Abstract
Purpose
To evaluate the effects of single-nucleotide polymorphisms (SNPs) in microRNA-related genes on clinical outcomes in colorectal cancer (CRC) patients receiving first-line fluoropyrimidine-based chemotherapy.
Experimental Design
Forty-one SNPs in 26 microRNA-related genes were genotyped in 1,097 CRC patients recruited at the University of Texas MD Anderson Cancer Center. Patients were enrolled between 1990 and 2008 and last follow-up was in 2010. The associations between genotypes and recurrence-free survival (RFS), progression-free survival (PFS), and overall survival (OS) stratified by clinical stage were analyzed in 741 newly diagnosed patients (diagnosed within 1 year) and replicated the findings in additional 356 patients.
Results
In patients with stage III disease, mir608:rs4919510 was associated with increased risk for both recurrence (HR=2.72; 95%CI, 1.38 to 5.33) and death (HR=3.53; 95%CI, 1.42 to 8.73). The associations were confirmed in the replication set and the combined HR for training and replication sets was 1.94 (95% CI, 1.31 to 2.86) for recurrence and 2.35 (95%CI, 1.40 to 3.93) for death, respectively. The mir219-1:rs213210 showed consistent association with death in the training set (HR=3.86; 95%CI, 1.33 to11.22), the replication set (HR = 3.33; 95% CI, 1.39 to 7.98) and combined dataset (HR = 3.53; 95% CI, 1.80 to 6.95). In combined analysis of these two SNPs, patients carrying the variant genotypes at both sites exhibited a 5.6 fold increased risk of death.
Conclusion
Genetic polymorphisms in the microRNA pathway may predict prognosis in stage III CRC patients treated with fluoropyrimidine-based chemotherapy.
Keywords: colorectal cancer, polymorphism, microRNA, chemotherapy, recurrence, survival
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second-leading cause of cancer-related deaths in Western countries (1). Stage-I disease usually can be cured by surgery alone, but patients with high-risk stage-II, -III, or -IV disease are recommended for chemotherapy in the adjuvant or palliative setting (1-3). 5-Fluorouracil (5-FU) is the most frequently used chemotherapeutic agent for treating CRC patients. However, a substantial portion of CRC patients do not benefit from 5-FU based treatment regimens, whereas treatment-related toxicity may be severe and dose-limiting (4, 5). One strategy to improve the efficacy and reduce toxicity is to determine subgroups of patients who would have a poor or favorable prognosis after 5-FU-based chemotherapy, which may allow personalized treatment of CRC patients.
MicroRNA (miRNA) is a growing family of 21- to 23-nucleotide, endogenous, single-stranded non-coding RNAs that post-transcriptionally regulate the expression of target mRNA transcripts. A series of biogenesis steps yield mature miRNAs from pri-miRNA to pre-miRNA (6) (7) (8). Given that impaired miRNA processing can result in a substantial decrease in mature miRNA levels(9) and many miRNAs target mRNA transcripts involved in cell proliferation, differentiation, and apoptosis (10), it is conceivable that single-nucleotide polymorphisms (SNPs) in genes of miRNA and of the miRNA biogenesis pathway may have effects on cancer risk, prognosis, and treatment response. SNPs are inherited genetic variations, can be easily assayed by high-throughput and reliable technology, is capable of affecting gene expression or protein function, and therefore, are alternative or complementary markers to tissue based biomarkers. Several genome-wide association studies (GWAS) have identified at least 16 CRC susceptibility SNPs (11-14). However, due to the large sample size requirement and the heterogeneity of tumors and treatment, GWAS of clinical outcomes of CRC or any other cancers has not been successfully conducted yet. Candidate gene approach is still the main approach in association studies of clinical outcomes. Several SNPs in the miRNA-related genes have been associated with the risk of cancers(15-17). A pre-miRNA SNP (rs11614913) in miR-196a2 was found to be associated with poor survival in individuals with non-small-cell lung cancer (18). A SNP in miR26a-1 was associated with disease response to irinotecan-based chemotherapy in patients with metastatic CRC (19). However, another study in Korean patients with surgically resected CRC did not observe any association between the polymorphisms in microRNA biogenesis-pathway and clinical outcomes (20). To further dissect these issues, we have performed a comprehensive study with regard to genetic variations in microRNA-related genes and clinical outcomes of CRC patients.
Materials and Methods
Study Population and Data Collection
1097 histologically confirmed colorectal adenocarcinoma patients were recruited at the University of Texas MD Anderson Cancer Center between January 1990 and June 2008, with follow-up to January 2010. Among them, there were 741 newly diagnosed patients (diagnosed within 1 year), which was analyzed as a training set in this study. The remaining 356 patients had a long history (diagnosed >1 year) of CRC prior to recruitment and was used as a replication set. A self-administered questionnaire was used for all patients to collect epidemiological data including demographical characteristics, tobacco use, family history of cancer and medical history. We reviewed patients’ medical records to collect clinical information including date of diagnosis, performance status, clinical stage, tumor location, histological grade, pathological stage, treatment, and tumor recurrence/ progression. Information on vital status was obtained from the medical records and Social Security Death Index (SSDI). Each patient donated 10 to 20-mL blood sample. All study participants signed an informed consent form, and the study was approved by the Institutional Review Board of MD Anderson Cancer Center.
SNP Selection and Genotyping
The selection of genes and SNPs has been described previously (16). Briefly, after an extensive mining of the International HapMap Project database (http://www.hapmap.org) (21), dbSNP database (http://www.ncbi.nlm.nih.gov/SNP) (22) and miRBase registry (http://microrna.sanger.ac.uk/)(23) for SNPs in microRNA pathway genes, we identified 41 potential functional polymorphisms: 24 SNPs in 11 genes related to miRNA biogenesis, 7 SNPs in 7 pre-miRNAs or mature miRNAs, and 10 SNPs in 8 pri-miRNAs. DNA was isolated from the peripheral blood samples using a QIAampDNA extraction kit (Qiagen, Valencia, California). SNP genotyping was done using the SNPlex assay, according to the manufacturer's instructions (Applied Biosystems, Foster City, CA). Internal quality controls and negative controls were used to ensure genotyping accuracy, and 5% of all samples were randomly selected and genotyped in duplicate, with 100% concordance. The average call rate for the SNPs was 98.8%. Two SNPs in pri-miRNAs (rs107822 and rs10425222) were excluded from further analysis due to the call rate <95%.
