceae) in group III, Eustoma (Gentianacea) and Gladiolus in group IV, Trifolium (Fabaceae) and Vicia in group V; Lupinus in groups VI, VII and VIII; and Pisum (Fabaceae) in group IX. Thus, original host species represented in groups I, II and IV (the former general group) were from diverse origins, but those in the other groups were not. Therefore, phylogenetic group names based on natural hosts no longer seemed appropriate.
Recombination is one of the major means by which plant virus evolution and the emergence of new viruses or virus strains occurs [17]–[21]. There is evidence for high levels of recombination within the Potyviridae in particular [22]–[26]. Wylie and Jones [9] suggested that recombination played an important role in host specialization of BYMV following plant domestication. This suggestion was based on their analysis of seven complete genomes and 64 coat protein (CP) gene sequences. This predicted their general group to be ancestral in 12 out of 19 firm or tentative recombination patterns. However, recombination has been found to reduce host specificity and broaden natural host ranges, such as occurred with the emergence of Maize streak virus as an agricultural pathogen in Africa [21], [27], [28]. Therefore, given the subsequent availability of many more whole BYMV genomes and an increase in the numbers of phylogenetic groupings [16], the suggested role of recombination in the evolution of host specialization of BYMV warranted further analysis.
This research investigated the role that recombination plays in the evolution of BYMV. It examined the hypotheses that (i) recombination is associated with the expansion of natural host ranges in the three groups that contain isolates originally from both monocots and dicots (generalists), and that (ii) groups with narrow natural host ranges (specialists) might now be expanding their natural host ranges due to intermingling of strains formerly isolated from each other within crop domestication centers, resulting in recombination events creating groups with broader natural host ranges. To address these hypotheses, we undertook recombination analyses of 33 complete BYMV coding regions and two of the closely related Clover yellow vein virus (CYVV). These analyses included one CYVV and 13 BYMV genomes obtained as part of research on BPS [16]. As potyviruses frequently undergo recombination (see above), wherever possible, whole genome sequences should be used for recombination analysis. Therefore, our research did not include recombination analysis of BYMV and CYVV CP genes, despite many more CP sequences being available on Genbank. To determine if recombination was playing a role in their symptom expression, our research also examined the example of infection with BYMV causing BPS (late infection) or systemic necrosis (early infection) in L. angustifolius plants [6], [29].
Materials and Methods
Thirty-three complete or nearly complete BYMV genomes and two CYVV genomes were retrieved from Genbank (Table 1). They were trimmed to the length of their coding regions, and aligned by Clustal W in MEGA 5.2.1 prior to analysis for recombination [30]. The RDP4 package [31] was used to detect recombination between them. Default parameters were used for the seven programs implemented within RDP: RDP [32], GENECONV [33], Bootscan [34], MaxChi [35], Chimaera [36], 3Seq [37] and SiScan [38] which included using a Bonferroni corrected P value cutoff of 0.05. A recombination pattern was considered to be a firm event, and genuine evidence of actual recombination, if detected by four or more of these programs, and anything less than four programs was not considered [9], [24].
Table 1. Bean yellow mosaic virus and Clover yellow vein virus genomes analyzed for recombination.
