Abstract
Insects use taste to evaluate food, hosts, and mates. Drosophila has many “orphan” taste neurons that express no known taste receptors. The Ionotropic Receptor (IR) superfamily is best known for its role in olfaction, but virtually nothing is known about a clade of ~35 members, the IR20a clade. Here, a comprehensive analysis of this clade reveals expression in all taste organs of the fly. Some members are expressed in orphan taste neurons, whereas others are coexpressed with bitter- or sugar-sensing Gustatory receptor (Gr) genes. Analysis of the closely related IR52c and IR52d genes reveals signatures of adaptive evolution, roles in male mating behavior, and sexually dimorphic expression in neurons of the male foreleg, which contacts females during courtship. These neurons are activated by conspecific females and contact a neural circuit for sexual behavior. Together, these results greatly expand the repertoire of candidate taste and pheromone receptors in the fly.
Introduction
Taste, or contact chemosensation, refers to the detection of chemicals through direct contact with a solid or liquid substrate. Insects rely on taste to detect a remarkable diversity of compounds, thus allowing them to evaluate potential nutrients, toxins, egg-laying sites, mates, pathogens, prey and predators (Blum, 1996; Liman et al., 2014). The far-ranging roles of taste in insect ecology raise possibilities for translating new understanding of taste into new means of controlling insect agricultural pests and vectors of disease (van der Goes van Naters and Carlson, 2006).
The fruit fly Drosophila melanogaster contains taste neurons in its mouthparts, legs, pharynx and wings (Stocker, 1994). These neurons are found in sensilla, which are cuticular compartments with a single pore for tastants to enter. Physiological recordings from taste sensilla in Drosophila and other insects have revealed responses of taste neurons to sugars, salts, bitter compounds, water and a large diversity of other tastants (Liman et al., 2014).
Much remains to be learned about the molecular underpinnings of these taste responses. A variety of candidate receptors have been identified in taste neurons, ranging from Gustatory receptors (Grs) and Pickpocket/Degenerin-Epithelial Sodium Channels (Ppk/DEG-ENaCs) to Transient receptor potential (Trp) channels (Liman et al., 2014; Lu et al., 2012; Thistle et al., 2012; Toda et al., 2012). More recently, a few members of the Ionotropic Receptor (IR) superfamily - which was initially found by virtue of expression in olfactory neurons – were found to be expressed in taste neurons (Benton et al., 2009; Croset et al., 2010; Zhang et al., 2013). Nonetheless, receptors of these diverse classes have been mapped to only a fraction of taste neurons in the fly. There remain many taste neurons to which no known taste receptors have been assigned and which can be considered orphan taste neurons.
Here we show that a virtually unexamined clade of ~35 genes within the IR superfamily in D. melanogaster is expressed in taste neurons. We examine their expression by analyzing GAL4 drivers representing nearly all members of the clade. Collectively, they are expressed in almost all taste sensilla of the fly. Two of these genes, IR52c and IR52d, are coexpressed in sensilla of the male foreleg, which makes contact with females during sexual behavior. These genes are expressed in sexually dimorphic taste neurons in which no other known taste receptors are expressed. The genes show signatures of adaptive evolution, and genetic analysis reveals roles in male sexual behavior. The neurons in which they are expressed are activated by exposure to D. melanogaster females, but not to females of a sibling species or to males. These neurons send projections to the CNS that form potential synaptic contacts with fruitless+ neurons, which act in sexual behavior. Taken together, this work substantially expands the inventory of candidate taste receptors in Drosophila. It also identifies receptors and neurons likely to act in the detection of mating partners in the fly.
Results
A large clade of candidate chemoreceptors expressed in taste organs
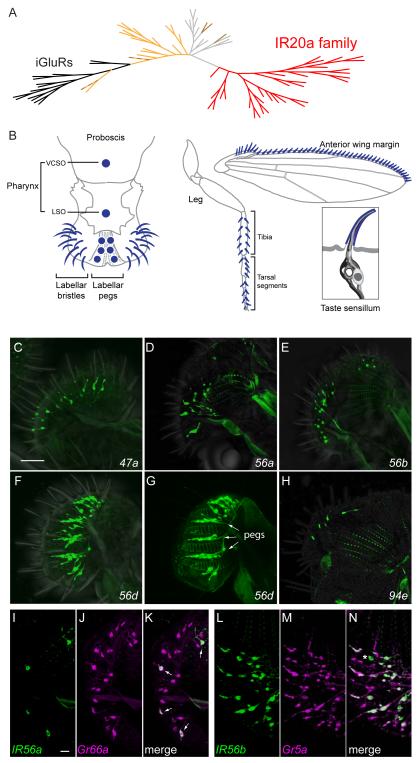
In an in silico screen for novel chemoreceptors in D. melanogaster, a screen that yielded Odorant receptor (Or) and Gustatory receptor (Gr) genes (Clyne et al., 2000; Clyne et al., 1999), we also identified another candidate chemoreceptor gene that belonged to a group of ~35 related genes. Initial RT-PCR experiments with several of these genes suggested they were expressed in taste neurons: transcripts were detected in the proboscis and legs, which are taste organs, but were absent in Poxn70 mutants that lack taste neurons (Figure S1A). We initiated a systematic analysis of the expression of nearly all of the 35 genes by constructing GAL4 transcriptional reporters for them, and we detected their expression in taste neurons as detailed below. During this analysis, others identified a superfamily of ~60 genes designated Ionotropic Receptor genes (IRs) because of their distant homology to ionotropic glutamate receptor genes (iGluRs); 17 of the genes were expressed in the antenna, an olfactory organ (Benton et al., 2009; Croset et al., 2010). Since the 35 genes that we identified form a previously uncharacterized clade within the IR superfamily, we refer to them here as the IR20a clade, after the member with the numerically lowest cytological position (Figure 1A).
Figure 1.
The IR20a clade and members that are expressed in the labellum.
(A) Phylogenetic tree of IRs and iGluRs. The IR20a clade is represented in red and the iGluRs in black. IRs that are expressed in antenna are indicated in light orange (Silbering et al., 2011). Brown branches indicate IRs that have been found to be expressed only in taste sensilla; dashed brown branches indicate IRs expressed in both taste and olfactory sensilla (Croset et al., 2010; Zhang et al., 2013). The remaining IRs are represented in light gray. The alignment includes 33 IR20a clade members; IR56e and IR60f were omitted because they are short truncated polypeptides. See also Figure S1.
(B) Taste sensilla (dark blue) distributed in the pharynx (VCSO and LSO), labellum, leg and anterior wing margin. In addition, a typical taste sensillum is shown here with four different types of taste neurons (support cells are omitted for simplicity). Adapted from elsewhere (Dahanukar et al., 2007; Yarmolinsky et al., 2009).
(C-H) IR-GAL4 drivers that label cells in taste bristles (C-F, H, GFP) and taste pegs (G, GFP, arrows) on the labellum. (G) Labellum visualized at the level of the taste pegs, which lie between the pseudotrachea (autofluorescent ladder-like structures).
(I-K) IR56a-GAL4 (green, GFP) is expressed in a subset of Gr66a-RFP-expressing neurons (magenta) in the labellum. Small arrows indicate co-expression of both reporters.
(L-N) Almost all IR56b-GAL4-labeled (green, GFP) neurons are co-labeled by Gr5a-LexA (magenta, tandem-Tomato), except for a small proportion that is only labeled by IR56b-GAL4 (*). Images are z-projections of confocal images. Scale bars represent 40 μm in C-H and 10 μm in I-N. See also Table S1 and Figure S2.
The IR20a clade genes are more distantly related to iGluRs than the rest of the IR superfamily (Figure 1A). Of 35 IR20a clade genes, seven contain premature stop codons or deletions and may be pseudogenes. The remaining 28 genes encode proteins with three predicted transmembranes domains, as do other IRs and iGluRs. Predicted proteins of the IR20a clade are highly divergent: their mean sequence identity is 16.3% + 0.3%, consistent with the high levels of divergence observed in chemoreceptor families (Croset et al., 2010). Comparison of members of the IR20a clade with the IRs expressed in the antenna and the remaining IRs suggests that each of the three groups of IRs harbor sequence motifs that are unique to each group, mainly within the predicted ligand-binding domains (Figure S1B and legend)(Benton et al., 2009). These sequence relationships are consistent with the possibility that IRs of different clades bind different classes of ligands.
In our systematic GAL4 expression analysis, we examined 30 of the genes: all 28 genes encoding presumptive full-length proteins and 2 putative pseudogenes (Table S1). To maximize the fidelity of these transcriptional reporters, we fused both the 5′ and 3′ flanking regions of the IR20a genes to the 5′ and 3′ ends of GAL4, respectively, in most cases (the average construct size, excluding GAL4 sequences, was 8.3±0.6 kb). Using these GAL4 constructs to drive GFP expression, we examined expression in the labellum, legs, pharynx, and anterior wing margin, regions that contain numerous taste sensilla (Figure 1B).
