SUMMARY
Mutations of SDCCAG8 are associated with nephronophthisis and Bardet-Biedl Syndrome, as well as schizophrenia; however, the function of SDCCAG8 remains largely unknown. Here, we show that SDCCAG8 regulates centrosomal accumulation of pericentriolar material and neuronal polarization and migration in the developing mouse cortex. Sdccag8 expression is selectively elevated in newborn neurons prior to their commencement of radial locomotion, and suppression of this expression by short-hairpin RNAs or a loss-of-function allele impairs centrosomal recruitment of γ-tubulin and pericentrin, interferes with microtubule organization, decouples the centrosome and the nucleus, and disrupts neuronal migration. Moreover, SDCCAG8 interacts and co-traffics with pericentriolar material 1 (PCM1), a centriolar satellite protein crucial for targeting proteins to the centrosome. Expression of SDCCAG8 carrying a human mutation causes neuronal migration defects. These results reveal a critical role for SDCCAG8 in controlling centrosomal properties and function, and provide insights into the basis of neurological defects linked to SDCCAG8 mutations.
INTRODUCTION
The mammalian cortex consists of an extraordinary number of neurons organized into a six-layered laminar structure, and is essential for all higher-order brain functions. The development of the cortex begins with the proliferation of neuroepithelial cells that gives rise to radial glial progenitors (RGPs) in the ventricular zone (VZ). The extensive division of RGPs is responsible for producing nearly all cortical excitatory neurons either directly or indirectly through transit amplifying progenitors, such as intermediate progenitors (IPs) (also called basal progenitors) and short neural precursors (Anthony et al., 2004; Englund et al., 2005; Haubensak et al., 2004; Heins et al., 2002; Malatesta et al., 2000; Miyata et al., 2004; Noctor et al., 2001; Noctor et al., 2004; Stancik et al., 2010). Newborn neurons then migrate radially along the radial glial fibers of RGPs to constitute the future cortex (Hatten, 1990; Rakic, 1972). Effective migration of neurons over long distances from their birthplace is essential for the proper formation and function of the cortex, and defects in this migration process have been associated with various brain disorders, such as lissencephaly, epilepsy, mental retardation, and schizophrenia (Ayala et al., 2007; Kerjan and Gleeson, 2007; Kwan et al., 2012; Metin et al., 2008; Ross and Walsh, 2001; Valiente and Marin, 2010).
Previous studies suggest that centrosomal integrity and function is crucial for the mammalian brain development (Higginbotham and Gleeson, 2007; Kuijpers and Hoogenraad, 2011). As the major microtubule organizing center (MTOC) in animal cells, the centrosome influences most if not all MT-dependent processes, including cell shape, polarity and motility, as well as mitotic spindle formation and cell division (Bettencourt-Dias and Glover, 2007; Bornens, 2012; Luders and Stearns, 2007). It also serves as the basal body for ciliogenesis. In the developing cortex, both progenitor cell division and neuronal migration appear to be dependent on the centrosome. An increasing number of centrosomal proteins have been genetically linked to microcephaly largely resulting from defects in cortical neurogenesis, underscoring the importance of the centrosome in progenitor cell division (Higginbotham and Gleeson, 2007; Nigg and Raff, 2009). The centrosome has also been shown to lead the way during glial-guided neuronal migration (Tsai and Gleeson, 2005), the predominant mode of neuronal migration in the developing cortex (Rakic, 1972). In most radially migrating cortical neurons, the centrosome is localized ahead of the nucleus, facing the leading process (Cooper, 2013; Tsai and Gleeson, 2005). Live imaging analysis showed that during saltatory nucleokinesis of migrating neurons, the centrosome first moves into the leading process, followed by forward displacement of the nucleus, suggesting a coordinated ‘two–stroke’ movement of the centrosome and the nucleus during neuronal locomotion (Edmondson and Hatten, 1987; Nadarajah et al., 2001; Solecki et al., 2004; Tsai et al., 2007). This coordination between the centrosome and the nucleus depends on the MT network that couples these two organelles, including the perinuclear cage that surrounds the nucleus (Rivas and Hatten, 1995; Tsai and Gleeson, 2005). However, how this MT-based coupling is regulated in radially migrating neurons remains largely unclear.
The capacity of the centrosome to organize MTs is thought to be critical for coordinated movement of the centrosome and the nucleus, and neuronal migration (Kuijpers and Hoogenraad, 2011). The centrosome is composed of two orthogonally arranged centrioles surrounded by a complex matrix of proteins known as the pericentriolar material (PCM) (Bettencourt-Dias and Glover, 2007; Bornens, 2012; Luders and Stearns, 2007). The PCM contains essential proteins responsible for MT nucleation and anchorage, including γ-tubulin (γ-TUB) and pericentrin (PCNT) (Doxsey et al., 1994; Stearns et al., 1991; Zheng et al., 1995). In fact, the PCM is a dynamic structure, and the exchange and transport of PCM is regulated to support MTOC activity and function of the centrosome. For example, in mitotic cells, the amount of γ-TUB at the centrosome increases about 3–5 fold at G2/M phase and this accompanies a strong increase in MT nucleating activity essential for mitotic spindle assembly (Khodjakov and Rieder, 1999). Given its critical role in facilitating cell division, the mitosis-specific PCM recruitment process has been extensively studied (Barr et al., 2004). In contrast, little is known about the regulation of PCM in post-mitotic cells for the fulfillment of centrosomal function.
Centriolar satellites, the highly motile granules concentrated near the centrosome and dispersed across the cytoplasm, have been thought to be crucial for protein trafficking to the centrosome and communication between the centrosome and the surrounding cytoplasm (Kubo et al., 1999). The hallmark protein involved in controlling PCM dynamics through centriolar satellites is pericentriolar material 1 (PCM1) (Kubo et al., 1999). It is a central scaffold of centriolar satellites and removal of PCM1 results in disappearance of the satellite structures near the centrosome, reduction in protein recruitment to the centrosome, and subsequent defect in MT organization at the centrosome (Dammermann and Merdes, 2002). Moreover, it forms a complex with p150glued, a subunit of the dynein/dynactin motor complex, to transport cargo towards the centrosome along the MTs (Kim et al., 2004). Together, these results suggest an important role for PCM1 and centriolar satellites in regulating the capacity of the centrosome to organize MTs.
Serologically defined colon cancer antigen 8 (SDCCAG8) has recently been linked to nephronophthisis (NPHP) (Otto et al., 2010), as well as Bardet-Biedl Syndrome (BBS) (Billingsley et al., 2012; Schaefer et al., 2011) (thus also called NPHP10 and BBS16). It was originally cloned as a stably localized centrosomal protein of unknown function (Kenedy et al., 2003). Candidate exon capture analysis and homozygosity mapping studies identified it as the cause of retinal-renal ciliopathy with diverse clinical phenotypes (Otto et al., 2010). Besides renal and retinal abnormalities, patients with SDCCAG8 mutations often exhibit mental retardation, cognitive impairment and seizures (Otto et al., 2010; Schaefer et al., 2011). Additionally, a genome-wide association study reported SDCCAG8 to be associated with an elevated risk of schizophrenia (Hamshere et al., 2012). These observations indicate an important role for SDCCAG8 in brain development and function. In this study, we found that SDCCAG8 is selectively up-regulated in newborn cortical neurons poised for radial migration. It forms a complex with PCM1 and regulates the recruitment of PCM, including γ-TUB and PCNT, to the centrosome to effect strong MTOC activity. Perturbations of SDCCAG8 expression or function disrupt MT organization and impair coupling between the centrosome and the nucleus, and their coordinated migration. Moreover, expression of a human SDCCAG8 mutant causes neuronal migration defects. Together, these results suggest that SDCCAG8 regulates the centrosomal accumulation of PCM and neuronal migration in the developing cortex, which may underlie the neurological defects associated with human SDCCAG8 mutations.
RESULTS
Selective elevation of SDCCAG8 expression in nascent cortical neurons
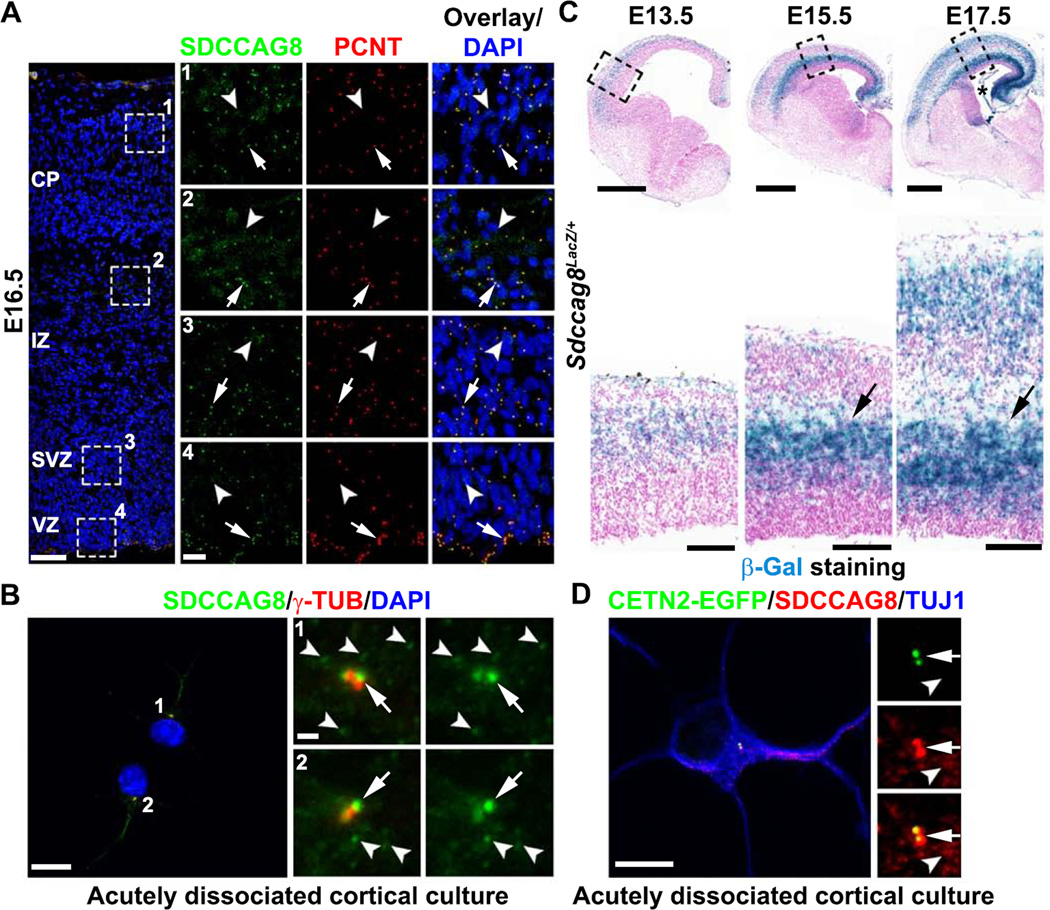
To reveal the function of SDCCAG8 in mammalian cortical development, we examined its expression and subcellular localization in the developing mouse cortex. We stained embryonic day 16.5 (E16.5) mouse brain sections with antibodies against SDCCAG8 and PCNT, a well-characterized centrosomal marker. SDCCAG8 exhibited a punctate expression pattern and co-localized with PCNT (Figures 1A and S1A, arrows), suggesting that SDCCAG8 is localized to the centrosome in the developing cortex. We also noticed small SDCCAG8 puncta away from the centrosome (Figures 1A and S1A, arrowheads). To further characterize the subcellular distribution of SDCCAG8, we prepared acutely dissociated cortical cultures and stained them with the antibodies against SDCCAG8 and γ-TUB, another well-characterized centrosomal marker. At each centrosome labeled by γ-TUB, SDCCAG8 existed as a pair of dots (Figure 1B, arrows), suggesting that it is localized to the centrioles, as previously shown in other cell types (Kenedy et al., 2003; Otto et al., 2010). Besides the centriolar localization, SDCCAG8 was also found as small puncta surrounding the centrosome and dispersed in the cytoplasm (Figure 1B, arrowheads).
