Highlights
-
•
PDIL1;1 efficiently catalyzed both disulfide bond formation and disulfide bond reduction.
-
•
Two redox-active sites of PDIL1;1 were involved in disulfide reduction.
-
•
Disulfide reduction activity of PDIL1;1 increased with increasing GSH concentration.
Abbreviations: ER, endoplasmic reticulum; ERO1, ER oxidoreductin 1; GSH, glutathione; GSSG, glutathione disulfide; PDI, protein disulfide isomerase; TRX, thioredoxin
Keywords: Disulfide bond, Glutathione, PDI, Protein storage organelle, Rice
Abstract
Protein disulfide isomerases (PDIs), a family of thiol-disulfide oxidoreductases that are ubiquitous in all eukaryotes, are the principal catalysts for disulfide bond formation. Here, we investigated three rice (Oryza sativa) PDI family members (PDIL1;1, PDIL1;4, and PDIL2;3) and found that PDIL1;1 exhibited the highest catalytic activity for both disulfide bond formation and disulfide bond reduction. The activity of PDIL1;1-catalyzed disulfide bond reduction, in which two redox-active sites were involved, was enhanced by increasing the glutathione concentration. These results suggest that PDIL1;1 plays primary roles in both disulfide bond formation and disulfide bond reduction, which allow for redox control of protein quality and packaging.
1. Introduction
The developing endosperm of rice (Oryza sativa) synthesizes large amounts of structurally and physicochemically diverse proteins, which serve as nutritional reserves for germinating seedlings. Disulfide bonds, which covalently link pairs of Cys residues and impart thermodynamic stability to proteins [1], permit redox control of protein packaging into and releasing from storage organelles. Only natively folded proteins with correct patterns of disulfide bonds can be transported from the endoplasmic reticulum (ER) to protein storage organelles [2]. For example, highly hydrophobic prolamins are packed into ER-derived protein body (designated PB-I) within the ER by a polymerization process involving the formation of intermolecular disulfide bonds [3], and proglutelins, which are composed of acidic and basic subunits, acquire intramolecular disulfide bonds before being transported from the ER to protein storage vacuole-derived protein body (designated PB-II) [4].
The ER contains enzymes that mediate the formation of disulfide bonds in proteins undergoing the folding process. Protein disulfide isomerases (PDIs), a family of thiol-disulfide oxidoreductases that are ubiquitous in all eukaryotes, are the principal catalysts for disulfide bond formation [5]. Members of the PDI family contain two thioredoxin (TRX)-fold redox-active domains (designated a and a′), each of which possesses a pair of Cys residues (Cys-X-X-Cys) that are involved in thiol-disulfide exchange reactions. The activity of PDIs depends on the redox state of the Cys residues, which can exist either in an oxidized (disulfide) form, which gains electrons for the formation of disulfide bonds in substrate proteins, or in a reduced (dithiol) form, which donates electrons for the reduction of disulfide bonds [6]. Non-native disulfide bonds frequently form between incorrectly paired Cys residues in folding intermediates [7], and reduction of these incorrect Cys pairings is essential for conversion of the non-native disulfide bonds to native ones.
The genomes of rice, Arabidopsis (Arabidopsis thaliana), and maize (Zea mays, Zm) each encode at least seven PDI proteins predicted to have two redox-active Cys pairs: PDIL1;1–1;4 and PDIL2;1–2;3 [8]. ZmPDIL1;1, ZmPDIL1;3, and ZmPDIL2;3, whose rice orthologues are PDIL1;1, PDIL1;4, and PDIL2;3, respectively, are expressed at much higher levels than other ZmPDILs in the endosperm [8]. Various lines of evidence have shown that specific members of the PDI family are involved in oxidative folding, processing, and assembly of storage proteins [9–12]. The active-site Cys residues of reduced PDI are oxidized by the flavoenzyme ERO1, which transfers electrons from PDI to molecular oxygen, a source of oxidizing power [13,14]. In rice endosperm cells, knockdown of ERO1 promotes aggregation of proglutelins through non-native intermolecular disulfide bonds [15]. In the ERO1-knockdown cells, which exhibit unfolded protein response-related induction of BiP (binding immunoglobulin protein), the level of PDIL1;1 decreases, the level of PDIL2;3 increases, and the level of PDIL1;4 remains unaltered relative to the corresponding levels in wild-type cells [15]. Knockout of PDIL1;1 also causes formation of aggregates containing proglutelins through non-native intermolecular disulfide bonds [15]. These findings raise the question of how the ER of endosperm cells establishes the pathways dedicated to the reduction of incorrect Cys pairings and subsequent formation of native disulfide bonds.
