Abstract
RNA is believed to have been the first reservoir of genetic information, but despite its ancient history, RNA continues to fascinate and is only now beginning to be understood in its entire variety and communication modality. New discoveries include the pseudogene RNA network regulating PTEN transcription and translation and the identification of circular RNAs as a new class of competing endogenous RNA molecules that sequester microRNAs to suppress their function.
For decades, RNA was considered to be merely a ‘carrier’ of genetic information, but in the past 15 years unexpected discoveries in the epigenetic landscape, from microRNAs (miRNAs) to pseudogenes and the recently described long noncoding RNAs (lncRNAs), have completely revolutionized this simplistic view. miRNAs are small RNA molecules of 20–22 nucleotides that post-transcriptionally negatively modulate the translation and stability of target RNA molecules containing miRNA-responsive elements (MREs) in their sequence1. LncRNAs and pseudogenes are longer and surprisingly more numerous than protein-encoding genes and are extremely versatile in their mechanism of action2,3.
Although one miRNA can potentially regulate hundreds of different mRNAs (each one in turn containing several MREs) the majority of these transcripts are actively expressed and translated, which supports the existence of several mechanisms that counteract miRNA regulation to achieve homeostasis. Probably the most ancestral and intuitive example among these regulatory mechanisms is the cross-talk and functional competition among different RNA molecules, as epitomized by the interplay between miRNAs and competing endogenous RNAs (ceRNAs)4 (described below).
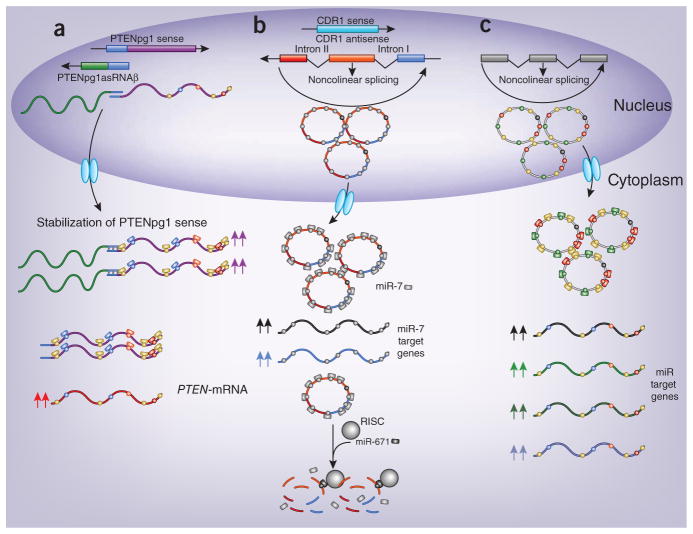
Three recent papers have now shed light on an intertwined system of interaction between noncoding RNA molecules and mRNAs from protein-coding genes. Johnsson et al.5 have identified a pseudogene network able to regulate phosphatase and tensin homolog (PTEN) at both the transcriptional and the post-transcriptional level. The PTEN pseudogene (PTENpg1) locus encodes three different lncRNA molecules: two functional antisense RNAs (asRNAs) and one sense PTENpg1. PTENpg1 asRNAα acts in trans, localizes to the PTEN promoter and inhibits PTEN transcription by recruiting epigenetic repressor complexes, whereas PTENpg1 asRNAβ, which is partially complementary to PTENpg1 sense, promotes stabilization of PTENpg1 sense by binding its 5′ end. PTENpg1 sense was previously shown to act as a ceRNA for PTEN6 through its ability to compete for several miRNAs that also target PTEN. Thereby, the stabilization of PTENpg1 sense by PTENpg1 asRNAβ can regulate its ceRNA activity6 (Fig. 1a), perhaps in a tissue-specific manner, which further highlights the regulatory activity on antisense and pseudogene RNAs species.
Figure 1.
New RNA species counteract miRNA function. (a) PTENpg1 asRNAβ is partially complementary to the first exon of PTENpg1 sense and promotes its stabilization (purple arrows). Thereby, PTENpg1 sense increases PTEN-mRNA abundance (red arrows) by sequestering miRNAs that also target PTEN. (b) circRNA CDR1 as (ciRS-7) arises from head-to-tail splicing of its precursor. CircRNA CDR1 as (ciRS-7) contains ~70 MREs for miR-7 and increases the expression of miR-7 target genes (black and blue arrows) by sequestering miR-7. CircRNA CDR1 as (ciRS-7) is under miR-671 regulation. miR-671 is almost perfectly complementary to CDR1 as (ciRS-7), and, although circularization protects from canonical nucleases, this is not sufficient to counteract AGO2 slicer activity in RISC. (c) circRNAs containing several distinct MREs can sequester different miRNA families and increase the expression of all genes (colored arrows) under miRNA regulation. MREs and miRNAs are indicated as color-coded circles and rectangles, respectively, on the RNA molecules.