Figure 1. SDCCAG8 expression and localization in the developing cortex.
(A) Representative images of E16.5 cortices stained for SDCCAG8 (green) and PCNT (red), a centrosomal marker, and DAPI (blue). High magnification images of the representative areas (broken lines) in the CP (1), IZ (2), SVZ (3), and VZ (4) are shown to the right. Note the localization of SDCCAG8 at the centrosome (arrows) as well as the small puncta in the cytoplasm (arrowheads). Scale bars: 50 µm and 10 µm. (B) Representative images of two acutely dissociated culture cells (1 and 2) from E15.5 cortices stained for SDCCAG8 (green) and γ-TUB (red), another centrosomal marker. High magnification images of the centrosomal region are shown to the right. Note the localization of SDCCAG8 as two dots at the centrosome (arrows), as well as the cytoplasmic puncta (arrowheads). Scale bars: 10 µm and 1 µm. (C) Representative images of brain sections of Sdccag8Lacz/+ mice at E13.5, E15.5, and E17.5 stained for β-Gal (blue) and counter-stained with Nuclear Fast Red (magenta). High magnification images of the cortices (broken lines) are shown at the bottom. Note the selective elevation of Sdccag8 expression around the IZ (arrows), where newborn neurons reside prior to their radial migration to the CP. The asterisk indicates the expression of Sdccag8 in the choroid plexus. Scale bars: 500 µm and 100 µm. (D) Representative images of acutely dissociated cortical neurons expressing CETN2-EGFP (green) stained for SDCCAG8 (red) and TUJ1, an immature neuronal marker (blue). Note the localization of SDCCAG8 at the centrioles (arrows) and the cytoplasmic puncta (arrowheads). Scale bars: 10 µm and 2 µm.
We next characterized the developmental expression profile of SDCCAG8 by exploiting a gene-trap mouse line, in which the LacZ gene was inserted into the Sdccag8 gene locus. Using β-galactosidase (β-Gal) as a reporter of Sdccag8 expression, we found that during the peak phase of cortical neurogenesis and neuronal migration (E12.5-E18.5), Sdccag8 was selectively expressed at high levels around the intermediate zone (IZ) (Figure 1C, arrows), where newborn neurons temporarily reside prior to their radial migration to the cortical plate (CP) – the future cortex. In comparison, the expression of Sdccag8 was low in the VZ and the subventricular zone (SVZ) (Figures 1C, S1C and S1D), where RGPs and IPs are located, respectively. These results suggest that, while SDCCAG8 is present in RGPs and IPs, its expression is selectively up-regulated in postmitotic neurons right before they commence radial migration into the CP. Interestingly, while the level of centrosomal SDCCAG8 in the IZ was not significantly different from that in other regions (Figures 1A and S1A, arrows), the level of cytoplasmic SDCCAG8 as small puncta was higher in the IZ (Figures 1A and S1A, arrowheads). In acutely dissociated cortical neurons identified by the neuronal marker TUJ1, SDCCAG8 was localized to the centrioles labeled by CENTRIN2-EGFP (CETN2-EGFP) (Figure 1D, arrows), as well as the cytoplasmic puncta (Figure 1D, arrowheads). Notably, Sdccag8 was also expressed in the choroid plexus (Figure 1C, asterisk).
SDCCAG8 depletion affects neuronal distribution
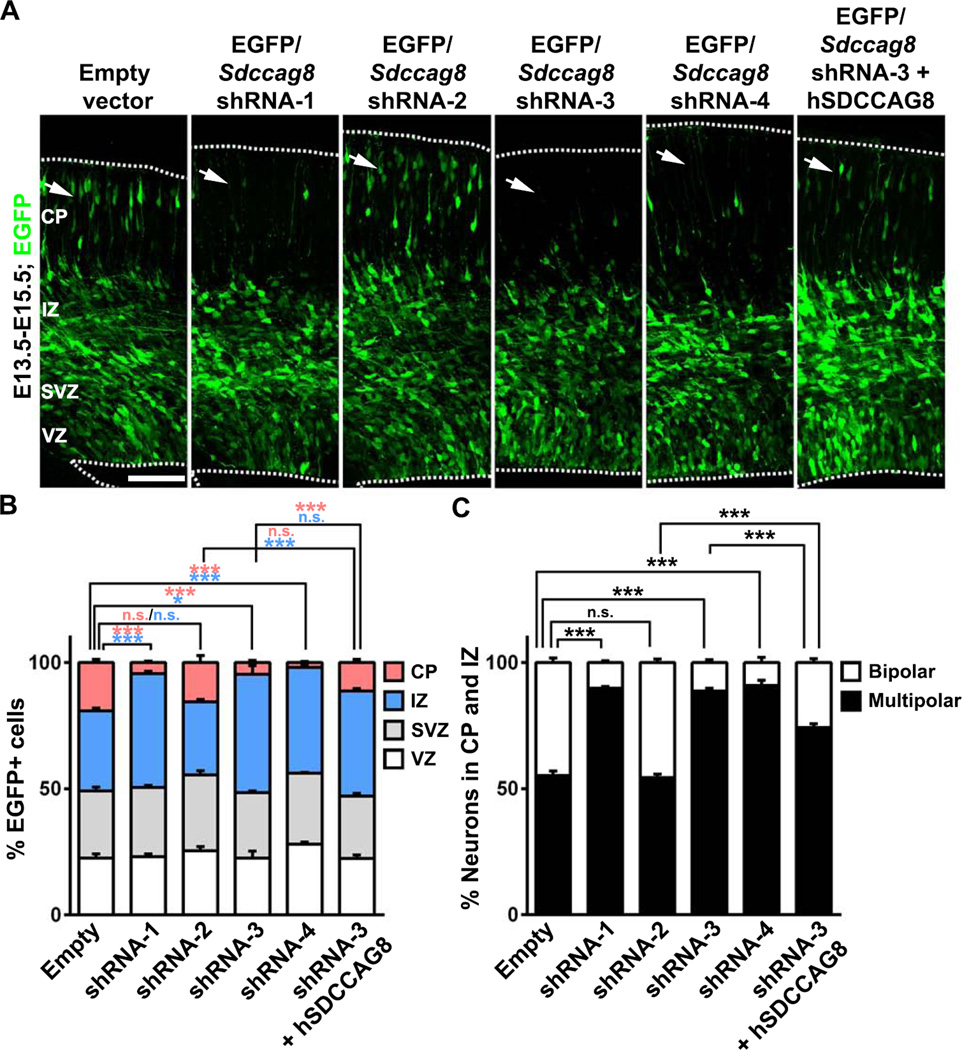
The preferential up-regulation of SDCCAG8 in newborn neurons in the IZ suggests that it may play a functional role. To address this, we developed short hairpin RNAs (shRNAs) against Sdccag8 to suppress its expression. Of four Sdccag8 shRNAs (1 to 4), three (1, 3 and 4) showed effective suppression of SDCCAG8 expression in COS7 cells (Figures S2A) and in the developing cortex (Figure S2B). We then introduced these Sdccag8 shRNAs together with enhanced green fluorescent protein in a bicistronic plasmid, i.e. EGFP/Sdccag8 shRNA, into the developing mouse cortex at E13.5 using in utero electroporation to examine their effect on cortical development. When examined at E15.5, we found that in cortices expressing the empty plasmid with no shRNA, many transfected EGFP-expressing cells were found in the CP (Figure 2A left, arrow). In contrast, in cortices expressing Sdccag8 shRNA-1, −3, or −4, but not −2, EGFP-expressing cells were rarely found in the CP (Figure 2A right, arrows).
Figure 2. Depletion of SDCCAG8 disrupts neuronal distribution in the developing cortex.
(A) Representative images of E15.5 cortices electroporated with EGFP/empty vector control, Sdccag8 shRNA1-4, or Sdccag8 shRNA-3 together with shRNA-resistant human SDCCAG8 (hSDCCAG8) at E13.5. Note a lack of transfected cells expressing EGFP (green) in the CP (arrows) in cortices expressing Sdccag8 shRNA-1, −3 and −4, and the partial rescue of this defect by co-expression of shRNA-resistant hSDCCAG8. Broken lines indicate the pial surface and the VZ surface. Scale bar: 100 µm (B) Quantification of the percentage of EGFP-expressing (EGFP+) cells in different regions of the developing cortex. Data are presented as mean ± s.e.m. (Empty vector, n = 926 cells; shRNA-1, n = 791 cells; shRNA-2, n = 989 cells; shRNA-3, n = 1,150 cells; shRNA-4, n = 1,444 cells; shRNA-3 + hSDCCAG8, n = 1,305 cells; 5 brains for each condition). ***, p<0.001; *, p<0.05; n.s., not significant. (C) Quantification of the percentage of EGFP+ cells in the IZ and the CP with a bipolar or multipolar morphology. Data are presented as mean ± s.e.m. (Empty vector, n = 1,283 cells; shRNA-1, n = 773 cells; shRNA-2, n = 1,267 cells; shRNA-3, n = 1,071 cells; shRNA-4, n = 867 cells; shRNA-3 + hSDCCAG8, n = 765 cells; 5 brains for each condition). ***, p<0.001. n.s., not significant.
We quantified the fraction of EGFP-expressing cells in different regions of the developing cortex. Compared to the empty plasmid control, the fraction of EGFP-expressing cells in the CP was drastically reduced, whereas the fraction in the IZ was concomitantly increased, in cortices expressing Sdccag8 shRNA-1, −3 and −4, but not −2 (Figure 2B). There was no significant difference in the VZ or the SVZ. Consistent with this, we observed no obvious changes in the fraction of EGFP-expressing cells that expressed PAX6 (Figures S3A and S3B), an RGP marker, or TBR2 (Figures S3C and S3D), an IP marker (Englund et al., 2005). In addition, there was no change in neuronal production, as revealed by the fraction of EGFP-expressing cells that expressed TUJ1, an immature neuronal marker (Figures S3E and S3F). Together, these results suggest that depletion of SDCCAG8 does not obviously affect progenitors or the production of neurons, but changes the distribution of neurons in the IZ and the CP. This selective effect on postmitotic neurons likely reflects the preferential up-regulation of Sdccag8 expression in nascent neurons, which renders them sensitive to Sdccag8 shRNAs.
Two additional lines of evidence confirmed the specificity of Sdccag8 shRNAs. First, the effects of Sdccag8 shRNAs were tightly correlated with their efficacy in suppressing SDCCAG8 expression. Sdccag8 shRNA-1, −3 and −4 strongly suppressed SDCCAG8 expression and caused an obvious phenotype, whereas Sdccag8 shRNA-2 failed to suppress SDCCAG8 expression and did not cause any phenotype (Figures 2A, 2B, S2A and S2B). In the following experiments, we used shRNA-1 or −3 to suppress SDCCAG8 expression and shRNA-2 as a control. Second, the defective neuronal distribution in the CP was partially rescued by co-expression of the shRNA-resistant human SDCCAG8 (hSDCCAG8, Figures 2A, 2B and S2C).
