Abstract
Mitochondrial Ca2+ homeostasis plays a key role in the regulation of aerobic metabolism and cell survival1, but the molecular identity of the Ca2+ channel, the mitochondrial calcium uniporter2, was still unknown. We have identified in silico a protein (denominated MCU) that shares tissue distribution with MICU1, a recently characterized uniporter regulator3, coexists with uniporter activity in phylogeny and includes two trasmembrane domains in the sequence. siRNA silencing of MCU in HeLa cells drastically reduced mitochondrial Ca2+ uptake. MCU overexpression doubled the [Ca2+]mt rise evoked by IP3-generating agonists, thus significantly buffering the cytosolic elevation. The purified MCU protein exhibited channel activity in planar lipid bilayers, with electrophysiological properties and inhibitor sensitivity of the uniporter. A mutant MCU, in which two negatively-charged residues of the putative pore forming region were replaced, had no channel activity and reduced agonist-dependent [Ca2+]mt transients when overexpressed in HeLa cells. Overall, these data demonstrate that the identified 40 kDa protein is the channel responsible for Ruthenium Red-sensitive mitochondrial Ca2+ uptake, thus providing molecular basis for this process of utmost physiological and pathological relevance.
The past two decades have seen the re-appraisal of the key role of mitochondria in decoding the highly pleiotropic Ca2+ signals evoked by physiological and pathological stimuli4. Mitochondria undergo rapid changes in matrix Ca2+ concentration ([Ca2+]mt) upon cell stimulation, because their low affinity uptake systems are exposed to microdomains of high [Ca2+] in proximity of ER or plasma membrane Ca2+ channels5-9. In turn, [Ca2+]mt increases upregulate aerobic metabolism10-11 and sensitize mitochondria to apoptotic challenges, favouring the release of caspase cofactors12-13. Mitochondrial Ca2+ buffering also shapes the amplitude and spatio-temporal patterns of cytosolic Ca2+ concentration ([Ca2+]cyt) increases14-16. The properties of the Ca2+ transporter are still largely those characterized in the 60s: i) electrogenic transport (hence the name calcium “uniporter”), then shown to be a highly selective channel17, ii) sensitivity to Ruthenium Red and, iii) low affinity for the cation1-2,18. Its molecular identity, however, has remained elusive.
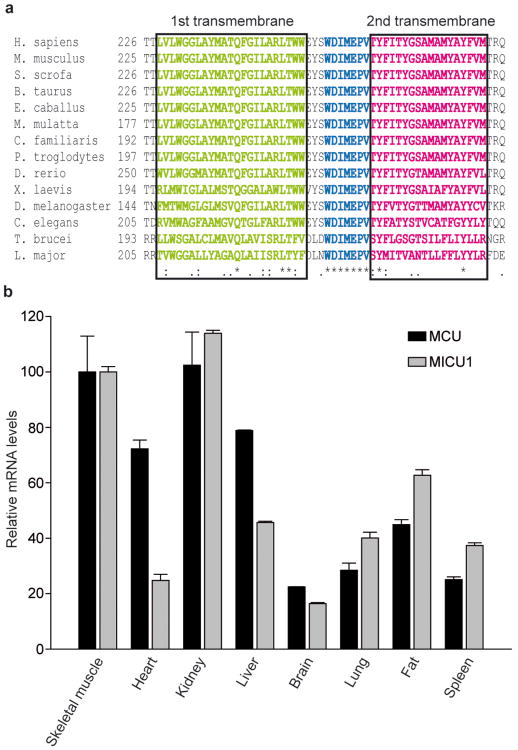
Recently, a uniporter regulator (named MICU1), necessary for rapid mitochondrial Ca2+ uptake in intact cells, was identified in silico in the MitoCarta database3. We thus looked in the same database for the actual transporter, i.e. an integral inner membrane protein fulfilling the criteria for being the bona fide MCU. Among 529 candidates with ubiquitous expression in mammalian tissues, we restricted the analysis to 89 proteins with 2 or more predicted transmembrane domains (TMD) in the primary sequence. Of these, 20 were absent in Saccaromyces cerevisiae (lacking a RuR-sensitive mitochondrial Ca2+ uptake route19), and, among those, 14 were significantly conserved also in kinetoplastids (Trypanosoma and Leishmania), in which rapid, uncoupler-sensitive Ca2+ fluxes to mitochondria were measured20-21. This list (Supplementary Table 1) included 13 proteins with either already characterized functions (complex I subunits, assembly regulators, etc.) or poor alignment scores. The remaining protein (NP_001028431) includes a highly conserved domain comprising two transmembrane regions and an intervening loop enriched in acidic residues (Fig. 1a), suggestive of a possible common role in cation permeation. This domain was also detected by Hidden Markov Model-based analysis of evolutionary conserved proteins (Pfam PF04678 and Panther PTHR1346 domains, with undefined function), further supporting the significance of the hit. We thus considered this protein a putative mitochondrial Ca2+ channel, named it MCU (“mitochondrial calcium uniporter”) and characterized it in detail. Tissue expression profiling by quantitative Real Time PCR showed ubiquitous presence in all investigated tissues (Fig. 1b), in agreement with a housekeeping role, with expression levels correlating with those of MICU1.
Figure 1. MCU includes two highly conserved transmembrane domains and is ubiquitously expressed in mammals, similarly to its putative regulator MICU1.
a, Alignment of the putative transmembrane domain and pore region of MCU proteins from 14 different species. b, Quantitative Real Time PCR analysis of mouse tissues. mRNA extraction and quantitative PCR as described in the methods section. Expression levels are normalized to skeletal muscle, and presented as means ± S.D. (n = 3).