The mouthparts of the fly include two labellar lobes, whose lateral and medial surfaces contain taste bristles and taste pegs, respectively (Falk et al., 1976). GAL4 drivers representing five of the genes, IR47a, IR56a, IR56b, IR56d, and IR94e, show expression in the labellar bristles, and IR56d-GAL4 is also expressed in taste pegs (Figure 1C-H). Labellar bristles have stereotyped positions and electrophysiological response profiles towards tastants (Figure S2A)(Weiss et al., 2011), and we identified the sensilla expressing these GAL4 drivers. The five drivers are expressed in different subsets of sensilla (Figure, S2B). IR56a-GAL4 was distinguished from the other drivers in that it is expressed in sensilla that contain bitter neurons but is excluded from L-type sensilla, which do not. We therefore carried out a double-label experiment with Gr66a-RFP, a marker of bitter neurons in the labellum. We found that IR56a-GAL4 is in fact expressed in a subset of Gr66a-RFP+ cells (Figure 1 I-K). By contrast, many labellar neurons that express IR56b-GAL4 and IR56d-GAL4 also express Gr5a-LexA, a marker of sugar neurons (Figure 1 L-N and Figures S2 C-E). These data suggest that IR56a functions in bitter neurons, and IR56b and IR56d function in sugar neurons; one possibility is that they detect or modulate the detection of aversive and attractive compounds, respectively.
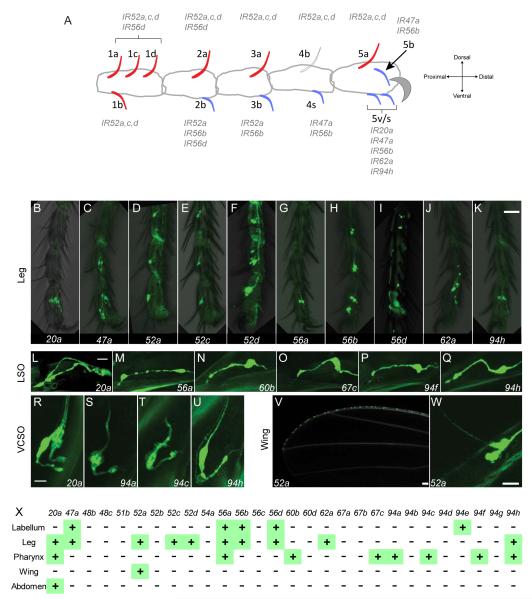
Legs contain taste sensilla on the tibia and, distal to it, on five tarsal segments. GAL4 drivers representing 10 of the genes are expressed in subsets of these leg sensilla. Two drivers, IR52a-GAL4 and IR56d-GAL4, are expressed in sensilla of both tibia and tarsal segments, while eight are expressed exclusively in tarsi (Figure 2A-K). We mapped the IR20a clade genes to specific sensilla on the tarsal segments and found that seven of the 10 are expressed in sensilla that respond to food components such as sugars, salts and bitter compounds (Figure 2A and S2F, G)(Ling et al., 2014). Accordingly, these sensilla (2b, 3b, 4s, 5v/s and 5b) also express Gr genes that are associated with the detection of both sugars and bitter compounds (Ling et al., 2014). These six sensilla are largely ventrally-oriented and likely make contact with underlying food sources.
Figure 2.
IR20a clade members are expressed in the legs, pharynx and wing margin.
(A) Spatially stereotyped taste sensilla on the female foreleg and the drivers expressed in them. Blue sensilla respond to food-related tastants, such as sugars, salt and bitter compounds; red sensilla do not (Ling et al., 2014; Meunier et al., 2000). Sensillum 4b is not colored due to variation in its response to sugars. Sensilla 5v and 5s are morphologically similar and adjacent to each other; as the present analysis does not provide sufficient resolution to distinguish these two sensilla, we have assigned the drivers expressed in either one to “5v/s”. Male forelegs contain additional sensilla close to 1a-d, 2a, 3a, and 4b. Midlegs and hindlegs are sexually monomorphic.
(B-K) Drivers that label cells in taste sensilla in legs.
(L-U) Drivers that label cells in the LSO 7th sensillum (L-Q) and VCSO (R-U) along the pharynx (Gendre et al., 2004).
(V-W) IR52a-GAL4 labels cells on the wing margin.
(X) Summary of expression in taste organs. Not shown are 7 putative pseudogenes. Scale bars represent 30 μm in B-K, 10 μm in L-Q, 10 μm in R-U, 50 μm in V, and 20 μm in W. See also Figure S2.
Intriguingly, GAL4 drivers representing IR52a, IR52c, IR52d, and IR56d are expressed in leg sensilla that have neither been found to respond to food components nor to express Gr genes (Ling et al., 2014). Most of these sensilla are dorsally-oriented and are less likely to make contact with food sources. Among these genes, IR52c and IR52d are expressed in forelegs but not midlegs or hindlegs, whereas IR52a is expressed in all legs, as indicated by both GAL4 and RT-PCR analysis (Figure S2G, K). The restriction of expression to forelegs is of special interest because male forelegs play a role in courtship behavior towards females and in the prevention of courtship behavior toward other species (Cook, 1977; Fan et al., 2013).
The pharynx contains taste sensilla that are likely to detect compounds in partially digested food (Miller, 1950). We found expression of eight GAL4 drivers in the pharynx (Figure 2L-U): Six are expressed in the labral sense organ (LSO, Figure 1B and 2 L-Q), and four are expressed in the ventral cibarial sense organ (VCSO, Figure 1B and 2R-U). Of the eight pharyngeal drivers, five are expressed in the pharynx but not in other taste tissues examined (Figure 2X). The expression of some of these drivers is strikingly sparse: IR60b-, IR67c-, IR94a- and IR94f-GAL4 each labeled only two neurons in the entire fly, both pharyngeal neurons; while such sparse expression suggests specialized functions (Joseph and Heberlein, 2012), we acknowledge the possibility of expression in other cells at levels below our detection limits.
The anterior wing margin contains curved taste bristles whose response profiles have received little attention and whose behavioral role has been enigmatic (Stocker, 1994). We found expression of IR52a-GAL4 in many of these taste sensilla (Figure 2V, W). The expression was observed in neurons that extend dendrites into these bristles, as is characteristic of taste neurons but not mechanosensory neurons. Some neurons of the wing margin have been found previously to express pickpocket25 (ppk25), a DEG/ENaC channel. Double-label analysis showed that ppk25-GAL4 and IR52a-LEXA are expressed in distinct populations of cells on the wing margin, suggesting that IR52a is expressed in orphan neurons of the wing (Figure S2L-N).
Finally, a driver representing IR20a is expressed in multi-dendritic neurons of the abdominal wall, in addition to its expression in legs and the pharynx (Figure S2O). Several Gr-GAL4 drivers have been shown to be expressed in similar neurons, although no taste function has been ascribed to them (Park and Kwon, 2011).
In summary, we have provided evidence that 16 members of the IR20a clade are expressed in adult taste sensilla: five in labellar sensilla, ten in leg sensilla, eight in pharyngeal sensilla, and one in wing sensilla (Figure 2X). While we have verified the expression of several IRs with RT-PCR (Figure S1A), we have been unable to detect them by in situ hybridization, presumably because their expression levels are below our detection threshold (not shown; see also (Benton et al., 2009)). Drivers of many of these genes are expressed in multiple organs, while other drivers are expressed sparsely in a single organ. Some taste sensilla express multiple IR-GAL4 drivers and some express drivers of both IR and Gr genes. IR52a, IR52c, IR52d, and IR56d are expressed in leg sensilla that neither respond to food-related tastants nor express Gr genes.
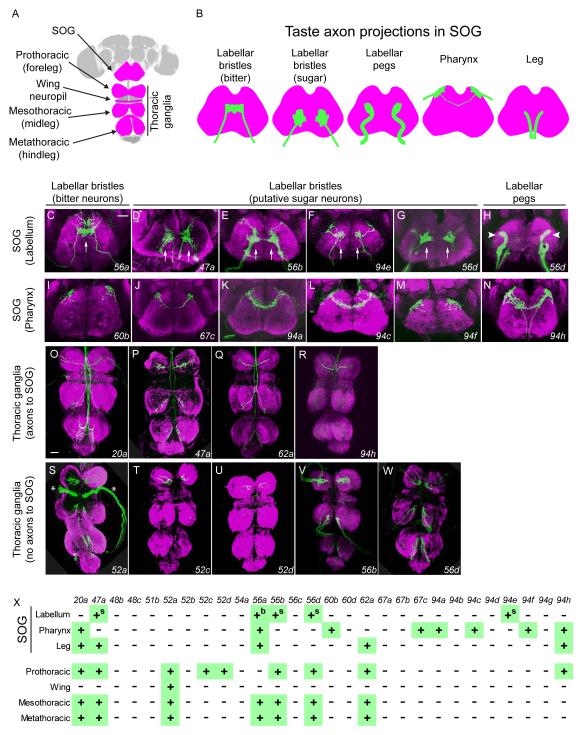
IR20a-clade-expressing neurons send axons to taste centers in the CNS
We next asked whether the IR-GAL4-expressing cells send projections into taste centers in the CNS. Projections of taste neurons in the CNS depend on the location of the tissue from which they originate and the taste quality that they subserve, e.g. bitter or sweet (Figure 3A, B)(Boll and Noll, 2002; Inoshita and Tanimura, 2006; Wang et al., 2004). Taste neurons from the labellum and pharynx send axons to the suboesophageal ganglion (SOG)(Figure 3B). Taste neurons from the legs and wing margin project to specific thoracic ganglia in the ventral nerve cord (Figure 3A). A subset of taste neurons in the legs also sends axon terminals to the posterior SOG after passing through the thoracic ganglia (Figure 3B).
Figure 3.
IR20a clade GAL4 drivers label axons that target taste centers in the CNS.
(A) CNS of Drosophila, with regions contacted by taste axons highlighted in magenta. The SOG receives projections from the labellum, pharynx, and legs. The pro-, meso-, metathoracic and wing ganglia receive projections from the forelegs, midlegs, hindlegs and wing margins, respectively.