In this study, we evaluated the catalytic activities of three rice PDI family members, PDIL1;1, PDIL1;4, and PDIL2;3, for the formation and reduction of disulfide bonds. PDIL1;1 showed the highest catalytic activity for both the formation and the reduction of disulfide bonds. PDIL1;1-catalyzed reduction of disulfide bonds was facilitated by increasing the concentration of glutathione (GSH), which constitutes the principal redox buffer in the ER and plays a role in the reductive pathway [16,17]. The physiological role of PDIL1;1 in the endosperm ER is discussed.
2. Materials and methods
2.1. Construction of expression plasmids
Expression plasmids containing DNA fragments encoding the predicted mature-sized rice PDIL1;1 (GenBank: AK068268; Glu26−Leu512) and PDIL2;3 (GenBank: AK062254; Ser19−Leu441) were prepared as described previously [12]. The DNA fragment encoding the predicted mature-sized PDIL1;4 (GenBank: AK071514; Ser23−Leu563) was amplified from the full-length cDNA by PCR with a set of primers (5′-ataggatcctccgacgacgacctcga-3′ and 5′-tatctcgagtctagagagcgtcgttgc-3′) and inserted into pGEX-6P-3 vector (GE Healthcare) at the appropriate sites. Cys-to-Ala substitutions were introduced into the PDIL1;1 construct by means of a QuikChange II site-directed mutagenesis kit (Agilent Technologies) and the following sets of primers: Cys72Ala, 5′-ccgtggtgtggacacgccaagaagctcgctcc-3′ and 5′-ggagcgagcttcttggcgtgtccacaccacgg-3′; Cys414Ala, 5′-ccatggtgcggacacgccaagaagctggctcc-3′ and 5′-ggagccagcttcttggcgtgtccgcaccatgg-3′. The desired mutations were confirmed by DNA sequencing.
2.2. Expression and purification of recombinant PDILs
The expression plasmids were used to transform Escherichia coli BL21(DE3)pLysS cells (Novagen). The cells were grown for 3 h at 37 °C in Luria–Bertani medium containing carbenicillin (50 μg/mL), and isopropyl-β-d-thiogalactopyranoside was then added to a final concentration of 0.5 mM. After further propagation for 12 h at 25 °C, the cells were harvested by centrifugation at 5,000g for 5 min and were lysed in phosphate-buffered saline (pH 7.3) in the presence of lysozyme (1 mg/mL) and 1 mM phenylmethanesulfonyl fluoride. After centrifugation at 16,000g for 20 min, the supernatant was used to bind glutathione S-transferase-tagged recombinant PDIL proteins to glutathione sepharose 4B (GE Healthcare), and then the tag was removed with protease in 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, and 1 mM ethylenediaminetetraacetic acid (EDTA), in accordance with the manufacturer’s instructions. The yields of recombinant proteins after purification were as follows: approximately 5 mg/L bacterial culture for PDIL1;1 and PDIL1;4 and approximately 2 mg/L bacterial culture for PDIL2;3.
2.3. Assays for RNase oxidative refolding and insulin disulfide reduction activities
RNase oxidative refolding activity was measured by means of RNase-catalyzed hydrolysis of cytidine 2′,3′-cyclic monophosphate (cCMP; Sigma–Aldrich) [18,19]. Reduced and denatured RNase (10 μM, Sigma–Aldrich), prepared as described previously [12], was incubated with each recombinant PDIL (1 μM) in 100 mM Tris–HCl (pH 8.0) containing 1 mM GSH, 0.2 mM glutathione disulfide (GSSG), and 2 mM EDTA at 28 °C for the times indicated in the figures, and the reactions were stopped by the addition of N-ethylmaleimide (final concentration, 2 mM). After the addition of 2 mM cCMP, hydrolysis of cCMP resulting from the restoration of RNase activity was measured as an increase in absorbance at 296 nm by means of a spectrophotometer (UV-1800, Shimadzu).