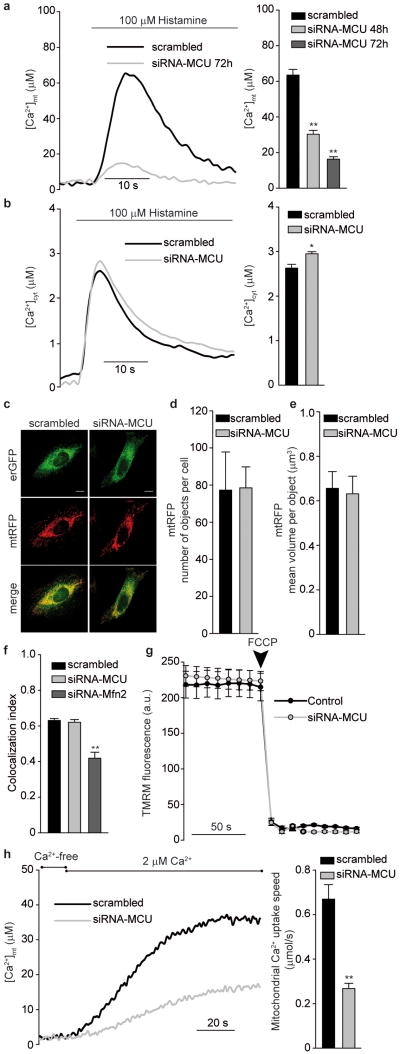
To verify the role of MCU in mitochondrial Ca2+ handling, siRNA oligonucleotides were synthesized and tested (Supplementary Fig. 1), Ca2+ measurements were then carried out with aequorin-based mitochondrial and cytosolic Ca2+ probes (mtAEQ and cytAEQ) (Fig. 2a and Supplementary Fig. 1). MCU-silenced and control HeLa cells, perfused with modified Krebs-Ringer buffer supplemented with 1 mM CaCl2 (KRB), were challenged, where indicated, with 100 μM histamine, an IP3-generating agonist causing Ca2+ release from the ER. In control cells, the [Ca2+]mt rise evoked by histamine stimulation was ∼63 μM (Fig. 2a). In MCU-silenced cells mitochondrial Ca2+ accumulation was drastically reduced (peak value ∼30 μM and ∼16 μM 48 h and 72 h after silencing, respectively). The drastic reduction was specific of mitochondria, as [Ca2+]cyt rises were almost unaffected (rather, a small but significant increase was detected, possibly due to reduced mitochondrial buffering) (Fig. 2b). Overexpression of MICU1 did dot rescue the [Ca2+]mt reduction in MCU-silenced cells, and had a marginal effect in control cells (Supplementary Fig. 2). Experiments were then carried out to exclude that the observed effect was secondary to changes in mitochondrial morphology (i.e. fragmentation, swelling or redistribution away from the ER) or in the driving force for Ca2+ accumulation (i.e. a collapse of mitochondrial membrane potential, ΔΨ). On the former aspect, mitochondria and ER were labeled in living cells by co-expressing mtRFP and erGFP (Fig. 2c and Supplementary Fig. 3). mtRFP imaging showed the typical three-dimensional interconnected network, with no difference in mitochondrial number and volume between control and MCU-silenced cells (Fig. 2d and e). Also the ER showed a similar distribution in control and MCU-silenced cells, and no difference was detected in the number of mtRFP/erGFP fluorescence overlaps, whereas in cells in which the ER/mitochondria tether mitofusin 2 (Mfn2)22 was silenced a marked reduction was observed (Fig. 2f and Supplementary Fig. 3). As to the driving force for Ca2+uptake, no difference was detected in the loading of the fluorescent dye tetramethylrhodamine methyl ester (TMRM) (Fig. 2g), thus ruling out a ΔΨ drop in MCU-silenced cells. In brief, mitochondria simply appear to have an intrinsically reduced ability of accumulating Ca2+. Direct evidence was obtained by permeabilizing MCU-silenced and control cells in intracellular buffer (IB), containing 100 μM EGTA (Ca2+-free) and then imposing an EGTA-buffered fixed [Ca2+] of 2 μM (2 μM Ca2+). Under those conditions, in MCU-silenced cells Ca2+ uptake was initiated at a speed that was three-fold lower than controls (Fig. 2h).
Figure 2. MCU silencing strongly inhibits mitochondrial Ca2+ uptake without causing morphological rearrangement or changes in the electrochemical gradient.
a, [Ca2+]mt and b, [Ca2+]cyt measurements in MCU-silenced cells. c, Fluorescence images of mtRFP- and erGFP-labeled mitochondria and ER, respectively. d and e, Mitochondrial number and volume, as deduced by calculating object number (d) and size (e). f, ER/mitochondria colocalization, estimated by Pearson's correlation coefficient. g, TMRM fluorescence measurements. h, [Ca2+]mt measurements in permeabilized cells. In this and following figures, experiments are representative of >5 trials, conditions are in the methods section, and statistics in Supplementary Table 2. * p<0.05, ** p<0.001
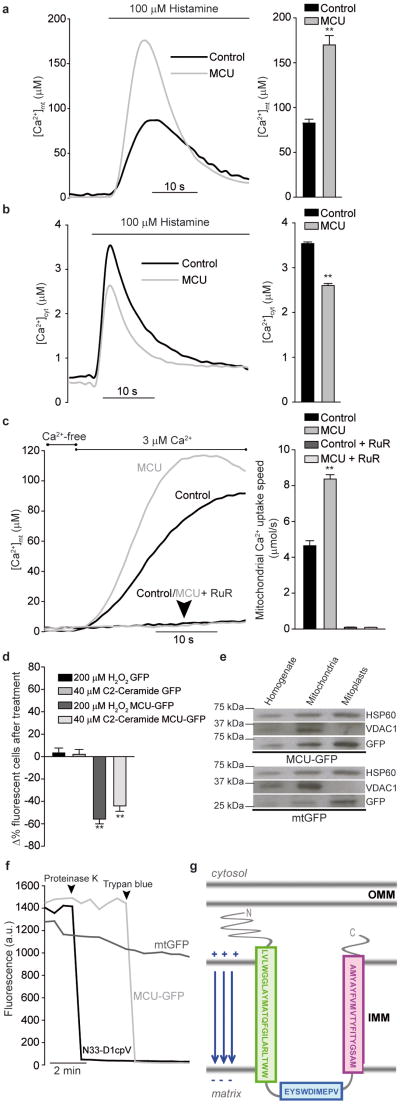
To demonstrate that MCU promotes Ca2+ uptake, overexpression experiments were carried out. The full-length cDNA of the protein, obtained by RT-PCR from skeletal muscle RNA, was identical to that deposited in the NCBI databank (NM_001033259). The cDNA was cloned in pcDNA3.1 and co-expressed with aequorin probes. MCU-overexpressing cells showed a dramatic increase in the histamine-evoked [Ca2+]mt rise (+ 106%, ∼169 μM in MCU-overexpressing vs. ∼82 μM in control cells, Fig. 3a). The greater mitochondrial response was not secondary to alterations of the cytosolic response. Rather, a significant reduction in the [Ca2+]cyt transient was observed, most likely due to increased Ca2+ clearance by mitochondria (Fig. 3b). Enhanced mitochondrial Ca2+ uptake was confirmed also upon capacitative Ca2+ influx (Supplementary Fig. 4) and in permeabilized cells (Fig. 3c). As in Fig. 2h, mitochondrial Ca2+ uptake was initiated by switching the medium, after digitonin permabilization, from IB/EGTA (Ca2+-free) to IB/Ca2+ (3 μM Ca2+). MCU-overexpressing cells showed a faster rate of Ca2+ uptake and reached a higher plateau level (Figure 3c). Finally, we verified whether the increased intrinsic ability of mitochondria to accumulate Ca2+ correlated with sensitivity to apoptotic challenges (Fig. 3d). Microscopy counts of cell viability after treatment with C2-ceramide or H2O2 showed that MCU-expressing cells were more efficiently killed, thus confirming the notion that mitochondrial Ca2+ loading synergizes with these apoptotic stimuli12,23.
