Abstract
Despite its many favorable properties as a sample support for biological electron microscopy, graphene is not widely used because its hydrophobicity precludes reliable protein deposition. We describe a method to modify graphene using a low-energy hydrogen plasma, which reduces hydrophobicity without degrading the graphene lattice. We show that the use of plasma-treated graphene enables better control of protein distribution in ice for electron cryo-microscopy and improved image quality by reducing radiation-induced sample motion.
Improved electron microscopes, more stable cryo-stages and direct electron detectors with high quantum efficiency have recently brought about a revolution in biological electron microscopy (EM), enabling near atomic resolution 3D structure determination for several molecular complexes1,2. Even so, specimen preparation methods have remained largely unchanged since they were initially developed for electron cryo-microscopy (cryo-EM) almost three decades ago3.
Thin films of amorphous carbon (am-C) are typically used as EM supports (grids). Am-C films are often semiconducting, and their conductivity and mechanical strength (with a Young’s modulus of 0.2-200 GPa) can be reduced by several orders of magnitude depending on the conditions during their production4. This makes am-C and the ice it supports prone to surface charging and radiation-induced chemical and physical changes5, which lead to blurring of particle images and degradation of information content2,6,7. Since conditions are not well controlled during the fabrication of most am-C support membranes, there is a large variation in their performance; microscope time and samples are often wasted on poor substrates. In contrast, graphene8, an atomically thin, mechanically robust conductor, comprises a near ideal substrate for imaging nanoscale specimens by EM. Suspended graphene conducts charge ballistically over the sub-micron distances that span a hole used for imaging molecules in ice8, which should reduce the buildup of surface charge during electron beam exposure. Further, graphene’s in-plane mechanical strength (Young’s modulus 1 TPa)9 is the highest ever measured for any material.
Despite its potential advantages, the ease of large-scale manufacture using chemical vapor deposition (CVD), and some promising preliminary work on using it as a substrate for biological molecules10-13, graphene has not been widely adopted for use with biological specimens. There are two reasons for this: graphene is hydrophobic, which precludes the deposition of proteins from aqueous solutions, and it is susceptible to surface contamination during manufacturing, handling and storage. Previous attempts to use graphene as an EM support required either harsh solvents that are incompatible with most proteins or conversion of the graphene to graphene oxide11,12. Graphene oxide is less than ideal since: (1) it contributes nearly as much background signal as thin am-C [11], (2) it is often an insulator making it less able to neutralize accumulated surface charge14, and (3) it has decreased mechanical strength, making it less stable than graphene.
Recently, investigators showed that it is possible to convert graphene to its fully hydrogenated form, graphane, using extended hydrogen plasma treatments15. Here we demonstrate that ≦ 5% hydrogenation of graphene with a low-energy hydrogen plasma can address the two problems described above: removal of surface contamination and making graphene suitably hydrophilic for use with proteins.
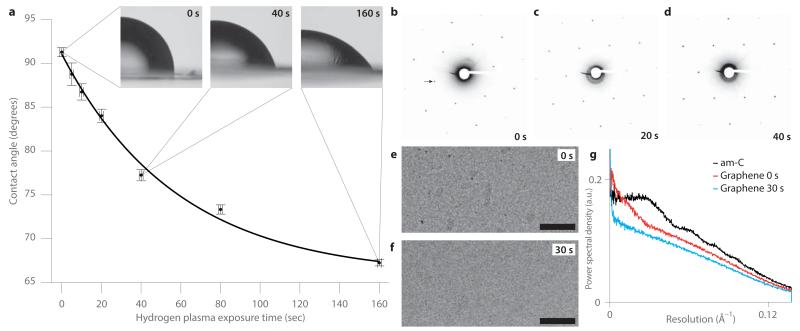
A low-energy, pure hydrogen plasma contains H, H+, H2+, H3+ and free electrons16. We hypothesized that the primary chemical reaction of graphene with hydrogen plasma is hydrogenation via the reaction: sp2C + H ⇄ sp3CH. We assessed the change in hydrophobicity of a graphene surface in response to a hydrogen plasma, by measuring the static contact angle between water, air and graphene (Fig. 1a). With increasing plasma dose, the contact angle decreased exponentially from a value of 91 ± 0.5° to a saturation value of 66 ± 1.3° (std. err.). This corresponds to a reduction in the graphene-water interfacial energy of 0.19 ± 0.02 eV/nm2 (Supplementary Note 1). To monitor the conversion to graphane, we measured the change in the lattice constant15 after plasma exposure using electron diffraction (Fig. 1b-d and Supplementary Fig. 1). After an 80 second dose of plasma treatment, less than 20% of the carbon-carbon sp2 bonds were converted to carbon-hydrogen sp3. Furthermore, the sharp peaks in the diffractograms after plasma treatment (Fig. 1c-d) demonstrated that the underlying graphene lattice is preserved.