| Accession number | Sequence ID | Phylogenetic grouping | Locationb | Orginal isolate host | Original host typec | Genome Reference |
| HG970860 | PN83Aa | I | WA, Australia | Lupinus angustifolius | IC, D | [16] |
| HG970861 | PN80Aa | I | WA, Australia | L. angustifolius | IC, D | [16] |
| HG970852 | GB17A | I | WA, Australia | L. angustifolius | IC, D | [16] |
| FJ492961 | Fr | I | South Korea | Freesia sp. | IC, D | unpublished |
| HG970847 | MD1 | I | WA, Australia | L. cosentinii | NW, D | [16] |
| JX173278 | KP2 | I | WA, Australia | Diuris magnifica | N, M | [42] |
| HG970851 | SP1 | I | WA, Australia | L. angustifolius | IC, D | [16] |
| HG970865 | AR93Ca | I | WA, Australia | L. angustifolius | IC, D | [16] |
| HG970869 | NG1 | I | WA, Australia | L. angustifolius | IC, D | [16] |
| JX156423 | SW3.2 | II | WA, Australia | Diuris sp. | N, M | [42] |
| HG970850 | MD7 | II | WA, Australia | L. cosentinii | NW, D | [16] |
| HG970863 | AR87Ca | II | WA, Australia | L. angustifolius | IC, D | [16] |
| HG970855 | LMBNN | II | WA, Australia | L. angustifolius | IC, D | [16] |
| HG970858 | ES55Ca | II | WA, Australia | L. angustifolius | IC, D | [16] |
| HG970854 | GB32Aa | II | WA, Australia | L. angustifolius | IC, D | [16] |
| HG970859 | ES11A | II | WA, Australia | L. angustifolius | IC, D | [16] |
| AB079886 | M11 | III | Japan | Gladiolus hybrida | IC, M | [43] |
| AB079887 | IbG | III | Japan | Gladiolus hybrida | IC, M | [43] |
| AB439729 | Gla | III | Hokkaido, Japan | Gladiolus hybrida | IC, M | [44] |
| AB079888 | GB2 | IV | Japan | - | - | unpublished |
| D83749 | MBGP | IV | Japan | - | - | [45] |
| NC003492 | MB4 | IV | Japan | - | - | [45] |
| AB439730 | G1 | IV | Japan | Gladiolus hybrida | IC, M | [44] |
| AM884180 | Lisianthus | IV | Taiwan | Eustoma russellianum | IC, D | unpublished |
| AY192568 | GDD | IV | USA | Gladiolus sp. | IC, M | [46] |
| AB439732 | 92-1 | V | Japan | Trifolium pratense | IC, D | [44] |
| U47033 | S | V | SA, Australia | Vicia faba | IC, D | [47] |
| HG970866 | LP | VI | WA, Australia | L. pilosus | IC, D | [16] |
| HG970868 | LPexFB | VI | WA, Australia | V. faba | IC, D | [16] |
| AB439731 | 90-2 | VII | Japan | V. faba | IC, D | [44] |
| HG970867 | FB | VII | WA, Australia | V. faba | IC, D | [16] |
| DQ641248 | WLMV | VIII | Idaho, USA | L. albus | IC, D | [48], [49] |
| AB373203 | CS | IX | Japan | Pisum sativum | IC, D | unpublished |
| NC003536 | CYVV | n/a | Japan | Phaseolus vulgaris | IC, D | [50] |
| HG970870 | CYVV AUS | n/a | NSW, Australia | T. repens | IC, D | [16] |
Indicates the sample originally came from a L. angustifolius plant with black pod syndrome.
NSW, New South Wales; SA, South Australia; WA, Western Australia.
Host types: D, dicot; IC, introduced cultivated plant; M, monocot; N, native plant; NW, naturalized weed.