Figure 3. MCU overexpression increases mitochondrial Ca2+ accumulation in intact and permeabilized cells, buffers cytosolic [Ca2+] rises, and sensitizes to apoptotic simuli; GFP-tagged MCU demonstrates mitochondrial localization and suggests a putative membrane topology.
a, [Ca2+]mt and b, [Ca2+]cyt measurements in MCU-overexpressing cells. c, [Ca2+]mt measurements in permeabilized cells. d, Cell viability upon apoptotic challenge. e, Subcellular fractionation of MCU-GFP- and mtGFP-expressing cells and Western blotting for GFP, matrix (HSP60) and OMM (VDAC1) markers. f, GFP proteinase K degradation and fluorescence quenching by Trypan blue in permeabilized cells. g, Schematic representation of the predicted MCU topology. RuR, Ruthenium Red. ** p<0.001.
We then investigated the subcellular distribution of MCU. For this purpose, tagged versions of MCU were generated (MCU-GFP and MCU-Flag), that had the same effect on [Ca2+]mt as MCU (Supplementary Fig. 5). GFP fluorescence and immunocytochemistry labeling with anti-Flag antibody completely overlapped with mitochondrial markers (Supplementary Fig. 6). Then, intra-mitochondrial distribution and topology was investigated. HeLa cells were transfected with MCU-GFP (or the matrix probe mtGFP) and harvested. The mitochondrial fraction was obtained by differential centrifugation and either blotted directly, or subjected to osmotic swelling, thus obtaining the mitoplast fraction devoid of the outer membrane. Both MCU and mtGFP were progressively more enriched in the mitochondrial and mitoplast fractions (Fig. 3e). Mitochondrial localization was also confirmed by immuno-EM of HeLa cells expressing MCU-Flag (Supplementary Fig. 7). To prove the inner membrane localization and get information on the topology of MCU, we carried out an experiment in digitonin-permeabilized cells, in which GFP fluorescence was visualized before and after treatment with proteinase K or Trypan blue, a fluorescence quencher that crosses the outer, but not the inner mitochondrial membrane9 (Fig. 3f). Proteinase K had no effect on MCU-GFP fluorescence (but abrogated fluorescence of the cytosolic, OMM-anchored N33D1cpV probe, confirming that MCU is not on the OMM), whereas Trypan blue abrogated MCU-GFP (but not mtGFP) fluorescence (Fig. 3f), thus demonstrating that MCU is in the IMM, with the C-terminus most likely located in the intermembrane space (Fig. 3g).
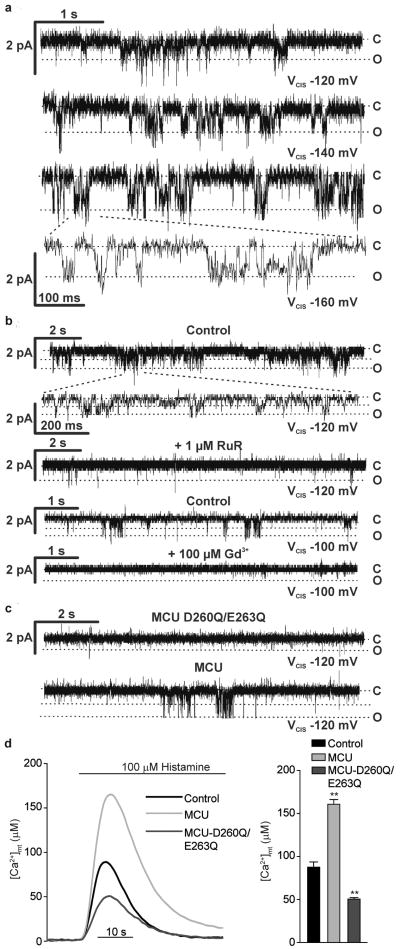
Finally, we investigated the channel activity of the purified protein reconstituted in a planar lipid bilayer. For this purpose, a His-tagged MCU was generated and expressed in two different heterologous expression systems, E. coli and wheat-germ cell-free transcription/translation. The purified protein (Supplementary Fig. 8) was inserted into lipid bilayers, and its electrophysiological activity was assessed in a medium containing only Ca2+ as cation. With both preparations, we measured channel activity, with the properties previously reported for the uniporter17,24: conductance of 6-7 pS (in the negative voltage range applied in the cis compartment), fast kinetics, low probability of opening at low voltages (Po), Po increase with voltage, and inhibition by Ruthenium Red and the lanthanide Gadolinium (Gd3+) (Fig. 4a and b and Supplementary Fig. 9). Then, to get evidence of cation permeation across the putative pore-forming domain, we mutated into glutamines two negatively charged residues of the conserved region (D260Q, E263Q) and assessed the electrophysiological properties of the mutant protein (MCUD260Q,E263Q) produced in vitro. MCUD260Q,E263Q failed to give rise to Ca2+-permeable channel activity in bilayer experiments, while in the same membrane the subsequent addition of MCU initiated channel activity with the characteristics described above (Fig. 4c). MCUD260Q,E263Q, when overexpressed in HeLa cells, showed the same membrane topology as MCU (Supplementary Fig. 10) and caused a marked reduction in histamine-induced [Ca2+]mt rises, both compared to MCU-overexpressing and control cells (Fig. 4d). MCUD260Q,E263Q thus acts as a dominant-negative mutant, most likely by either inhibiting MCU channel activity within an oligomer or competing for docking sites or essential regulators.