Statistical Analysis
The study endpoints were overall survival (OS), progression-free survival (PFS), and recurrence-free survival (RFS). OS was calculated from pathologic diagnosis to death regardless of the cause. PFS was defined as the time from pathologic diagnosis until disease progression or death, whichever occurred first . RFS was defined as the time from the date of pathologic diagnosis until first recurrence or death from any cause. Statistical analyses were performed using STATA software (version 10, STATA Corporation, College Station, TX). Cox's proportional hazards model was used to estimate hazard ratios (HRs) and their 95% confidence intervals (CIs) for the multivariate survival analyses in the training set and the replication set. The analyses were adjusted for age, gender, ethnicity, clinical stage, histologic grade, and if necessary, treatment modality and chemotherapy regimen. The Benjamini-Hochberg False Discovery Rate (FDR) method was used to control for multiple comparisons with a threshold of q=0.05 (24). The Cochrane Q statistics test was used for the assessment of heterogeneity. If the result of the Q test was not significant, the fixed-effects model was chosen. Otherwise, the pooled HR was estimated using the random effects model. Associations between genotypes and OS, PFS, and RFS were estimated using the Kaplan-Meier method, and statistical significance was determined using the log-rank test. We also evaluated the combined effects of the SNPs by the number of genotypes identified from the main effects analysis of single SNPs. All P-values were two-sided and a P ≤ 0.05 was considered statistically significant.
Results
Patient Characteristics
Table 1 shows selected demographic, tumor, and treatment characteristics in 1097 CRC patients included in this study. Of the 741 patients in training set, the median patient age was 58 years (range, 19–97 years). The mean age (year ± SD) for stage I, II, III, and IV patients was 58.5±1.5, 59.1±1.0, 58.5±0.84, and 55.2±0.77, respectively. The median follow-up time was 32.5 months. There were 41 recurrences (22.7%) and 31 deaths (17.1%) among 181 patients with stage II disease, and 60 recurrences (27.8%) and 41 deaths (19.0%) among 216 patients with stage III disease, but median RFS and OS had not been reached during the follow-up time. Among 277 patients with stage IV disease, 148 patients (53.4%) had progression and 147 (53.1%) were dead, with median PFS of 18.2 months and median OS of 36.1 months. Age (P=0.03) and clinical stage (P=2×10-17) were significantly associated with RFS and PFS, while clinical stage (P=2×10-25) and histological grade (P=0.0006) correlated with OS. In addition, those who underwent curative surgery or received fluoropyrimidine-based chemotherapy had better clinical outcome.
Table 1.
Selected demographic and clinical characteristics of CRC patients
| Training set (N=741) |
Replication set (N=356) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | Recurrence /progression |
No recurrence /progression |
P Value |
Dead | Alive | P value |
Recurrence /progression |
No recurrence /progression |
P value |
Dead | Alive | P value |
| Age, mean (SD) | 55.9(13.2) | 58.1(12.7) | 0.03 | 57.5(13.9) | 57.3(12.6) | 0.86 | 56.2(12.5) | 53.8(13.5) | 0.25 | 56.7(125) | 54.75(12.7) | 0.16 |
| Gender, N (%) | ||||||||||||
| Male | 161(63.4) | 297(61.1) | 147(66.5) | 312(60.0) | 188(59.7) | 17(42.5) | 128(58.7) | 78(56.5) | ||||
| Female | 93(36.6) | 189(38.9) | 0.55 | 74(33.5) | 208(40.0) | 0.09 | 127(40.3) | 23(57.5) | 0.04 | 90(41.3) | 60(43.5) | 0.68 |
| Race, N (%) | ||||||||||||
| Caucasian | 204(80.6) | 407(83.9) | 182(82.7) | 429(82.7) | 263(83.5) | 31(77.5) | 179(82.1) | 116(84.1) | ||||
| African-American | 23(9.1) | 39(8.0) | 20(9.1) | 42(8.1) | 19(6.0) | 3(7.5) | 15(6.9) | 7(5.1) | ||||
| Others | 26(10.3) | 39(8.0) | 0.50 | 18(8.2) | 48(9.2) | 0.83 | 33(10.5) | 6(15.0) | 0.63 | 24(11.0) | 15(10.9) | 0.78 |
| Tumor location, N (%) | ||||||||||||
| Proximal | 66(26.4) | 156(32.2) | 61(28.0) | 161(31.1) | 88(28.9) | 12(30.0) | 63(30.3) | 37(27.0) | ||||
| Distal | 75(30.0) | 126(26.0) | 72(33.0) | 129(25.0) | 111(36.5) | 14(35.0) | 71(34.1) | 55(40.1) | ||||
| Rectal | 109(43.6) | 202(41.7) | 0.23 | 85(39.0) | 227(43.9) | 0.08 | 105(34.5) | 14(35.0) | 0.98 | 74(35.6) | 45(32.8) | 0.52 |
| Stage, N (%) | ||||||||||||
| Stage I | 5(2.0) | 61(12.6) | 2(0.9) | 64(12.3) | 37( 11.7) | 1(2.5) | 23(10.6) | 15(10.9) | ||||
| Stage II | 41(16.1) | 140(28.8) | 31(14.0) | 150(28.8) | 78(24.8) | 9(22.5) | 43(19.7) | 45(32.6) | ||||
| Stage III | 60(23.6) | 156(32.1) | 41(18.6) | 175(33.7) | 108(34.3) | 7(17.5) | 70(32.1) | 45(32.6) | ||||
| Stage IV | 148(58.3) | 129(26.5) | 2×10−17 | 147(66.5) | 131(25.2) | 1×10−25 | 92(29.2) | 23(57.5) | 0.002 | 82(37.6) | 33(23.9) | 0.01 |
| Histology grade, N (%) | ||||||||||||
