Abstract
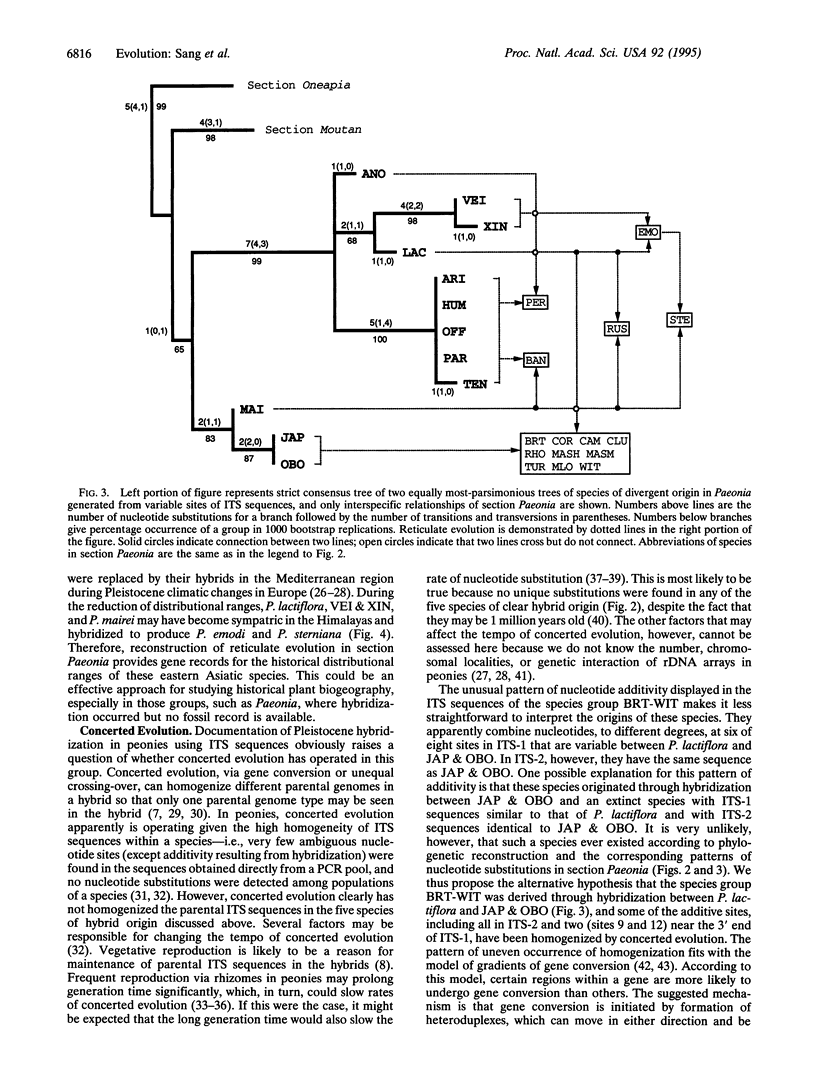
The internal transcribed spacers (ITS) of nuclear ribosomal DNA of 33 species of genus Paeonia (Paeoniaceae) were sequenced. In section Paeonia, different patterns of nucleotide additivity were detected in 14 diploid and tetraploid species at sites that are variable in the other 12 species of the section, suggesting that reticulate evolution has occurred. Phylogenetic relationships of species that do not show additivity, and thus ostensibly were not derived through hybridization, were reconstructed by parsimony analysis. The taxa presumably derived through reticulate evolution were then added to the phylogenetic tree according to additivity from putative parents. The study provides an example of successfully using ITS sequences to reconstruct reticulate evolution in plants and further demonstrates that the sequence data could be highly informative and accurate for detecting hybridization. Maintenance of parental sequences in the species of hybrid origin is likely due to slowing of concerted evolution caused by the long generation time of peonies. The partial and uneven homogenization of parental sequences displayed in nine species of putative hybrid origin may have resulted from gradients of gene conversion. The documented hybridizations may have occurred since the Pleistocene glaciations. The species of hybrid origin and their putative parents are now distantly allopatric. Reconstruction of reticulate evolution with sequence data, therefore, provides gene records for distributional histories of some of the parental species.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Coen E., Strachan T., Dover G. Dynamics of concerted evolution of ribosomal DNA and histone gene families in the melanogaster species subgroup of Drosophila. J Mol Biol. 1982 Jun 15;158(1):17–35. doi: 10.1016/0022-2836(82)90448-x. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Eickbush T. H., Burke W. D. The silkmoth late chorion locus. II. Gradients of gene conversion in two paired multigene families. J Mol Biol. 1986 Aug 5;190(3):357–366. doi: 10.1016/0022-2836(86)90007-0. [DOI] [PubMed] [Google Scholar]
- Gaut B. S., Muse S. V., Clark W. D., Clegg M. T. Relative rates of nucleotide substitution at the rbcL locus of monocotyledonous plants. J Mol Evol. 1992 Oct;35(4):292–303. doi: 10.1007/BF00161167. [DOI] [PubMed] [Google Scholar]
- Gunnersen J. M., Crawford R. J., Tregear G. W. Expression of the relaxin gene in rat tissues. Mol Cell Endocrinol. 1995 Apr 28;110(1-2):55–64. doi: 10.1016/0303-7207(95)03516-a. [DOI] [PubMed] [Google Scholar]
- Hess J. F., Schmid C. W., Shen C. K. A gradient of sequence divergence in the human adult alpha-globin duplication units. Science. 1984 Oct 5;226(4670):67–70. doi: 10.1126/science.6474190. [DOI] [PubMed] [Google Scholar]
- Hillis D. M., Dixon M. T. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol. 1991 Dec;66(4):411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- Hillis D. M., Moritz C., Porter C. A., Baker R. J. Evidence for biased gene conversion in concerted evolution of ribosomal DNA. Science. 1991 Jan 18;251(4991):308–310. doi: 10.1126/science.1987647. [DOI] [PubMed] [Google Scholar]
- Li W. H., Tanimura M., Sharp P. M. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J Mol Evol. 1987;25(4):330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- Nagylaki T., Petes T. D. Intrachromosomal gene conversion and the maintenance of sequence homogeneity among repeated genes. Genetics. 1982 Feb;100(2):315–337. doi: 10.1093/genetics/100.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., Dover G. A. Population genetics of multigene families that are dispersed into two or more chromosomes. Proc Natl Acad Sci U S A. 1983 Jul;80(13):4079–4083. doi: 10.1073/pnas.80.13.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J. F., Schnabel A., Seelanan T. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc Natl Acad Sci U S A. 1995 Jan 3;92(1):280–284. doi: 10.1073/pnas.92.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]