Figure 4. The purified MCU protein exhibits channel activity in lipid bilayers, with the properties of the uniporter; MCUD260Q,E263Q shows no channel activity and reduces [Ca2+]mt transients in cells.
a-c Electrophysiological recordings of purified MCU or MCUD260Q,E263Q, produced in E. coli (a and b) or in vitro (c) and reconstituted in planar lipid bilayers. a, MCU traces at different applied voltages; b, MCU traces before and after addition of RuR or Gd3+; c, MCUD260Q,E263Q traces. After 10 minutes recording with no current, MCU was added and channel activity was detected after 1-2 minutes (lower trace). d, [Ca2+]mt measurements in MCUD260Q,E263Q-expressing cells.
In conclusion, we have identified a 40 kDa protein, which fulfils the requirements for being the long searched mitochondrial calcium uniporter: it contains two transmembrane domains and exhibits channel activity in vitro with the previously characterized properties of the uniporter, it is localized in the inner membrane and, finally, it dramatically enhances mitochondrial Ca2+ uptake upon overexpression. The availability of molecular information opens the mitochondrial checkpoint in cellular Ca2+ signaling to a deeper understanding. Informative animal models can now be generated and new drugs developed to influence processes regulated by mitochondrial Ca2+ signals, such as aerobic metabolism and cell death.
Full methods (on-line only)
Bioinformatics screening
The MitoCarta database was screened for ubiquitously expressed proteins (expressed in at least 12 out of 14 tissues analyzed, obtaining 529 candidates) and proteins containing 2 or more TMD were selected by using TMHMM algorithm2. 89 out of 1098 proteins present in MitoCarta fulfilled these requirements. Sequences of these candidates were retrieved and aligned with the Saccaromyces cerevisiae proteome by a standard Gonnet matrix to exclude homologues. All alignments with a P value < 0.001 were excluded. 20 out of 89 remained and were subsequently aligned with kinetoplastids proteomes using the TriTrypDB server. 14 out of 20 showed significant scores. These candidates were manually aligned through different species to look for putative highly-conserved channel forming domains.
RNA extraction and Gene Expression Analyses
For the expression analysis of MCU and MICU1 in mouse tissues, adult male C57BL/6 mice (28-30 g) were used. Skeletal muscles (tibialis anterior), heart, brain, spleen, lung, liver, kidney and visceral fat were excised form 3 age-matched animals. Total RNA was purified using SV Total RNA Isolation kit (Promega) following manufacturer instructions. The RNA was quantified and controlled for its quality using the RNA 6000 LabChip kit (Agilent Technologies) in conjunction with an Agilent Bioanalyzer 2001. An equal amount of total RNA from the three animals was pooled together for each organ, complementary DNA was generated with a cDNA synthesis kit (SuperScript VILO; Invitrogen) and analyzed by quantitative Real Time PCR using the SYBR green chemistry (Bio-Rad). The primers were designed and analyzed with Primer3. Identity of the amplicons was confirmed by their dissociation profiles and gel analysis. Quantitative PCR standard curves were constructed by using serial dilutions of pooled cDNAs of the analyzed samples, using at least 4 dilution points and the efficiency of all primer sets was between 95 and 105%. Real-time was performed using Primer sequences were as follows:
MICU1-fw GTCGAACTCTCGGACCATGT
MICU1-rv CAAAGTCCCAGGCAGTTTCT
The primers amplify a fragment of 199 bp.
MCU-fw AAAGGAGCCAAAAAGTCACG
MCU-rv AACGGCGTGAGTTACAAACA
The primers amplify a fragment of 200 bp.
For the expression analysis of MCU silencing total RNA was purified from HeLa cells transfected with scrambled, siRNA-MCU#1 and siRNA-MCU#2 for 48 hours following the standard Trizol protocol. The RNA was quantified, controlled for its quality and retrotranscribed as described above. Complementary DNA was analyzed by quantitative Real Time PCR using the SYBR Green chemistry (Bio-Rad). All data were normalized to GAPDH expression. The oligonucleotide primers specific for MCU are the same as above. The oligonucleotide primers specific for GAPDH are the following:
GADPH-fw CACCATCTTCCAGGAGCGAG
GADPH-rv CCTTCTCCATGGTGGTGAAGAC
The primers amplify a fragment of 101 bp.
Constructs and siRNA
Mouse MCU (NM_001033259) was amplified from mouse skeletal muscle cDNA by PCR using the following primers:
- For the cloning in pEGFP-N1:
fw: 5′-GAATTCGCCACCATGGCGGCCGCCGCAGGTAG-3′
rv: 5′-GGATCCACTTCCTTTTCTCCGATCTGTCG-3′
The PCR fragment was cloned into EcoRI and BamHI sites in pEGFP-N1 (Clontech).
- For the cloning in pcDNA3.1:
fw: 5′-GGTACCGCCACCATGGCGGCCGCCGCAGGTAG-3′
rv: 5′-GAATTCTCATTCCTTTTCTCCGATCTGTC-3′
The PCR fragment was cloned into KpnI and EcoRI sites in pcDNA3.1 (Invitrogen).
- For the cloning of MCU-Flag in pcDNA3.1:
fw: 5′-GGTACCGCCACCATGGCGGCCGCCGCAGGTAG-3′
rv: 5′-GGAATTCTCACTTATCGTCGTCATCCTTGTAATCTTCCTTTTCTCCGATCTGTC-3′
The PCR fragment was cloned into KpnI and EcoRI sites in pcDNA3.1 (Invitrogen).