| Well-differentiated | 9(3.6) | 25(5.2) | 6(2.8) | 28(5.5) | 10(3.3) | 0(0.0) | 6(2.8) | 4(2.9) | ||||
| Moderate-differentiated | 192(76.5) | 377(78.7) | 157(72.0) | 413(80.5) | 237(77.2) | 33(84.6) | 158(74.9) | 113(83.1) | ||||
| Poorly-differentiated | 50(19.9) | 77(16.1) | 0.29 | 55(25.2) | 72(14.0) | 0.0006 | 60(19.5) | 6(15.4) | 0.40 | 47(22.3) | 19(14.0) | 0.16 |
| Primary tumor curative surgery | ||||||||||||
| Yes | 158(62.2) | 388(79.8) | 113(51.1) | 433(83.3) | 272(86.3) | 22(55.0) | 176(80.7) | 119(86.2) | ||||
| No | 96(37.8) | 98(20.2) | 2×10−7 | 108(48.9) | 87(16.7) | 1×10−19 | 43(13.7) | 18(45.0) | 7×10−7 | 42(19.3) | 19(13.8) | 0.18 |
| Fluoropyrimidine-based chemotherapy | ||||||||||||
| Yes | 214(84.3) | 318(65.4) | 174(78.7) | 359(69.0) | 239(75.9) | 37(92.5) | 176(80.7) | 101(73.2) | ||||
| No | 40(15.7) | 168(34.6) | 6×10−8 | 47(21.3) | 161(31.0) | 0.007 | 76(24.1) | 3(7.5) | 0.02 | 42(19.3) | 37(26.8) | 0.10 |
There were 356 patients included in replication set. The mean age for stage I, II, III, and IV patients was 62.3±2.0, 59.1±1.0, 58.5±0.84, and 55.2±0.77, respectively. The median OS for the four stages was 63.2, 79.6, 71.8, and 36.6 months, respectively. These patients were diagnosed outside of MD Anderson Cancer Center over a year ago and came to MD Anderson Cancer Center for treatment due to potential tumor recurrence or progression; therefore, the recurrence, progression and death rates were higher than newly diagnosed patients (Table 1).
Training Set
We assessed the association of each individual SNP with OS, PFS, and RFS using a multivariate Cox model, adjusting for age, gender, ethnicity, smoking status, tumor site and histologic grade, and stratified by stages (Table 2). Among them, GEMIN4:rs7813 and rs910925 were in strong linkage disequilibrium (r2=0.99), thus we only included rs7813 in the subsequent analysis.
Table 2.
SNPs associated with outcomes of CRC patients receiving 5-FU based chemotherapy stratified by stages
| Training set |
Replication set |
Pooled analysis |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Model# | HR(95%CI) | P value* | q value | HR(95%CI) | P value* | HR(95%CI) | P value | P for Hetero.† | MAF†† |
| Recurrence: | |||||||||||
| Stage II | |||||||||||
| RAN | rs14035 | add | 2.32(1.28-4.21) | 0.005 | 0.06 | 1.00(0.56-1.78) | 1.00 | 1.52(0.67-3.46) | 0.32 | 0.05 | 12% |
| mir373 | rs12983273 | add | 2.48(1.19-5.14) | 0.02 | 0.07 | 1.22(0.63-2.37) | 0.55 | 1.68(1.03-2.74) | 0.04 | 0.16 | 13% |
| Drosha | rs6877842 | dom | 0.29(0.09-0.92) | 0.04 | 0.16 | 1.05(0.52-2.10) | 0.89 | 0.75(0.41-1.36) | 0.34 | 0.06 | 18% |
| Stage III | |||||||||||
| mir608 | rs4919510 | dom | 2.72(1.38-5.33) | 0.004 | 0.04 | 1.64(1.02-2.63) | 0.04 | 1.94(1.31-2.86) | 0.0008 | 0.22 | 17% |
| GEMIN3 | rs197412 | add | 2.07(1.31-3.27) | 0.002 | 0.03 | 1.07(0.72-1.57) | 0.74 | 1.47(0.72-2.81) | 0.24 | 0.03 | 10% |
| XPO5 | rs11077 | dom | 0.38(0.19-0.78) | 0.008 | 0.04 | 1.29(0.79-2.10) | 0.31 | 0.72(0.22-2.38) | 0.59 | 0.005 | 40% |
| GEMIN4 | rs2740348 | dom | 2.56(1.25-5.24) | 0.01 | 0.04 | 0.86(0.50-1.50) | 0.61 | 1.45(0.50-4.21) | 0.50 | 0.02 | 18% |
| GEMIN3 | rs197388 | add | 1.82(1.08-3.05) | 0.02 | 0.07 | 0.99(0.57-1.70) | 0.96 | 1.36(0.94-1.99) | 0.11 | 0.11 | 29% |
| GEMIN4 | rs7813 | add | 1.62(1.06-2.49) | 0.03 | 0.07 | 1.25(0.84-1.87) | 0.27 | 1.41(1.05-1.89) | 0.02 | 0.39 | 14% |
| Progression | |||||||||||
| Stage IV | |||||||||||
| let7f-2 | rs17276588 | dom | 3.12(1.61-6.06) | 0.0008 | 0.008 | 1.69(0.72-4.01) | 0.23 | 2.48(1.47-4.19) | 0.0007 | 0.28 | 2% |
| mir30c-1 | rs16827546 | dom | 2.12(1.22-3.67) | 0.007 | 0.04 | 1.12(0.50-2.51) | 0.78 | 1.73(1.10-2.73) | 0.02 | 0.20 | 4% |
| Drosha | rs6877842 | dom | 1.61(1.07-2.43) | 0.02 | 0.08 | 0.97(0.58-1.62) | 0.89 | 1.32(0.96-1.82) | 0.09 | 0.13 | 18% |
| DICER | rs13078 | dom | 1.51(1.01-2.24) | 0.04 | 0.12 | 0.74(0.45-1.21) | 0.23 | 1.07(0.53-2.16) | 0.41 | 0.03 | 14% |
| Survival: | |||||||||||
| Stage II | |||||||||||
| AGO2 | rs4961280 | rec | 5.49(1.39-21.74) | 0.02 | 0.18 | 4.61(0.73-29.16) | 0.10 | 5.16(1.71-15.53) | 0.004 | 0.84 | 13% |
| Stage III | |||||||||||
| mir608 | rs4919510 | dom | 3.53(1.42-8.73) | 0.006 | 0.05 | 1.93(1.03-3.62) | 0.04 | 2.35(1.40-3.93) | 0.001 | 0.28 | 17% |
| mir219-1 | rs213210 | dom | 3.86(1.33-11.22) | 0.01 | 0.06 | 3.33(1.39-7.98) | 0.007 | 3.53(1.80-6.95) | 0.0003 | 0.83 | 6% |
| mir604 | rs2368392 | rec | 5.20(1.55-17.50) | 0.007 | 0.05 | 0.51(0.17-1.50) | 0.22 | 1.62(0.16-15.95) | 0.68 | 0.005 | 25% |
| Stage IV | |||||||||||
| DICER | rs13078 | dom | 1.66(1.09-2.52) | 0.02 | 0.21 | 0.87(0.51-1.48) | 0.61 | 1.30(0.93-1.80) | 0.12 | 0.06 | 14% |
| TRBP | rs784567 | rec | 1.59(1.03-2.43) | 0.04 | 0.31 | 0.76(0.42-1.38) | 0.37 | 1.13(0.65-2.31) | 0.52 | 0.05 | 4% |
Adjusted for age, gender, ethnicity, smoking status, tumor site, histologic grade, and treatment.