Figure 1. Low-energy hydrogen plasma treatment renders graphene hydrophilic and removes contamination.
(a) Plots of the air-water-graphene contact angle versus exposure time, with insets containing examples of optical micrographs used to measure the angles. The curve is an exponential fit to the data with rate constant of 1/58 seconds. Error bars are the std. err. at each plasma dose for the 3-5 measurements in y and the estimated accuracy of the exposure time, ± 1 second, in x. (b-d) Selected area electron diffraction patterns for the same suspended graphene sample: before hydrogen plasma exposure (b), after 20 seconds (c) and after 40 seconds (d). Arrow points to the 0-110 reflection at 2.14 Å and sets the scale for all three diffractograms. The change in the lattice constant for c & d relative to b is less than the error in the measurement, ≈0.9% (Supplementary Fig. 1). (e-f) Electron micrographs of suspended graphene before (e) and after (f) 30 second hydrogen plasma treatment respectively. Scale bars are 1000 Å, nominal defocus is −2.0 μm and fluence (electron dose) is 25 e−/Å2. (g) Power spectral density in each complete micrograph (Supplementary Fig. 2), normalized to the total image intensity, for 28 ± 1.4 Å thick am-C (Supplementary Fig. 2e), graphene before plasma treatment (e), and graphene after plasma treatment (f).
We studied graphene’s properties as a substrate for cryo-EM before and after plasma treatment as compared to am-C. When suspended graphene is prepared using typical transfer methods, it is usually contaminated with adsorbents (Fig. 1e). Exposing the graphene to 30 seconds of hydrogen plasma removed most of the surface contamination (Fig. 1f) and decreased the background signal level in the image (Fig. 1g). Compared to a thin (28 ± 1.4 Å) layer of am-C (Supplementary Fig. 2), the background signal of the treated graphene lattice was much lower and featureless (Fig. 1g).
Hydrogen plasma treatment likely removes surface contamination much faster than it modifies graphene, since hydrocarbon contaminants are easily broken down and volatilized by the energetic plasma. Since the graphene sp2 bond is highly resistant to chemical modification, it is only slowly reduced by hydrogen in the plasma. Hydrogen species in the 10-15 eV range used here have insufficient energy to directly remove carbon atoms from their sp2 bonds (21 eV)17, eliminating sputtering damage to the lattice.