Insulin disulfide reduction assays [20] were performed using dithiothreitol (DTT) or GSH as an electron donor. Insulin (125 μM, Sigma–Aldrich) was incubated with each recombinant PDIL (0.5 μM) in 100 mM phosphate buffer (pH 8.0) containing 2 mM EDTA at 28 °C for the times indicated in the figures, either in the presence of 160 μM DTT or in the presence of 0.2 mM GSSG and GSH (1, 2, or 4 mM). The increase in turbidity accompanying reduction of the intramolecular disulfide bonds of insulin was monitored at 650 nm by means of a spectrophotometer (UV-1800, Shimadzu).
3. Results
3.1. Catalytic activities of recombinant PDIL1;1, PDIL1;4, and PDIL2;3 for disulfide bond formation and reduction
We evaluated the catalytic activities of recombinant PDIL1;1, PDIL1;4, and PDIL2;3, all of which contain two TRX-like domains with a Cys-X-X-Cys motif, but in a different arrangement (Fig. 1A); specifically, we compared their activities for disulfide bond formation by using reduced, denatured RNase as a substrate in the presence of a glutathione redox buffer. PDIL1;1 showed the highest catalytic activity for oxidative RNase refolding, followed by PDIL1;4 (Fig. 1B). The catalytic activity of PDIL2;3 was relatively low (Fig. 1B), as reported previously [12].
Fig. 1.
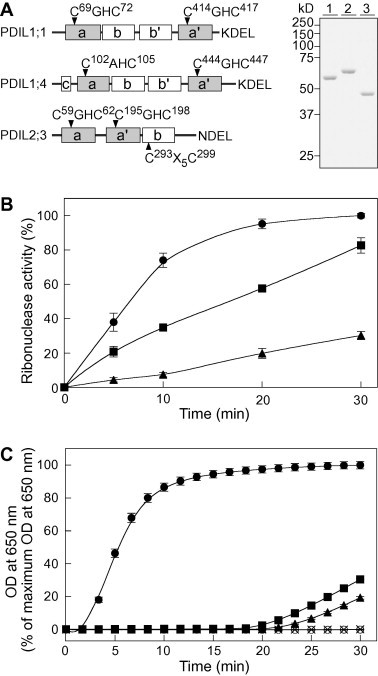
Disulfide bond formation and reduction activities of recombinant PDIL1;1, PDIL1;4, and PDIL2;3. (A) Schematic illustration of rice PDIL1;1, PDIL1;4, and PDIL2;3 (left panel) and SDS–PAGE of the recombinant PDILs (right panel). Redox-active TRX domains a and a′ (gray boxes; Cys residues indicated by arrowheads) and redox-inactive TRX domains b and b′ (white boxes) were predicted by NCBI conserved domain searches (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). The acid N-terminal domain (c, white box) for PDIL1;4 and the structural disulfide CX5C (arrowhead) for PDIL2;3 are also indicated. Purified PDIL1;1 (Glu26−Leu512, lane 1), PDIL1;4 (Ser23−Leu563, lane 2), and PDIL2;3 (Ser19−Leu441, lane 3) were separated by SDS–PAGE with 10% (w/v) acrylamide and stained with Coomassie Brilliant Blue. The predicted molecular masses of recombinant PDIL1;1, PDIL1;4, and PDIL2;3 are 54, 60, and 46 kD, respectively. (B) Oxidative protein-refolding activities of the PDILs. Reduced, denatured RNase was refolded in the presence of PDIL1;1 (●), PDIL1;4 (■), or PDIL2;3 (▴) and GSSG and GSH. The oxidative RNase-refolding activities of the PDILs were assayed by monitoring the RNase-catalyzed hydrolysis of cCMP (as indicated by an increase in the absorbance at 296 nm). The data are presented as percentages of the highest activity (means ± SD of three independent determinations). (C) Disulfide bond reduction activities of the PDILs. Insulin was incubated in the presence of PDIL1;1 (●), PDIL1;4 (■), or PDIL2;3 (▴) using DTT as an electron donor, and the increase in turbidity accompanying reduction of the insulin disulfide bonds by the PDILs was monitored at 650 nm for 30 min, as described in the Materials and Methods. DTT (160 μM) in the absence of PDIL1;1 (×) and PDIL1;1 (0.5 μM) in the absence of DTT (○) served as negative controls. The data (OD at 650 nm) are presented as percentages of the maximum OD at 650 nm observed (means ± SD of three independent determinations).