Genetic model of inheritance: dom- dominant model, rec- recessive model, add- additive mode.
Cochran's Q statistic to test for heterogeneity between studies.
MAF: minor allele frequency
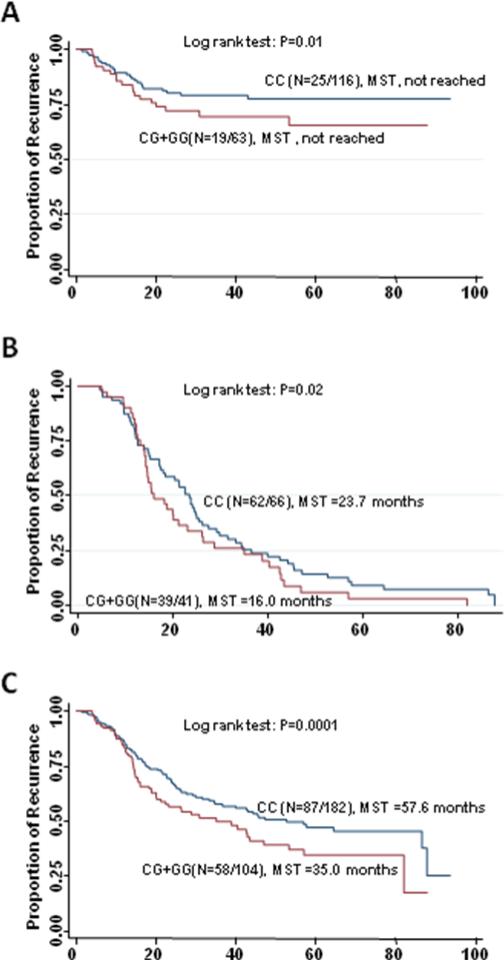
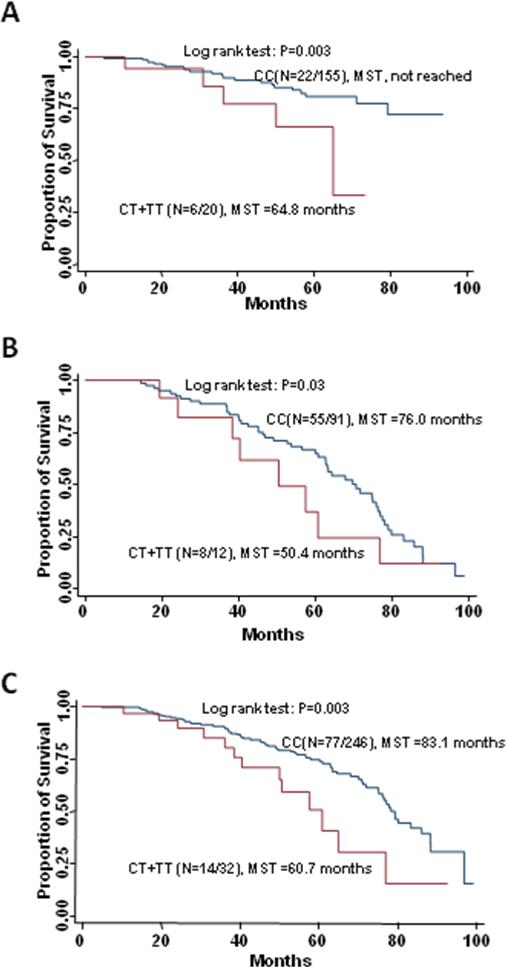
Due to the excellent prognosis and rare events of recurrence and death, we did not carry out analysis of stage I patients. Among 117 patients with stage II disease who received 5-FU based chemotherapy, the most significant association with recurrence was conferred by the variant allele of RAN:rs14035 in a dose-dependent manner (per allele HR=2.32; 95%CI, 1.28 to 4.21;P=0.005, q=0.06). The homozygous variant genotype of AGO2:rs4961280 was significantly associated with an increased risk of death (HR=5.49; 95%CI, 1.39 to 21.74; P=0.02, q=0.18). Among 179 patients with stage III disease who received 5-FU based chemotherapy, the variant-containing genotypes of mir608:rs4919510 exhibited increased risks of both recurrence (HR=2.72; 95%CI, 1.38 to 5.33; P=0.004; q=0.03) and death (HR=3.53; 95%CI, 1.42 to 8.73; P=0.006; q=0.05). Moreover, these patients had a significantly shorter RFS (104.3 versus >117.8 months, log-rank P = 0.01; Figure 1A) and OS (125.5 vs. >117.8 months, log-rank P = 0.01) compared with those with the wild-type genotype. The GEMIN3: rs197412 and XPO5:rs11077 had highly significant associations with recurrence (HR=2.07, 95% CI, 1.31 to 3.27; P=0.002; q=0.03; and HR = 0.38, 95% CI, 0.19 to 0.78; P=0.002; q=0.03, respectively), while mir604:rs2368392 and mir219-1:rs213210 showed highly significant associations with death (HR=5.20; 95%CI, 1.55 to17.50; P=0.007; q=0.05; and HR=3.86; 95%CI, 1.33 to11.22; P=0.01; q=0.06, respectively). This increase in risk of death resulted in a decreased OS for those with the variant-containing genotypes of mir219-1:rs213210 compared to patients carrying the homozygous wild type genotype (64.8 vs. >117.8 months, log-rank P = 0.003; Figure 2A). Among 218 patients with stage IV disease, the effects of the variant-containing genotypes of let-7f-2:rs17276588 and mir30c-1:rs16827546 on progression were significant after adjusting for multiple comparison (HR=3.12; 95%CI, 1.61 to 6.06; P=0.0008, q=0.008; and HR=2.12; 95%CI, 1.22 to 3.67; P=0.007; q=0.04, respectively). Two SNPs, Drosha:rs13078 (HR = 1.66; 95% CI, 1.09 to 2.52; P=0.02) and TRBP:rs784567 (HR = 1.59; 95% CI, 1.03 to 2.43; P=0.04), were associated with the risk of death, but lost significance after adjusting for multiple comparison.