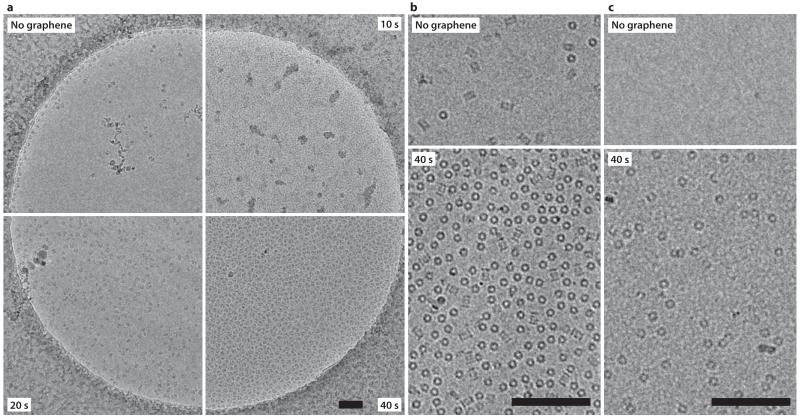
Having shown that we can control the hydrophobicity of graphene, we then tested hydrogen plasma-treated graphene as a substrate for cryo-EM using purified ribosomes. We transferred monolayer graphene to holey carbon EM grids, subjected the grids to various doses of hydrogen plasma, and used them to prepare vitrified ice samples containing 70S ribosomes. On grids without graphene, very few ribosomes were visible in the ice (~60 particles/μm2), as most were attracted to the surface of the am-C and the edges of the holes (Fig. 2a, Supplementary Fig. 3). Using the same concentration, freezing and blotting conditions with graphene grids treated with 10 seconds of hydrogen plasma, only small patches of vitreous ice and clumped ribosomes were visible (Fig. 2a). This indicated incomplete wetting and was consistent with the small change in contact angle at this dose. After 20 seconds of hydrogen plasma, the graphene surface was more uniformly wet and the ice quality was greatly improved, demonstrating that ribosomes adsorbed to the surface (~600 particles/μm2). Increasing the plasma dose to 40 seconds led to very densely packed ribosomes (~1900 particles/μm2). Selected area diffractograms acquired after each image confirmed the vitreous nature of the ice and verified the presence of the graphene layer (Supplementary Fig. 4). Overall, our results demonstrate a monotonic relationship between hydrogen plasma dose and surface particle density, thereby allowing the experimenter to use graphene to tune particle distribution.
Figure 2. Dose dependent adsorption of proteins on hydrogen plasma-treated graphene.
(a) Electron cryo-micrographs of 70S ribosomes in vitrified ice at 80 K. Upper left quadrant is a standard grid treated with a 10 s hydrogen plasma dose. Other three quadrants show grids covered with monolayer graphene, and treated with 10, 20 and 40 s of hydrogen plasma. All other sample concentration, blotting, vitrification and imaging conditions are the same for all four grids. Scale bar is 1000 Å. (b-c) Electron micrographs of 20S proteasome (b) or apoferritin (c) molecules on graphene treated with 40 s of hydrogen plasma (lower panels) and molecules in unsupported ice from an adjacent region of the same grid (upper panels). Scale bars are 1000 Å and magnification is the same for (b) and (c).
To demonstrate that adsorption to partially hydrogenated graphene is not specific to the 70S ribosome, we also tested the 80S ribosome (Supplementary Fig. 5), the 20S proteasome (2b, Supplementary Fig. 6a) and apoferritin (Fig. 2c, Supplementary Fig. 6b). 80S ribosomes have a 10-fold higher density on graphene grids, compared to grids without an additional support. The density of proteasome particles on the graphene surface is 6-fold higher than in the unsupported ice and is likely near saturation due to steric effects. Apoferritin showed a >500-fold increase in particle surface density on graphene.
To study cryo-EM image quality on graphene, we determined the three dimensional structure of the 70S ribosome from three micrographs on graphene (Supplementary Fig. 5). Using 2061 individual particle images, we obtained a reconstructed density map at 19 Å resolution (Supplementary Fig. 7). Next we calculated a reconstruction from 20,050 80S particles on graphene (Supplementary Fig. 5), which reconstructed to a resolution of 5.0 Å(Supplementary Fig. 7). Using these data, we assessed the quality of the images on graphene. Imperfections in particle images are often characterized using an empirical model of short-range atomic motion (B-factors), which provides a measure of the image quality. We observed an ~35% reduction in the B-factor of 80S ribosomes on graphene compared to am-C, which is indicative of the improvement in the information content of each particle image.