Results
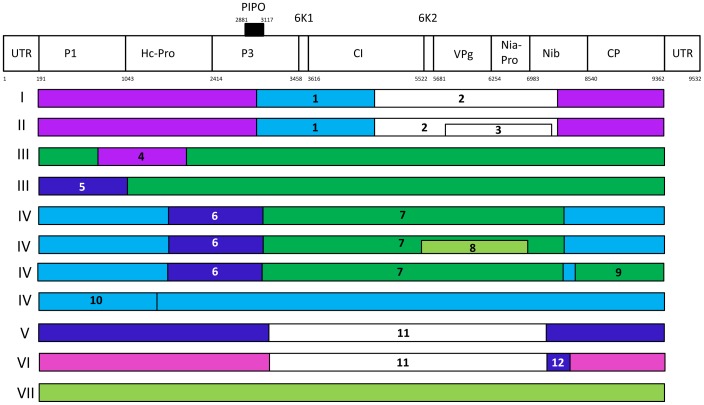
When the complete coding regions of 33 BYMV and two CYVV isolates were analyzed, 12 firm recombination events were identified (Table 2, Fig. 1). The 16 sequences within phylogenetic groups I and II all had two recombination events across their P3, 6K1, CI, 6K2, VPg, Nia-Pro and Nib genes (events 1 and 2). The parental sequences for event 1 were from groups IV and VII. With event 2, one was unknown and the other from group IV. The seven sequences within group II also contained another, event 3 which occurred across the VPg, Nia-Pro and Nib genes. It had one unknown parental sequence and one from group I. Two of the sequences from group III (MB11 and IbG) contained event 4, which was across the P1 and Hc-Pro genes. It had parental sequences from groups II and V. The third sequence from group III (Gla) contained event 5, located in the P1 gene. Its parental sequences were from groups V and IV. Four of the sequences from group IV (MBGP, G1, Lisianthus and GB2) contained recombination events 6 and 7. Event 6 was located across the Hc-Pro and P3 genes and event 7 across the region from P3 to Nib. The parental sequences for events 6 and 7 were groups V and II, and III and VII, respectively. The sequence from GB2 contained an extra event across the region from CI to Nia-Pro. Its parental sequences were from groups VII and IV. The sequence Lisianthus had another event across the Nib and CP genes, and its parental sequences were from groups III and IV. The remaining sequence from group IV (GDD) contained event 10, located across the P1 and Hc-Pro genes with parental sequences from groups IV and I. Event 11 was found in both group V and VI sequences, and stretched from the P3 to the Nib regions. Parental sequences for event 11 were an unknown and group III. Group VI sequences (LP and LPexFB) had an additional event (event 12) in the Nib region with parental sequences from group V and an unknown sequence. There was no evidence of recombination in sequences from groups VII to IX, or in the CYVV sequences. The greatest P-values across all 12 recombination events ranged from 6.701×10−7 to 1.968×10−160.
Table 2. Recombination events in the coding regions of 33 Bean yellow mosaic virus and 2 Clover yellow vein virus genomes.
| Event | Phylogenetic groupinga | Recombinant sequences | Programs detected byb | Start position in genomec | Genes affected | Parental sequencesd | Parental phylogenetic group | P-valuee |
| 1 | I, II | PN83A, PN80A, GB17A, Fr, MD1, KP2, SP1, AR93C, NG1, SW3.2, MD7, AR87C, LMBNN, ES55C, GB32A, ES11A | R, G, B, M, C, S, 3 | 2947–3089 | P3, PIPO | GI×90-2 | IV×VII | 1.30×10−78 (3f) |
| 2 | I, II | PN83A, PN80A, GB17A, Fr, MD1, KP2, SP1, AR93C, NG1, SW3.2, MD7, AR87C, LMBNN, ES55C, GB32A, ES11A | R, G, B, M, C, S, 3 | 5203–5457 | CI | unknown × MBGP | unknown × IV | 3.130×10−61 (G) |
| 3 | II | SW3.2, MD7, AR87C, LMBNN, ES55C, GB32A, ES11A | R, G, B, M, C, S, 3 | 5816–5829 | VPg | unknown × AR93C | unknown ×I | 2.447×10−44 (S) |
| 4 | III | MB11, IbG | R, G, B, M, C, S, 3 | 1073–1099 | Hc-Pro | SW3.2 ×S | II×V | 3.623×10−67 (G) |
| 5 | III | Gla | R, G, B, M, C, S, 3 | 1–191 (undetermined) | 5′UTR-P1 | S× MBGP | V×IV | 7.042×10−110 (G) |
| 6 | IV | MBGP, G1, Lisianthus, GB2 | R, G, B, M, C, S, 3 | 1721–1929 | Hc-Pro | S× ES11A | V×II | 1.495×10−30 (G) |
| 7 | IV | MBGP, G1, Lisianthus, GB2 | R, G, B, M, C, S | 2231–2318 | Hc-Pro | M11 ×90-2 | III×VII | 1.773×10−55 (S) |
| 8 | IV | GB2 | R, G, B, M, C, S, 3 | 5506–5556 | CI-6K2 | 90-2×G1 | VII×IV | 1.968×10−160 (G) |
| 9 | IV | Lisianthus | R, G, B, M, C, S, 3 | 8336 | Nib | IbG×G1 | III×IV | 1.661×10−79 (S) |
| 10 | IV | GDD | R, G, B, M, C, S, 3 | 1–191 (undetermined) | 5′UTR-P1 | MBGP × PN83A | IV×I | 7.249×10−40 (S) |
| 11 | V, VI | LP, LPexFB, S, 92-1 | R, G, B, M, C, S, 3 | 3236–3306 | P3 | unknown × M11 | unknown ×III | 3.051×10−47 (G) |
| 12 | VI | Lp, LPexFB | R, B, M, C | 7588–7872 | Nib | 92-1× unknown | V× unknown | 6.701×10−7 (M) |
Phylogenetic grouping determined by Kehoe et al. (2014b).