Figure 1.
Kaplan-Meier recurrence-free survival curves of patients with stage III disease receiving fluoropyrimidine-based chemotherapy by genotypes of mir608:rs4919510. (A) training set, (B) replication set, and (C) combined set.
Figure 2.
Kaplan-Meier overall survival curves of patients with stage III disease receiving fluoropyrimidine-based chemotherapy by genotypes of mir219-1:rs213210. (A) training set, (B) replication set, and (C) combined set
Replication Set
Genotype data was analyzed in 53 stage II, 108 stage III, and 110 stage IV patients. As shown in Table 3, the significant associations between mir608:rs4919510 and risk of both recurrence and death and the association between mir219-1:rs213210 and risk of death were confirmed in stage III patients in the replication set. Patients with the variant allele-containing genotypes of the mir608:rs4919510 showed increased risks of recurrence (HR=1.64; 95% CI, 1.02 to 2.63; P=0.04) and death (HR=1.93; 95% CI, 1.03 to 3.62; P=0.04) in the replication set. The pooled analysis HR was 1.94 (95% CI, 1.31 to 2.86; P=0.0008) for recurrence and 2.35 (95%CI, 1.40 to 3.93; P=0.001) for death. Furthermore, compared with patients carrying the wild-type genotype, those with the variant-containing genotypes had a decrease in RFS in replication set (16.0 vs.23.7 months, log-rank P = 0.02; Figure 1B) and combined dataset (35.0 vs. 57.6 months, log-rank P = 0.0001; Figure 1C), respectively. The increase in risk of death also resulted in a decrease in OS in combined dataset (74.8 vs.103.7 months, log-rank P = 0.0009, data not shown).
Table 3.
Association of mir608:rs4919510 and mir219-1:rs213210 with recurrence and survival in stage III patients receiving fluoropyrimidine-based chemotherapy
| Training set |
Replication set |
Pooled analysis |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Event/No event | HR (95%CI) | P* | MST† | Log rank p | Event/No event | HR (95%CI) | P* | MST† | Log rank P | HR (95%CI) | P* | MST† | Log rank P |
| Recurrence: | ||||||||||||||
| mir608:rs4919510 | ||||||||||||||
| CC | 25/91 | Reference | >117.8 | 62/4 | Reference | 24.5 | Reference | 87.8 | ||||||
| CG+GG | 19/44 | 2.72 (1.38-5.33) | .004 | 104.3 | .01 | 39/2 | 1.64 (1.02-2.63) | .04 | 15.4 | .04 | 1.65 (1.13- 2.41) | .01 | 28.7 | .0001 |
| Survival: | ||||||||||||||
| mir608:rs4919510 | ||||||||||||||
| CC | 16/100 | Reference | >117.8 | 41/25 | Reference | 79.7 | Reference | 103.7 | ||||||
| CG+GG | 12/51 | 3.53 (1.42-8.73) | .006 | 125.5 | .01 | 24/17 | 1.93 (1.03-3.62) | .04 | 62.7 | .08 | 1.96 (1.19- 3.21) | .008 | 74.8 | .0009 |
| mir219-1:rs213210 | ||||||||||||||
| CC | 22/133 | Reference | >117.8 | 55/36 | Reference | 76.4 | Reference | 101.8 | ||||||
| CT+TT | 6/14 | 3.86 (1.33-11.22) | .01 | 64.8 | .003 | 8/4 | 3.33 (1.39-7.98) | .007 | 50.4 | .03 | 3.22 (1.70- 6.10) | .0003 | 62.7 | .003 |
Adjusted for age, gender, ethnicity, smoking status, tumor site, histologic grade, and treatment
MST, median event-free survival times in months
The variant-containing genotypes of the mir219-1:rs213210 showed consistent association with death in replication set (HR = 3.33; 95% CI, 1.39 to 7.98; P=0.007) and combined dataset (HR = 3.53; 95% CI, 1.80 to 6.95; P=0.0003). These patients also had a significantly shorter OS than those with the wild-type genotype in replication set (50.4 vs.76.0 months, log-rank P = 0.03; Figure 2B) and combined dataset (60.7 vs. 83.1 months, log-rank P = 0.003; Figure 2C).