Knowledge of the endogenous competition between noncoding RNAs recently expanded with the discovery of a new, highly prevalent class of conserved RNA molecules called circular RNAs (circRNAs or ciRS)7,8. The majority of circRNAs overlap with coding genes, often at the protein-coding portion of the mRNAs, and arise from circularized splicing of the reverse ends (in head-to-tail manner) of one or more exons (Fig. 1b). The circRNA CDR1 as (also known as ciRS-7) is an extraordinary example of a competing endogenous RNA species9,10 (Fig. 1b). This brain-enriched circRNA contains more than 70 miR-7 MREs and forms complexes with Argonaute (AGO) protein, the catalytic core of the RNA-induced silencing complex (RISC). CDR1 as (ciRS-7) is coexpressed with miR-7 in the brain and is colocalized in the P bodies, which suggests that miR-7 compartmentalizes CDR1 as (ciRS-7) to this organelle9. Silencing of CDR1 as (ciRS-7) in HEK-293 cells results in downregulation of miR-7 target genes as well as other genes, whereas morpholino knockdown of miR-7 and ectopic expression of CDR1 as (ciRS-7) are both associated with morphological defects in the midbrain9,10.
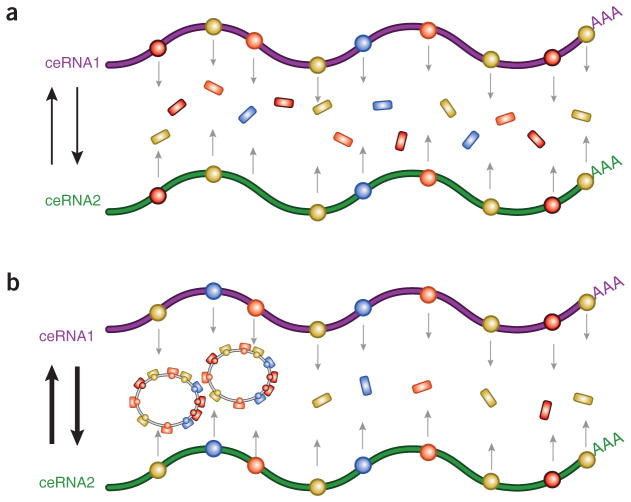
Astonishingly, it has become recently apparent that ~80% or our genome is biochemically functional and 70% or more is actively transcribed3. This indicates that the total number of noncoding RNA genes vastly outnumbers that of the coding genome, with pseudogenes and lncRNAs among the most numerous species. But algorithms able to predict functional motifs of noncoding RNAs are still lacking in contrast to those of protein-coding genes. Base pair complementarity, however, represents a general property of all RNA molecules. On this basis, we have previously proposed that RNA species can communicate with one another by using the MREs of miRNAs as letters of a new RNA language, whereby RNA species sharing the same subset of MREs can regulate their relative abundance by competing for miRNAs4 (Fig. 2a). The direct ceRNA cross-talk between PTEN and PTENpg1 sense represents one such example. The new findings by Johnsson et al.5 now add another layer to this regulatory crosstalk whereby PTENpg1 asRNAβ actively contributes to this ceRNA network by stabilizing PTENpg1 sense (Fig. 1a).
Figure 2.
circRNAs enhance ceRNAs cross-talk. (a) ceRNA1 and ceRNA2 regulate each other’s levels by competing for the same pool of miRNAs. However, ceRNAs are still under negative regulation by abundant miRNAs. (b) circRNAs sharing MREs in common with ceRNA1 and ceRNA2 sequester excess miRNAs and in turn enhance ceRNA cross-talk and abundance. Thereby, the intertwined network among miRNAs, circRNAs and ceRNAs regulates cellular homeostasis, whereas its dysregulation may contribute to diseases such as cancer and neurodegenerative disorders. MREs and miRNAs are indicated as color-coded circles and rectangles, respectively, on the RNA molecules.
Intriguingly, circRNAs are now emerging as potent ceRNA molecules. CircRNAs are expressed in a tissue-specific manner, are stable and conserved and may have regulatory roles during differentiation and evolution. Hundreds of human genes express circRNA isoforms in amounts comparable to those of their linear counterparts. If we consider that the majority of these arise from coding gene exons8, we speculate that circRNAs containing MREs shared by their linear counterparts might enable and modulate ceRNA cross-talk and ceRNA networks by reducing miRNA pressure on protein-coding RNAs in a tissue- or cell-specific manner, hence acting as circ-ceRNAs. As PTENpg1 sense enhances PTEN expression by miRNA sequestration, circRNAs arising from the protein-coding gene—or the pseudogene—could protect the mRNA through the same mechanism (Fig. 1c).