- For the cloning of MCU in pET-28A(+):
fw: 5′-AGGATCCATGGCGGCCGCCGCAGGTAG-3′
rv: 5′-ACTCGAGTCATTCCTTTTCTCCGATCT-3′
The PCR fragment was cloned into BamHI and XhoI sites in pET-28A(+) (Novagen).
- For the cloning of MCU deleted of the mitochondrial targeting sequence (aa 1-54) (MCUΔMTS) in pET-28A(+):
fw: 5′-CATATGGCTTCCTGGCAGAGCGTGGG-3′
rv: 5′-CTCGAGTCATTCCTTTTCTCCGATCT-3′
The PCR fragment was cloned into NdeI and XhoI sites in pET-28A(+) (Novagen).
- For the cloning of MCU in pIVEX 1.3 WG:
fw: 5′-ACATATGGCGGCCGCCGCAGGTAGATC-3′
rv: 5′-TCTCGAGTTCCTTTTCTCCGATCTGTC-3′
The PCR fragment was cloned into NdeI and XhoI sites in pIVEX 1.3 WG (Roche).
- The generation of the pcDNA3.1-MCUD260Q,E263Q-Flag and pEGFP-N1-MCUD260Q,E263Q mutant was performed by mutagenesis PCR using the wild type pcDNA3.1-MCU-Flag and pEGFP-N1-MCU vectors as template and the mutagenesis primer:
5′-CTGGTGGGAGTACTCGTGGCAAATCATGCAACCCGTCACCTACTTCATCAC-3′.
Mouse MICU1 (NM_144822) was amplified from mouse skeletal muscle cDNA by PCR using the following primers:
- For the cloning of MICU1-HA in pcDNA3.1:
fw: 5′-CGGATCCGCCACCATGTTTCGTCTTAACACCCT-3′
rv: 3′- GCTCGAGTCACAGGGAAGCGTAGTCAGGCACATCGTAGGGGTATTTGGGCAGAGCAAAGTCCC-5′
The PCR fragment was cloned into BamHI and XhoI sites in pcDNA3.1 (Invitrogen).
- For the cloning of MICU1 in pEGFP-N1:
fw: 5′-CCTCGAGATGTTTCGTCTTAACACCCT-3′
rv: 5′-CGGATCCCGTTTGGGCAGAGCAAAGTCCC-3′
The PCR fragment was cloned into XhoI and BamHI sites in pEGFP-N1 (Clontech).
- To silence MCU specific siRNA were designed:
siRNA-MCU#1: nucleotide 899-917 of the corresponding mRNA
5′- GCCAGAGACAGACAAUACUtt-3′
3′-ttCGGUCUCUGUCUGUUAUGA -5′
siRNA-MCU#2: nucleotide 360-378 of the corresponding mRNA
5′- GGGAAUUGACAGAGUUGCUtt-3′
3′-ttCCCUUAACUGUCUCAACGA -5′
The non-targeting siRNA (scrambled) is the following:
5′- GCCUAAGAACGACAAAUCAtt-3′
3′-ttCGGAUUCUUGCUGUUUAGU -5′
Cell culture and transfection
In all the experiments HeLa cells were used. Cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Euroclone), supplemented with 10% fetal bovine serum (FBS) (Euroclone) and transfected with a standard calcium-phosphate procedure. For aequorin measurements, the cells were seeded 24 hours before transfection onto 13 mm glass coverslips and allowed to grow to 50% confluence before transfection. For morphologic analyses cells were seeded 24 hours before transfection onto 24 mm glass coverslips and allowed to grow to 50% confluence before transfection, unless otherwise specified.
Aequorin measurements
HeLa cells grown on 13 mm round glass coverslips at 50% confluence were transfected with the cytosolic (cytAEQ) or the low-affinity mitochondrial (mtAEQmut, referred in the text as mtAEQ) probe (as previously described3) together with the indicated siRNA or plasmid. pcDNA3.1 was used as control unless otherwise indicated. The coverslip with the cells was incubated with 5 μM coelenterazine for 1-2 hours in KRB (Krebs-Ringer modified buffer: 125 mM NaCl, 5 mM KCl, 1 mM Na3PO4, 1 mM MgSO4, 5.5 mM glucose, 20 mM HEPES, pH 7.4, 37° C) supplemented with 1mM CaCl2, and then transferred to the perfusion chamber. All aequorin measurements were carried out in KRB. Agonists and other drugs were added to the same medium, as specified in the text. The experiments were terminated by lysing the cells with 100 μM digitonin in a hypotonic Ca2+-rich solution (10 mM CaCl2 in H2O), thus discharging the remaining aequorin pool. The light signal was collected and calibrated into [Ca2+] values by an algorithm based on the Ca2+ response curve of aequorin at physiological conditions of pH, [Mg2+] and ionic strength, as previously described3. Representative traces are shown in the figures whereas the full dataset is included in the Supplementary Table S2. In the experiments with permeabilized cells, a buffer mimicking the cytosolic ionic composition, (intracellular buffer [IB]) was employed: 130 mM KCl, 10 mM NaCl, 2 mM K2HPO4, 5mM succinid acid, 5 mM malic acid, 1 mM MgCl, 20 mM HEPES, 1 mM pyruvate, 0.5 mM ATP and 0.1 mM ADP (pH 7 at 37°C). IB was supplemented with either 100 μM EGTA (IB/EGTA) or a 2 mM EGTA and 2 mM HEEDTA-buffered [Ca2+] of 2 or 3 μM (IB/Ca2+), calculated with Chelator software4. HeLa cells were permeabilized by a 1 min perfusion with 50 μM digitonin (added to IB/EGTA) during luminescence measurements. Mitochondrial Ca2+ uptake speed was calculated as the 1st derivative by using the SLOPE excel function and smoothed for 3 time points. The higher value reached during Ca2+ addition represents the maximal Ca2+ uptake speed. [Ca2+]mt following capacitative Ca2+ influx was measured by perfusing HeLa cells with the SERCA blocker cyclopiazonic acid (CPA, 20 μM) in a KRB solution, containing no Ca2+ and 100 μM EGTA. In this protocol, mitochondrial Ca2+ uptake were evoked by adding 2 mM CaCl2 to the medium. All of the results are expressed as means ± S.E.M., and Student's t test was used for the statistic. All the materials were from Sigma Aldrich unless specified.