We next examined the disulfide bond reduction activities of these PDILs using insulin as a substrate in the presence of DTT. PDIL1;1 almost immediately showed a marked increase in turbidity owing to rapid reduction of the insulin disulfide bonds (Fig. 1C, solid circles; the calculated maximum rate for PDIL1;1 was 0.0027 ± 0.0001 ΔA650 s−1). In contrast, for both PDIL1;4 and PDIL2;3, turbidity did not start to increase for approximately 20 min, and the rates of increase were lower than the rate for PDIL1;1 (Fig. 1C, solid squares and triangles). These results indicate that PDIL1;1 efficiently catalyzed reduction of the insulin disulfide bonds. No significant gain in turbidity was measurable during the 30-min assay period when 160 μM DTT was used as an electron donor in the absence of PDIL1;1 (Fig. 1C, crosses); nor was there any measurable increase in turbidity with PDIL1;1 in the absence of DTT (Fig. 1C, open circles).
3.2. Effects of Cys-to-Ala mutation on disulfide bond reduction activity of PDIL1;1
PDIL1;1 has two Cys pairs: Cys69GHCys72 and Cys414GHCys417 (Fig. 1A, left panel), both of which are involved in oxidative RNase refolding [12]. We produced three variants of PDIL1;1, in which the second Cys residue in each Cys pair was mutated to an Ala residue in the a domain (Cys72Ala), in the a′ domain (Cys417Ala), or in both domains (Cys72Ala/Cys417Ala). Using insulin as a substrate, we examined the effects of these mutations on the disulfide bond reduction activity of the mutants. Mutation in either the a domain (Cys72Ala) or the a′ domain (Cys417Ala) delayed the appearance of turbidity accompanying reduction of the insulin disulfide bonds compared with the wild-type (Fig. 2). Mutation in both redox-active sites (Cys72Ala/Cys417Ala) led to a slower increase in turbidity than did mutation in a single redox-active site (Cys72Ala or Cys417Ala) (Fig. 2). These results suggest that both redox-active sites of PDIL1;1 are involved in reduction of the insulin disulfide bonds.
Fig. 2.
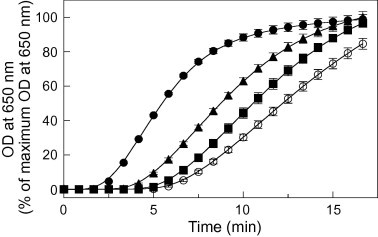
Comparison of disulfide bond reduction activities of wild-type and mutated PDIL1;1. Insulin was incubated with DTT as an electron donor in the presence of the wild-type PDIL1;1 (●) and three PDIL1;1 mutants: Cys72Ala mutant (■), Cys417Ala mutant (▴), and Cys72Ala/Cys417Ala mutant (○). The increase in turbidity accompanying reduction of the insulin by the PDILs was monitored at 650 nm. The data (OD at 650 nm) are presented as percentages of the maximum OD at 650 nm observed (means ± SD of three independent determinations).
3.3. Effects of GSH concentration on disulfide bond reduction activity of PDIL1;1
We examined how varying the GSH concentration affected the disulfide bond reduction activity of PDIL1;1. PDIL1;1-catalyzed reduction of the insulin disulfide bonds was measured in a redox buffer containing GSH at a concentration of 1, 2, or 4 mM in the presence of GSSG at a fixed concentration (0.2 mM). Using GSH as an electron donor, PDIL1;1 catalyzed reduction of the insulin disulfide bonds, and insulin reduction activity clearly increased with increasing GSH concentration (Fig. 3). This result suggests that the availability of a reducing equivalent, GSH, profoundly affects the catalytic activity of PDIL1;1 for disulfide bond reduction. There was no significant increase in turbidity in the absence of PDIL1;1 at a GSH concentration of 4 mM during the 20-min assay period (Fig. 3, open squares).
Fig. 3.
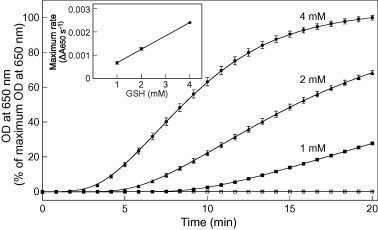
Effects of GSH concentration on PDIL1;1-catalyzed reduction of disulfide bonds. Insulin was incubated in the presence of PDIL1;1, GSSG (0.2 mM), and GSH at 1 mM (■), 2 mM (▴), or 4 mM (●); the increase in turbidity accompanying reduction of the insulin disulfide bonds by PDIL1;1 was monitored at 650 nm for 20 min. GSH (4 mM) and GSSG (0.2 mM) in the absence of PDIL1;1 served as negative controls (□). The data (OD at 650 nm) are presented as percentages of the maximum OD at 650 nm observed (means ± SD of three independent determinations). Inset: maximum rates of insulin disulfide reduction (tangents to the steepest part of the curve) were calculated from the increase in absorbance at 650 nm per second and are plotted against GSH concentration (means ± SD of three independent determinations).