R, RDP; G, GENECONV; B, Bootscan; M, Maxchi; C, Chimaera; S, SiScan; 3, 3Seq.
Numbers represent nucleotide position in the genome.
Source of recombinant fragment. Minor parent is listed first, followed by the major parent.
The P-value is the greatest value for the event in question.
The program which detected the greatest P-value.
Figure 1. Recombination events between the coding regions of 33 Bean yellow mosaic virus (BYMV) and two from Clover yellow vein virus (ClYVV) genomes.
The locations of genes in the BYMV genome are indicated by the diagram at the top of the Figure. Twelve recombination events were found, labeled 1–12. Each recombination event correlates with the event column in Table 2. Each color represents a phylogenetic group, apart from purple, which represents two such groups, I and II. The phylogenetic groupings of Kehoe et al. 2014a are indicated at the left hand side of the picture. The colour of each event refers to the phylogenetic grouping of its predicted parental sequences, which are detailed within Table 2. The white colour represents events whose parental sequences are unknown. The sequences analyzed were: HG970847, HG970851, HG970852, HG970860, HG970861, HG970865, HG970869, FJ492961 and JX173278 (Phylogenetic group I); HG970850, HG97054, HG970855, HG970858, HG970859, HG970863 and JX156423 (II); AB079886, AB079887 and AB439729 (III); D83749, AM884180 and AY192568 (IV); AB439732 and U47033 (V); HG970866 and HG970868 (VI); AB439731 and HG970867 (VII); DQ641248 (VIII); AB373203 (IX); NC003536 and HG970870 (ClYVV). No recombination events were detected in sequences from phylogenetic groups VII–IX or within ClYVV, but a sequence from group VII is suggested as a parental sequence for one of those from group IV.
Six of the sequences analysed from groups I and II (PN83A, PN80A, AR93C, Ar87C, ES55C and GB32A) were BYMV isolates from L. angustifolius plants with BPS, but there was no recombination event specific to these sequences. This was also the case with three isolates (GB17A, NG1 and ES11A) from L. angustifolius plants with systemic necrosis within groups I and II.
Discussion
Our research found extensive recombination amongst diverse BYMV genome sequences which is likely to have significant evolutionary implications for the virus. It revealed the presence of extensive recombination within three BYMV phylogenetic groups that include both monocots and dicots as natural hosts, supporting the suggestion that recombination leads to broadening of natural host ranges. It therefore provides evidence for the hypothesis that recombination is responsible for the wide natural host ranges of the BYMV groups that invade both dicots and monocots. It also found recombination events in three BYMV phylogenetic groups with narrow natural host ranges indicating they might now have the potential to broaden their natural host ranges. It therefore provides support for the hypothesis that groups with narrow natural host ranges might now be expanding their natural host ranges due to intermingling of strains formerly isolated from each other within crop domestication centers, resulting in recombination events and broader natural host ranges. Such a scenario would occur as a result of recombination within mixed infections between previously isolated groups. Thus, past expansion of international trade in plants and plant products would have brought BYMV isolates that evolved in isolated crop domestication centers into contact with each other resulting in recombination. These results have broader implications concerning the likely role of recombination in the evolution of plant viruses in general, especially where a distinction exists between specialist and generalist virus groups. Our research also found no indication that recombination is playing a role in producing isolates causing BPS or systemic necrosis in L. angustifolius plants.