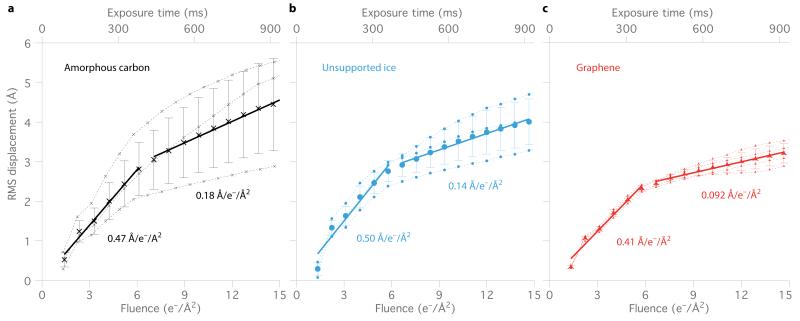
To investigate the origins of the improved image quality, we analyzed movie data of individual particles acquired using a direct electron detector for 80S ribosomes on three substrates: am-C, unsupported ice and graphene (Fig. 3). We used the individual frames of the movies to determine the trajectory of each particle and then calculated the root mean squared (RMS) ensemble displacement for each frame for thousands of particles on each test grid, and repeated the measurements for multiple grids (Fig. 3 and Supplementary Fig. 8). We found that 80S ribosome particles in ice irradiated with high-energy electrons exhibited two distinct phases of motion, an initial fast phase from 0 to ~6 e-/Å2, followed by a slower phase. Relative to conventional grids, the speed of first and second phases were 10-20% and 30-50% reduced on graphene, respectively, which likely accounts for the improved image quality on graphene. We note the striking differences in radiation-induced movement from grid to grid for conventional grids. While some conventional grids performed similarly to graphene (Fig. 3 and Supplementary Fig. 8), there is a much larger variation from one grid to the next, which is likely due to uncontrolled differences in their conductivity and mechanical properties. The well-defined and reproducible mechanical and electrical properties of graphene likely account for its more consistent performance.
Figure 3. Reduced motion of proteins on graphene substrates: speed plots.
The average 80S ribosome displacement from its initial position is plotted vs. time/ electron fluence for data collected in ice: supported by a continuous layer of am-C (a), without any support layer (b) and supported on a graphene substrate (c). Each point (dotted lines) represents the root mean squared (RMS) displacement of thousands of particles from a single grid (see Methods), whose positions were measured using a five-frame running average under constant electron beam irradiation (300 keV; 16 e−/Å2/s). Solid lines are the linear fits to the two phases of motion, with the slopes (ensemble particle speed) as indicated. Error bars are the standard error of the mean of the replicate experiments (3 separate grids for a,b 4 for c). All plots have the same scale.
In summary, we have demonstrated that a low-energy hydrogen plasma can non-destructively modify graphene hydrophobicity. Surprisingly, even the addition of one hydrogen atom for every 20 carbon atoms renders graphene hydrophilic and induces protein adsorption. We propose a simple model (Supplementary Note 2) to understand this large change in interfacial energy (~7 kT/nm2) and its saturation with only partial hydrogenation of the lattice18.
A major problem with conventional cryo-EM grids is the lack of precise control of the distribution of proteins within a thin layer of vitreous ice. During blotting and vitrification, proteins often segregate to the air/water interface or to other surfaces present. The use of hydrogen plasma-treated graphene enables better control of particle surface density independent of other sample preparation variables (concentration, humidity, blot time, etc.). Additionally, the specific adsorptive properties of the hydrogen treated graphene surface allow the use of much lower concentrations of protein, e.g., 150 times less for the case of apoferritin and 10 times less for 80S ribosomes.