Mitochondrial and ER morphology analysis
Images for investigating mitochondrial morphology and ER/mitochondrial contact sites were taken on a Leica TCS-SP5-II equipped with a PlanApo 100x/1.4 N.A. objective. For all images, pinhole was set to 1 airy unit, pixel size was 75 nm and a Z-stack was acquired for the whole depth of the cell by sampling at 130 nm in the Z plane. 488 nm Ar-laser line was used to excite GFP or AlexaFluor488 and its signal collected in the 492-537 nm range, while RFP (and MitoTracker Red and AlexaFluor546) fluorescence was excited by the 543 nm HeNe laser and its emission was collected in the 555-700 nm range. For each image, PMT gain was slightly adjusted in order to maximize signal and avoid saturation. Images were then all analyzed with the Fiji image processing package based on ImageJ. Mitochondrial morphology was calculated with the 3D Object Counter plugin, while colocalization was evaluated with the JaCoP plugin5. The colocalization index is represented by Pearson's coefficient calculated following Costes randomization (100 cycles) and automatic threshold calculation6.
Immunofluorescence
HeLa cells were grown on 24 mm coverslips and transfected with MCU-Flag encoding plasmid when 50% confluent. After 24 hours, cells were washed with PBS, fixed in 4% formaldehyde for 10 minutes and quenched with 50 mM NH4Cl in PBS. Cells were permeabilized for 10 minutes with 0.1% Triton X-100 in PBS and blocked in PBS containing 2% BSA and 0.05% Triton X-100 for 1 hour. Cells were then incubated with primary antibodies (anti-HSP60 and anti-Flag) for 3 hours at room temperature and washed 3 times with 0.1% Triton X-100 in PBS. The appropriate isotype matched, AlexaFluor conjugated secondary antibodies (Invitrogen) were used and coverslips were mounted with ProLong Gold Antifade reagent (Invitrogen). Images were taken on a Leica SP5-II as detailed above.
Fractionation and Western blotting
108 HeLa cells were homogenized with a Dounce homogenizer (100 strokes) and centrifuged at 600×g to remove entire cells. The supernatant was then centrifuged at 8000×g to pellet the crude mitochondrial fraction. Mitoplasts were obtained by osmotic swelling by incubating mitochondrial fraction in 20 mM HEPES for 20 minutes. Proteins were quantified using the BCA Protein Assay Kit (Pierce) following the manufacturer instructions. 10 μg of protein were separated by SDS-PAGE, transferred onto nitrocellulose membranes (GE Healthcare) and probed using the following antibodies: anti-VDAC1(Abnova), anti-HSP60 (Santa Cruz) and anti-GFP (Abcam). Isotype matched, horseradish peroxidase conjugated secondary antibodies (BioRad) were used followed by detection by chemiluminescence (GE Healthcare).
For testing the siRNAs efficacy, HeLa cells were grown on 10 cm petri dishes and transfected when 30% confluent. 8 μg of MCU-Flag encoding plasmid was co-transfected with scrambled, siRNA-MCU#1 or siRNA-MCU#2. After 72h cells were washed, harvested and lysed in RIPA buffer (150 mM NaCl, 25 mM TRIS, 1% Triton-X100, 0.5% Na-deoxycholate and 0.1% SDS, pH = 8) added with proteases and phosphatases inhibitor cocktails (Roche). Proteins were quantified with BCA method and 25 μg of each sample were loaded on a Novex NuPage Bis-Tris 4-12% precast gel (Invitrogen), transferred onto nitrocellulose membranes and probed with an anti-β– Tubulin (Sigma) and anti-Flag (Sigma) antibodies. Isotype matched, horseradish peroxidase conjugated secondary antibodies (BioRad) were used followed by detection by chemiluminescence (GE Healthcare).
Measurements of mitochondrial ΔΨ
Mitochondrial ΔΨ was measured by loading cells with 20 nM tetramethyl rhodamine methyl ester (TMRM, Invitrogen) for 30 minutes at 37°C. Images were taken on a inverted microscope (Zeiss Axiovert 100) equipped with a PlanFluar 40×/1.3 N.A. objective, a Photometrics Coolsnap ES and a LED-based illumination system (Cairn OptoLed II). TMRM excitation was performed at 560 nm and emission was collected through a 590-650 nm bandpass filter. Images were taken every 10 seconds with a fixed 200 ms exposure time. 10 μM FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone), an uncoupler of oxidative phosphorylation, was added after 12 acquisitions to completely collapse the electrical gradient established by the respiratory chain (ΔΨ).
Trypan blue quenching experiment
After transfection with mtGFP, MCU-GFP or N33D1cpV, cells were analyzed on spinning disk imaging system composed of an inverted microscope (Zeiss Axiovert 200) equipped with a PlanApo 63×/1.4 N.A. objective, a BD-CARVII confocal head, a Photometrics Cascade 512B camera, a piezoelectric Z motor (Phisik instruments) and a 300 watt Xenon bulb (Sutter Instruments). At each time point, a Z-stack was taken (z-step: 500 nm) using standard EGFP filterset (Semrock GFP-A-Basic-000) and the best focus plane was selected for subsequent analysis. After 1 minute permeabilization in IB containing 50 μM digitonin, cells were incubated in 1 ml of IB, first treated with 4 units of proteinase K and then with 0.005% trypan blue. Best focus images of each time point were background corrected and mitochondrial mean fluorescence intensity was measured with MetaMorph software (Molecular devices).
Cell death experiment
Cell sensitivity to apoptotic stimuli was evaluated as previously described7. HeLa cells grown on 24 mm round glass coverslips at 30% confluence were transfected with MCU-GFP or GFP alone. The effect on cell fate was evaluated by applying an apoptotic challenge (40 μM C2-ceramide or 200 μM H2O2) and comparing the survival of transfected and non-transfected cells. In these experiments, the percentage of GFP-positive cells was calculated before and after applying an apoptotic stimulus (C2-ceramide or H2O2). In mock-transfected cells, although the total number of cells is reduced after cell death induction, the apparent transfection efficiency was maintained (i.e. transfected and non-transfected cells have the same sensitivity to the apoptotic stimulus and thus die to the same extent). However, when cells are transfected with a construct influencing their sensitivity to apoptosis, this will be reflected by a change in the fraction of fluorescent cells, i.e. in the “apparent” transfection efficiency. Thus, protection from apoptosis results into an apparent increase of transfection, whereas a decrease reflects a higher sensitivity to apoptosis. Data are reported as the mean percentage change in the apparent transfection efficiency after apoptotic challenge compared to vehicle-treated cells. Cells were extensively washed with PBS, stained with DAPI and two images per field (blue and green fluorescence) were taken at 20× magnification (mean transfection efficiency were roughly 30% for both GFP and MCU-GFP). At least 10 fields per coverslip were randomly imaged and counted. Data presented are the sum of at least two different coverslip per experimental condition carried out in three different independent experiments.