Besides the distribution change, we noticed that SDCCAG8 depletion affected the morphology of neurons in the CP and the IZ. Compared to the control, the fraction of neurons with a bipolar morphology was greatly reduced in cortices expressing effective Sdccag8 shRNAs (Figures 2A and 2C). The morphologic switch from multipolar to bipolar has been thought to reflect neuronal polarization (Barnes and Polleux, 2009) and is likely critical for neuronal migration (LoTurco and Bai, 2006). Notably, the defect in this morphological switch was partly overcome by E18.5, as many neurons in the CP and the IZ became bipolar (Figures S4A and S4C). Related to this, there was no obvious deficit in axon specification of dissociated cortical neurons in culture expressing Sdccag8 shRNA (Figures S4D and S4E). However, the distribution of neurons in E18.5 cortices remained severely disrupted (Figures S4A, S4B, S4F and S4H), while the laminar fate specification of neurons were not affected, as revealed by the expression of CUX1 and CTIP2, two well-characterized neuronal markers for the superficial and deep layer neurons, respectively (Greig et al., 2013; Kwan et al., 2012) (Figures S4F and S4G). In addition, SDCCAG8 depletion did not obviously affect the formation of primary cilia by RGPs in the VZ, as revealed by the expression of a well-established ciliary marker ARL13B (Caspary et al., 2007) (Figure S5). Together, these results suggest a key function of SDCCAG8 in regulating neuronal migration in the developing cortex.
SDCCAG8 depletion impairs neuronal migration
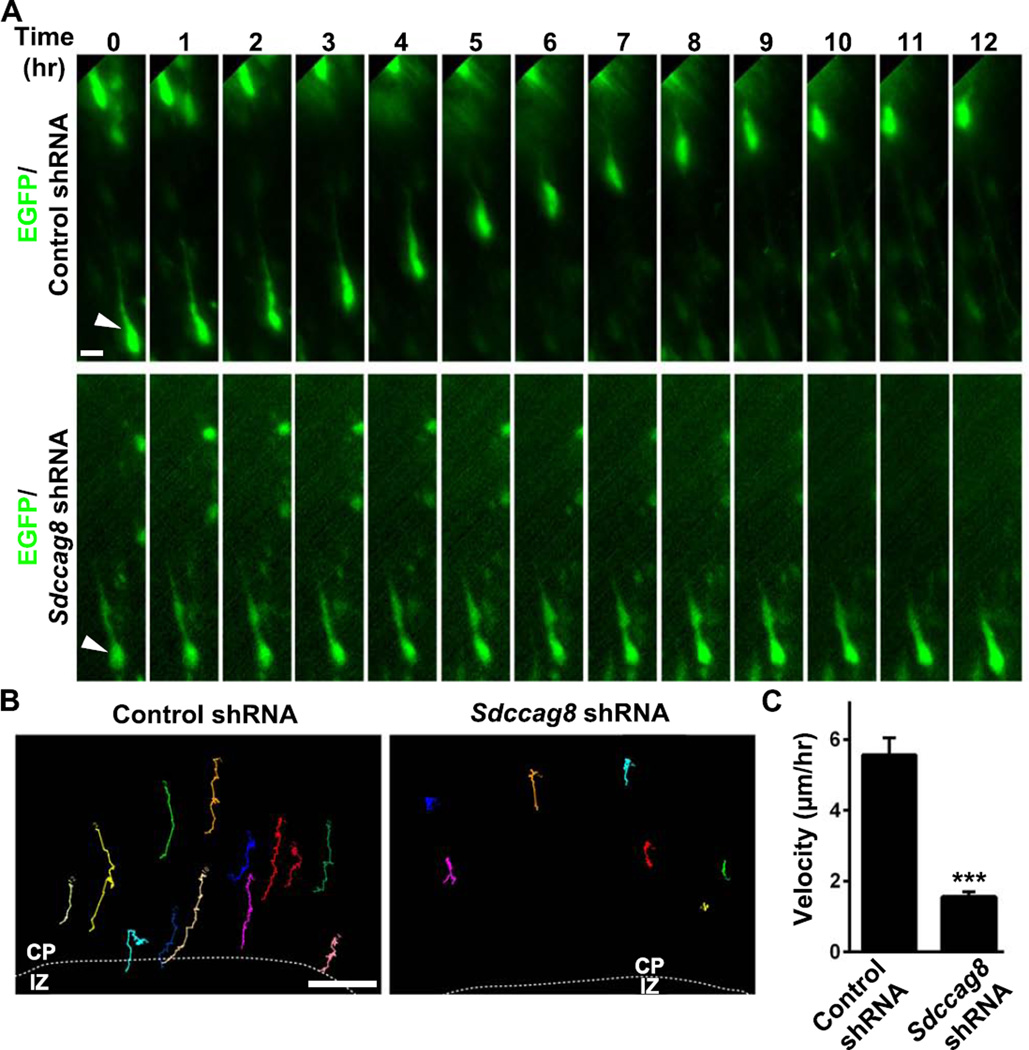
To define the function of SDCCAG8 in regulating neuronal migration, we performed live imaging experiments to track the migration of neurons expressing control or Sdccag8 shRNA. We introduced control or Sdccag8 shRNA together with EGFP into the developing cortex at E13.5 and prepared organotypic cortical slice cultures at E16.5. Slices harboring sparsely labeled neurons in the CP were subjected to time lapse imaging analysis (Figure 3). As expected, bipolar cells expressing EGFP/Control shRNA migrated efficiently through the upper IZ and the CP towards the pial surface (Figures 3A top and 3B left; Movie S1). However, bipolar cells expressing EGFP/Sdccag8 shRNA appeared mostly stationary and rarely migrated over long distances while maintaining a bipolar morphology (Figures 3A bottom and 3B right; Movie S2). Consequently, the velocity of neuronal migration was drastically reduced (Figure 3C). These results clearly showed that SDCCAG8 depletion impairs neuronal migration in the developing cortex.
Figure 3. Depletion of SDCCAG8 impairs neuronal migration.
(A) Representative kymographs of bipolar migrating neurons expressing EGFP/Control or Sdccag8 shRNA (green). Arrows indicate the cell body of neurons. Scale bar: 10 µm. (B) Representative traces of bipolar neurons expressing Control or Sdccag8 shRNA. Broken lines indicate the border between the IZ and the CP. Scale bar: 20 µm. (C) Quantification of the migration velocity of bipolar neurons expressing Control or Sdccag8 shRNA. Data are presented as mean ± s.e.m. (Control, n = 44; Sdccag8, n = 66). ***, p<0.001.
SDCCAG8 depletion disrupts coordinated movement of the centrosome and the nucleus
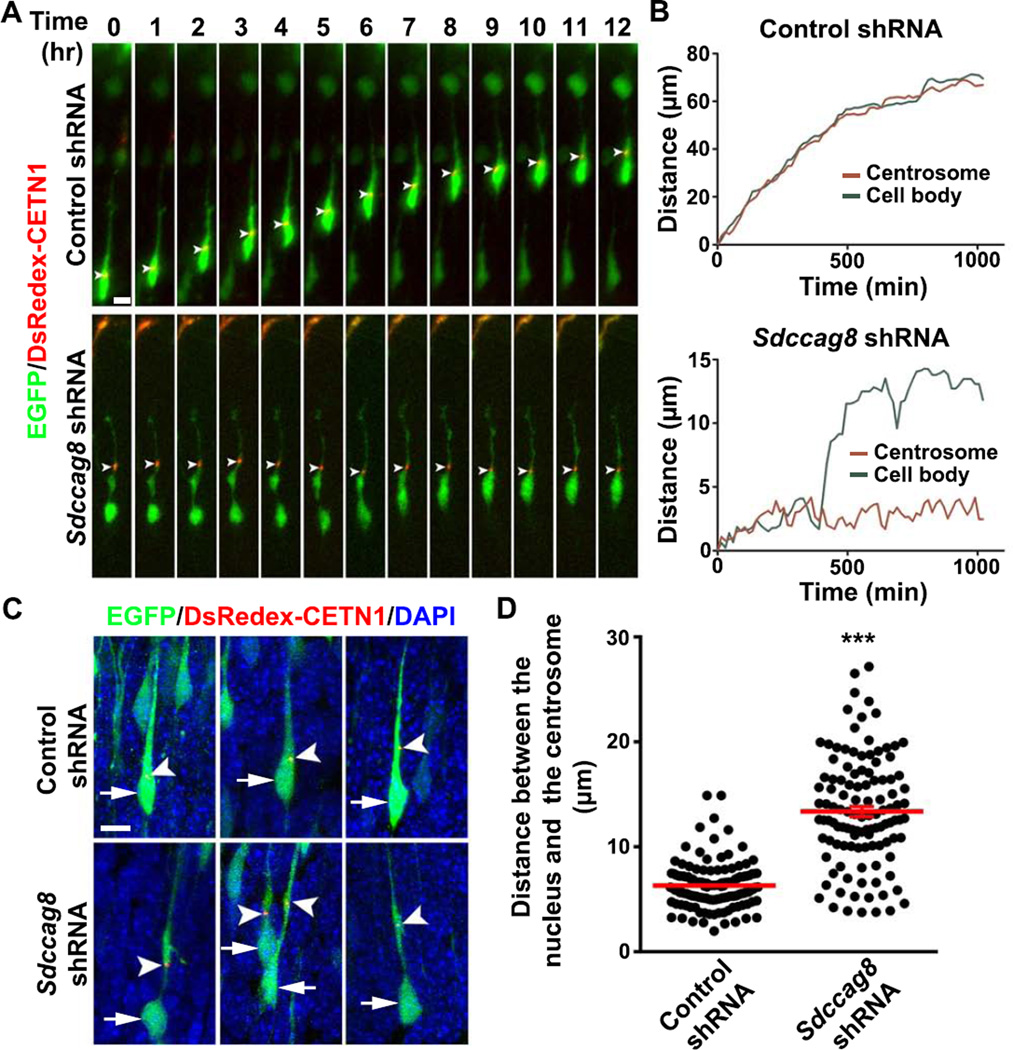
The predominant mode of neuronal radial migration in the developing cortex is glial-guided ‘two-stroke’ locomotion, in which the centrosome advances into a dilated region in the leading process, followed by nuclear translocation (Ayala et al., 2007; Cooper, 2013; Hatten, 1990; Rakic, 1971; Tsai and Gleeson, 2005). A key feature of such migration is the coordinated movement between the nucleus and the centrosome. To further pinpoint the function of SDCCAG8, we performed live imaging analysis of the movement of the centrosome as well as the cell body of migrating bipolar neurons in organotypic cortical slice cultures. We introduced a plasmid carrying Centrin1 fused with DsRedexpress (DsRedex-CETN1) that reliably labels the centrosome (Wang et al., 2009) together with EGFP/Control or Sdccag8 shRNA into the developing cortex. As expected, in migrating bipolar neurons expressing Control shRNA, we observed a progressive advance of the centrosome into the leading process, which was tightly coordinated with nuclear translocation (Figures 4A and 4B, top; Movie S3). As a result, the nucleus always remained relatively close to the centrosome (Figures 4C top and 4D).
Figure 4. Depletion of SDCCAG8 impairs coordinated movement of the centrosome and the nucleus.
(A) Representative kymographs of bipolar migrating neurons expressing DsRedex-CETN1 (red) to label the centrosome and EGFP/Control (top) or Sdccag8 (bottom) shRNA. Arrowheads indicate the position of the centrosome. Scale bar: 5 µm. (B) Traces of the cell bodies (green) and the centrosomes (red) in migrating neurons in (A). (C) Representative images of bipolar neurons in E16.5 cortices electroporated with DsRedex-CETN1 (red) and EGFP/Control or Sdccag8 shRNA (green) at E13.5 and stained with DAPI (blue). Arrows indicate the cell bodies and arrowheads indicate the centrosomes labeled by DsRedex-CETN1. Scale bar: 10 µm. (D) Quantification of the distance between the centrosome and the nucleus (µm) in bipolar neurons. Each dot represents a neuron and red lines represent mean ± s.e.m. (Control, n = 131; Sdccag8, n = 114). ***, p<0.001.