Another problem in cryo-EM is specimen-dependent degradation of image quality7; this can be improved in two ways using hydrogen plasma-treated graphene. First, graphene is effectively invisible at resolutions used in structural biology (>2.1 Å) leading to reduced background noise in the images compared to am-C. Second, graphene generally reduces electron radiation-induced particle movement (image blurring) and thus increases the information in each image. We speculate that such movement is caused by a complicated buildup of charge and stress on the specimen19,20, which likely includes the creation of large electric fields across regions of the irradiated area, electrical breakdown of the ice layer in response to these fields, density changes within the ice due to radiolysis and the mechanical release of stresses in the ice created during vitrification. Further work is needed to elucidate the detailed dynamics and underlying mechanisms of these particle motions.
Hydrogen plasma-treated graphene offers a reproducible and tunable surface for the adsorption of proteins. We envision that the preparation of biological specimens for cryo-EM will move from a trial-and-error art to systematic screening of surface conditions by using a range of plasma treatments. We also expect that low-energy plasma treatment of graphene-based sensors will enable the detection and characterization of specific biological molecules and complexes.
METHODS
Graphene synthesis
Graphene was synthesized by chemical vapor deposition (CVD) on copper foil substrates21,22. Briefly, an ~18 cm2 section of 25 μm copper foil (Alfa Aesar# 13382) was placed in the 25 mm diameter quartz tube of a dry-pumped CVD furnace, evacuated to <20 mTorr, and then exposed to a continuous flow of Hydrogen (99.999%) gas at a flow of 20 sccm, bringing the pressure in the reaction tube to 500 mTorr. The temperature of the oven was elevated under thermocouple control to 1000 °C over the course of ~15 minutes. Once the temperature was reached, an additional 20 sccm of methane (99.999%) was added to the reaction chamber for a duration of 15 minutes, bringing the total pressure to 700 mTorr. After methane addition was over, the heating was turned off and the reaction tube was slowly cooled to ambient temperature under continuing hydrogen flow over a period of 2 hours. Once cool, the hydrogen flow was stopped, the chamber vented with dry nitrogen and the graphene on copper foil was removed from the growth tube and stored in a critically-cleaned Fluorware wafer container inside a clean, low-humidity storage box until use.
Hydrogen plasma treatment and contact angle measurement
To measure the graphene water contact angle, individual 3.2 mm diameter disks were punched from the graphene on copper foils using a custom-made disk punch. Disks were initially cleaned by submersing them for 10 seconds in CMOS grade isopropyl alcohol (Sigma) and then after the residual solvent evaporated, were placed on a nitric-acid cleaned glass slide inside a commercial plasma reaction chamber (Fischione Model 1070) where the grids were located 15 ± 1 cm from the edge of the RF coils. The design of this instrument is such that the acceleration of ions across the plasma sheath is minimized so high-energy species are not expected to contribute to reactions at the graphene surface23. The source of hydrogen was a high-purity electrolysis hydrogen generator (Dominik Hunter model 20H-MD). Controls for the zero time exposure dose included those with and without various solvent cleaning treatments; we found the contact angle was the same for all to within experimental error. Each disk was mounted in an optical microscope (Zeiss Axiophot), and a 1 μL droplet of 18 MΩ (5.5 × 10−8 S/cm) deionized water was applied and immediately (within 5 seconds) upon withdrawal of the pipette tip, imaged using a calibrated digital camera attached to the microscope (Zeiss ERc5s). We measured the droplet evaporation rate (1.4 nL/sec) and found it contributed a negligible change in angle (<0.5°) during the delay between water application and image acquisition. Each image was subsequently analyzed to obtain the contact angle by graphically measuring the angle between the substrate plane and the tangent to the droplet at the point it met the surface. This measurement was performed 3-5 times for each of the plasma doses (n = 5, 3, 5, 5, 4, 5, 4); the values were then normalized by dividing by the ratio of the droplet volume measured in the image to the value for the zero plasma dose. We found this normalization was required to correct for the error in pipetting a volume this small. The multiple measurements were then averaged, and the standard error of the mean taken as the error in angle at each dose. Finally, we confirmed the presence of graphene covering the punched disks by subsequently transferring the graphene to EM grids as described below, and then imaging them with an electron microscope. We note that the contact angle of 91° for intrinsic graphene (Fig. 1a) is well within the wide range of reported values24. Contact angles in the 60-80° range are typically used for protein deposition on standard am-C substrates25. The rate constant of the exponential fit was 1/58 seconds.
