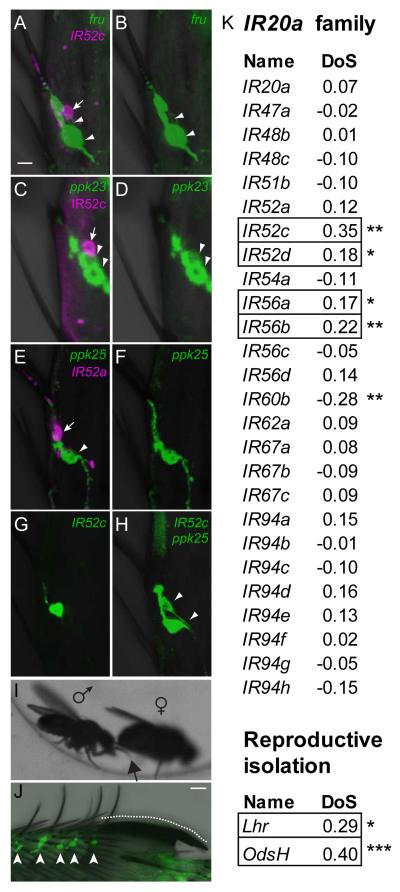
Figure 6.
IR52c+ neurons are adjacent to fru+ neurons in dorsal sensilla and members of IR20a clade, including IR52c and IR52d, show signatures of adaptive evolution.
(A-B) IR52c-GAL4 (magenta, tandem-Tomato, arrow) labels a neuron, which is adjacent to but distinct from a neuron labeled by fru-LexA (green, GFP, arrowheads).
(C-D) anti-IR52c antibody (magenta, arrow) labels a neuron adjacent to but distinct from two which are labeled by ppk23-GAL4 (green, GFP, arrowheads).
(E-F) IR52a-LEXA (magenta, tandem-Tomato, arrow) labels a neuron that is distinct from that labeled by ppk25-GAL4 (green, GFP, arrowhead).
(G-H) GFP driven by IR52c-GAL4 labels one neuron (G), while a combination of IR52c-GAL4 and ppk25-GAL4 labels two neurons (H), indicating that these two drivers express in different cells (H, arrowheads). Neurons in sensilla 5a of male T5 tarsal segments are shown in A-H. Scale bar in A represents 5 μm in A-H.
(I) Contact of the dorsal surface of the male foreleg (arrow) with the female abdomen. See also Movie S1.
(J) A high density of IR52c-GAL4 expressing neurons (arrowheads) close to the sex comb (dotted line). Scale bar represents 10 μm.
(K) Some members of the IR20a clade show signatures of adaptive evolution.
Four of 26 members of the D. melanogaster IR20a clade show signatures of adaptive evolution, when compared with their orthologs in D. simulans using the McDonald-Kreitman test (MKT). MKT looks for evidence of natural selection by comparing variation within species to the divergence between species. DoS (Direction of Selection) indicates adaptive evolution when positive (boxed) and purifying selection when negative (Stoletzki and Eyre-Walker, 2011). The Lhr and OdsH genes, which are responsible for hybrid incompatibility between D. melanogaster and D. simulans, serve as references for DoS values for adaptive evolution. Data were derived from the popDrowser website (Ramia et al., 2011). Data for IR52b and IR60d were not available, and those for 7 putative pseudogenes are not included. “*” denotes P<0.05 and “**” denotes P<0.01.