In stark contrast, in bipolar neurons expressing Sdccag8 shRNA, the centrosome appeared mostly stationary in the leading process with the nucleus lagging distantly behind for a long period of time (Figures 4A and 4B, bottom; Movie S4). While the nucleus eventually moved close to the centrosome, we rarely observed a complete two-stroke locomotion of these neurons over the entire course of imaging. Consequently, the distance between the nucleus and the centrosome became highly variable and was substantially increased compared to that in control neurons (Figures 4C bottom and 4D). It is worth noting that the centrosome in neurons expressing Sdccag8 shRNA was often found around the dilated region in the leading process (Figure 4C). We also observed some forward movement of the centrosome in neurons expressing Sdccag8 shRNA (Figure S6A). Therefore, the defect in migration of neurons lacking SDCCAG8 appears to be largely due to the uncoordinated movement of the nucleus and the centrosome.
SDCCAG8 depletion impairs PCM recruitment and MT link between the centrosome and the nucleus
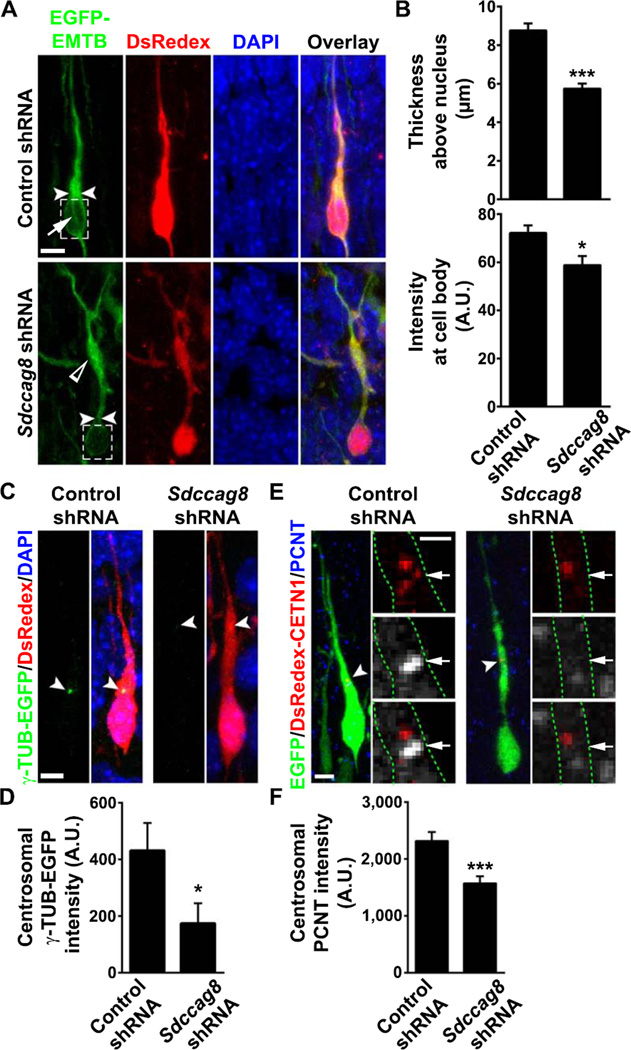
Previous studies showed that a perinuclear cage of MTs is critical for the tight coupling between the nucleus and the centrosome, and their coordinated movement during neuronal locomotion (Rivas and Hatten, 1995). To reveal MT organization in migrating neurons, we utilized the MT binding domain of ensconsin (EMTB) fused to EGFP, EGFP-EMTB, a well-characterized fluorescent probe to visualize MTs (Faire et al., 1999). We introduced EGFP-EMTB together with DsRedex/Control or Sdccag8 shRNA into the developing cortex and examined the distribution of EGFP-EMTB in migrating neurons. In neurons expressing Control shRNA, abundant MTs were found in the leading process, especially above the nucleus (Figure 5A top, arrowheads). Moreover, we frequently found MT bundles extending around the nucleus (Figure 5A top, arrow), likely representing the perinuclear cage. In contrast, MTs in neurons expressing Sdccag8 shRNA were mostly found in the dilated region of the leading process (Figure 5A bottom, open arrowhead) where the centrosome was located (Figure 4C bottom), but not enriched above the nucleus (Figure 5A bottom, arrowheads). We measured the width of EGFP-EMTB-labeled MTs above the nucleus and found that it was substantially reduced in neurons expressing Sdccag8 shRNA compared to control (Figure 5B top). Furthermore, the MTs around the nucleus, as measured by the EGFP-EMTB fluorescence intensity at the cell body (Figure 5A broken lines), were significantly decreased (Figure 5B bottom). Consistent with this loss of MTs around the nucleus that forcefully couple the centrosome and the nucleus in bipolar migrating neurons, we found that the shape of the nucleus was also altered (Figures S6B and S6C). While the nuclei in control neurons were mostly spindle-shaped, they became more rounded in neurons expressing Sdccag8 shRNA, as reflected by a decrease in the ratio of the long to short axis of the nuclei. Together, these results suggest that SDCCAG8 depletion disrupts the MT organization that couples the centrosome and the nucleus in migrating bipolar neurons.
Figure 5. Depletion of SDCCAG8 impairs PCM accumulation and MT organization between the centrosome and the nucleus.
(A) Representative images of bipolar neurons expressing EGFP-EMTB (green) that labels MTs and DsRedex/Control or Sdccag8 shRNA (red) stained for DAPI (blue). Arrowheads indicate the thickness of EGFP-EMTB-labeled MTs above the nucleus and broken lines highlight MTs around the nucleus. The arrow indicates MTs that form the perinuclear cage. The open arrowhead indicates the accumulation of MTs in the dilated region of the leading process some distance away from the nucleus in cells expressing Sdccag8 shRNA. Scale bar: 10 µm (B) Quantification of the thickness of the EGFP-EMTB-labeled MTs above the nucleus (top) and its intensity around the nucleus (bottom). Data are presented as mean ± s.e.m. (Control, n = 24; Sdccag8, n = 28). A.U., arbitrary unit. *, p<0.05; ***, p<0.001. (C) Representative images of bipolar neurons expressing γ-TUB-EGFP (green) and DsRedex/Control or Sdccag8 shRNA stained with DAPI (blue). Arrowheads indicate the centrosomes where γ-TUB-EGFP accumulates. Scale bar: 5 µm. (D) Quantification of the γ-TUB-EGFP focus intensity at the centrosome. Data are presented as mean ± s.e.m. (Control, n = 54; Sdccag8, n = 43). *, p<0.05 (E) Representative images of bipolar neurons expressing DsRedex-CETN1 (red) and EGFP/Control or Sdccag8 shRNA (green) stained for PCNT (blue or white). Broken lines indicate the cell boundary. Arrowheads and arrows indicate the centrosomes labeled by DsRedex-CETN1. Scale bars: 5 µm and 2 µm. (F) Quantification of the PCNT intensity at the centrosome. Data are presented as mean ± s.e.m. (Control, n = 106; Sdccag8, n = 100). ***, p<0.001.
The centrosome functions as the major MTOC. The aberrant MT organization in neurons expressing Sdccag8 shRNA indicates potential defects of the centrosome. Given that the PCM at the centrosome is essential for MT organization (Doxsey et al., 1994; Stearns et al., 1991; Zheng et al., 1995), we examined the centrosomal accumulation of the PCM by analyzing two classic PCM proteins, γ-TUB and PCNT. We introduced γ-TUB fused with EGFP (Khodjakov and Rieder, 1999), γ-TUB-EGFP, together with DsRedex/Control or Sdccag8 shRNA into the developing cortex and examined the localization of γ-TUB-EGFP in migrating bipolar neurons. In neurons expressing control shRNA, a prominent focus of γ-TUB-EGFP was frequently observed at the location where the centrosome typically resided (Figures 5C left, arrowheads, and 5D); however, γ-TUB-EGFP foci were barely observed in neurons expressing Sdccag8 shRNA (Figures 5C right, arrowheads, and 5D). These results suggest that SDCCAG8 depletion disrupts the centrosomal accumulation of γ-TUB.
We next examined the effect of SDCCAG8 depletion on the centrosomal accumulation of endogenous PCNT. The centrosome was labeled by DsRedex-CENT1 and endogenous PCNT was revealed by immunofluorescence. Robust PCNT puncta were frequently found at the centrosome in neurons expressing control shRNA (Figures 5E left arrows, and 5F), but were less prominent in neurons expressing Sdccag8 shRNA (Figures 5E right arrows, and 5F), suggesting that SDCCAG8 depletion impairs the centrosomal accumulation of PCNT. Notably, expression of Sdccag8 shRNA did not affect the expression level of γ-TUB and PCNT (Figure S6D). Together, these results demonstrate that SDCCAG8 regulates the centrosomal recruitment of PCM, which is essential for the MTOC activity of the centrosome and the proper MT organization that couples the centrosome and the nucleus in migrating neurons.
SDCCAG8 co-localizes and co-traffics with PCM1
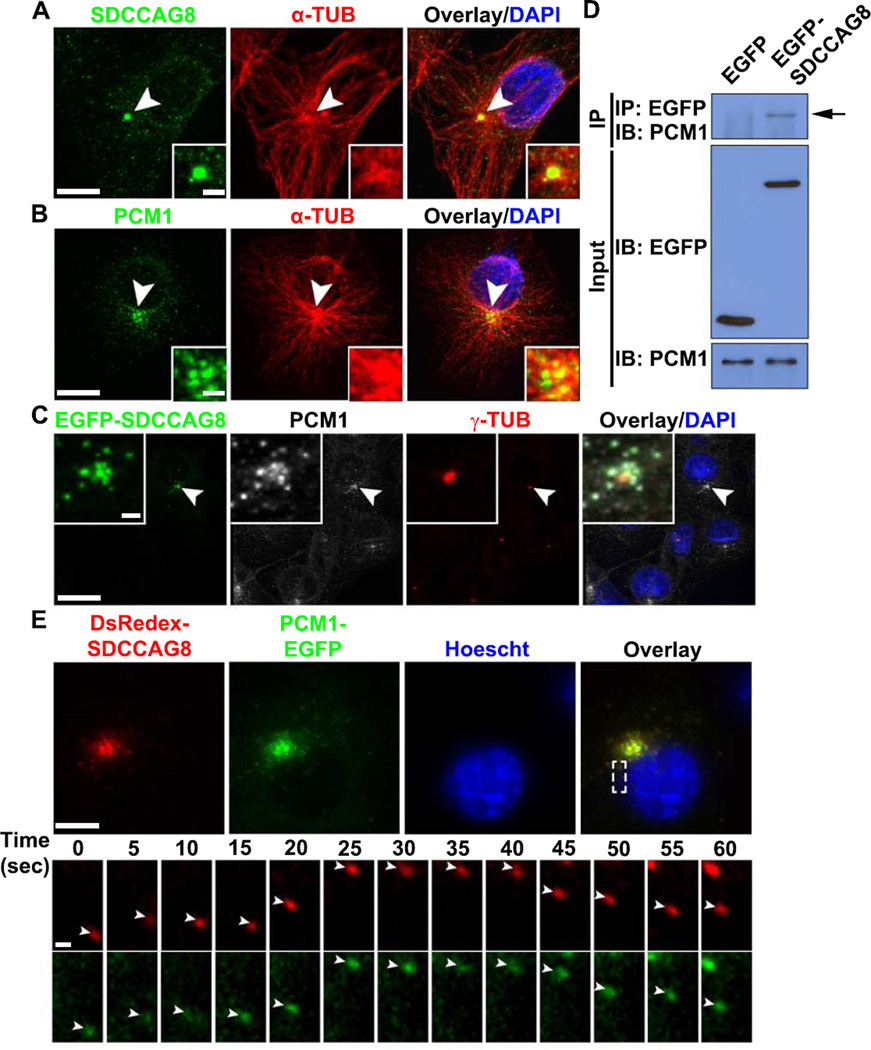
The recruitment of PCM to the centrosome has often been associated with small granules that are transported along MTs known as centriolar satellites (Dammermann and Merdes, 2002; Kubo et al., 1999). While SDCCAG8 was localized to the centrioles (Figures 1A, 1B, 1D and S1A, arrows), it was also found as small puncta surrounding the centrosome as well as in the cytoplasm (Figures 1A, 1B, 1D and S1A, arrowheads). To test whether these small SDCCAG8 puncta represent centriolar satellites along MTs, we examined the distribution of SDCCAG8 in COS7 cells, which provide better spatial resolution of MT organization. We found that SDCCAG8 in COS7 cells exhibited a similar expression pattern as neurons (Figure 6A). In addition to the centrosomal localization, SDCCAG8 puncta were found to be distributed along MTs around the centrosome and throughout the cell. This expression pattern was reminiscent of PCM1 (Figure 6B), a well-characterized centriolar satellite protein that plays a central role in targeting proteins to the centrosome (Dammermann and Merdes, 2002; Kubo et al., 1999). Moreover, we found that SDCCAG8 tagged with EGFP, which exhibited the same distribution pattern as endogenous SDCCAG8, co-localized with PCM1 (Figure 6C) and co-immunoprecipitated with PCM1 (Figure 6D). These results suggest that SDCCAG8 forms a complex with PCM1 at centriolar satellites.