We do not know exactly the actual competing capability of the majority of circRNAs and pseudogenes, but if they are evolutionarily selected to protect their cognate coding mRNAs, they may represent a powerful weapon against excess miRNAs and hence enhance ceRNA cross-talk, which occurs in permissive molecular environments11 (Fig. 2b). Memczac et al.10 and Hansen et al.9 have described CDR1 as (ciRS-7) as a hot spot for miR-7, but this circRNA is also a miR-671 target. CircRNAs can potentially include several different MREs and use them to ‘talk’ with many other genes in a combinatory manner. Importantly, different miRNAs can exert distinct effects on the same circRNA; whereas miR-7 titers away CDR1 as (ciRS-7) and promotes its compartmentalization in the P bodies, miR-671 binding results in its degradation12. This implies that, beyond their intrinsic stability, the amount of each circRNA may be finely tuned by specific cellular mechanisms that are still largely unknown. Moreover, turnover and degradation of circRNAs and other ceRNAs could also play a key part in the regulation of the ceRNA language. For instance, the degradation of a circRNA loaded with multiple miRNAs would release the burden of bound miRNAs, with a deep impact on overall ceRNA activity, by affecting the translation of tens or even hundreds of ceRNA transcripts, perhaps in localized fashion10.
As the biogenesis of circRNAs remains largely unknown, this will become another area of intense investigation. Noncolinear splicing is probably one of the most reliable mechanisms for generating circRNAs; however whether this is actively regulated remains to be elucidated. The high conservation of circRNAs makes it possible to study their function in vivo during development as well as in diseases such as cancer, and their stability (due to their circular form) makes the spread of their ceRNA activity throughout different tissues possible. CircRNAs could be easily collected from bodily fluids and might represent new biomarkers in diagnosis as well as in response to specific therapies. Conversely, ectopic injection of therapeutic circRNAs (able, for example, to counteract oncogenic miRNAs) could overcome the problem of therapeutic delivery of small RNA molecules. Although we cannot rule out other mechanisms of action for circRNAs, it is worth noting that CDR1 as (ciRS-7), with more than 70 MREs, does behave as a potent ceRNA. Other circRNAs and pseudogenes could contain multiple subsets of MREs, and the recent discovery of thousands of aberrant chimeric transcripts in prostate cancer, associated not with DNA rearrangements but instead with noncanonical splicing13, suggests that other potential ceRNA molecules with unforeseen biological impacts remain to be discovered. In taking advantage of next-generation sequencing technologies and sophisticated algorithms, however, the challenge remains to determine the functional significance of all this information and decipher the complexity of the intertwined relationship between the coding and noncoding genome.
Acknowledgments
We thank F. Bersani and Y. Tay for helpful discussions and comments and T. Garvey for critical editing of the manuscript. Our work is supported by the US National Institutes of Health and National Cancer Institute. R.T. is supported by a Marie Curie International Outgoing Fellowship for Career Development (project DECODER 273518). C.L. is supported by a Fondazione Marche and Italian Scientists and Scholars in North America Foundation postdoctoral fellowship.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Contributor Information
Riccardo Taulli, Cancer Genetics Program, Beth Israel Deaconess Cancer Center, Departments of Medicine and Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA. Department of Oncology, University of Turin School of Medicine, Turin, Italy.
Cristian Loretelli, Cancer Genetics Program, Beth Israel Deaconess Cancer Center, Departments of Medicine and Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Pier Paolo Pandolfi, Email: ppandolf@bidmc.harvard.edu, Cancer Genetics Program, Beth Israel Deaconess Cancer Center, Departments of Medicine and Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
References
- 1.Bartel DP. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guttman M, Rinn JL. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djebali S, et al. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salmena L. Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnsson P, et al. Nat Struct Mol Biol. 2013;20:440–446. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poliseno L, et al. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salzman J. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeck WR. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen TB, et al. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 10.Memczak S, et al. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 11.Ala U, et al. Proc Natl Acad Sci USA. 2013 Mar 27; doi: 10.1073/pnas.1222509110. published online. [DOI] [Google Scholar]
- 12.Hansen TB, et al. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kannan K. Proc Natl Acad Sci USA. 2011;108:9172–9177. doi: 10.1073/pnas.1100489108. [DOI] [PMC free article] [PubMed] [Google Scholar]