(B) Axonal projections (green) of taste neurons in the SOG. Axons lie in different planes along the z-axis: axons from labellar pegs and pharyngeal sensilla are anterior; axons from labellar bristles are in middle; axons from legs are posterior. Axons from the pharynx cross the midline in some cases (dotted line).
(C-H) Axons from labellar taste neurons. (C) Single arrow indicates axons of bitter neurons crossing the midline. (D-G, arrows) Axons are concentrated on both sides of midline, in a pattern resembling that of sugar neuron projections. In H, the arrowheads indicate axons that resemble those from labellar pegs. D includes projections from the legs.
(I-N) Axons from pharyngeal taste neurons. N includes projections from the legs.
(O-R) Axons that pass through the thoracic ganglia and project towards the SOG.
(S-W) Axons that terminate within the thoracic ganglia. Asterisks in S indicate projections into the wing neuropil labeled by IR52a-GAL4.
All images are from females. Z-projections of axons expressing GFP are in green and the synaptic marker Bruchpilot is in magenta. Scale bars in C and O represent 30 μm for C-N, and O-W, respectively.
(X) Summary of axonal projection patterns of neurons expressing GAL4 drivers. “b” and “s” indicate axons that resemble those of bitter and sugar neurons, respectively. IR56a-GAL4 labels axon projections consistent with its peripheral expression, in addition to other neurons (not shown).
All five GAL4 constructs that label the labellum label projections in the SOG, supporting the conclusion that they are expressed in neurons. Of the IR-GAL4 drivers that are expressed in the labellum, one, IR56a-GAL4, labels projections in the central region of the SOG with the terminals concentrated at the midline, as observed for bitter taste neurons (Figure 3B, C). The projection patterns of IR47a-, IR56b-, IR56d-, and IR94e-GAL4 resemble those of sugar neurons: they are concentrated in the regions flanking the midline, with little or no midline crossing (Figure 3B, D-G). The projection patterns of neurons labeled by IR56a-GAL4 and IR56b-GAL4 are consistent with their co-expression with markers of bitter- and sugar-sensing Gr drivers, respectively, in the periphery (Figure 1I-N).
IR56d-GAL4 labels two distinct types of axon terminals in the SOG, visible in different focal planes (Figure 3G-H). One type, in the central region (arrows), likely represents projections from labellar bristles, while the other type, in the lateral-anterior region, likely represents projections from pegs (Inoshita and Tanimura, 2006). These patterns are consistent with the expression of IR56d-GAL4 in the periphery (Figure 1F-G). The projections in the central region resemble those of sugar-sensing labellar neurons, which is consistent with the co-labeling of labellar neurons by drivers of IR56d and Gr5a, a sugar-sensing Gr (Figure S2 C-E; for projections of IR47a-GAL4 and IR94e-GAL4 see Fig. S2 H-J and legend)(Dahanukar et al., 2001).
Drivers expressed in the pharynx - representing IR60b, IR67c, IR94a, IR94c, IR94f, and IR94h - label projections in the dorso-anterior SOG (Figures 3I-N). The projections of the IR60b and IR94f drivers differ from the others in that they showed no projections towards the midline.
The drivers expressed in the legs label projections in the thoracic ganglia. Moreover, the map of expression in forelegs, midlegs, and hindlegs (Figure S2G) matches the map of projections in the prothoracic, mesothoracic and metathoracic ganglia, respectively (Figure 3O-X). Some leg projections pass through the thoracic ganglia to extend to the SOG (IR20a-, IR47a-, IR62a- and IR94h-GAL4; Figure 3O-R), while others appear to terminate in the thoracic ganglia without projecting to the SOG (IR52a-, IR52c-, IR52d-, IR56b-, and IR56d-GAL4)(Figure 3S-W). These two categories of leg projection patterns seem likely to represent distinct neural circuits, although we acknowledge that it is possible to miss faintly labeled leg projections into the SOG among the latter group of drivers. None of the drivers label axon projections in the antennal lobe, where olfactory receptor neurons (ORNs) terminate, indicating that none of them are expressed in ORNs. In summary, the labeling of axons in taste centers of the CNS by these IR-GAL4 drivers matches well with their peripheral expression patterns and supports their expression in taste neurons (Figure 2X and 3X).
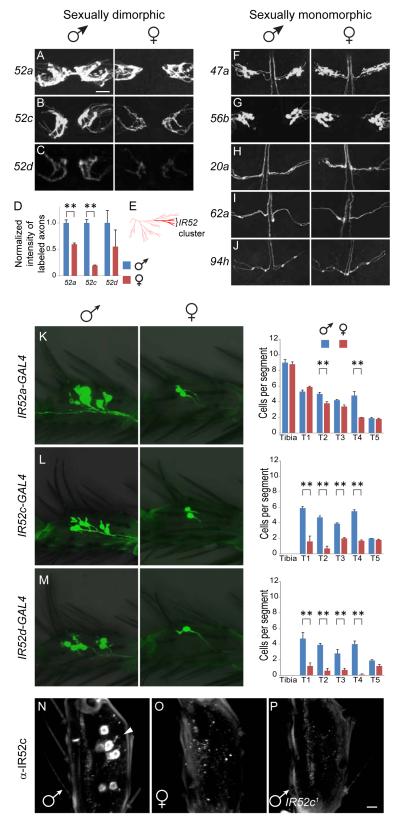
Sexual dimorphism and structural differences among axonal projections
Sexual dimorphisms were found for some taste axons projecting from the forelegs, which act in male courtship behavior (Figure 4A-B). Two kinds of sexual dimorphism were observed in the prothoracic ganglia. First, axons labeled by IR52a-GAL4 cross the midline in a commissure in males, but not in females (Figure 4A). Second, GFP labeling is stronger in males than females in axons labeled by IR52a- and IR52c-GAL4 (Figure 4 A, B, D). Interestingly, IR52a, c, and d are closely related in sequence and are arranged in a tandem cluster along with IR52b in the D. melanogaster reference genome (Figure 4E). We did not observe expression with an IR52b-GAL4 driver and thus focused on the other three genes. In contrast to IR52a-, and IR52c-GAL4 drivers, no sexual dimorphism was observed in five other drivers that send projections to the prothoracic ganglion (Figure 4F-J).
Figure 4.
Some IR20a clade genes show sexually dimorphic expression.
(A-D) Axons in the prothoracic ganglia labeled by GAL4 drivers of IR52a and c are sexually dimorphic. Mean expression levels of IR52d-GAL4 were higher in males as well; however the levels were relatively weak, the signal-to-noise ratios were lower and the variance was higher. “**” denotes p<0.01 (one-way ANOVA); n=7-8.
(E) Phylogenetic tree of IRs and iGluRs highlighting position of the IR52 cluster, which includes IR52a, IR52c and IR52d (bright red). The remainder of the IR20a clade is in pink.
(F-J) Axons labeled by drivers of IR47a, IR56b, IR20a, IR62a and IR94h are sexually monomorphic.
(K-M) Sexually dimorphic expression of IR52a-, IR52c-, and IR52d-GAL4 drivers in forelegs. The numbers of cells expressing GFP in each distal foreleg segment are indicated. In (A-C, K-M), GAL4 lines were aged for 4-6 weeks to ensure high penetrance of GFP expression. “**” denotes p<0.01 (one-way ANOVA, n = 9-15).
(N-P) In flies younger than 10 days post-eclosion, anti-IR52c labels cells in forelegs of males (N), but not females (O). The antibody did not label midlegs or hindlegs (not shown). (P) Antibody labeling is specific, as it did not label the forelegs of IR52c1 males. Tarsal segment T4 is shown in K-P. Scale bar in A represents 20 μm in all panels, except that scale bar in P represents 5 μm in N-P.
The dimorphism observed with the antibody is stronger than that observed with the driver, perhaps because the driver expression was examined in aged flies. See also Figure S3.
Axons labeled by drivers of IR52a, c, and d have varicosities, likely representing neuronal synapses, distributed in a C-shaped pattern in the prothoracic ganglia (Figure 4A-C). In contrast, axons labeled by other foreleg-expressing GAL4 drivers either have varicosities in a different spatial pattern (IR47a, IR56b, Figure 4F,G), or do not show varicosities (IR20a, IR62a, IR94h, Figure 4H-J). Axons labeled by the latter group of drivers may form synapses in the prothoracic ganglia without obvious varicosities. Nonetheless, these differences suggest that different populations of leg neurons have different connectivities in the CNS.
Sexually dimorphic expression of the IR52 cluster in legs
The sexual dimorphism in CNS projections labeled by IR52a- and IR52c-GAL4 prompted us to ask whether expression is sexually dimorphic in forelegs. We found that IR52a-GAL4 is expressed in comparable numbers of cells in males and females in the tibia and in most tarsal segments, although somewhat more cells were labeled in the second and fourth tarsal segments of males than females (Figure 4K). However, IR52c-and IR52d-GAL4 were expressed in many more cells in the male foreleg than the female foreleg (Figure 4L-M). Thus the sexual dimorphism in intensity of labeling of axon terminals by IR52a- and IR52c-GAL4 are consistent with their labeling patterns in the periphery.