Graphene grid preparation
To create suspended monolayer graphene EM grids, we use a method based on one first described by Regan et al.26 and further developed in Ref. 10. We started by cleaning commercial holey carbon on 300 mesh gold grids (Quantifoil Au 300 1.2/1.3) by using anti-capillary tweezers to immerse them individually in chloroform, acetone and isopropyl alcohol for ~15 seconds each (Sigma-Aldrich, ultra-high purity semiconductor grade solvents). We found this helped to remove any residual photoresist and large surface contaminants remaining on the Quantifoil films after manufacture. After blotting dry in air, we then mounted the grids in a custom-made stainless steel suspension holder inside the plasma reactor mentioned above. The chamber was evacuated to <10−5 Torr and then ultra high purity argon and oxygen (BOC 99.9999%) were admitted in a mass ratio of 9:1 to a steady state pressure of 21 mTorr. The auto-tuned RF plasma was sparked at 40 Watts (<3 W reverse power) and applied for the specified time, in this case 10-30 s for cleaning the grid. Quantifoil grids were then used immediately for graphene transfer.
To cover the Quantifoil grid with a graphene monolayer, a 3.2 mm disk of graphene on copper was cut from a larger foil using a custom made mechanical punch. The disk was immersed in acetone, and isopropanol (Sigma, CMOS grade) for 10-15 seconds each and blotted dry prior to use. The plasma cleaned Quantifoil grid was then applied, carbon side down, to the disk. An optical microscope (Zeiss Axiophot) was used to inspect the grid “sandwich” making sure that both the grid and the disk were flat, free of particulate contaminants and in good contact with each other before the next step. Then 7 μL of CMOS grade isopropyl alcohol was added to the top of the grid, and the droplet was allowed to dry in air. The receding meniscus of the alcohol pulled the carbon film of the Quantifoil in contact with the graphene surface, which was verified by the change in the color of the reflected light from the surface.
Next the grid-disk sandwich was floated in ~50 mL of buffered FeCl3 (Sigma) in a crystallization dish for 20 min. The grid was then transferred with a flamed Pt loop to 32% HCl (Sigma CMOS grade) for 5 min, then 10% HCl for 5 min followed by 3 rinses in 18 MΩ deionized water. After the final water step, the grid was transferred using the loop to a piece of filter paper (Whatmann #1) in an acid-washed, glass petri dish and stored in a low-humidity box until use. All steps of the graphene transfer were monitored using an optical microscope. Grids were discarded if there were any defects.
Diffraction studies of hydrogen treated suspended graphene
Selected area diffraction studies of suspended graphene were performed before and after hydrogen plasma exposure in the following way: First, CVD-grown graphene was transferred to a pre-cleaned Quantifoil grid as described above. The grid was mounted in a single-tilt holder (FEI) that had been cleaned with a 75%/25% Ar/O2 plasma at 50 W for 5 min. The grid was imaged with 300 keV electrons in a FEI Tecnai F30 microscope whose residual column pressure was nominally 88 nTorr (measured at the closest ion pump) and with an anti-contamination shield surrounding the sample cooled by liquid nitrogen. Selected area diffractograms were collected on a liquid cooled 2K×2K CCD camera (Tietz F224HD) using a fluence of ~30 e−/Å2/s, a nominal camera length of 690 mm, exposure time of 1 second, and a 10 μm diameter selected area aperture which corresponded to an interrogated area of 0.30 μm2 at the sample. The actual camera length was calibrated using the 111 to 311 lattice reflections of a thin film of polycrystalline aluminum. After the first diffractogram was collected, the sample was removed from the column and immediately transported in a sealed, carefully cleaned container while still mounted in the holder to the plasma chamber, exposed to pure hydrogen plasma (with the same conditions as above) while mounted in the holder, and then immediately returned to the electron microscope for collection of the second data set. During repeated experiments using this transfer process we saw no evidence of contamination of the graphene sample. The systematic error in the measurement limits the precision to about 0.9% (Supplementary Fig. 1). Final diffractograms were inverted to black on white to improve contrast when printed.