Our results resemble those of Wylie and Jones [9] in that the recombination patterns found were similar. However, the dataset from our whole genome analysis was much larger (35 compared to their eight) and revealed four additional firm recombination events. Overall, we detected 12 such events across 33 BYMV and two CYVV genomes, whereas they detected eight events across seven BYMV and one CYVV genome. Their study also identified three tentative recombination events involving BYMV genomes from group IV and an unknown parent within the 3′ region of the CYVV genome. In contrast, our analysis, which excluded tentative recombination events, did not reveal any firm events involving either of the two CYVV sequences as a parent. The use of more whole genome sequences gives us greater confidence in the results.
Our results showed eight recombination events within the former general group, now groups I, II and IV (two or three events per genome), and five amongst the former specialist groups where groups III and VI had two events each and group V had one event. Groups VII–IX had no recombination events. Our findings therefore showed that the groups with the most recombination had the broadest natural host ranges that included both monocots and dicots (I, II, IV). They also found recombination within groups III, V and VI (formerly specialist groups) thereby giving them the potential to broaden their natural host ranges and thus regeneralize. However, caution is required over our interpretation as groups V–IX were only represented by one or two genomes each, so there are likely to be as yet undetected recombination events. Likewise, the limited numbers of sequences in groups V–IX also make deductions difficult regarding (i) the parents of these sequences, or (ii) the roles of these sequences as parents in other recombination events generally. Also, one of the specialist phylogenetic groups based on CP genes reported by Wylie et al. [13] was their canna group. Isolates from this group were unrepresented by complete genome sequences, so they could not be evaluated.
All three recombination events present in BYMV groups I and II encompass the P3, PIPO, CI and VPg regions of the genome. These regions are responsible for pathogenicity, virus long distance movement, virulence determinance towards potyvirus resistance, replication and protein-protein interactions [8], [11], [12]. However, the recombination events we detected were in isolates originally collected from symptomatic plants in field, glasshouse and experimental situations, so pathogenicity was the only characteristic that could be related to recombination. Moreover, not all viral recombinants will necessarily give rise to viable, fit variants. The nature of potyviruses is such that functions of some genes overlap with others [8]. Recombinant fitness is determined by (i) the degree to which intragenome interactions are disrupted by the event, and (ii) the divergence between the exchanged sequences, where the higher the divergence, the greater the probability that intragenome disruption will occur [39]. Recombinant virus strains or isolates with disrupted intragenome interactions are likely to be removed by negative or purifying selection, e.g. as found within the Geminivirdae [39]. Thus, the recombination events detected in our analysis do not reflect overall BYMV recombination rate.
Most of the complete BYMV genomes available for analysis were from Australia or Japan, so there is little scope for deductions based on geography. With the exception of one from a Freesia spp. in South Korea, all isolates with genomes that fit into groups I and II were collected from south-western Australia. Moreover, there are also BYMV isolates from Australia in three other groups (V, VI and VII). These findings reinforce the suggestion that BYMV arrived in Australia on at least five different occasions and that international trade, for example of bulbs and seeds, is likely responsible for the worldwide distribution of BYMV [13], [40].
It appears unlikely that any of the recombination events detected in groups I and II (events 1, 2 and 3) were responsible for the emergence of BPS as a significant disease of L. angustifolius caused by BYMV. No recombination event was specific to the six BYMV isolates originally from plants with BPS. Moreover, they did not differ from the four BYMV isolates originally from L. angustifolius plants with systemic necrosis, and one other from a plant with a susceptible reaction (non-necrotic symptoms) [6], [29], [41]. Furthermore, recombination analysis did not distinguish sequences of these L. angustifolius isolates from those of any other hosts in groups I or II.