Figure 6. SDCCAG8 interacts, co-localizes and co-traffics with PCM1.
(A, B) Representative images of COS7 cells stained for SDCCAG8 (A) or PCM1 (B) (green), α-TUB (red) and with DAPI (blue). Arrowheads indicate the centrosome. High magnification images of the centrosomal region are shown in the inset. Scale bars: 10 µm and 2.5 µm. (C) Representative images of COS7 cells expressing a low level of EGFP-SDCCAG8 (green) stained for PCM1 (white), γ-TUB (red), and with DAPI (blue). Arrowheads indicate the centrosome and high magnification images of the centrosomal region are shown in the inset. Scale bars: 20 µm and 2 µm. (D) Interaction between EGFP-SDCCAG8 with PCM1 detected by co-immunoprecipitation in COS7 cells. (E) Representative images of live COS7 cells expressing DsRedex-SDCCAG8 and PCM1-EGFP stained with Hoescht (blue). High magnification kymographs of one region (broken lines) are shown at the bottom. Note that DsRedex-SDCCAG8 and PCM1-EGFP puncta (arrowheads) co-traffic. Scale bars: 10 µm and 1 µm.
To test whether SDCCAG8 indeed traffics together with PCM1, we introduced DsRedex-tagged SDCCAG8, DsRedex-SDCCAG8, and EGFP-tagged PCM1, PCM1-EGFP, into COS7 cells, and performed live imaging experiments (Figure 6E). Both DsRedex-SDCCAG8 and PCM1-EGFP formed small puncta around the centrosome and throughout the cytoplasm, and co-localized well with each other (Figure 6E top). Moreover, SDCCAG8-DsRedex and PCM1-EGFP puncta moved together rapidly in the cell (Figure 6E bottom; Movie S5). These results demonstrate that SDCCAG8 and PCM1co-localize and co-traffic together.
SDCCAG8 over-expression delocalizes PCM1 and impairs centrosomal PCM recruitment and neuronal migration
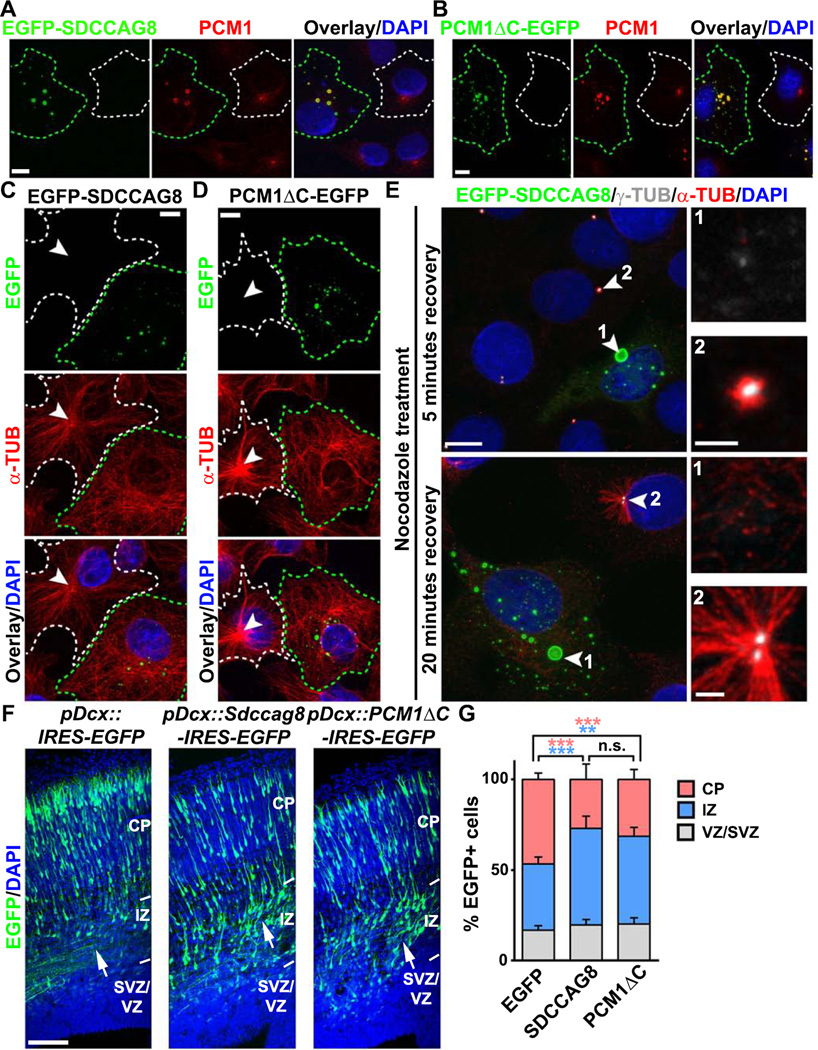
SDCCAG8 contains multiple coiled-coil domains (Otto et al., 2010). Similar to other coiled-coil domain-containing proteins that have a strong propensity for oligomerization (Burkhard et al., 2001), over-expression of SDCCAG8 led to the formation of ectopic aggregates (Figure 7A, green). Consistent with a tight association between SDCCAG8 and PCM1, we found that PCM1 was strongly recruited to the ectopic SDCCAG8 aggregates (Figure 7A, red). This delocalization of PCM1 was phenotypically similar to the expression of the carboxyl-terminal truncation mutant form of PCM1, PCM1ΔC-EGFP, which aggregated and sequestered endogenous PCM1 (Figure 7B, green cell), as previously shown (Dammermann and Merdes, 2002). Moreover, similar to PCM1ΔC expression, SDCCAG8 over-expression impaired the accumulation of PCM, including both γ-TUB and PCNT, at the centrosome (Figures S7A and S7B). Consequently, the MT organization was disrupted (Figures 7C and 7D). While control cells contained a well focused MT network, radiating from a single MTOC at the centrosome (broken white lines, arrowheads), cells over-expressing SDCCAG8 or PCM1ΔC contained mostly randomly distributed MTs throughout the cytoplasm (broken green lines).
Figure 7. SDCCAG8 over-expression disrupts PCM1 accumulation, MT organization and neuronal migration.
(A, B) Representative images of COS7 cells expressing EGFP-SDCCAG8 (A) or PCM1ΔC-EGFP (B) (green) stained for endogenous PCM1 (red) and with DAPI (blue). Green broken lines indicate an EGFP-SDCCAG8- or PCM1ΔC-EGFP-expressing cell and white broken lines indicate a nearby non-transfected cell. Scale bars: 10µm. (C, D) Representative images of COS7 cells expressing EGFP-SDCCAG8 (C) or PCM1ΔC-EGFP (D) (green) stained for α-TUB (red) and with DAPI (blue). Green broken lines indicate an EGFP-SDCCAG8- or PCM1ΔC-EGFP-expressing cell and white broken lines indicate a nearby non-transfected cell. Arrowheads indicate the radial organization of MTs emanating from the centrosome. Scale bars: 10µm. (E) Representative images of COS7 cells expressing EGFP-SDCCAG8 (green, cell 1) recovering from nocodazole treatment stained for γ-TUB (white) and α-TUB (red), and with DAPI (blue). Arrowheads indicate the centrosomes. High magnification images of the centrosomal region are shown to the right. Scale bars: 10 µm, 2 µm, and 2 µm. (F) Representative images of E16.5 cortices electroporated with EGFP (left), SDCCAG8-IRES-EGFP (middle), or PCM1ΔC-IRES-EGFP (right) (green) under the control of a doublecortin (Dcx) promoter at E13.5 and stained with DAPI (blue). Arrows indicate the accumulation of cells expressing SDCCAG8 or PCM1ΔC in the IZ. IRES stands for internal ribosome entry site. Scale bar: 100 µm. (G) Quantification of the distribution of EGFP+ cells in different regions of the developing cortex. Data are presented as mean ± s.e.m. (EGFP, n = 2,407 cells; SDCCAG8, n = 1,703 cells; PCM1ΔC, n = 1,763 cells; five brains for each condition). ***, p<0.001; **, p<0.01; n.s., not significant.
To test whether this is due to defective MT nucleation and anchorage at the centrosome, we performed an MT re-growth assay (Figure 7E). While control cells showed robust initial growth of small MT asters at the centrosome harboring a high level of γ-TUB (cell 2, arrowhead), cells over-expressing SDCCAG8 did not (cell 1, arrowhead). Within 20 minutes, an exuberant radial MT organization was obvious in control cells (cell 2, arrowhead), whereas only scarce MTs were found to be randomly distributed in the cytoplasm in cells over-expressing SDCCAG8 (cell 1, arrowhead). Together, these results suggest that SDCCAG8 over-expression delocalizes PCM1, disrupts centrosomal accumulation of PCM, and impairs MT organization at the centrosome. Notably, no MTs were found to be nucleated or anchored at the protein aggregates formed by SDCCAG8, suggesting that proteins segregated to these structures were not competent to nucleate or anchor MTs by themselves.