We then raised an antibody against a peptide specific for IR52c and performed quantitative RT-PCR. The antibody labeled cells in the forelegs of 5-day-old males, but not 5-day-old females (Figure 4N, O). Labeling was observed only in females that were >10d old, and in these older females staining was weak and sporadic (not shown). The specificity of the antibody was confirmed in tests with a mutant containing a transposon in the IR52c ORF; the mutant leg did not show labeling (Figure 4P). We also confirmed the fidelity of the IR52c-GAL4 driver by showing co-labeling of neurons in male forelegs by the driver and the anti-IR52c antibody (Figure S3A-C). Consistent with the anti-IR52c antibody staining, quantitative RT-PCR revealed a corresponding sexual dimorphism in mRNA transcript levels in the forelegs (Figure S3D). IR52c transcripts could not be detected in midlegs, hindlegs, head, thorax or abdomen (Figure S2K, S3E). Similarly, the male genitalia did not show labeling with IR52c-GAL4 (Figure S3E inset). Therefore, within the limits of our detection methods, IR52c expression is only detected in the forelegs. The male-biased expression of IR52c occurs at the levels of transcript and protein abundance, in addition to the number of cells expressing this gene.
Expression of the IR52 cluster in male sensilla that are likely to contact females
We next compared the expression of IR52a-GAL4, IR52c, and IR52d-GAL4 to each other and to other molecular markers in male forelegs. First we asked whether they are coexpressed. Since the ORFs of IR52a-d fall within a tight cluster of ~11 kb with short intergenic regions, it seemed plausible that they shared some common transcriptional enhancers. Double-labeling with anti-IR52c antibody and IR52d-GAL4 revealed complete, or nearly complete, coexpression in the forelegs (Figure S3F-H), the only legs in which they are expressed. Anti-IR52c and IR52a-GAL4 revealed substantial, although not complete coexpression in dorsal sensilla of forelegs (Figure S3I-K); moreover, instead of being restricted to forelegs, IR52a drivers are expressed in dorsal sensilla of all legs, and a small fraction of cells expressing IR52a drivers are in ventral sensilla that do not express IR52c (Figure 2A).
Two broadly expressed IR genes that are not members of the IR20a clade, IR25a and IR76b, have been previously detected in taste organs (Croset et al., 2010; Zhang et al., 2013). We found that anti-IR52c stains cells that are labeled by drivers of IR25a and IR76b (Figure S3L-Q). The coexpression of IR52c with drivers of IR52a and IR52d, and of IR25a and IR76b, raises the possibility of heteromultimerization of IR20a clade members, reminiscent of antennal IRs which form heteromeric odor receptor complexes with IR25a and IR76b (Abuin et al., 2011).
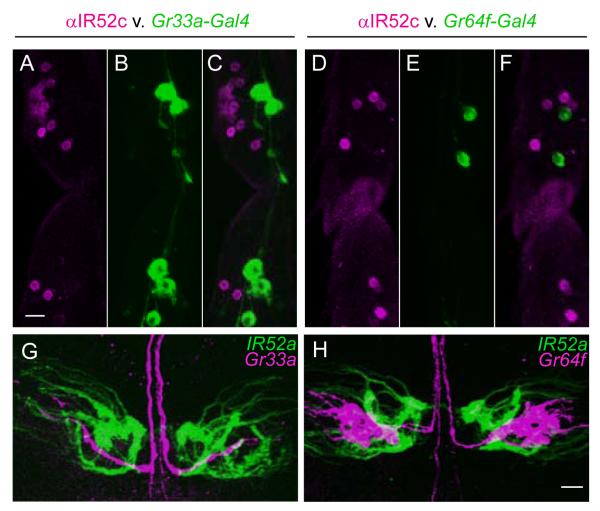
As noted above, IR52a-, IR52c-, and IR52d-GAL4 expression is observed in foreleg sensilla that have a different orientation (red sensilla in Figure 2A) than those shown to respond to food-related tastants (blue sensilla). Double-label experiments confirmed that IR52c is not expressed in the same sensilla as a marker of bitter-sensing neurons, Gr33a-GAL4, or a marker of sugar-sensing neurons, Gr64f-GAL4 (Dahanukar et al., 2007; Moon et al., 2009) (Figure 5A-F). Given that IR52c and IR52d-GAL4 are co-expressed, the latter is most likely not expressed in food-sensing neurons.
Figure 5.
Neurons expressing IR52c and IR52a-GAL4 are distinct from neurons expressing markers of bitter or sugar neurons.
(A-F) IR52c is not expressed in bitter or sugar neurons. In male forelegs, neurons labeled by anti-IR52c antibody (magenta, A, C, D, F) in dorsally-oriented taste sensilla are distinct from neurons in ventral sensilla expressing Gr33a-GAL4 (green, B, C) and Gr64f-GAL4 (green, E, F). Scale bar represents 10 μm in A-F.
(G, H) Comparison of axonal projections of foreleg taste neurons expressing IR52a-GAL4 (green) with bitter neurons expressing Gr33a-GAL4 (magenta, G), and sugar neurons expressing Gr64f-GAL4 (magenta, H). Scale bar represents 10 μm in G, H. See also Figure S3.
A distinction between IR52a-expressing neurons and bitter or sugar neurons was clear in the CNS: we constructed an IR52a-LEXA fusion and found that neurons that express it send projections that do not overlap with those of bitter neurons expressing Gr33a-GAL4 (Figure 5G) or of sugar neurons expressing Gr64f-GAL4 (Figure 5H). The lack of overlap suggests that IR52a-expressing neurons form synapses with different higher order neurons than do bitter or sugar neurons and activate different circuits.
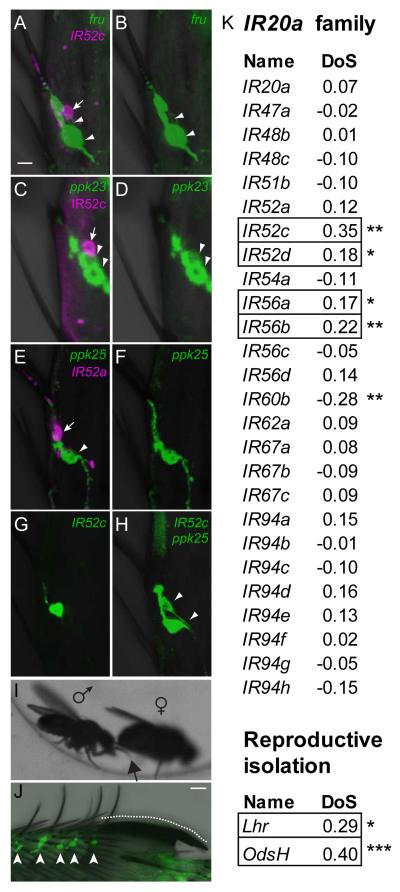
The sexually dimorphic expression of IR52a-GAL4, IR52c, and IR52d-GAL4, along with their expression in sensilla that do not respond to typical food tastants, suggested the possibility of a role for the genes in mating behavior. The transcription factor encoded by fruitless (fru) has previously been shown to be expressed in foreleg taste neurons associated with male sexual behavior (Toda et al., 2012). Interestingly, we found that IR52c-GAL4 neurons and fru-LEXA neurons are mutually exclusive (Figure 6A-B). In some cases, IR52c-GAL4 and fru-LEXA label adjacent neurons that innervate the same sensilla. Consistent with the results above, cells labeled by either anti-IR52c, IR52a-LEXA or IR52c-Gal4 are adjacent to, but distinct from those labeled by GAL4 drivers of ppk23 or ppk25, ion channel genes expressed in subsets of fru+ taste neurons (Figure 6C-H) (Lu et al., 2012; Starostina et al., 2012; Thistle et al., 2012; Toda et al., 2012). In addition, IR52c+ neurons are distinct from those labeled by a GAL4 driver of Gr32a, which is required for the inhibition of male-male and inter-species courtship (Figure S3R-T) (Fan et al., 2013; Miyamoto and Amrein, 2008). Given that the doublesex gene is required for the sexual dimorphism in numbers of taste sensilla on the forelegs (Mellert et al., 2012), we performed a developmental lineage tracing experiment and found that at least some IR52c+ neurons are descended from the dsx+ cell lineage (Figure S3U-W). Taken together, the simplest interpretation of our results is that IR52c+ neurons represent a novel class of sexually dimorphic taste neurons in the leg.
Figure 6.
IR52c+ neurons are adjacent to fru+ neurons in dorsal sensilla and members of IR20a clade, including IR52c and IR52d, show signatures of adaptive evolution.
(A-B) IR52c-GAL4 (magenta, tandem-Tomato, arrow) labels a neuron, which is adjacent to but distinct from a neuron labeled by fru-LexA (green, GFP, arrowheads).
(C-D) anti-IR52c antibody (magenta, arrow) labels a neuron adjacent to but distinct from two which are labeled by ppk23-GAL4 (green, GFP, arrowheads).
(E-F) IR52a-LEXA (magenta, tandem-Tomato, arrow) labels a neuron that is distinct from that labeled by ppk25-GAL4 (green, GFP, arrowhead).
(G-H) GFP driven by IR52c-GAL4 labels one neuron (G), while a combination of IR52c-GAL4 and ppk25-GAL4 labels two neurons (H), indicating that these two drivers express in different cells (H, arrowheads). Neurons in sensilla 5a of male T5 tarsal segments are shown in A-H. Scale bar in A represents 5 μm in A-H.
(I) Contact of the dorsal surface of the male foreleg (arrow) with the female abdomen. See also Movie S1.
(J) A high density of IR52c-GAL4 expressing neurons (arrowheads) close to the sex comb (dotted line). Scale bar represents 10 μm.
(K) Some members of the IR20a clade show signatures of adaptive evolution.