4. Discussion
During seed development and germination, the redox state of storage proteins changes dramatically; during seed development, the acquisition of disulfide bonds facilitates packaging of the proteins into storage organelles, and during germination, conversion of the oxidized forms back to the reduced forms increases their proteolytic susceptibility and thus the availability of nitrogen and sulfur [21]. The h-type TRX, which is reduced by NADPH-dependent TRX reductase, plays specific and critical roles in the reduction of disulfide-bonded storage proteins during germination [22]. Previous studies showed that PDIL1;1 is essential for the formation of disulfide bonds in storage proteins, such as proglutelins, during seed development [10,15]. Our current results suggest that PDIL1;1 plays primary roles not only in transferring oxidizing equivalents to substrate proteins but also in transferring reducing equivalents to incorrectly paired Cys residues and that the PDIL1;1-dependent reduction system contributes to the quality control of storage proteins in the ER of the developing endosperm.
The oxidative pathway involving ERO1 (a de novo disulfide-generating enzyme) and PDI (a disulfide-transferring enzyme) has been extensively studied in eukaryotes (reviewed in [6,23]). Human ERO1α-based oxidation of PDI involves intramolecular transfer of electrons from the reduced Cys residues in the a domain of PDI to the disulfide in the a′ domain and subsequent intermolecular transfer of electrons from the reduced Cys residues in the a′ domain of PDI to the shuttle disulfide of ERO1α [24]. Although the ER is a more oxidizing environment than the cytosol (the GSH/GSSG ratio ranges from 1:1 to 3:1 in the ER and from 30:1 to 100:1 in the cytosol) [16], the ER contains a high concentration of GSH (estimated to be 9 mM [25]), which can be considered to be the most likely source of reducing equivalents for PDI in the ER. Recently, the PDI-mediated reductive pathways have been elucidated. ERp57, which is a member of the mammalian PDI family and has an abb′a′ (b and b′ designate redox-inactive TRX-fold domains) domain organization, plays a direct role in the isomerization of non-native disulfide bonds in certain glycoproteins [26,27]. Mammalian ERdj5, which possesses a J domain and six TRX domains (including the two C-terminal TRX domains involved in disulfide bond reduction), cleaves non-native disulfide bonds to facilitate degradation of misfolded glycoproteins recognized by EDEM1 [28,29] and to ensure correct folding of the substrate, which is the low-density lipoprotein receptor [30]. Analysis of the crystal structure of yeast PDI (Pdi1p; abb′a′ domain organization) has revealed that the two redox active sites face each other across a wide hydrophobic surface in the central cleft; this advantageous arrangement allows PDI to effectively interact with substrates and catalyze the formation and reduction of their disulfide bonds [31]. Although PDIL1;1 and PDIL1;4 share the abb′a′ domain organization (Fig. 1A), only PDIL1;1 shows high activity for the reduction of disulfide bonds when insulin is used as a substrate (Fig. 1C), suggesting that PDIL1;1 and PDIL1;4 have evolved to acquire distinct functions in the endosperm cells of rice.