Having found that SDCCAG8 over-expression impairs PCM recruitment and centrosomal MTOC activity, we next examined its effect on neuronal migration. When expressed in dissociated cortical neurons in culture, SDCCAG8 tagged with FLAG colocalized with PCM1 (Figure S7C). Previous studies have shown that PCM1-mediated PCM recruitment is important for the behavior of RGPs (Ge et al., 2010). To avoid potential alterations in neurogenesis, we selectively expressed SDCCΔG8 or PCM1AC together with EGFP in postmitotic neurons using a doublecortin (Dcx) promoter (Franco et al., 2011). Compared to control neurons expressing EGFP alone, neurons expressing EGFP/SDCCAG8 or EGFP/PCM1ΔC exhibited a strong migration phenotype (Figures 7F, 7G and S7D–F), similar to that of neurons expressing Sdccag8 shRNA (Figures 2A and 2B) or Pcm1 shRNA (de Anda et al., 2010; Kamiya et al., 2008). While a majority of control neurons exited the IZ and migrated to the CP, a large number of neurons expressing SDCCΔG8 or PCM1AC remained in the IZ (Figures 7F arrows and S7D–F). These results suggest that disruptions of PCM recruitment to the centrosome and MT organization in migrating neurons by over-expression of SDCCAG8 or PCM1ΔC impair neuronal migration.
Expression of human SDCCAG8 truncation mutant disrupts neuronal migration
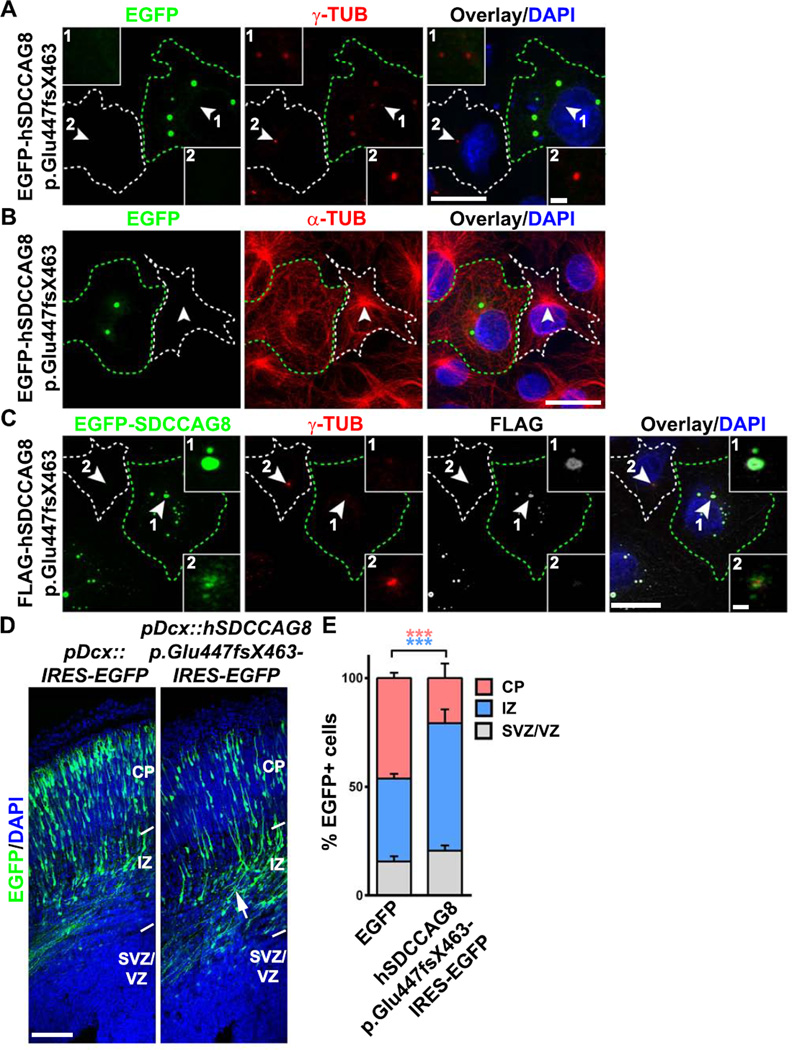
Mutations of SDCCAG8 have been identified from a cohort of human patients afflicted with NPHP, as well as seizure and mental retardation (Otto et al., 2010). To gain more insight, we analyzed one particular truncating mutation that causes mental retardation, hSDCCAG8 p.Glu447fsX463, in which a single base G is inserted between nucleotides 1339–1340, resulting in a reading frame shift and protein truncation. We found that this mutation disrupted the interaction between hSDCCAG8 and PCM1 (Figures S8A and S8B) and failed to rescue the neuronal migration defect caused by Sdccag8 shRNA (Figures 2, S8C and S8D). When expressed in COS7 cells, hSDCCAG8 p.Glu447fsX463 formed aggregates and disrupted centrosomal accumulation of PCM protein y-TUB (Figure 8A) and MT organization (Figure 8B). Given that hSDCCAG8 p.Glu447fsX463 did not interact with PCM1 (Figures S8A and S8B), we tested the possibility that this mutant protein exerted its effect through interaction with and sequestration of wild type full length SDCCAG8, which in turn disrupted centrosomal PCM recruitment and MTOC activity. Consistent with this, we found that hSDCCAG8 p.Glu447fxX463 recruited full length SDCCAG8 to the aggregates and impaired centrosomal accumulation of γ-TUB (Figures 8C and S8E). Moreover, expression of this mutant human truncated form of SDCCAG8 disrupted neuronal migration in the developing cortex (Figures 8D and 8E). These results further support the function of SDCCAG8 in regulating centrosomal accumulation of PCM and neuronal migration, and suggest that defective neuronal migration in the cortex likely contributes to the brain defect in human patients with SDCCAG8 mutations.
Figure 8. Expression of human SDCCAG8 truncation mutant disrupts PCM recruitment, MT organization and neuronal migration.
(A, B) Representative images of COS7 cells expressing EGFP-hSDCCAG8 p.Glu447fsx463 (green) stained for γ-TUB (A) or α-TUB (red) and with DAPI (blue). Arrowheads indicate the centrosome. Green broken lines indicate the cell expressing EGFP-hSDCCAG8 p.Glu447fsx463 (cell 1) and white broken lines indicate a nearby non-transfected cell (cell 2). High magnification images of the centrosomal region are shown in the inset. Scale bars: 20 µm and 2 µm. (C) Representative images of COS7 cells expressing FLAG-hSDCCAG8 p.Glu447fsx463 (white) and EGFP-SDCCAG8 (green) stained for γ-TUB (red) and with DAPI (blue). Arrowheads indicate the centrosomes. Green broken lines indicate the transfected cell (cell 1) and white broken lines indicate the nearby non-transfected cell (cell 2). High magnification images of the centrosomal region are shown in the inset. Scale bars: 20 µm and 2 µm. (D) Representative images of E16.5 cortices electroporated with EGFP (left) or hSDCCAG8 p.Glu447fsx463/EGFP (right) (green) under the control of the Dcx promoter at E13.5 stained with DAPI (blue). Arrow indicates the accumulation of cells expressing hSDCCAG8 p.Glu447fsx463 in the IZ. Scale bar: 100 µm. (E) Quantification of the distribution of EGFP+ cells in different regions of the developing cortex. Data are presented as mean ± s.e.m. (EGFP, n = 2,241 cells; hSDCCAG8 p.Glu447fsX463, n = 1,826 cells; five brains for each condition). ***, p<0.001.
Sdccag8 knockout impairs neuronal migration in the cortex
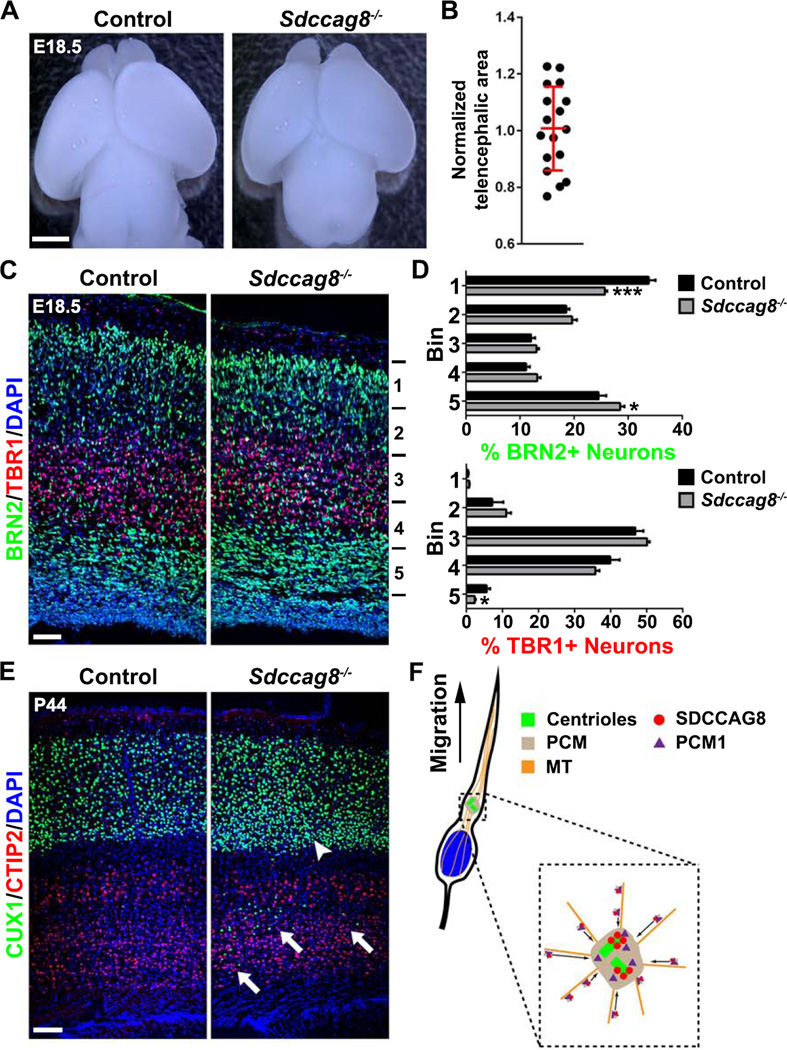
To further define the function of SDCCAG8 in regulating neuronal migration, we generated a loss-of-function mouse allele of Sdccag8, as confirmed by Western blot (Figure S9A) and immunohistochemistry (Figure S9B). While wild type (Sdccag8+/+) and heterozygous (Sdccag8+/−) animals survived to adulthood with no obvious deficits, homozygous (Sdccag8−/−) animals mostly died after birth with obvious defects of cleft palate and polydactyly (Figure S9C). We observed one Sdccag8−/− animal that survived to postnatal day (P) 44 with no clear cleft palate but polydactyly, and was sacrificed due to malocclusion. Notably, compared to littermate wild type or heterozygous controls, there was no obvious microcephaly in Sdccag8-/- animals at E18.5 (Figures 9A and 9B), suggesting cortical neurogenesis is grossly normal in mice lacking SDCCAG8. In addition, we observed no obvious defects in primary cilia formation by RGPs at the VZ surface (Figures S9E–G). A few (4 out of 18) Sdccag8−/− animals exhibited an enlarged lateral ventricle (Figure S9D), which might be due to abnormalities of primary cilia in the choroid plexus (Figures S9H and S9I, arrows), where SDCCAG8 was abundantly expressed (Figure 1C, asterisk).
Figure 9. Sdccag8 knockout impairs neuronal migration in the cortex.
(A) Representative whole mount images of E18.5 Control (Sdccag8+/+ or Sdccag8+/−) and Sdccag8−/− brains. Scale bar: 2 mm. (B) Quantification of relative telencephalic area of Sdccag8−/− brains cortices to the littermate controls. Red lines represents mean ± s.e.m (n = 17). (C) Representative images of E18.5 Control (Sdccag8+/−) and Sdccag8−/− cortices stained for BRN2 (green) and TBR1 (red), two specific markers for the superficial and deep layer neurons, and with DAPI (blue). Scale bar: 50 µm. (D) Quantification of the percentage of BRN2+ and TBR1+ neurons in different bins of the cortex (Control, n = 7 brains; Sdccag8−/−, n = 6 brains). ***, p<0.001; *, p<0.05. (E) Images of P44 Control and Sdccag8−/− cortices stained for CUX1 (green) and CTIP2 (red), the respective superficial and deep layer neuronal markers, and with DAPI (blue). Note the presence of substantial CUX1+ neurons near the bottom of the cortex (arrows) and the relative high density of CUX1+ neurons at the bottom of the superficial layers (arrowhead), indicating a neuronal migration defect. Scale bar: 100 µm. (F) A model illustrating the role of SDCCAG8 in regulating PCM recruitment and neuronal migration in the developing cortex.