As more whole genomes sequences are submitted to databases, particularly from regions of the world in which BYMV specialist groups may have originated, or where crop domestication has occurred, the picture should become clearer and we will be better able to answer the question for BYMV – to specialize or not to specialize?
Acknowledgments
We thank E. Gajda, S. Vincent and M. Banovic for glasshouse and laboratory support.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All data used for the analysis and subsequent findings in this paper are freely available on public databases (both Genbank and ENA) and the accession numbers are provided in Table 1 in the manuscript. Some of the data are also from the companion paper (pone.0104580) which was accepted with minor revisions at the same time as this one.
Funding Statement
This research was funded by an Australian Postgraduate Award (APA), and an Australian Grains Research and Development Corporation (GRDC) Studentship, Project number GRS10039. It was undertaken using the facilities at the Department of Agriculture and Food Western Australia. This study forms part of a PhD project by the first author at the University of Western Australia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bos L (1970a) Bean yellow mosaic virus. Association of Applied Biologists. Descriptions of Plant Viruses No.40.
- 2. Bos L (1970b) The identification of three new viruses isolated from Wisteria and Pisum in the Netherlands, and the problem of variation within the potato virus Y group. Neth J Pl Path 76: 8–46. [Google Scholar]
- 3. Jones RAC, McLean GD (1989) Virus diseases of lupins. Ann Appl Biol 114: 609–637. [Google Scholar]
- 4.Edwardson JR, Christie RG (1991) CRC handbook of viruses infecting legumes CRC Press, Boca Raton, FL. [Google Scholar]
- 5.Buichell BJ (2008) Narrow-leafed lupin breeding in Australia – where to from here? In Palta JA, Berger JB (editors). 2008. rLupins for health and wealthwealthn in Austr2007) 12th International Lupin Conference, 14–18 September 2008, Fremantle, Western Australia. International Lupin Association, Canterbury, New Zealand.
- 6. Kehoe MA, Buirchell BJ, Coutts BA, Jones RAC (2014) Black pod syndrome of Lupinus angustifolius is caused by late infection with Bean yellow mosaic virus . Plant Dis 98: 739–745. [DOI] [PubMed] [Google Scholar]
- 7. Berlandier FA, Thackray DJ, Jones RAC, Latham LJ, Cartwright L (1997) Determining the relative roles of different aphid species as vectors of cucumber mosaic and bean yellow mosaic viruses in lupins. Ann Appl Biol 131: 297–314. [Google Scholar]
- 8. Urcuqui-Inchima S, Haenni A, Bernardi F (2001) Potyvirus proteins: a wealth of functions. Virus Research 74: 157–175. [DOI] [PubMed] [Google Scholar]
- 9. Wylie SJ, Jones RAC (2009) Role of recombination in the evolution of host specialization within Bean yellow mosaic virus . Phytopathology 99: 512–518. [DOI] [PubMed] [Google Scholar]
- 10. Chung BY-W, Miller WA, Atkins JF, Firth AE (2008) An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci 105: 5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wen R-H, Hajimorad MR (2010) Mutational analysis of the putative pipo of soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virol 400: 1–7. [DOI] [PubMed] [Google Scholar]
- 12. Choi SH, Hagiwara-Komoda Y, Nakahara KS, Atsumi G, Shimada R, et al. (2013) Quantitative and Qualitative involvement of P3N-PIPO in overcoming recessive resistance against Clover yellow vein virus in pea carrying the cyv1 gene. J Virol 87: 7326–7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wylie SJ, Coutts BA, Jones MGK, Jones RAC (2008) Phylogenetic analysis of Bean yellow mosaic virus isolates from four continents: relationship between the seven groups found and their hosts and origins. Plant Dis 92: 1596–1603. [DOI] [PubMed] [Google Scholar]