We have recently demonstrated that PDIL1;1 and PDIL2;3, which are localized in the ER lumen and at the surface of PB-I, respectively, constitute critical and different pathways for disulfide bond formation in the endosperm cells of rice [12,15]: PDIL1;1 plays an essential role in the formation of disulfide bonds in proglutelins, as described above; whereas PDIL2;3 plays an important role in PB-I development by promoting the intermolecular disulfide bond-linked assembly of Cys-rich prolamins into the core of PB-I. However, although knockdown of PDIL2;3 does not impair the formation of disulfide bonds in PB-II-targeted storage proteins, such as proglutelins, knockout of PDIL1;1 leads to the formation of large amounts of aggregates containing proglutelins through non-native intermolecular disulfide bonds and impairs the normal development of both PB-II and PB-I [10,12,15]. These findings suggest that the PDIL1;1-dependent pathways are also required for the PDIL2;3-dependent pathway to function in the development of PB-I. However, the mechanism for coordination of the functions of PDIL1;1 and PDIL2;3 has not been uncovered. The mammalian ER has been suggested to possess an oxidative system in which PDI (a PDIL1;1 orthologue) cooperatively increases fidelity in the rapid but promiscuous formation of disulfide bonds by P5 (a PDIL2;3 orthologue), which is preferentially oxidized by peroxiredoxin 4, another disulfide-generating enzyme [32]. PDIL1;1 shows markedly high catalytic activity for disulfide bond reduction using GSH as an electron donor (Fig. 1C and 3). This result opens up the possibility that when disulfide bond formation results in incorrect pairings of Cys residues, PDIL1;1 greatly facilitates the prompt reduction of incorrect disulfide bonds in the ER of rice endosperm. Recently, it has been shown that after being oxidized directly by ERO1α, PDI receives electrons preferentially from other PDI family members, including P5 and ERp57 [33]. Further study is needed to determine whether PDIL1;1 transfers oxidizing equivalents to PDIL2;3 in the ER of rice endosperm.
Acknowledgements
This work was supported by a Grant-in-Aid for Young Scientists (23780101) from the Japan Society for the Promotion of Science, by a Grant-in-Aid for Scientific Research (26450116) from the Japan Society for the Promotion of Science, and by the Ministry of Education, Culture, Sports, Science and Technology of Japan. Y.O. and Y.K. designed and performed research; Y.O. and Y.K. analyzed data; Y.O. wrote the paper.
References
- 1.Fass D. Disulfide bonding in protein biophysics. Ann. Rev. Biophys. 2012;41:63–79. doi: 10.1146/annurev-biophys-050511-102321. [DOI] [PubMed] [Google Scholar]
- 2.Onda Y. Oxidative protein-folding systems in plant cells. Int. J. Cell Biol. 2013;2013:1–15. doi: 10.1155/2013/585431. 585431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogawa M., Kumamaru T., Satoh H., Iwata N., Omura T., Kasai Z., Tanaka K. Purification of protein body-I of rice seed and its polypeptide composition. Plant Cell Physiol. 1987;28:1517–1527. [Google Scholar]
- 4.Yamagata H., Sugimoto T., Tanaka K., Kasai Z. Biosynthesis of storage proteins in developing rice seeds. Plant Physiol. 1982;70:1094–1100. doi: 10.1104/pp.70.4.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freedman R.B., Hirst T.R., Tuite M.F. Protein disulphide isomerase: building bridges in protein folding. Trends Biochem. Sci. 1994;19:331–336. doi: 10.1016/0968-0004(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 6.Hatahet F., Ruddock L.W. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid. Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 7.Jansens A., van Duijn E., Braakman I. Coordinated nonvectorial folding in a newly synthesized multidomain protein. Science. 2002;298:2401–2403. doi: 10.1126/science.1078376. [DOI] [PubMed] [Google Scholar]
- 8.Houston N.L., Fan C.Z., Xiang Q.Y., Schulze J.M., Jung R., Boston R.S. Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 2005;137:762–778. doi: 10.1104/pp.104.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C.P., Larkins B.A. Expression of protein disulfide isomerase is elevated in the endosperm of the maize floury-2 mutant. Plant Mol. Biol. 1996;30:873–882. doi: 10.1007/BF00020800. [DOI] [PubMed] [Google Scholar]
- 10.Takemoto Y., Coughlan S.J., Okita T.W., Satoh H., Ogawa M., Kumamaru T. The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol. 2002;128:1212–1222. doi: 10.1104/pp.010624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamauchi S., Wadahama H., Iwasaki K., Nakamoto Y., Nishizawa K., Ishimoto M., Kawada T., Urade R. Molecular cloning and characterization of two soybean protein disulfide isomerases as molecular chaperones for seed storage proteins. FEBS J. 2008;275:2644–2658. doi: 10.1111/j.1742-4658.2008.06412.x. [DOI] [PubMed] [Google Scholar]