We next assessed neuronal migration in the developing cortices of Sdccag8−/− animals and their littermate controls. We stained E18.5 brain sections with no obviously enlarged ventricle for BRN2 and TBR1, two specific neuronal markers for the superficial and deep layer neurons (Greig et al., 2013; Kwan et al., 2012), and examined the distribution of these two neuronal populations in the cortex (Figures 9C and 9D). Interestingly, compared to the control, there were significantly less BRN2-expressing neurons at the top of the cortex, but substantially more BRN2-expressing neurons near the bottom of the cortex, in Sdccag8−/− animals. In addition, there were less TBR1-expressing neurons near the bottom of the cortex. These results suggest that SDCCAG8 removal impairs neuronal migration in the developing cortex. Consistent with this, we found that in the P44 Sdccag8−/− cortex there were substantially more CUX1-expressing neurons in the deep layers (Figure 9E arrows), as well as in the lower part of the superficial layers (Figure 9E arrowhead), than that in the littermate control cortex. Together, these results clearly demonstrate that SDCCAG8 is required for proper neuronal migration in the cortex.
DISCUSSION
In this study, we showed that SDCCAG8 regulates centrosomal recruitment of PCM, which supports the MTOC activity of the centrosome, the MT-mediated coupling between the centrosome and the nucleus, and neuronal migration in the developing cortex (Figure 9F). This regulation is achieved through the interaction between SDCCAG8 and PCM1, a principal centriolar satellite protein involved in targeting proteins to the centrosome. Either depletion or over-expression of SDCCAG8 disrupted centrosomal recruitment of PCM and cortical neuronal migration.
The function of the centrosome depends on its MT nucleation and organization activity, which is largely carried out by the PCM (Doxsey et al., 1994; Stearns et al., 1991; Zheng et al., 1995). Therefore, the abundance of PCM at the centrosome is often tightly regulated. For example, the mitotic kinase Polo-like kinase 1 (Plk1)-dependent recruitment of PCM to mitotic centrosomes in late G2 cells is important for bipolar spindle formation and cytokinesis (Barr et al., 2004; Wang et al., 2011). We found that SDCCAG8 is present in both progenitor cells and postmitotic neurons of the developing cortex. However, the expression of SDCCAG8 is preferentially up-regulated, especially at the centriolar satellites, in postmitotic neurons in the IZ that are poised for persistent radial migration. Moreover, removal of SDCCAG8 impaired neuronal migration. Notably, there is no microcephaly in Sdccag8−/− mice, although we could not exclude any subtle changes in progenitors and neurogenesis. Mutations of a number of centrosomal proteins have been associated with microcephaly (Gilmore and Walsh, 2013; Higginbotham and Gleeson, 2007; Nigg and Raff, 2009), while other centrosomal protein mutations are linked to ‘ciliopathies’ and perhaps mental retardation but no microcephaly (Oh and Katsanis, 2012). These differences in phenotypic manifestations may be related to distinct functions of the corresponding proteins at the centrosome. Our results suggest that an important function of SDCCAG8 is to regulate PCM recruitment in postmitotic neurons to support their migration. Regulation of PCM at the centrosome in mitotic and non-mitotic cells probably involves distinct mechanisms. It will be interesting to test whether SDCCAG8 and PCM1 together represent a general molecular pathway in regulating PCM abundance in non-mitotic cells that demand a strong MTOC activity.
Neuronal migration in the developing cortex is complex. Newborn neurons actively change their morphology and mode of movement to complete the process. After a brief initial radial migration from the site of origin to the SVZ, they adopt a multipolar morphology and largely pause in the SVZ or the bottom of the IZ (Kriegstein and Noctor, 2004). Then, they become bipolar with a leading process pointing towards the pia and a trailing process pointing away, and migrate rapidly and persistently in a radial direction along radial glial fibers to reach their final position. The nature of this pause in migration prior to effective radial migration is not well understood. One possibility is to allow neurons to acquire the necessary properties for rapid glial-guided locomotion. Our data on the expression and function of SDCCAG8 suggest that PCM recruitment and strong MTOC activity at the centrosome may be a key part of this preparation process.
It is largely accepted that glial-guided neuronal locomotion depends on the centrosome (Cooper, 2013; Tsai and Gleeson, 2005), which carries out the major MTOC activity. Likely in response to migratory cues detected by the leading process, the centrosome first moves into a dilated region of the leading process. Subsequently, the nucleus moves towards the centrosome to complete a ‘two-stroke’ migration step. This coordinated movement between the centrosome and the nucleus depends on the tight coupling between these two organelles, which is at least partly mediated by a special MT-based structure, the perinuclear cage (Rivas and Hatten, 1995). The formation and maintenance of the perinuclear cage in rapidly migrating bipolar neurons likely requires a strong MTOC activity. On one hand, to be mobile, the centrosome may not be constantly attached to the nucleus. On the other hand, the centrosome and the nucleus need to be coupled to effectively coordinate their movement. Interestingly, SDCCAG8 is selectively up-regulated in nascent neurons right before the onset of glial-guided radial locomotion. It promotes PCM recruitment, thereby enhancing the MTOC activity of the centrosome. Disruptions of SDCCAG8 expression and/or function perturbed MT organization, decoupled the centrosome and the nucleus, and impaired their coordinated movement. It appears that the forward movement of the centrosome is not strongly affected, as the centrosome was found in the dilated region of the leading process, similar to that in control neurons. However, the nucleus lags far behind – even though our live imaging experiments showed that the nucleus could eventually translocate to meet the centrosome, likely driven by the force generated by residual MTs and their associated motor proteins, as well as the actin cytoskeleton and myosin motors (Schaar and McConnell, 2005; Solecki et al., 2009; Tsai et al., 2007; Vallee et al., 2009).
Consistent with a tight regulation of the coupling between the centrosome and the nucleus in rapidly migrating neurons, an increasing number of proteins have been found to control the process. They include MT and MT-based motor-associating proteins, such as doublecortin (DCX), Lissencephaly 1 (LIS1), NUDE-like protein (NDEL1) and focal adhesion kinase (FAK) (Deuel et al., 2006; Shu et al., 2004; Solecki et al., 2009; Tanaka et al., 2004; Tsai et al., 2005; Xie et al., 2003), the polarity proteins (Solecki et al., 2004), and the nuclear envelope proteins (Zhang et al., 2009). Notably, in some instances, the centrosome has been shown to move behind the nucleus (Distel et al., 2010; Sakakibara et al., 2013; Umeshima et al., 2007). We found that the vast majority of radially migrating, bipolar cortical neurons move with the centrosome leading the nucleus, indicating that radial migration with the centrosome behind the nucleus seems to be the minor rather than the rule in the developing cortex. The centrosome and its MTOC activity have also been implicated in the switch from multipolar to bipolar morphology (de Anda et al., 2010). We found that disruption of SDCCAG8 expression and/or function did not prevent, but clearly delayed, this morphological transition.
Mutations of SDCCAG8 have been found in human patients with NPHP and/or BBS (Billingsley et al., 2012; Otto et al., 2010; Schaefer et al., 2011). Besides retinal and renal dysfunction, the patients are characterized by mental retardation and seizures. NPHP and BBS are both broadly classified as ciliopathies (Hurd and Hildebrandt, 2011). We found that disruptions of SDCCAG8 expression and/or function did not obviously affect ciliogenesis in the developing cortex. While we could not completely rule out any subtle changes in the cilia that may lead to brain phenotypes, our study suggested that mutations of SDCCAG8 impair centrosomal property and function, MT organization and consequently neuronal migration in the developing cortex. Abnormal cortical neuron migration has been previously linked to the neurological defects associated with BBS, as in the case of BBS4 and BBS1 (Ishizuka et al., 2011; Kamiya et al., 2008). Similar to SDCCAG8, BBS4 associates with PCM1 and regulates recruitment of PCM and microtubule organization (Kim et al., 2004). Another centrosomal protein implicated in neuronal migration is the schizophrenia-associated protein disrupted-in-schizophrenia 1 (DISC1) (Kamiya et al., 2005; Kamiya et al., 2008), which also interacts with PCM1 and is involved in recruitment of PCM to the centrosome (Miyoshi et al., 2004; Shimizu et al., 2008). Interestingly, a recent report linked SDCCAG8 with schizophrenia (Hamshere et al., 2012). Both DISC1 and PCM1 have been previously linked to schizophrenia. Therefore, SDCCAG8, PCM1, DISC1 and BBS4 may define a key molecular pathway underlying mental defects associated with NPHP, BBS and certain forms of schizophrenia.
EXPERIMENTAL PROCEDURES
Mice, Plasmids and In Utero Electroporation
In utero electroporation experiments were performed using CD-1 mice (Charles River) as previously described (Bultje et al., 2009) (see Supplementary Experimental Procedures). Sdccag8LacZ/+ mice containing a LacZ cassette inserted in intron 1 of the Sdccag8 locus were provided by Dr. Hildebrandt (Children’s Hospital, Boston). Mice carrying CETN2-EGFP (Higginbotham et al., 2004) were obtained from The Jackson Laboratory (CB6-Tg(CAG-EGFP/CETN2)34Jgg/J). Sdccag8−/− mice was generated using the embryonic stem cell line (EPD0581-2-D04) obtained from the International Knockout Mouse Consortium (IKMC). Plasmids were described in Supplementary Experimental Procedures. All procedures for animal handling and usage were approved by the institutional research animal resource center (RARC).
Immunohistochemistry and Imaging
Mouse brain sections were prepared and immunohistochemistry were carried out as previously described (Bultje et al., 2009). Confocal images were acquired using an Olympus FV1000 confocal microscope and analyzed with Fluoview (Olympus), Volocity (Perkin Elmer), Image J (NIH), and Photoshop (Adobe). Quantifications of cell distribution, telencephalic area and fluorescence intensity of γ-TUB-EGFP, PCNT and EGFP-EMTB are described in Supplementary Experimental Procedures. Data are presented as the mean ± s.e.m. and statistical significance was assessed using a two-tailed Student’s t-test.
Cell Culture, Western Blot and Immunoprecipitation
COS7 cells were cultured and transfected as previously described (Bultje et al., 2009). Dissociated cortical cultures were prepared as previously described (Shi et al., 2003). Immunostaining was carried out and cells on coverslips were mounted on glass slides using Fluoromount G mounting media (Electron Microscopy Sciences), and imaged using an inverted microscope equipped with epifluorescent illumination and a cooled CCD camera (Axio Observer, Zeiss). Images were analyzed with Axiovision (Zeiss) and Photoshop.
For live imaging analysis in COS7 cells, cells were maintained at physiological conditions using a stage top incubation system mounted on an epi-fluorescence microscope (Axio Observer, Zeiss). Before imaging, the cells were exposed to NucBlue Live ReadyProbe (Life Technologies) to stain DNA. Images were acquired at the rate of 1 frame every ∼2.5 seconds.
Western blotting and immunoprecipitation reactions were performed using standard protocols (see Supplementary Experimental Procedures).