Immunogold electron microscopy
HeLa cells were transfected with control (pcDNA3.1) or MCU-Flag plasmids. 24 hours after transfection cells were fixed in 4% formaldehyde and 0.1% glutaraldehyde in 0.1M cacodylate buffer. Dehydratation was carried out by subsequent incubation with increasing ethanol concentrations (20, 40, 60, 80 and 100%). Cells were then infiltrated and embedded in LRW resin and 100 nm slices were obtained. For immunogold staining slices were first blocked in PBS containing 2% BSA and 0.05% Triton X-100 and then incubated for 1 hour with anti-Flag (Sigma) antibody. After 5 washes with PBS, slices were incubated for 1 hour with an isotype matched 10 nm gold particles conjugated secondary antibody (Sigma). After 5 washes with PBS and 1 wash with H2O, slices were contrasted with uranyl acetate for 15 minutes and lead citrate for 6 minutes. Slices observation and imaging were carried out on a Tecnai G2 TEM.
Protein expression and purification
E.coli
MCU and MCUΔMTS were cloned into pET-28A(+) vector, as described above. Competent cells were transformed with the indicated constructs. Induction was performed at 20°C (O.D.600 0.4) with 0.35 mM IPTG for 24 hours. Following sonication in Buffer A (50 mM Na-phosphate, pH 7.4, 300 mM NaCl + protease inhibitor cocktail), membraneous fraction was collected by centrifugation (10 min 12000×g) and solubilized with Decy-β-D-maltopyranoside (2.5%) for 3 hours at room temperature. The non-solubilized material was removed by centrifugation and the supernatant was loaded on a Nickel chromatography column (HIS-Select Nickel affinity gel, Sigma). Column was washed with buffer A and the protein was eluted with imidazole gradient (50-200 mM). Fractions of 500 μl were collected and dialyzed against buffer A without NaCl. A concentration of 0.025% of detergent was maintained throughout the purification.
In vitro
MCU was cloned into pIVEX 1.3 WG, as described above. In vitro expression was performed by using RTS™100 Wheat Germ CECF Kit (Roche). After expression, the reaction mix was solubilized with 2 % Triton X100 for 90 minutes at 30° C under shaking.
Gel electrophoresis
SDS-PAGE was perfomed using 6 M urea and standard protocols. 30 μl of eluted fractions and 1 μl of the reaction mix/lane were loaded.
Electrophysiology
Electrophysiology experiments were carried out as previously described8-9. A Warner Instruments (Hamden, CT, USA) electrophysiological planar bilayer apparatus was used. Bilayers of approximately 150-200 pF capacity were prepared using azolectin in decane containing 1% chloroform (Sigma, St. Louis, Mo.) across a 250 μm hole in a polystirene cuvette. Azolectin was partially purified by precipitation with cold acetone from a chloroform solution. The inside of the cuvette constituted the trans compartment. The standard experimental medium was 100 mM CaCl2 (or Ca-gluconate), 10 mM HEPES/pH 7.2. The lipid membrane was built under symmetric ionic conditions and both chambers contained 3 ml of solution. The contents of both chambers were stirred by magnetic bars when desired. Connections to the electrodes were provided by agar bridges. Purified proteins were added to the cis side. Control experiments with empty membrane or with detergents used for the purification showed no activity. All voltages reported are those of the cis chamber, zero being assigned by convention to the trans (grounded) side. Currents are considered as positive when carried by cations flowing from the cis to the trans compartment. A BC-525C unit and headstage were used to control parameters and amplify signals. Output was recorded with a 10 kHz bandwidth on videotape using a Medical Systems (New York) PCM-2 interface. Data were acquired at 2 kHz, filtered at 500 Hz and analyzed offline using the pClamp program set (Axon Instruments, Union City, CA, USA). Conductance was determined by averaging the measured amplitudes of single channel events (n ≥ 50) at various applied voltages.
Statistical analysis of data
Statistical data are presented as mean ± S.E.M., unless otherwise speficied. Significance was calculated by Student's t test, and correlation analysis was performed with the SigmaPlot 11.0 software (Systat Software Inc.).
Supplementary Material
Acknowledgments
We thank M. Zoratti for help in the analysis of electrophysiological data, G. Merli for carrying out some of the experiments and P. Bernardi, T. Pozzan and L. Cendron for helpful discussions and for the N33D1cpV expression plasmid (T.P.). This research was supported by grants from the Italian Ministry of Education, University and Research, European Commission (FP7 “MyoAGE”, no. 223576), NIH (Grant #1P01AG025532-01A1), Cariparo Foundation (Padua), the Italian Association for Cancer Research (AIRC) and Telethon-Italy (GPP1005).
Footnotes
Author Contributions. D.D.S. performed bioinformatic analysis, Ca2+ measurements and morphological analysis of organelles. A.R. performed molecular biology and gene expression analysis. D.D.S. and A.R. contributed equally to the study. E.T. expressed and purified the protein in heterologous systems; I.S. performed and analyzed the electrophysiology experiments; R.R. discussed the results and wrote the paper.
Author Information. The authors declare no conflict of interest.