- 12.Onda Y., Nagamine A., Sakurai M., Kumamaru T., Ogawa M., Kawagoe Y. Distinct roles of protein disulfide isomerase and P5 sulfhydryl oxidoreductases in multiple pathways for oxidation of structurally diverse storage proteins in rice. Plant Cell. 2011;23:210–223. doi: 10.1105/tpc.110.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross E., Kastner D.B., Kaiser C.A., Fass D. Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell. 2004;117:601–610. doi: 10.1016/s0092-8674(04)00418-0. [DOI] [PubMed] [Google Scholar]
- 14.Gross E., Sevier C.S., Heldman N., Vitu E., Bentzur M., Kaiser C.A., Thorpe C., Fass D. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc. Natl. Acad. Sci. U.S.A. 2006;103:299–304. doi: 10.1073/pnas.0506448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onda Y., Kumamaru T., Kawagoe Y. ER membrane-localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proc. Natl. Acad. Sci. U.S.A. 2009;106:14156–14161. doi: 10.1073/pnas.0904429106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang C., Sinskey A.J., Lodish H.F. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 17.Cuozzo J.W., Kaiser C.A. Competition between glutathione and protein thiols for disulphide-bond formation. Nat. Cell Biol. 1999;1:130–135. doi: 10.1038/11047. [DOI] [PubMed] [Google Scholar]
- 18.Lyles M.M., Gilbert H.F. Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: dependence of the rate on the composition of the redox buffer. Biochemistry. 1991;30:613–619. doi: 10.1021/bi00217a004. [DOI] [PubMed] [Google Scholar]
- 19.Rancy P.C., Thorpe C. Oxidative protein folding in vitro: a study of the cooperation between quiescin-sulfhydryl oxidase and protein disulfide isomerase. Biochemistry. 2008;47:12047–12056. doi: 10.1021/bi801604x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 1979;254:9627–9632. [PubMed] [Google Scholar]
- 21.Buchanan B.B., Balmer Y. Redox regulation: a broadening horizon. Ann. Rev. Plant. Biol. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- 22.Kobrehel K., Wong J.H., Balogh A., Kiss F., Yee B.C., Buchanan B.B. Specific reduction of wheat storage proteins by thioredoxin h. Plant Physiol. 1992;99:919–924. doi: 10.1104/pp.99.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sevier C.S., Kaiser C.A. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim. Biophys. Acta. 2008;1783:549–556. doi: 10.1016/j.bbamcr.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 24.Araki K., Nagata K. Functional in vitro analysis of the ERO1 protein and protein-disulfide isomerase pathway. J. Biol. Chem. 2011;286:32705–32712. doi: 10.1074/jbc.M111.227181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bass R., Ruddock L.W., Klappa P., Freedman R.B. A major fraction of endoplasmic reticulum-located glutathione is present as mixed disulfides with protein. J. Biol. Chem. 2004;279:5257–5262. doi: 10.1074/jbc.M304951200. [DOI] [PubMed] [Google Scholar]
- 26.Jessop C.E., Bulleid N.J. Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 2004;279:55341–55347. doi: 10.1074/jbc.M411409200. [DOI] [PubMed] [Google Scholar]
- 27.Jessop C.E., Chakravarthi S., Garbi N., Hammerling G.J., Lovell S., Bulleid N.J. ERp57 is essential for efficient folding of glycoproteins sharing common structural domains. EMBO J. 2007;26:28–40. doi: 10.1038/sj.emboj.7601505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D.Y., Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- 29.Hagiwara M. Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5. Mol. Cell. 2011;41:432–444. doi: 10.1016/j.molcel.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 30.Oka O.B., Pringle M.A., Schopp I.M., Braakman I., Bulleid N.J. ERdj5 is the ER reductase that catalyzes the removal of non-native disulfides and correct folding of the LDL receptor. Mol. Cell. 2013;50:793–804. doi: 10.1016/j.molcel.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian G., Xiang S., Noiva R., Lennarz W.J., Schindelin H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell. 2006;124:61–73. doi: 10.1016/j.cell.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 32.Sato Y. Synergistic cooperation of PDI family members in peroxiredoxin 4-driven oxidative protein folding. Sci. Rep. 2013;3:2456. doi: 10.1038/srep02456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Araki K., Iemura S., Kamiya Y., Ron D., Kato K., Natsume T., Nagata K. Ero1-α and PDIs constitute a hierarchical electron transfer network of endoplasmic reticulum oxidoreductases. J. Cell Biol. 2013;202:861–874. doi: 10.1083/jcb.201303027. [DOI] [PMC free article] [PubMed] [Google Scholar]