Organotypic Slice Culture and Live Imaging
Organotypic cortical slice cultures were prepared as previously described (Bultje et al., 2009). For live imaging analysis, slices were maintained at physiological conditions with a stage-top microscope incubator, and Z-stack images were acquired on an inverted laser scanning confocal microscope (Zeiss 5Live) every 15 minutes for 16–18 hours.
Supplementary Material
HIGHLIGHTS.
SDCCAG8 is up-regulated in nascent cortical neurons and regulates their migration.
SDCCAG8 regulates PCM recruitment at the centrosome and MT organization.
SDCCAG8 interacts and co-traffics with centriolar satellite protein PCM1.
Expression of human SDCCAG8 mutant disrupts cortical neuronal migration.
ACKNOWLEDGEMENTS
We thank Dr. Kathryn V. Anderson, Dr. Meng-Fu Bryan Tsou, Yvette Chin and Ildiko Lily Erdy for critical reading of the manuscript. We thank Dr. Andreas Merdes (Universite de Toulouse, France) for the PCM1 antibody, Dr. Ulrich Mueller (The Scripps Research Institute) for the pDcx-IRES-EGFP plasmid, Dr. Chloe Bulinski (Columbia University) for the pEGFP-EMTB plasmid and Dr. Alexey Khodjakov (The Wadsworth Center) for the pEGFP-γ-TUB plasmid. We thank the Molecular Cytology Core Facility for technical support and the Shi Lab members for their input and comments on the manuscript. This work was supported by grants from the National Institutes of Health (R01DA024681 and R01NS085004 to S.-H.S; 1K99DK099434 to R.A.; DK068306 and RC4-DK090917 to F.H.) and the Simons Foundation (to S.-H.S). F.H. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Barnes AP, Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu Rev Neurosci. 2009;32:347–381. doi: 10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- Billingsley G, Vincent A, Deveault C, Heon E. Mutational analysis of SDCCAG8 in Bardet-Biedl syndrome patients with renal involvement and absent polydactyly. Ophthalmic Genet. 2012;33:150–154. doi: 10.3109/13816810.2012.689411. [DOI] [PubMed] [Google Scholar]
- Bornens M. The centrosome in cells and organisms. Science. 2012;335:422–426. doi: 10.1126/science.1209037. [DOI] [PubMed] [Google Scholar]
- Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein AR, Shi SH. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Cooper JA. Cell biology in neuroscience: mechanisms of cell migration in the nervous system. J Cell Biol. 2013;202:725–734. doi: 10.1083/jcb.201305021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Anda FC, Meletis K, Ge X, Rei D, Tsai LH. Centrosome motility is essential for initial axon formation in the neocortex. J Neurosci. 2010;30:10391–10406. doi: 10.1523/JNEUROSCI.0381-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuel TA, Liu JS, Corbo JC, Yoo SY, Rorke-Adams LB, Walsh CA. Genetic interactions between doublecortin and doublecortin-like kinase in neuronal migration and axon outgrowth. Neuron. 2006;49:41–53. doi: 10.1016/j.neuron.2005.10.038. [DOI] [PubMed] [Google Scholar]
- Distel M, Hocking JC, Volkmann K, Koster RW. The centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in vivo. J Cell Biol. 2010;191:875–890. doi: 10.1083/jcb.201004154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- Edmondson JC, Hatten ME. Glial-guided granule neuron migration in vitro: a high-resolution time-lapse video microscopic study. J Neurosci. 1987;7:1928–1934. doi: 10.1523/JNEUROSCI.07-06-01928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faire K, Waterman-Storer CM, Gruber D, Masson D, Salmon ED, Bulinski JC. E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics. J Cell Sci. 1999;112(Pt 23):4243–4255. doi: 10.1242/jcs.112.23.4243. [DOI] [PubMed] [Google Scholar]
- Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Muller U. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron. 2011;69:482–497. doi: 10.1016/j.neuron.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Frank CL, Calderon de Anda F, Tsai LH. Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron. 2010;65:191–203. doi: 10.1016/j.neuron.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore EC, Walsh CA. Genetic causes of microcephaly and lessons for neuronal development. Wiley interdisciplinary reviews Developmental biology. 2013;2:461–478. doi: 10.1002/wdev.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamshere ML, Walters JT, Smith R, Richards AL, Green E, Grozeva D, Jones I, Forty L, Jones L, Gordon-Smith K, et al. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME. Riding the glial monorail: a common mechanism for glial-guided neuronal migration in different regions of the developing mammalian brain. Trends Neurosci. 1990;13:179–184. doi: 10.1016/0166-2236(90)90044-b. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, Hack MA, Chapouton P, Barde YA, Gotz M. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- Higginbotham H, Bielas S, Tanaka T, Gleeson JG. Transgenic mouse line with green-fluorescent protein-labeled Centrin 2 allows visualization of the centrosome in living cells. Transgenic Res. 2004;13:155–164. doi: 10.1023/b:trag.0000026071.41735.8e. [DOI] [PubMed] [Google Scholar]
- Higginbotham HR, Gleeson JG. The centrosome in neuronal development. Trends Neurosci. 2007;30:276–283. doi: 10.1016/j.tins.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Hurd TW, Hildebrandt F. Mechanisms of nephronophthisis and related ciliopathies. Nephron Experimental nephrology. 2011;118:e9–14. doi: 10.1159/000320888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka K, Kamiya A, Oh EC, Kanki H, Seshadri S, Robinson JF, Murdoch H, Dunlop AJ, Kubo K, Furukori K, et al. DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature. 2011;473:92–96. doi: 10.1038/nature09859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, Sawamura N, Park U, Kudo C, Okawa M, et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Tan PL, Kubo K, Engelhard C, Ishizuka K, Kubo A, Tsukita S, Pulver AE, Nakajima K, Cascella NG, et al. Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses. Arch Gen Psychiatry. 2008;65:996–1006. doi: 10.1001/archpsyc.65.9.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenedy AA, Cohen KJ, Loveys DA, Kato GJ, Dang CV. Identification and characterization of the novel centrosome-associated protein CCCAP. Gene. 2003;303:35–46. doi: 10.1016/s0378-1119(02)01141-1. [DOI] [PubMed] [Google Scholar]
- Kerjan G, Gleeson JG. Genetic mechanisms underlying abnormal neuronal migration in classical lissencephaly. Trends Genet. 2007;23:623–630. doi: 10.1016/j.tig.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Badano JL, Sibold S, Esmail MA, Hill J, Hoskins BE, Leitch CC, Venner K, Ansley SJ, Ross AJ, et al. The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat Genet. 2004;36:462–470. doi: 10.1038/ng1352. [DOI] [PubMed] [Google Scholar]
- Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers M, Hoogenraad CC. Centrosomes, microtubules and neuronal development. Mol Cell Neurosci. 2011;48:349–358. doi: 10.1016/j.mcn.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Kwan KY, Sestan N, Anton ES. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 2012;139:1535–1546. doi: 10.1242/dev.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoTurco JJ, Bai J. The multipolar stage and disruptions in neuronal migration. Trends Neurosci. 2006;29:407–413. doi: 10.1016/j.tins.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Luders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- Metin C, Vallee RB, Rakic P, Bhide PG. Modes and mishaps of neuronal migration in the mammalian brain. J Neurosci. 2008;28:11746–11752. doi: 10.1523/JNEUROSCI.3860-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Asanuma M, Miyazaki I, Diaz-Corrales FJ, Katayama T, Tohyama M, Ogawa N. DISC1 localizes to the centrosome by binding to kendrin. Biochem Biophys Res Commun. 2004;317:1195–1199. doi: 10.1016/j.bbrc.2004.03.163. [DOI] [PubMed] [Google Scholar]
- Nadarajah B, Brunstrom J, Grutzendler J, Wong R, Pearlman A. Two modes of radial migration in early development of the cerebral cortex. Nat Neurosci. 2001;4:143–150. doi: 10.1038/83967. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Noctor S, Flint A, Weissman T, Dammerman R, Kriegstein A. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noctor S, Martínez-Cerdeño V, Ivic L, Kriegstein A. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Oh EC, Katsanis N. Cilia in vertebrate development and disease. Development. 2012;139:443–448. doi: 10.1242/dev.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto EA, Hurd TW, Airik R, Chaki M, Zhou W, Stoetzel C, Patil SB, Levy S, Ghosh AK, Murga-Zamalloa CA, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Guidance of neurons migrating to the fetal monkey neocortex. Brain Res. 1971;33:471–476. doi: 10.1016/0006-8993(71)90119-3. [DOI] [PubMed] [Google Scholar]
- Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol. 1972;145:61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- Rivas RJ, Hatten ME. Motility and cytoskeletal organization of migrating cerebellar granule neurons. J Neurosci. 1995;15:981–989. doi: 10.1523/JNEUROSCI.15-02-00981.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ME, Walsh CA. Human brain malformations and their lessons for neuronal migration. Annu Rev Neurosci. 2001;24:1041–1070. doi: 10.1146/annurev.neuro.24.1.1041. [DOI] [PubMed] [Google Scholar]
- Sakakibara A, Sato T, Ando R, Noguchi N, Masaoka M, Miyata T. Dynamics of Centrosome Translocation and Microtubule Organization in Neocortical Neurons during Distinct Modes of Polarization. Cereb Cortex. 2013 doi: 10.1093/cercor/bhs411. [DOI] [PubMed] [Google Scholar]
- Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proc Natl Acad Sci U S A. 2005;102:13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer E, Zaloszyc A, Lauer J, Durand M, Stutzmann F, Perdomo-Trujillo Y, Redin C, Bennouna Greene V, Toutain A, Perrin L, et al. Mutations in SDCCAG8/NPHP10 Cause Bardet-Biedl Syndrome and Are Associated with Penetrant Renal Disease and Absent Polydactyly. Molecular syndromology. 2011;1:273–281. doi: 10.1159/000331268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Matsuzaki S, Hattori T, Kumamoto N, Miyoshi K, Katayama T, Tohyama M. DISC1-kendrin interaction is involved in centrosomal microtubule network formation. Biochem Biophys Res Commun. 2008;377:1051–1056. doi: 10.1016/j.bbrc.2008.10.100. [DOI] [PubMed] [Google Scholar]
- Shu T, Ayala R, Nguyen MD, Xie Z, Gleeson JG, Tsai LH. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44:263–277. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Trivedi N, Govek EE, Kerekes RA, Gleason SS, Hatten ME. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 2009;63:63–80. doi: 10.1016/j.neuron.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancik EK, Navarro-Quiroga I, Sellke R, Haydar TF. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J Neurosci. 2010;30:7028–7036. doi: 10.1523/JNEUROSCI.6131-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Evans L, Kirschner M. Gamma-tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Tsai JW, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. The Journal of cell biology. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LH, Gleeson JG. Nucleokinesis in neuronal migration. Neuron. 2005;46:383–388. doi: 10.1016/j.neuron.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Umeshima H, Hirano T, Kengaku M. Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons. Proc Natl Acad Sci U S A. 2007;104:16182–16187. doi: 10.1073/pnas.0708047104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente M, Marin O. Neuronal migration mechanisms in development and disease. Curr Opin Neurobiol. 2010;20:68–78. doi: 10.1016/j.conb.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Vallee RB, Seale GE, Tsai JW. Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol. 2009;19:347–355. doi: 10.1016/j.tcb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WJ, Soni RK, Uryu K, Tsou MF. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J Cell Biol. 2011;193:727–739. doi: 10.1083/jcb.201101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell. 2003;114:469–482. doi: 10.1016/s0092-8674(03)00605-6. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.