Four of 26 members of the D. melanogaster IR20a clade show signatures of adaptive evolution, when compared with their orthologs in D. simulans using the McDonald-Kreitman test (MKT). MKT looks for evidence of natural selection by comparing variation within species to the divergence between species. DoS (Direction of Selection) indicates adaptive evolution when positive (boxed) and purifying selection when negative (Stoletzki and Eyre-Walker, 2011). The Lhr and OdsH genes, which are responsible for hybrid incompatibility between D. melanogaster and D. simulans, serve as references for DoS values for adaptive evolution. Data were derived from the popDrowser website (Ramia et al., 2011). Data for IR52b and IR60d were not available, and those for 7 putative pseudogenes are not included. “*” denotes P<0.05 and “**” denotes P<0.01.
We captured high-speed video recordings of mating behavior to determine whether sensilla on the male forelegs expressing IR52a-GAL4, IR52c, and IR52d-GAL4 are likely to make contact with females. In all of 13 pairs of flies examined, the males contacted the female cuticle with the dorsal surface of the forelegs during courtship (Figure 6I and Movie S1) (Cook, 1977). Male foreleg sensilla expressing IR52a, IR52c, and IR52d are thus well-positioned to contact the female during mating behavior.
We note finally that there is a high density of sensilla expressing GAL4 drivers of IR52a, IR52c, and IR52d in close proximity to the sex comb, which is on the dorsal aspect of tarsal segment T1 of the male foreleg (Figure 6J and data not shown). The sex comb is used by the male to grasp regions near the female genitalia during copulation attempts (Cook, 1977). Thus, during mating behavior, IR52c+ sensilla are likely to come in contact with regions of the female near the genitalia in addition to other regions.
IR52c and IR52d show signatures of adaptive evolution
As sex-related genes have been proposed to evolve more rapidly than other genes, (Civetta and Singh, 1998), we asked whether sexually dimorphic IRs show interesting evolutionary signatures by examining a genome-wide comparisons between D. melanogaster and a sibling species. A database of polymorphisms within a natural population of D. melanogaster (Mackay et al., 2012) has recently , made it possible in principle to distinguish adaptive evolution from neutral variation, by comparing variation within D. melanogaster to the divergence between D. melanogaster and D. simulans, using the McDonald-Kreitman test (Stoletzki and Eyre-Walker, 2011).
Interestingly, IR52c and IR52d, along with 2 other IR20a clade genes, show signatures of adaptive evolution: they exhibit a statistically significant, positive Direction of Selection (DoS) value, reminiscent of genes involved in reproductive isolation between Drosophila species, such as Lhr and OdsH (Figure 6K) (Brideau et al., 2006; Ramia et al., 2011; Ting et al., 1998). Moreover, the coexpression of IR52c and IR52d in sexually dimorphic foreleg sensilla suggests the possibility of coevolution of the two genes.
The IR20a clade overall appears to be evolving rapidly: 12 of its members lie in four gene clusters that underwent expansions and contractions in gene number during the evolution of Drosophila species (Croset et al., 2010). IR52c and IR52d lie in one of these dynamic clusters.
Roles for IR52c and IR52d in male sexual behavior
The high DoS values of IR52c and IR52d, coupled with their male-biased patterns of expression, prompted us to ask whether these two genes act in male sexual behavior. We generated Δcd, a deletion mutant in which the ORFs of IR52c and IR52d are disrupted while the rest of the IR52 cluster is left intact (Figure S4A, B). To minimize genetic background effects, we placed the double mutant chromosome in a transheterozygous combination with a small deficiency, Df(2R)IR52a-d removing the entire IR52 cluster (Δcd/Df), and compared mutant and control genotypes in an outbred background (Supplemental Experimental Procedures; Figure S4A).
In D. melanogaster, copulation is preceded by an elaborate courtship ritual. The male fly detects the female fly in part through taste sensilla on his foreleg and labellum and then produces a courtship song by unilateral wing vibrations (Cook, 1977; Fan et al., 2013; Greenspan and Ferveur, 2000; Thistle et al., 2012). Various aspects of courtship behavior are thought to act cumulatively in back-and-forth exchanges of signals between two flies before culminating in copulation.
To analyze courtship behavior we developed and validated FlyVoyeur, an automated tracking algorithm that allows convenient quantitation of a variety of parameters of fly behavior, including rapid wing movements that are difficult to score manually (see Supplemental Experimental Procedures).
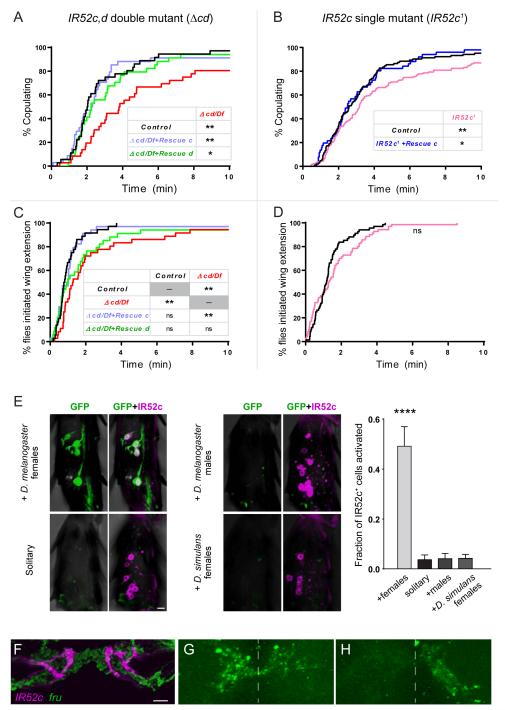
Δcd/Df males showed a prolongation of the time taken to achieve successful copulation (i.e. copulation latency, Figure 7A). For example, the t1/2 of copulation – the time at which half the males had begun copulating – was four minutes for the mutant but only two minutes for the control in our experimental paradigm. To test the possibility that the phenotype was due to a mutation at another site, we introduced into Δcd/Df males either of two constructs containing 13.9 kb of genomic DNA. One construct included IR52a, IR52b, and IR52c (Rescue c, Figure S4C); the second construct, inserted into the same genomic position as the first, contained IR52a, IR52b, and IR52d, but not IR52c (Rescue d, Figure S4C). The Rescue c construct conferred a restoration of the copulation latency of the double mutant to control levels, significantly different from the double mutant, while the Rescue d provided a weaker degree of rescue that is nonetheless significant (Figure 7A). The simplest interpretation of these results is that both IR52c and IR52d play roles in determining male copulation latencies, and that the roles of these genes are at least partially redundant.
Figure 7.
The roles of IR52c and IR52d in male sexual behavior.
(A) Disruption of both IR52c and IR52d in males delays copulation. The double mutant is a transheterozygous combination of Δcd, a ~2.5kb deletion that disrupts only IR52c and IR52d and a ~ 33kb deficiency removing the entire IR52 cluster (Figure S4A).
Introducing either genomic fragments with a wildtype copy of IR52c or IR52d rescued the copulation delay (Figure S4C).
(B) IR52c1 mutant males showed delayed copulation, which can be rescued with wildtype IR52c.
(C) Δcd/Df showed a delay in courtship initiation, which was rescued by introducing wildtype copy of IR52c, but not IR52d.
(D) IR52c1 mutant males do not show a significant delay in courtship initiation.
(A-D) Mutants and controls were in a Berlin-K/Canton-S outbred background (see also Figure S4 and Supplemental Experimental Procedures); small differences are nonetheless expected in the genetic backgrounds of the single mutant and the double mutant. n = 34-36 in (A, B), 40-131 in (C) and 68-70 in (D). Statistical tests were performed using survival analysis (log-rank test). “**” denotes p<0.01 and “*” denotes p<0.05.
(E) Activation of a calcium-sensitive immediate-early gene cassette (NFAT/CaLEXA) (Masuyama et al., 2012) occured in IR52c+ foreleg cells of males incubated with virgin females for three days, but not in those of solitary males (Supplemental Experimental Procedures). Males of the genotype IR52c-GAL4/LexAop-CD8-GFP-2A-CD8-GFP; 2x IR52c-GAL4/UAS-mLEXA-VP16-NFAT, LexAop-CD2-GFP were tested; IR52c-GAL4 drives the expression of the LEXA-NFAT fusion protein in IR52c+ cells. Calcium influx into these cells led to nuclear entry of the fusion protein and activated expression of GFP from the LEXA responsive promoters. The identity of GFP-expressing cells (green) was verified with anti-IR52c labeling staining (magenta). Male foreleg tarsal segments of males showed greater GFP expression in IR52c+ cells when exposed to D. melanogaster females than when kept solitary or than when exposed to other males or D. simulans virgin females (Kruskal-Wallis test followed by Dunn’s multiple comparison test; n = 20-26 tarsal segments [117-174 cells, 6.8±0.06 cells per segment]; “****” denotes p < 0.0001). Scale bar represents 5 μm. Tarsal segment 4 is shown here.
(F) Foreleg neurons expressing IR52c-GAL4 (magenta, RFP) and fru-LEXA (green, GFP) project to the prothoracic ganglia to form interdigitating terminals.
(G) Reconstitution of native GFP fluorescence was observed between IR52c+ and fru+ neurons across putative synpases using a modified GRASP method involving a synapse-targeting signal (Fan et al., 2013).
(H) Amputation of a single foreleg abolished GRASP signal on one side of the prothoracic ganglia while preserving the signal on the other side. Dotted lines in (H, I) indicate the midline. The orientations of the sections in (G) and (H) are not identical. Scale bar in (F) represents 10 μm in (F-H).