Vitrification and electron cryo-microscopy using hydrogen plasma-treated graphene grids
Frozen stocks of Thermus thermophilus 70S ribosomes27 or Saccharomyces cerevisiae 80S2 provided by the V. Ramakrishnan lab were thawed and diluted in 5 mM HEPES pH 7.5, 50 mM KCl, 10 mM NH4Cl, 10 mM Mg-acetate, 6 mM β-mercaptoethanol (70S) or in 3 mM HEPES-KOH pH 7.45, 6.6 mM Tris-acetate pH 7.2, 3 mM NH4Cl, 6.6 mM NH4-acetate, 48 mM K-acetate, 4 mM Mg-acetate, 2.4 mM DTT (80S), all the while being kept on ice. Ribosomes were used at 70 nM (70S) or 80 nM (80S) for graphene or carbon supported grids, and at 800 nM for unsupported Quantifoil grids. Horse spleen apoferritin (Sigma) and human 20S proteasomes (Enzo) were diluted to 100 nM in phosphate-buffered saline pH 7.4 (PBS). Grids were treated with a pure hydrogen plasma as described above, for the indicated times just prior to use. For the datasets used for 3D ribosome reconstructions and radiation-induced motion measurements, the hydrogen plasma exposure time was 20 seconds, just prior to use, and the dose for the apoferritin and proteasome was 40 seconds. 70S ribosome grids were placed in a cryoplunger (FEI Vitrobot IV) equilibrated to 4 °C and 100% relative humidity; 3 μL of sample was applied, allowed to incubate for 60 seconds, blotted with force setting −20 for 2 seconds, and then plunged into liquid ethane at just above its melting point. 80S ribosome, proteasome and apoferritin grids were placed in a nearly identical cryoplunger (FEI Vitrobot III) and prepared with empirically equivalent settings: equilibrated to 4 °C and 100% relative humidity; 3 μL of sample was applied, allowed to incubate for 15 seconds, blotted with force setting −2 mm for 5 seconds, and then plunged into liquid ethane at just above its melting point. Grids were then stored in liquid nitrogen until they were transferred to an electron microscope for imaging. Previously published cryo-EM studies of apoferritin28 required sample concentrations that were ~150 times greater (15 μM vs. 100 nM used here), presumably because most molecules are lost to other surfaces present during the blotting process.
For the 80S data in Supplementary Fig. 5, we made four individual grids with varying plasma dose. The first grid examined had 15 s hydrogen plasma treatment. It had a low density of particles, but was otherwise good. The second grid examined had 20 s hydrogen plasma treatment. The particle density was excellent and all our data were collected from this grid. The remaining grids were discarded without examination. For the data in Fig. 2 we prepared two Quantifoil grids and one graphene grid for each hydrogen plasma condition. All grids had suitable ice, except the one with 10 s hydrogen plasma treatment, which was too hydrophobic to wet properly. For the additional data in Fig. 3, we prepared 4 graphene grids, all of which had good ice and three were chosen for data collection. Thus, the failure rate for preparing cryo-specimens on graphene substrates is very low.
Vitrified 70S ribosomes were imaged with 300 keV electrons in a Polara (FEI) under very low-dose conditions (2.5 e−/Å2/s; <0.2 e −/Å2 pre-exposure in low-mag mode) at a nominal magnification of 23 kX at a temperature of 80-90 K. Exposures were one second on a CMOS direct-electron detector (FEI back-thinned Falcon II) where the calibrated pixels corresponded to (4.58 Å)2 at the sample and the frame rate was 17 frames per second.