- 14. Jones RAC (1993) Effects of cereal borders, admixture with cereals and plant density on the spread of bean yellow mosaic potyvirus into narrow-leafed lupins (Lupinus angustifolius). Ann. Appl. Biol 122: 501–518. [Google Scholar]
- 15. Jones RAC (2009) Plant virus emergence and evolution: origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control. Virus Res 141: 113–130. [DOI] [PubMed] [Google Scholar]
- 16.Kehoe MA, Buirchell BJ, Coutts BA, Jones RAC (2014a) Plant virology and next generation sequencing: experiences with a Potyvirus. PLoS ONE In Press. 10.1371/journal.pone.0104580. [DOI] [PMC free article] [PubMed]
- 17. Sztuba-Solinska J, Urbanowicz A, Figlerowicz M, Bujarski JJ (2011) RNA-RNA recombination in plant virus replication and evolution. Annu Rev Phytopathol 49: 415–443. [DOI] [PubMed] [Google Scholar]
- 18. Valli A, López-Moya J, Garcia JA (2007) Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae . J Gen Virol 88: 1016–1028. [DOI] [PubMed] [Google Scholar]
- 19. Roossinck MJ (2003) Plant RNA virus evolution. Curr Opin Micro 6: 406–409. [DOI] [PubMed] [Google Scholar]
- 20.Gibbs A, Gibbs M, Ohshima K, García-Arenal F (2008) More about plant virus evolution: past, present, and future. In: Domingo E, Parrish CR, Holland JJ, editors. Origin and Evolution of Viruses (Second edition), Elsevier Ltd. pp 229–250.
- 21. Varsani A, Shepherd DN, Monjane AL, Owor BE, Erdmann JB, et al. (2008) Recombination, decreased host specificity and increased mobility may have driven the emergence of maize streak virus as an agricultural pathogen. J. Gen. Virol 89: 2063–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chare ER, Holmes EC (2006) A phylogenetic survey of recombination frequency in plant RNA viruses. Arch Virol 151: 933–946. [DOI] [PubMed] [Google Scholar]
- 23. Revers F, Le Gall O, Candresse T, Le Romancer M, Dunez J (1996) Frequent occurrence of recombinant potyvirus isolates. J Gen Virol 77: 1953–1965. [DOI] [PubMed] [Google Scholar]
- 24. Ohshima K, Yamaguchi Y, Hirota R, Hamamoto T, Tomimura K, et al. (2002) Molecular evolution of Turnip mosaic virus: evidence of host adaptation, genetic recombination and geographical spread. J Gen Virol 83: 1511–1521. [DOI] [PubMed] [Google Scholar]
- 25. Karasev AV, Gray SM (2013) Genetic Diversity of Potato virus Y complex. Am. J. Potato Res 90: 7–13. [Google Scholar]
- 26. Tromas N, Zwart MP, Poulain M, Elena SF (2013) Estimation of the in vivo recombination rate for a plant RNA virus. J Gen Virol 95: 724–732. [DOI] [PubMed] [Google Scholar]
- 27. Van der Walt E, Rybicki EP, Varsani A, Plston JE, Billharz R, et al. (2009) Rapid host adaptation by extensive recombination. J. Gen. Virol 90: 734–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Monjane AL, van der Walt E, Varsani A, Rybicki EP, Martin DP (2011) Recombination hotspots and host susceptibility modulate the adaptive value of recombination during Maize streak virus evolution. BMV Evol. Biol 11: 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cheng Y, Jones RAC (2000) Biological properties of necrotic and non-necrotic strains of bean yellow mosaic virus in cool season grain legumes. Ann Appl Biol 136: 215–227. [Google Scholar]
- 30. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martin DP, Lemey P, Lott M, Moulton V, Posada D, et al. (2010) RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26: 2462–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martin D, Rybicki E (2000) RDP: detection of recombination amongst aligned sequences. Bioinformatics 16: 562–563. [DOI] [PubMed] [Google Scholar]
- 33. Padidam M, Sawyer S, Fauquet CM (1999) Possible emergence of new geminiviruses by frequent recombination. Virology 265: 218–225. [DOI] [PubMed] [Google Scholar]