To further examine the requirement of IR52c in sexual behavior, we tested IR52c1, a transposon insertion that disrupts IR52c while leaving the expression of other genes in the IR52 cluster intact (Figures 4P and S4A, D). We did not expect a strong phenotype in the copulation latency of the IR52c1 single mutant, since IR52c and IR52d appeared at least partially redundant. Nonetheless, IR52c1 showed a moderate but significant copulation delay phenotype that could be rescued by Rescue c (Figure 7B), providing further support for a role for IR52c in male copulation latency. We note that a transposon disrupting IR52e, another gene in the same cluster (see Extended Experimental Methods), caused no copulation delay, suggesting that the effect of disrupting IR52c is not generally applicable to all genes within the cluster (Figure S4A,D,J).
To further understand the copulation delay phenotype, we examined different aspects of courtship leading to copulation. Consistent with a defect in detecting potential mates, Δcd/Df males were slow to initiate their first unilateral wing extension to produce courtship songs, compared to control males (Figure 7C). This phenotype is fully restored by the Rescue c construct, suggesting a role for IR52c in courtship behavior. Introduction of Rescue d to Δcd/Df produced an intermediate phenotype that did not differ significantly from either control or Δcd/Df males. Δcd/Df males also showed reductions in the fraction of time spent in wing extension and frequency of licks, i.e. contacts between the male labellum and the female abdomen, relative to controls (Figure S4E-F). The Δcd/Df males showed normal consumption and preference for sucrose, indicating that the loss of IR52c and IR52d did not produce a general impairment of taste functions, and the mutant was normal in a test of locomotor function (Figure S4G). We did not detect with IR52c1 a significant delay in initiation of wing extension towards the female, nor did it show phenotypes in overall wing extension and lick frequency (Figure 7D, S4H-I). These results may reflect a partial redundancy of IR52c and its closest paralog, IR52d; the sensitivities of measurements of courtship and copulation may also differ.
IR52c+ neurons are activated by females and form putative synapses with fru+ neurons
The evidence that IR52c and IR52d act in male sexual behavior and their co-expression in sexually dimorphic foreleg taste neurons suggest that these neurons may sense potential mates. To test this possibility, we used a calcium-sensitive reporter cassette. This reporter cassette allows the targeted expression of a modified nuclear factor of activated T cells (NFAT) in IR52c+ cells. When calcium levels rise during neuronal activation, the modified NFAT enters the nucleus and induces GFP expression (Masuyama et al., 2012).
When male flies expressing the modified NFAT in IR52c+ cells were exposed to virgin females, we observed GFP expression in a large proportion of the cells (Figure 7E). In contrast, solitary males showed only a very small proportion of GFP-expressing cells; most legs showed no GFP expression in these control conditions (Figure 7E). We next exposed the male flies to other males and again found little if any GFP-labeling in IR52c+ cells. Finally, we exposed males to D. simulans virgin females and found very little labeling. The detection of calcium signals specifically after exposure to D. melanogaster females suggests that IR52c+ neurons are activated by the presence of conspecific females.
Next, we asked whether activation of the IR52c+ neurons influences courtship behavior, by ectopically expressing NaChBac, a bacterial sodium channel that increases the excitability of neurons and depolarizes them (Nitabach et al., 2006). Compared to controls, males that express NaChBac in IR52c+ neurons showed a decreased latency to initiation of wing extension (Figure S4K). The t1/2 of wing-extension – the time at which half the males had extended a wing – was less than half that required for each of two control lines (Figure S4K). This result suggests that activation of IR52c+ neurons promotes courtship.
Having found that IR52c+ neurons are activated by females and that activation of these neurons promotes courtship, we asked whether they interact with a neural circuit that governs sexual behavior. First, we examined whether the axonal projections of IR52c+ neurons lie in the vicinity of neurons expressing fru, which specifies a neural circuit for sexual behavior. Indeed, we observed that IR52c+ axon terminals interdigitate with neurites of fru+ neurons in the prothoracic ganglia (Figure 7F). Second, to determine whether there are putative synaptic contacts between IR52c+ and fru+ neurons, we performed an enhanced version of GFP reconstitution across synaptic partners (GRASP) in which the reconstitution of the native fluorescence of the split GFP protein is restricted to synapses (Fan et al., 2013). When we expressed the split GFP components in IR52c+ and fru+ neurons, we observed GRASP signals in the prothoracic ganglia (Figure 7G). Amputation of one foreleg of the flies abolished the GRASP signal in one side of the prothoracic ganglia, while preserving the signal from the side receiving input from the intact foreleg (Figure 7H). These results indicate that sites of putative synapses exist between IR52c+ and fru+ neurons in the central nervous system. Although we do not know the function of the particular subset of fru+ neurons contacting IR52c+ neurons, these data open the door for further investigation of the role of IR52c+ neurons in sexual behavior at the neural circuit level.
Discussion
A major class of candidate taste receptors
The IR20a clade represents a major addition to the repertoire of candidate taste receptors in Drosophila. The clade comprises ~35 genes, more than half the size of the Gr family of taste receptor genes. GAL4 drivers representing genes of this clade labeled taste neurons in all known taste organs, including the labellum, legs, pharynx, and wing. Different taste organs express distinct combinations of IR drivers, likely reflecting differences in the functions of these organs. The clade is likely to make wide-ranging contributions to taste function, and its identification opens diverse avenues for exploration of taste coding and behavior.
The study described here was initiated with an interest in discovering new receptors that detect chemical signals not sensed by other receptors, or that are expressed in orphan taste neurons to which no receptors had previously been mapped. Some of our IR drivers are in fact expressed in taste neurons to which no receptors had been mapped, thus “deorphanizing” these neurons; we estimate that on the order of one-fourth of orphan taste neurons express drivers of this clade. Among the receptors expressed in orphan taste neurons is IR52c, which shows signatures of adaptive evolution, is sexually dimorphic in its expression, and is required for normal copulation behavior. In addition, we have identified IR drivers that are coexpressed with Gr drivers, suggesting novel ways in which this clade may contribute to the taste code.
IR and Gr coexpression
We have found that some bitter-sensing neurons express drivers of both IR and Gr genes: IR56a and Gr66a. Likewise, some sugar-sensing neurons coexpress the driver of Gr5a with those of IR56b and IR56d. Thus the taste specificities of some individual taste neurons are likely to be conferred jointly by members of two entirely different classes of receptors.
If some taste neurons receive signals via two different classes of receptors, how do the neurons integrate the signals? In this light it will be interesting to test whether IRs and Grs interact within the neuron, and if so, whether such interactions modulate taste responses. Interactions between receptors of different classes have not been documented in chemosensory neurons, but there is ample precedent for such receptor cross-talk in central neurons (Lee et al., 2002). The multiplicity of IRs and Grs expressed in individual taste neurons expands the potential for combinatorial interactions within and perhaps between members of the two families.
Adaptive evolution within the IR20a clade
Signatures of adaptive evolution among the IR20a clade members could in principle reflect evolutionary responses to changes in the environment of D. melanogaster. Such environmental changes could necessitate the detection of new food sources or novel toxins or pathogens.
Alternatively, adaptive evolution may reflect speciation. For example, two genes that show adaptive evolution, Lhr and OdsH, are thought to act in reproductive isolation between the closely related species D. melanogaster and D. simulans (Figure 6K) (Brideau et al., 2006; Ting et al., 1998). Among chemoreceptor genes, four of the six odor receptors of the Or family that show signatures of adaptive evolution are expressed in antennal trichoid sensilla (Table S2). Neurons in trichoid sensilla respond to fly odors, some of which are known to function in sexual behavior (Ejima et al., 2007; Ramia et al., 2011; van der Goes van Naters and Carlson, 2007). Interestingly, both IR52c and IR52d show adaptive evolution (Figure 6K); these genes are co-expressed in the same foreleg neurons, and we have found evidence that they act in male mating behavior.
These results, taken together, suggest that signatures of adaptive evolution may in some cases reflect functions in mate detection or other aspects of sexual behavior. Accordingly, our results suggest how a relatively simple analysis of gene evolution with tools such as the publicly accessible popDrower database (http://popdrowser.uab.cat/fgb2/gbrowse/dgrp/) could be a powerful means of identifying new genes that mediate social interactions, sexual behavior, or speciation (Ramia et al., 2011).
Genes, cells and circuitry underlying chemoreception in sexual behavior
The IR52c, d double mutant males were slow in achieving copulation, a phenotype that may be partially attributed to their delay in initiating unilateral wing extension towards females. As wing extension occurs early during the courtship ritual (Manoli and Baker, 2004), our results suggest that IR52c and IR52d act in males in the detection of females. This notion is supported by the previous finding that contact between the male foreleg and the female abdomen induces wing extension and the generation of courtship song (Kohatsu et al., 2011).
Our results with the double and single mutant males suggest a partial redundancy between IR52c and IR52d. While both the IR52c, d double mutant and IR52c single mutant show delayed copulation, only the double mutants showed a delay in initiating wing extension. Sequence similarity and co-expression in foreleg neurons may allow IR52c and d to serve similar, partially redundant functions, and this may explain why the double mutant phenotype is stronger than that of the single mutant. Interestingly, re-introduction of wildtype IR52d into the double mutants produced a weak rescue of the copulation delay phenotype and no rescue of the delay in initiating wing extension; this contrasts with the rescue of both phenotypes by re-introducing wildtype IR52c (Figure 7 A, C). One possible explanation for this difference between IR52d and IR52c is that the expression level of IR52d is much lower than that of IR52c (qRT-PCR data in Figure S4B): the extremely low expression level of IR52d may have precluded a strong rescue of the double mutant phenotypes by the re-introduction of wildtype IR52d. Therefore, despite the sequence similarity between IR52c and IR52d, the large difference in expression levels of the two genes prevents a complete interchangeability between the two genes.