References
- 1.Szabadkai G, Duchen MR. Mitochondria: the hub of cellular Ca2+ signaling. Physiology (Bethesda) 2008;23:84–94. doi: 10.1152/physiol.00046.2007. [DOI] [PubMed] [Google Scholar]
- 2.Carafoli E. Historical review: mitochondria and calcium: ups and downs of an unusual relationship. Trends Biochem Sci. 2003;28:175–181. doi: 10.1016/S0968-0004(03)00053-7. [DOI] [PubMed] [Google Scholar]
- 3.Perocchi F, et al. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature. 2010;467:291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 5.Rizzuto R, Brini M, Murgia M, Pozzan T. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science. 1993;262:744–747. doi: 10.1126/science.8235595. [DOI] [PubMed] [Google Scholar]
- 6.Rizzuto R, et al. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 7.Csordas G, Thomas AP, Hajnoczky G. Quasi-synaptic calcium signal transmission between endoplasmic reticulum and mitochondria. EMBO J. 1999;18:96–108. doi: 10.1093/emboj/18.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Csordas G, et al. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol Cell. 2010;39:121–132. doi: 10.1016/j.molcel.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giacomello M, et al. Ca2+ hot spots on the mitochondrial surface are generated by Ca2+ mobilization from stores, but not by activation of store-operated Ca2+ channels. Mol Cell. 2010;38:280–290. doi: 10.1016/j.molcel.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Hajnoczky G, Robb-Gaspers LD, Seitz MB, Thomas AP. Decoding of cytosolic calcium oscillations in the mitochondria. Cell. 1995;82:415–424. doi: 10.1016/0092-8674(95)90430-1. [DOI] [PubMed] [Google Scholar]
- 11.Jouaville LS, Pinton P, Bastianutto C, Rutter GA, Rizzuto R. Regulation of mitochondrial ATP synthesis by calcium: evidence for a long-term metabolic priming. Proc Natl Acad Sci U S A. 1999;96:13807–13812. doi: 10.1073/pnas.96.24.13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinton P, et al. The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide-induced apoptosis: significance for the molecular mechanism of Bcl-2 action. EMBO J. 2001;20:2690–2701. doi: 10.1093/emboj/20.11.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pacher P, Hajnoczky G. Propagation of the apoptotic signal by mitochondrial waves. EMBO J. 2001;20:4107–4121. doi: 10.1093/emboj/20.15.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hajnoczky G, Hager R, Thomas AP. Mitochondria suppress local feedback activation of inositol 1,4, 5-trisphosphate receptors by Ca2+ J Biol Chem. 1999;274:14157–14162. doi: 10.1074/jbc.274.20.14157. [DOI] [PubMed] [Google Scholar]
- 15.Boitier E, Rea R, Duchen MR. Mitochondria exert a negative feedback on the propagation of intracellular Ca2+ waves in rat cortical astrocytes. J Cell Biol. 1999;145:795–808. doi: 10.1083/jcb.145.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tinel H, et al. Active mitochondria surrounding the pancreatic acinar granule region prevent spreading of inositol trisphosphate-evoked local cytosolic Ca(2+) signals. EMBO J. 1999;18:4999–5008. doi: 10.1093/emboj/18.18.4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirichok Y, Krapivinsky G, Clapham DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427:360–364. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- 18.Nicholls DG. Mitochondria and calcium signaling. Cell Calcium. 2005;38:311–317. doi: 10.1016/j.ceca.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Uribe S, Rangel P, Pardo JP. Interactions of calcium with yeast mitochondria. Cell Calcium. 1992;13:211–217. doi: 10.1016/0143-4160(92)90009-h. [DOI] [PubMed] [Google Scholar]
- 20.Xiong ZH, Ridgley EL, Enis D, Olness F, Ruben L. Selective transfer of calcium from an acidic compartment to the mitochondrion of Trypanosoma brucei. Measurements with targeted aequorins. J Biol Chem. 1997;272:31022–31028. doi: 10.1074/jbc.272.49.31022. [DOI] [PubMed] [Google Scholar]
- 21.Benaim G, Bermudez R, Urbina JA. Ca2+ transport in isolated mitochondrial vesicles from Leishmania braziliensis promastigotes. Mol Biochem Parasitol. 1990;39:61–68. doi: 10.1016/0166-6851(90)90008-a. [DOI] [PubMed] [Google Scholar]
- 22.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 23.Hajnoczky G, Csordas G, Madesh M, Pacher P. Control of apoptosis by IP(3) and ryanodine receptor driven calcium signals. Cell Calcium. 2000;28:349–363. doi: 10.1054/ceca.2000.0169. [DOI] [PubMed] [Google Scholar]
- 24.Michels G, et al. Regulation of the human cardiac mitochondrial Ca2+ uptake by 2 different voltage-gated Ca2+ channels. Circulation. 2009;119:2435–2443. doi: 10.1161/CIRCULATIONAHA.108.835389. [DOI] [PubMed] [Google Scholar]
- 25.Pinton P, Rimessi A, Romagnoli A, Prandini A, Rizzuto R. Biosensors for the detection of calcium and pH. Methods Cell Biol. 2007;80:297–325. doi: 10.1016/S0091-679X(06)80015-4. [DOI] [PubMed] [Google Scholar]
- 26.Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- 1.Kirichok Y, Krapivinsky G, Clapham DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427:360–364. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- 2.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 3.Pinton P, Rimessi A, Romagnoli A, Prandini A, Rizzuto R. Biosensors for the detection of calcium and pH. Methods Cell Biol. 2007;80:297–325. doi: 10.1016/S0091-679X(06)80015-4. [DOI] [PubMed] [Google Scholar]
- 4.Schoenmakers TJ, Visser GJ, Flik G, Theuvenet AP. CHELATOR: an improved method for computing metal ion concentrations in physiological solutions. Biotechniques. 1992;12:870–874. 876–879. [PubMed] [Google Scholar]
- 5.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 6.Costes SV, et al. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinton P, et al. The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide-induced apoptosis: significance for the molecular mechanism of Bcl-2 action. EMBO J. 2001;20:2690–2701. doi: 10.1093/emboj/20.11.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szabo I, Soddemann M, Leanza L, Zoratti M, Gulbins E. Single-point mutations of a lysine residue change function of Bax and Bcl-xL expressed in Bax- and Bak-less mouse embryonic fibroblasts: novel insights into the molecular mechanisms of Bax-induced apoptosis. Cell Death Differ. 2011;18:427–438. doi: 10.1038/cdd.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teardo E, et al. Characterization of a plant glutamate receptor activity. Cell Physiol Biochem. 2010;26:253–262. doi: 10.1159/000320525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.