- 34.Martin DP, Posada D, Crandall KA, Williamson C (2005) A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retrov. 21: , 98–102 [DOI] [PubMed] [Google Scholar]
- 35. Maynard Smith J (1992) Analyzing the mosaic structure of genes. J Mol Evol 34: 126–129. [DOI] [PubMed] [Google Scholar]
- 36. Posada D, Crandall KA (2001) Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. P Natl Acad Sci USA 98: 13757–13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boni MF, Posada D, Feldman MW (2007) An exact nanoparametric method for inferring mosaic structure in sequence triplets. Genetics 176: 1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gibbs MJ, Armstrong JS, Gibbs AJ (2000) Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16: 573–582. [DOI] [PubMed] [Google Scholar]
- 39. Martin DP, van der Walt E, Posada D, Rybicki EP (2005) The evolutionary value of recombination is constrained by genome modularity. PLoS Gen 1(4): e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gibbs AJ, Mackenzie AM, Wei K-J, Gibbs MJ (2008) The potyviruses of Australia. Arch Virol 153: 1411–1420. [DOI] [PubMed] [Google Scholar]
- 41. Cheng Y, Jones RAC (1999) Distribution and incidence of necrotic and non-necrotic strains of bean yellow mosaic virus in wild and crop lupins. Aust J Agric Res 50: 589–599. [Google Scholar]
- 42. Wylie SJ, Li H, Dixon KW, Richards H, Jones MGK (2013) Exotic and indigenous viruses infect wild populations and captive collections of temperate terrestrial orchids (Diuris species) in Australia. Virus Res 171: 22–32. [DOI] [PubMed] [Google Scholar]
- 43. Nakazono-Nagaoka E, Sato C, Kosaka Y, Natsuaki T (2004) Evaluation of cross-protection with an attenuated isolate of Bean yellow mosaic virus by differential detection of virus isolates using RT-PCR. J Gen Plant Pathol 70: 359–362. [Google Scholar]
- 44. Nakazono-Nagaoka E, Takahashi T, Shimizu T, Kosaka Y, Natsuaki T, et al. (2009) Cross-protection against Bean yellow mosaic virus (BYMV) and Clover yellow vein virus by attenuated BYMV isolate M11. Virology 99: 251–257. [DOI] [PubMed] [Google Scholar]
- 45. Nakamura S, Honkura R, Ugaki M, Ohshima M, Ohashi Y (1994) Nucleotide sequence of the 3′-terminal region of bean yellow mosaic virus RNA and resistance to viral infection in transgenic Nicotiana benthamiana expressing its coat protein gene. Ann Phytopathol Soc Jpn 60: 295–304. [Google Scholar]
- 46. Hammond J, Hammond RW (2003) The complete nucleotide sequence of isolate BYMV-GDD of Bean yellow mosaic virus and comparison to other potyviruses. Arch Virol 148: 2461–2470. [DOI] [PubMed] [Google Scholar]
- 47. Guyatt KJ, Proll DF, Menssen A, Davidson AD (1996) The complete nucleotide sequence of bean yellow mosaic potyvirus RNA. Arch Virol 141: 1231–1246. [DOI] [PubMed] [Google Scholar]
- 48. Hampton RO, Shukla DD, Jordan RL (1992) Comparative host range, serology, and coat protein peptide profiles of white lupin moaic virus. Phytopathology 82: 566–571. [Google Scholar]
- 49. Bruun-Rasmussen M, Møller IS, Tulinius G, Hansen JKR, Lund OS, et al. (2007) The same allele of translation initiation factor 4E mediates resistance against two Potyvirus spp. in Pisum sativum . Mol Plant Microbe In 20: 1075–1082. [DOI] [PubMed] [Google Scholar]
- 50. Takahashi Y, Takahashi T, Uyeda I (1997) A cDNA clone to Clover yellow vein potyvirus is highly infectious. Vir Genes 12: 235–243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All data used for the analysis and subsequent findings in this paper are freely available on public databases (both Genbank and ENA) and the accession numbers are provided in Table 1 in the manuscript. Some of the data are also from the companion paper (pone.0104580) which was accepted with minor revisions at the same time as this one.