Cumulative Effect
Since the variant genotypes of mir608:rs4919510 and mir219-1:rs213210 were confirmed in stage III patients receiving adjuvant fluoropyrimidine-based therapy, we next performed a joint analysis of these two SNPs. There was a significant trend for the increased risk of death with the increasing number of variant genotypes in training set (P for trend = 0.0001) and replication set (P for trend = 0.003). In the combined dataset, compared with patients without any variant genotype, patients carrying one variant genotype had a 2.51-fold increased risk of death (95% CI, 1.47 to 4.28; P =0.001), and the risk further increased to 5.6-fold (95% CI,1.91 to16.45; P= 0.002) for patients carrying two variant genotypes (P for trend = 5.25×10-5). Similar cumulative effect was also observed for recurrence in the training (P for trend=0.0006), replication (P for trend =0.01), and combined sets (P for trend =0.001). In the combined dataset, carrying one or two variant genotypes resulted in 1.67-fold (95%CI, 1.12-2.49; P=0.01) and 3.53-fold (95%CI, 1.51-8.26; P=0.004) increased risk of recurrence, respectively.
Discussion
Our results reveal that mir608:rs4919510 and mir219-1:rs213210, alone or in combination, showed consistent association with clinical outcome for patients with stage III disease in the training and replication set. To our knowledge, this is the largest study to date to investigate the role of genetic variations in miRNA pathway in clinical outcomes for CRC patients. Currently, tumor-node-metastasis (TNM) system remains the most commonly used criterion to predict prognosis and define the need for chemotherapy in CRC patients (25). Fluoropyrimidine-based chemotherapy was routinely used in stage III disease, but recurrence rates remained high.(26) Risk stratification based on germline genotype status would provide information for individual chemotherapy. In the present study, mir608:rs4919510 was consistently associated with both recurrence and survival. A previous study has demonstrated that the mir608:rs4919510 variant is associated with recurrence in patients with renal cell carcinoma (27). The SNP rs4919510 is located in the mature mir608 sequence and was predicted to bind to mir608 target sites within the ACDC (adiponectin and collagen domain containing), CD4 (CD4 antigen), GHR (growth hormone receptor), RXRB (retinoic X receptor beta), and TP53 (tumor protein p53) genes with lower free energies than the wild type allele (28). These targets could affect CRC clinical outcome, for example, previous investigations reported that stage III CRC patients with tumors carrying TP53 mutation benefited from 5-FU based postoperative chemotherapy (29) (30). Therefore, it is conceivable that the variant miR-608 could elicit different biological activities of these target molecules that could affect CRC clinical outcome. It has been reported that the variant allele of mir219-1:rs213210 was associated with susceptibility to developing esophageal cancer (31). We found that CRC patients carrying the variant genotypes of rs213210 had a worse survival, possibly through a mechanism of affecting the expression of mature mir219-1. The expression of mir219-1 was highly up-regulated in metastatic hepatocellular carcinoma and was also capable of predicting patient survival and recurrence (32). A previous study found that miR-219 is a target of the CLOCK and BMAL1 complex and exhibits robust circadian rhythms of expression (33), and another study showed that circadian clock protein BMAL1 coordinates in vivo tumor growth, S-phase progression, thymidylate synthase, and 5-FU therapeutic index (34). It is tempting to speculate that circadian clock related mechanism may partially explain the link between miR-219 and recurrence in stage III CRC patients receiving 5-FU based therapy. The functional consequence of mir219-1:rs213210 is not clear. Further studies are warranted to evaluate whether this SNP affects mir219-1 expression and processing, and the biological mechanisms underlying the association of this SNP and recurrence.
We observed a clear and significant trend towards increased recurrence and death risk as the number of variant genotypes of mir608:rs4919510 and mir219-1:rs213210 and patients carrying two variant genotypes had a 5.6-fold increased risk of death. These results suggest that the cumulative influence of multiple SNPs was able to further enhance the separation between the patients with stage III disease who would have unfavorable prognosis following 5-FU-based adjuvant chemotherapy.
The effect of GEMIN3:rs197412, XPO5:rs11077 and mir604:rs2368392 was also highly significant in stage III disease, though the results were not confirmed in the replication set. The variant allele of GEMIN3:rs197412 has been shown to be associated with susceptibility to developing premalignant oral lesions (35). XPO5:rs11077 was recently identified as a predictor of disease control rate for colon cancer patients treated with 5-fluorouracil and irinotecan (19). Polymorphisms affecting the expression of proteins involved in general miRNA biogenesis, such as GEMIN3 and XOP5, could alter global miRNA homeostasis and tissue-specific miRNA alterations (36-38). The global repression of miRNA maturation has been demonstrated to promote cellular transformation and tumorigenesis (9). The functional consequence of SNPs in pre-miRNA sequences, such as mir604:rs2368392, have already been suggested to affect the processing and levels of the mature miRNA(18)., Given that variability in expression levels of miRNA may influence tumor progression and sensitivity to a fluoropyrimidine agent (39) (40), these potential functional SNPs could emerge as potential predictors of prognosis and the efficacy of fluoropyrimidine.
For stage II diseases, RAN: rs14035 was associated with OS with high significance in patients receiving surgery and adjuvant fluoropyrimidine treatment. The key element in the nuclear export of pre-miRNAs is XPO5-RAN GTP-pre-miRNA heteroternary complex. The disruption in the pre-miRNA nucleo-cytoplasmic transport would impair the production of mature miRNAs in cancer cells (41). RAN is overexpressed in most cancer cell lines including colon cancer, suggesting its role in tumor transformation (42). Moreover, it was reported that RAN suppressed the activation of JNK and inhibited apoptosis induced by an anticancer drug (43). In patients with stage IV CRC, let7f-2:rs17276588 had significant predictive effect on progression in patients receiving fluoropyrimidine-based chemotherapy. This SNP remained significant after adjusting for multiple comparison in the training set and also showed the same direction in the replication set, although did not reach statistical significance. The let-7 family of miRNAs was found to regulate Ras activity by binding to the 3’-UTR of the human Ras gene (44). K-ras mutations have been associated with increased risk of relapse and death in CRC patients by the multi-center Colorectal Cancer Collaborative Group (RASCAL) study (45) (46). Nakajima et al. demonstrated an association between members of the let-7 family and responsiveness to the oral fluoroprimidine S-1 (47). Therefore, it is biologically plausible that SNPs in let7 family of miRNAs affect CRC prognosis and treatment response.