How do the foreleg neurons that co-express IR52c and IR52d contribute to male sexual behavior? We found evidence that IR52c+d+ neurons in male forelegs undergo activation in the presence of conspecific females, using an NFAT-based method (Masuyama et al., 2012). In contrast, extended exposure to conspecific males did not activate these neurons, suggesting that the IR52c+d+ neurons do not have a role in male-male social interactions in our experimental conditions. Similarly, D. simulans females did not activate these neurons. One simple interpretation is that IR52c+d+ neurons act in distinguishing potential mates from unproductive targets; this interpretation would be consistent with functional divergence of D. melanogaster IR52c and IR52d from their closest homologs in D. simulans, as suggested by the DoS values of these genes. We note that although D. melanogaster males do interact with conspecific males and D. simulans females (Wood and Ringo, 1980), it is possible that such interactions are not sufficiently intense to generate a signal in our experimental system (Fan et al., 2013; Miyamoto and Amrein, 2008), and we cannot exclude the possibility that IR52c+d+ neurons play a role in male-male or heterospecific interactions in more naturalistic conditions. Further investigation of these possibilities will help us understand the nature of the signal detected by IR52c+d+ neurons. The most important conclusion from our behavioral results is that IR52c+d+ foreleg neurons are activated by conspecific females, the reproductively relevant targets of the males. This conclusion is consistent with our observation that the ectopic expression of NaChBac in IR52c+d+ neurons promotes courtship. These results raise the question of how these neurons interact with the neural circuit that generates sexual behaviors.
fru governs the specification of much of the neural circuit that controls male sexual behavior (Kimura et al., 2008; Manoli et al., 2005; Stockinger et al., 2005). Interestingly, IR52c+ neurons do not express fru. However, our GRASP results suggest that IR52c+ neurons form putative synaptic contacts with fru+ neurons. These results provide an interesting parallel between IR52c+ and Gr32a+ leg neurons, neither of which express fru, but form putative synaptic contacts with fru+ neurons (Fan et al., 2013). Importantly, the observation of putative synaptic contacts between IR52c+ and fru+ neurons suggests a neural mechanism by which activation of IR52c+ neurons influences sexual behavior. While we do not know the identities of these fru+ neurons and their precise functions, this observation will serve as a foundation for future studies to further characterize these putative synapses at the functional level.
dsx governs the ontogenesis of a portion of the circuitry underlying male sexual behavior (Rideout et al., 2010; Robinett et al., 2010); this portion partially overlaps the circuitry specified by fru. We have found that at least some of the IR52c+ neurons may have descended from a cell lineage specified by dsx. These results are consistent with their locations in sexually dimorphic foreleg taste sensilla, which depend on dsx (Mellert et al., 2012).
Our results add a third gene family to two other families expressed in taste neurons, the Gr and ppk families, that have previously been implicated in male sexual behavior (Bray and Amrein, 2003; Fan et al., 2013; Lin et al., 2005; Lu et al., 2012; Miyamoto and Amrein, 2008; Moon et al., 2009; Starostina et al., 2012; Thistle et al., 2012; Toda et al., 2012; Watanabe et al., 2011). ppk25, ppk23, ppk29, Gr39a and Gr68a positively regulate courtship directed at female flies (but see (Ejima and Griffith, 2008), for Gr68a). The diversity of chemosensory genes acting in courtship stimulation may reflect the large number of putative pheromones in D. melanogaster females (Everaerts et al., 2010; Yew et al., 2009).
In this context, neither the IR52c,d double mutant, the ppk23, 29 double mutant, the ppk25 mutant nor the Gr39a mutant showed complete loss of male-female courtship (Lu et al., 2012; Starostina et al., 2012; Thistle et al., 2012; Toda et al., 2012; Watanabe et al., 2011). The phenotypes may be incomplete due to compensation from other sensory modalities such as vision and olfaction (Krstic et al., 2009) or to partial redundancy among these genes in the taste system. Interestingly, we have found evidence that IR52c is expressed in distinct cells from ppk23 and ppk25, suggesting that distinct cells on the male foreleg may have partially redundant roles in male sexual behavior. Similarly, in the context of male-male interaction, Gr32a+ and a subset of ppk23+ neurons share similar function in preventing males from courting other males, consistent with partial redundancy between distinct populations of taste neurons (Fan et al., 2013; Miyamoto and Amrein, 2008; Thistle et al., 2012; Toda et al., 2012).
Experimental Procedures
Transgenic constructs and expression analyses
To maximize the fidelity of the reporters, we flanked GAL4 with both the 5′ and 3′ flanking regions of the IR20a genes, in most cases (Table S1). Most drivers were inserted into common phiC31 integration sites (Supplemental Experimental Procedures). We analyzed the expression of 120 GAL4 lines, 2-10 lines for each gene, using UAS-mCD8-GFP (Lee and Luo, 1999). For selected genes, we confirmed their expression with RTPCR and immunohistochemistry (Supplemental Experimental Procedures).
Genetics, mating assays and automated scoring of behavior
IR52c1 is a Minos transposon, MB04402, inserted into the ORF of IR52c (BDGP Gene Disruption Project). Δcd was generated by imprecise excision of IR52c1, and Df(2R)IR52a-d was generated by FLP-mediated site-directed recombination. Details on genetics, mating assays and FlyVoyeur, the automated scoring algorithm, are in Supplemental Experimental Procedures.
CaLEXA (NFAT-based detection of neuronal calcium signal)
As detailed in Supplemental Experimental Procedures, a LEXA-NFAT fusion protein was expressed in IR52c+ cells to detect neuronal calcium signals, using a previously described construct (Masuyama et al., 2012).
GFP Reconstitution Across Synaptic Partners (GRASP)
The GRASP experiment was performed as described previously with modifications detailed in Supplemental Experimental Procedures (Fan et al., 2013). One component of the split GFP was targeted to the synapse through fusion with neurexin, while the other component was targeted to the plasma membrane through fusion with CD4 (Fan et al., 2013; Gordon and Scott, 2009).
Supplementary Material
Acknowledgments
We thank J. Wang, C. Montell, Y. Ben-Shahar, N. Shah, B. Baker, S. Goodwin, C. Pikielny, K. Scott, A. Dahanukar, J.-Y. Kwon, R. Prum, C. Clark and the Bloomington Stock Center for fly lines, vectors and equipment. This work was supported by grants from the NIH (to J.R.C.) and an NIH NRSA Fellowship to K.M. We are grateful to P. Clyne for his identification and initial characterization of these genes, and to S. Ritzenthaler and K. Foster for their early studies of these genes. We thank J. Chung for helping with behavioral analysis. We are grateful to C.-Y. Su for helpful comments on the manuscript and A. Keene for insightful comments on courtship behavior analysis. We thank Z. Berman and P. Graham for support.