Since the training and replication set split as we presented here may not be optimal due to the over-representation of recurrence and progression patients in the replication set, we also randomly split the entire dataset into a training and a replication set and re-performed all the analyses (Supplementary Table 1 and 2) and the results were comparable to the current split. For example, for survival, the mir608 SNP (P=0.019) and mir219-1 SNP (P=0.036) ranked the top two in P-values in the new training set. In the new validation set, mir219-1 SNP was the most significant SNP and mir608 SNP was borderline. In pooled analysis, the mir219-1 SNP ranked the first and the mir608 SNP ranked the third. Because we believe the original training set was cleaner and more optimal for initial screening, we presented the original results. Since our study is the first to comprehensively evaluate SNPs in miRNA pathways as prognostic factors for CRC patients receiving 5-FU based chemotherapy, the data of pooled analysis of all SNPs would provide valuable information to investigators in the field who may perform independent validation.
The major strength of our study is the large population size that allowed us to perform stage-stratified analyses in training and replication sets, which limited the confounding of tumor and treatment heterogeneity. The significant results, particularly mir608:rs4919510 and mir219-1:rs213210, remained significant after adjusting for multiple comparison and also were replicated in the replication set; therefore, these SNPs are not likely to be false positives. An additional strength is the comprehensive clinical data for each of these patients. The main limitation of this study is that the replication set was not newly diagnosed patients and those patients came to MD Anderson mainly due to potential recurrence or progression. Therefore, the relative small and heterozygous population in the replication set may result in false negatives.
In conclusion, we identified several highly plausible candidate SNPs in the miRNA pathway that are associated with clinical outcome in CRC patients and also observed significant cumulative effect of multiple SNPs in predicting prognosis in CRC patients receiving 5-FU-based chemotherapy. Further studies are needed to replicate these SNPs in independent populations, to functionally characterize the significant genetic variants, and to find the biological mechanisms underlying the associations.
Supplementary Material
Translational Relevance.
MicroRNA plays important roles in cellular proliferation, differentiation, and apoptosis. This study comprehensively evaluated the associations between single nucleotide polymorphisms (SNPs) in microRNA-related genes and clinical outcomes in colorectal cancer (CRC) patients receiving first-line fluoropyrimidine-based chemotherapy, and identified several biologically plausible individual predictors of recurrence and survival as well as cumulative effect in predicting patient outcomes. The validation and incorporation of the identified SNPs and interactions with the clinical variables may help identify CRC patients with distinct prognosis to allow for personalized cancer therapy.
Table 4.
Cumulative analysis of mir608:rs4919510 and mir219-1:rs213210 in stage III patients receiving fluoropyrimidine-based chemotherapy
| No. of Variant Genotypes | No recurrence, n (%) | Recurrence, n (%) | HR(95%CI) | P value* |
|---|---|---|---|---|
| Training set | ||||
| 0 | 78(80.41) | 19(19.59) | 1(reference) | |
| 1 | 52(71.23) | 21(28.77) | 2.97(1.40-6.28) | 0.004 |
| 2 | 3(60.00) | 2(40.00) | 10.48(2.02-54.47) | 0.005 |
| p for trend | 0.0006 | |||
| Replication set | ||||
| 0 | 3(5.36) | 53(94.64) | 1(reference) | |
| 1 | 1(2.50) | 39(97.50) | 1.63(0.98-2.70) | 0.06 |
| 2 | 1(16.67) | 5(83.33) | 3.41(1.21-9.58) | 0.02 |
| P for trend | 0.01 | |||
| Combined set | ||||
| 0 | 81(52.94) | 72(47.06) | 1(reference) | |
| 1 | 53(46.90) | 60(53.10) | 1.67(1.12-2.49) | 0.01 |
| 2 | 4(36.36) | 7(63.64) | 3.53(1.51-8.26) | 0.004 |
| P for trend | 0.001 | |||
| Alive, n (%) | Dead, n (%) | |||
| Training set | ||||
| 0 | 86(88.66) | 11(11.34) | 1(reference) | |
| 1 | 57(78.08) | 16(21.92) | 7.49(2.38-23.56) | 0.0006 |
| 2 | 4(80.00) | 1(20.00) | 14.98(1.36-164.94) | 0.03 |
| P for trend | 0.0001 | |||
| Replication set | ||||
| 0 | 21(37.50) | 35(62.50) | 1(reference) | |
| 1 | 17(42.50) | 23(57.50) | 2.04(1.04-4.01) | 0.04 |
| 2 | 2(33.33) | 4(66.67) | 7.34(2.03-26.52) | 0.002 |
| P for trend | 0.003 | |||
| Combined set | ||||
| 0 | 107(69.93) | 46(30.07) | 1(reference) | |
| 1 | 74(65.49) | 39(34.51) | 2.51(1.47-4.28) | 0.001 |
| 2 | 6(54.55) | 5(45.45) | 5.60(1.91-16.45) | 0.002 |
| P for trend | 5.25×10−5 | |||
Adjusted for age, gender, ethnicity, smoking status, tumor site, histologic grade, and treatment
Acknowledgments
Financial Support: This work was supported by a Multidisciplinary Research Program (MRP) grant on colorectal cancer from The University of Texas MD Anderson Cancer Center
References
- 1.Gravalos C, Garcia-Escobar I, Garcia-Alfonso P, Cassinello J, Malon D, Carrato A. Adjuvant chemotherapy for stages II, III and IV of colon cancer. Clin Transl Oncol. 2009;11:526–33. doi: 10.1007/s12094-009-0397-8. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham D, Atkin W, Lenz HJ, et al. Colorectal cancer. Lancet. 2010;375:1030–47. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 3.Marshall JL. Risk assessment in Stage II colorectal cancer. Oncology (Williston Park) 2010;24:9–13. [PubMed] [Google Scholar]
- 4.Raftery L, Goldberg RM. Optimal delivery of cytotoxic chemotherapy for colon cancer. Cancer J. 2010;16:214–9. doi: 10.1097/PPO.0b013e3181ddc5ac. [DOI] [PubMed] [Google Scholar]
- 5.Hoff PM, Saad ED, Costa F, et al. Literature Review and Practical Aspects on the Management of Oxaliplatin-Associated Toxicity. Clin Colorectal Cancer. 2011 doi: 10.1016/j.clcc.2011.10.004. epub. [DOI] [PubMed] [Google Scholar]
- 6.Sun G, Yan J, Noltner K, et al. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009;15:1640–51. doi: 10.1261/rna.1560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–12. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 8.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 9.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 10.Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27(Suppl 2):S52–7. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nature Genetics. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 12.Houlston RS, Webb E, Broderick P, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nature Genetics. 2008;40:1426–35. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houlston RS, Cheadle J, Dobbins SE, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nature Genetics. 2010;42:973–U89. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomlinson IP, Carvajal-Carmona LG, Dobbins SE, et al. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet. 2011;7:e1002105. doi: 10.1371/journal.pgen.1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horikawa Y, Wood CG, Yang H, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–62. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang H, Dinney CP, Ye Y, Zhu Y, Grossman HB, Wu X. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–7. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
- 17.Ye Y, Wang KK, Gu J, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila Pa) 2008;1:460–9. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Z, Chen J, Tian T, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–8. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boni V, Zarate R, Villa JC, et al. Role of primary miRNA polymorphic variants in metastatic colon cancer patients treated with 5-fluorouracil and irinotecan. Pharmacogenomics J. doi: 10.1038/tpj.2010.58. [DOI] [PubMed] [Google Scholar]
- 20.Lee HC, Kim JG, Chae YS, et al. Prognostic impact of microRNA-related gene polymorphisms on survival of patients with colorectal cancer. J Cancer Res Clin Oncol. 136:1073–8. doi: 10.1007/s00432-009-0754-6. [DOI] [PubMed] [Google Scholar]
- 21.The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 22.Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–9. [PubMed] [Google Scholar]
- 23.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 25.Smits KM, Cleven AH, Weijenberg MP, et al. Pharmacoepigenomics in colorectal cancer: a step forward in predicting prognosis and treatment response. Pharmacogenomics. 2008;9:1903–16. doi: 10.2217/14622416.9.12.1903. [DOI] [PubMed] [Google Scholar]
- 26.Mano MS, Duhoux F. Colon cancer: update on adjuvant therapy. Clin Colorectal Cancer. 2008;7:178–83. doi: 10.3816/CCC.2008.n.023. [DOI] [PubMed] [Google Scholar]
- 27.Lin J, Horikawa Y, Tamboli P, Clague J, Wood CG, Wu X. Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis. 2010;31:1805–12. doi: 10.1093/carcin/bgq168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landi D, Gemignani F, Barale R, Landi S. A catalog of polymorphisms falling in microRNA-binding regions of cancer genes. DNA Cell Biol. 2008;27:35–43. doi: 10.1089/dna.2007.0650. [DOI] [PubMed] [Google Scholar]
- 29.Godai TI, Suda T, Sugano N, et al. Identification of colorectal cancer patients with tumors carrying the TP53 mutation on the codon 72 proline allele that benefited most from 5-fluorouracil (5-FU) based postoperative chemotherapy. BMC Cancer. 2009;9:420. doi: 10.1186/1471-2407-9-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blandino G, Levine AJ, Oren M. Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene. 1999;18:477–85. doi: 10.1038/sj.onc.1202314. [DOI] [PubMed] [Google Scholar]
- 31.Ye Y, Wang KK, Gu J, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila) 2008;1:460–9. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 33.Cheng HY, Papp JW, Varlamova O, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–29. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood PA, Du-Quiton J, You S, Hrushesky WJ. Circadian clock coordinates cancer cell cycle progression, thymidylate synthase, and 5-fluorouracil therapeutic index. Mol Cancer Ther. 2006;5:2023–33. doi: 10.1158/1535-7163.MCT-06-0177. [DOI] [PubMed] [Google Scholar]
- 35.Clague J, Lippman SM, Yang H, et al. Genetic variation in MicroRNA genes and risk of oral premalignant lesions. Mol Carcinog. 2010;49:183–9. doi: 10.1002/mc.20588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10:399–416. doi: 10.2217/14622416.10.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faggad A, Budczies J, Tchernitsa O, et al. Prognostic significance of Dicer expression in ovarian cancer-link to global microRNA changes and oestrogen receptor expression. J Pathol. 2010;220:382–91. doi: 10.1002/path.2658. [DOI] [PubMed] [Google Scholar]
- 38.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boni V, Zarate R, Villa JC, et al. Role of primary miRNA polymorphic variants in metastatic colon cancer patients treated with 5-fluorouracil and irinotecan. Pharmacogenomics Journal. 2011;11:429–36. doi: 10.1038/tpj.2010.58. [DOI] [PubMed] [Google Scholar]
- 40.Svoboda M, Izakovicova Holla L, Sefr R, et al. Micro-RNAs miR125b and miR137 are frequently upregulated in response to capecitabine chemoradiotherapy of rectal cancer. Int J Oncol. 2008;33:541–7. [PubMed] [Google Scholar]
- 41.Melo SA, Moutinho C, Ropero S, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–15. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 42.Honma K, Takemasa I, Matoba R, et al. Screening of potential molecular targets for colorectal cancer therapy. Int J Gen Med. 2009;2:243–57. doi: 10.2147/ijgm.s5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woo IS, Jang HS, Eun SY, et al. Ran suppresses paclitaxel-induced apoptosis in human glioblastoma cells. Apoptosis. 2008;13:1223–31. doi: 10.1007/s10495-008-0247-0. [DOI] [PubMed] [Google Scholar]
- 44.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 45.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–84. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 46.Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001;85:692–6. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakajima G, Hayashi K, Xi Y, et al. Non-coding MicroRNAs hsa-let-7g and hsa-miR-181b are Associated with Chemoresponse to S-1 in Colon Cancer. Cancer Genomics Proteomics. 2006;3:317–24. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.