Footnotes
Supplemental Information Supplemental information includes Supplemental Experimental Procedures, five figures, two tables and one movie.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant Ionotropic Glutamate Receptors as Chemosensory Receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum MS. Semiochemical Parsimony in the Arthropoda. Annual Review of Entomology. 1996;41:353–374. doi: 10.1146/annurev.en.41.010196.002033. [DOI] [PubMed] [Google Scholar]
- Boll W, Noll M. The Drosophila Pox neuro gene: control of male courtship behavior and fertility as revealed by a complete dissection of all enhancers. Development. 2002;129:5667–5681. doi: 10.1242/dev.00157. [DOI] [PubMed] [Google Scholar]
- Bray S, Amrein H. A putative Drosophila pheromone receptor expressed in male-specific taste neurons is required for efficient courtship. Neuron. 2003;39:1019–1029. doi: 10.1016/s0896-6273(03)00542-7. [DOI] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science. 2006;314:1292–1295. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- Civetta A, Singh RS. Sex-related genes, directional sexual selection, and speciation. Mol Biol Evol. 1998;15:901–909. doi: 10.1093/oxfordjournals.molbev.a025994. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Carlson JR. Candidate taste receptors in Drosophila. Science. 2000;287:1830–1834. doi: 10.1126/science.287.5459.1830. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Cook RM. Behavioral role of the sexcombs in Drosophila melanogaster and Drosophila simulans. Behavior Genetics. 1977;7:349–357. doi: 10.1007/BF01077448. [DOI] [PubMed] [Google Scholar]
- Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahanukar A, Foster K, van der Goes van Naters WM, Carlson JR. A Gr receptor is required for response to the sugar trehalose in taste neurons of Drosophila. Nat Neurosci. 2001;4:1182–1186. doi: 10.1038/nn765. [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Lei YT, Kwon JY, Carlson JR. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007;56:503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejima A, Griffith LC. Courtship initiation is stimulated by acoustic signals in Drosophila melanogaster. PLoS ONE. 2008;3:e3246. doi: 10.1371/journal.pone.0003246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejima A, Smith BP, Lucas C, van der Goes van Naters W, Miller CJ, Carlson JR, Levine JD, Griffith LC. Generalization of courtship learning in Drosophila is mediated by cis-vaccenyl acetate. Curr Biol. 2007;17:599–605. doi: 10.1016/j.cub.2007.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everaerts C, Farine JP, Cobb M, Ferveur JF. Drosophila cuticular hydrocarbons revisited: mating status alters cuticular profiles. PLoS ONE. 2010;5:e9607. doi: 10.1371/journal.pone.0009607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk R, Bleiseravivi N, Atidia J. Labellar Taste Organs of Drosophila melanogaster. Journal of Morphology. 1976;150:327–341. doi: 10.1002/jmor.1051500206. [DOI] [PubMed] [Google Scholar]
- Fan P, Manoli DS, Ahmed OM, Chen Y, Agarwal N, Kwong S, Cai AG, Neitz J, Renslo A, Baker BS, et al. Genetic and neural mechanisms that inhibit Drosophila from mating with other species. Cell. 2013;154:89–102. doi: 10.1016/j.cell.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendre N, Luer K, Friche S, Grillenzoni N, Ramaekers A, Technau GM, Stocker RF. Integration of complex larval chemosensory organs into the adult nervous system of Drosophila. Development. 2004;131:83–92. doi: 10.1242/dev.00879. [DOI] [PubMed] [Google Scholar]
- Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan RJ, Ferveur JF. Courtship in Drosophila. Annu Rev Genet. 2000;34:205–232. doi: 10.1146/annurev.genet.34.1.205. [DOI] [PubMed] [Google Scholar]
- Inoshita T, Tanimura T. Cellular identification of water gustatory receptor neurons and their central projection pattern in Drosophila. Proc Natl Acad Sci U S A. 2006;103:1094–1099. doi: 10.1073/pnas.0502376103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph RM, Heberlein U. Tissue-specific Activation of a Single Gustatory Receptor Produces Opposing Behavioral Responses in Drosophila. Genetics. 2012;192:521–532. doi: 10.1534/genetics.112.142455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hachiya T, Koganezawa M, Tazawa T, Yamamoto D. Fruitless and doublesex coordinate to generate male-specific neurons that can initiate courtship. Neuron. 2008;59:759–769. doi: 10.1016/j.neuron.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Kohatsu S, Koganezawa M, Yamamoto D. Female Contact Activates Male-Specific Interneurons that Trigger Stereotypic Courtship Behavior in Drosophila. Neuron. 2011;69:498–508. doi: 10.1016/j.neuron.2010.12.017. [DOI] [PubMed] [Google Scholar]
- Krstic D, Boll W, Noll M. Sensory integration regulating male courtship behavior in Drosophila. PLoS ONE. 2009;4:e4457. doi: 10.1371/journal.pone.0004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FJS, Xue S, Pei L, Vukusic B, Chéry N, Wang Y, Wang YT, Niznik HB, Yu X.-m., Liu F. Dual Regulation of NMDA Receptor Functions by Direct Protein-Protein Interactions with the Dopamine D1 Receptor. Cell. 2002;111:219–230. doi: 10.1016/s0092-8674(02)00962-5. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Liman Emily R., Zhang Yali V., Montell C. Peripheral Coding of Taste. Neuron. 2014;81:984–1000. doi: 10.1016/j.neuron.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Mann KJ, Starostina E, Kinser RD, Pikielny CW. A Drosophila DEG/ENaC channel subunit is required for male response to female pheromones. Proc Natl Acad Sci U S A. 2005;102:12831–12836. doi: 10.1073/pnas.0506420102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F, Dahanukar A, Weiss LA, Kwon JY, Carlson JR. The Molecular and Cellular Basis of Taste Coding in the Legs of Drosophila. J Neurosci. 2014;34:7148–7164. doi: 10.1523/JNEUROSCI.0649-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Lamora A, Sun Y, Welsh MJ, Ben-Shahar Y. ppk23-Dependent Chemosensory Functions Contribute to Courtship Behavior in Drosophila melanogaster. PLoS Genet. 2012;8:e1002587. doi: 10.1371/journal.pgen.1002587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli DS, Baker BS. Median bundle neurons coordinate behaviours during Drosophila male courtship. Nature. 2004;430:564–569. doi: 10.1038/nature02713. [DOI] [PubMed] [Google Scholar]
- Manoli DS, Foss M, Villella A, Taylor BJ, Hall JC, Baker BS. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature. 2005;436:395–400. doi: 10.1038/nature03859. [DOI] [PubMed] [Google Scholar]
- Masuyama K, Zhang Y, Rao Y, Wang JW. Mapping neural circuits with activity-dependent nuclear import of a transcription factor. J Neurogenet. 2012;26:89–102. doi: 10.3109/01677063.2011.642910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellert DJ, Robinett CC, Baker BS. doublesex Functions Early and Late in Gustatory Sense Organ Development. PLoS ONE. 2012;7:e51489. doi: 10.1371/journal.pone.0051489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier N, Ferveur JF, Marion-Poll F. Sex-specific non-pheromonal taste receptors in Drosophila. Curr Biol. 2000;10:1583–1586. doi: 10.1016/s0960-9822(00)00860-5. [DOI] [PubMed] [Google Scholar]
- Miller A. The internal anatomy and histology of the imago of Drosophila melanogaster. In: Demerec M, editor. Biology of Drosophila. John Wiley & Sons; New York: 1950. pp. 420–534. [Google Scholar]
- Miyamoto T, Amrein H. Suppression of male courtship by a Drosophila pheromone receptor. Nat Neurosci. 2008;11:874–876. doi: 10.1038/nn.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SJ, Lee Y, Jiao Y, Montell C. A Drosophila gustatory receptor essential for aversive taste and inhibiting male-to-male courtship. Curr Biol. 2009;19:1623–1627. doi: 10.1016/j.cub.2009.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kwon JY. A systematic analysis of Drosophila gustatory receptor gene expression in abdominal neurons which project to the central nervous system. Mol Cells. 2011;32:375–381. doi: 10.1007/s10059-011-0128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramia M, Librado P, Casillas S, Rozas J, Barbadilla A. PopDrowser: the Population Drosophila Browser. Bioinformatics. 2011;28:595–596. doi: 10.1093/bioinformatics/btr691. [DOI] [PubMed] [Google Scholar]
- Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat Neurosci. 2010;13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Vaughan AG, Knapp JM, Baker BS. Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 2010;8:e1000365. doi: 10.1371/journal.pbio.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GS, Benton R. Complementary Function and Integrated Wiring of the Evolutionarily Distinct Drosophila Olfactory Subsystems. J Neurosci. 2011;31:13357–13375. doi: 10.1523/JNEUROSCI.2360-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starostina E, Liu T, Vijayan V, Zheng Z, Siwicki KK, Pikielny CW. A Drosophila DEG/ENaC Subunit Functions Specifically in Gustatory Neurons Required for Male Courtship Behavior. The Journal of Neuroscience. 2012;32:4665–4674. doi: 10.1523/JNEUROSCI.6178-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- Stockinger P, Kvitsiani D, Rotkopf S, Tirian L, Dickson BJ. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- Stoletzki N, Eyre-Walker A. Estimation of the neutrality index. Mol Biol Evol. 2011;28:63–70. doi: 10.1093/molbev/msq249. [DOI] [PubMed] [Google Scholar]
- Thistle R, Cameron P, Ghorayshi A, Dennison L, Scott K. Contact Chemoreceptors Mediate Male-Male Repulsion and Male-Female Attraction during Drosophila Courtship. Cell. 2012;149:1140–1151. doi: 10.1016/j.cell.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CT, Tsaur SC, Wu ML, Wu CI. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- Toda H, Zhao X, Dickson BJ. The Drosophila Female Aphrodisiac Pheromone Activates ppk23(+) Sensory Neurons to Elicit Male Courtship Behavior. Cell reports. 2012;1:599–607. doi: 10.1016/j.celrep.2012.05.007. [DOI] [PubMed] [Google Scholar]
- van der Goes van Naters W, Carlson JR. Insects as chemosensors of humans and crops. Nature. 2006;444:302–307. doi: 10.1038/nature05403. [DOI] [PubMed] [Google Scholar]
- van der Goes van Naters W, Carlson JR. Receptors and neurons for fly odors in Drosophila. Curr Biol. 2007;17:606–612. doi: 10.1016/j.cub.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Singhvi A, Kong P, Scott K. Taste representations in the Drosophila brain. Cell. 2004;117:981–991. doi: 10.1016/j.cell.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Toba G, Koganezawa M, Yamamoto D. Gr39a, a highly diversified gustatory receptor in Drosophila, has a role in sexual behavior. Behav Genet. 2011;41:746–753. doi: 10.1007/s10519-011-9461-6. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Dahanukar A, Kwon JY, Banerjee D, Carlson JR. The molecular and cellular basis of bitter taste in Drosophila. Neuron. 2011;69:258–272. doi: 10.1016/j.neuron.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D, Ringo JM. Male Mating Discrimination in Drosophila melanogaster, Drosophila simulans and Their Hybrids. Evolution. 1980;34:320–329. doi: 10.1111/j.1558-5646.1980.tb04820.x. [DOI] [PubMed] [Google Scholar]
- Yarmolinsky DA, Zuker CS, Ryba NJ. Common sense about taste: from mammals to insects. Cell. 2009;139:234–244. doi: 10.1016/j.cell.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yew JY, Dreisewerd K, Luftmann H, Muthing J, Pohlentz G, Kravitz EA. A new male sex pheromone and novel cuticular cues for chemical communication in Drosophila. Curr Biol. 2009;19:1245–1254. doi: 10.1016/j.cub.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YV, Ni J, Montell C. The Molecular Basis for Attractive Salt-Taste Coding in Drosophila. Science. 2013;340:1334–1338. doi: 10.1126/science.1234133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.