Vitrified 80S ribosomes were imaged with 300 keV electrons in a Titan Krios (FEI) under low-dose conditions (16 ± 3 e−/Å2/s; <0.2 e −/Å2 pre-exposure in low-mag mode) at a nominal magnification of 59 kX (Supplementary Fig. 5) or 47 kX (additional data in Supplementary Fig. 8) at a temperature of 80-90 K. Exposures were one second on a CMOS direct-electron detector (FEI back-thinned Falcon II) where the frame rate was 18 frames per second and calibrated pixels corresponded to (1.34 Å)2 or (1.745 Å)2 at the sample. The condenser lens and aperture settings were such that the electron beam was circularly symmetric about the center of the grid hole, and encompassed an area about 10-15% larger than the area of the suspended ice. Based on the symmetry of the interacting region, this illumination geometry minimizes the possible development of local electric fields and empirically gave the best images. All high-resolution data herein was collected using this illumination geometry. The first and last frames of the collection were discarded due to incomplete blanking of the beam. The region of the grid used for data collection had >95% single layer graphene coverage as verified by electron diffraction on about every fifth hole after image acquisition. A typical micrograph of 80S ribosomes on graphene is shown in Supplementary Fig. 9.
Single particle data analysis
For the 70S dataset, 2282 particles were picked from three micrographs using EMAN2’s boxer program with the semiautomated swarm method29. Next, the particles were preprocessed and extracted in 76 × 76 pixel boxes (scale is 4.58 Å/px) in Relion30, which included CTF fitting using CTFFIND331. After one round of 2D classification with 18 classes, 221 particles were discarded and the remaining 2061 were used for 3D refinement. The 3D refinement was carried out in Relion using an initial model generated from the 70S crystal structure32, low-pass filtered to 50 Å. The 3D refinement converged at an estimated angular accuracy of 3.6° and a resolution of 19.3 Å (0.143 gold-standard FSC). The final map was low-pass filtered to 19.3 Å. The angles assigned to each particle imaged in the final refinement iteration (rotation and tilt in Relion) were used to generate the plot of orientations. A combined PDB from 2WDK and 2WDL32 was fit to the map using UCSF Chimera’s rigid body fitting algorithm33. The 3D renderings of the maps and models were created in Chimera.
The 80S dataset on graphene shown in Supplementary Fig. 5 comprised 48386 particles picked from 401 micrographs again with EMAN2’s boxer program with the semiautomated swarm method. Micrographs were gain corrected after collection by dividing by the sum of all the images, normalized to its mean value. Particles were preprocessed and extracted in 328 × 328 pixel boxes with Relion, including CTF fitting using CTFFIND3. Eleven micrographs were rejected due to poor CTF fits. After 3 rounds of 2D classification, 20050 particles were kept for refinement in 3D. The vast majority of particles removed at this stage were individual 60S subunits. We used 20050 particles imaged on am-C using the same data collection strategy (fluence, exposure time, voltage, etc.), randomly selected from a previously published dataset2, for comparison. To facilitate comparison, we used the same initial model with the same coordinate orientation (EMDB-1780) 34, low-pass filtered to 60 Å. All other refinement parameters were the same for the two datasets. The 3D refinement used gold-standard FSC calculations to prevent over-fitting of the high resolution data, as implemented in Relion’s 3D autorefine algorithm35. FSC curves were calculated between the two halves of the data using the automated masking procedure in Relion post-process. Resolutions of the reconstructions cited in the text were determined using the gold-standard method with a 0.143 criterion36. B-factor changes cited in the text were calculated for linear fits to unsharpened, uncorrected Guinier plots between (10.0 Å)−2 and (5.2 Å)−2 for both datasets36.
Sample preparation and data collection for the additional datasets (Fig. 3) collected in unsupported ice (standard Quantifoil grid, no continuous carbon support layer), on am-C and on graphene were identical to the above. The numbers of particles used for comparison were: 20050 (graphene data from above), 8676, 9481 and 2715 for graphene; 55837, 15014 and 8807 for unsupported ice; and 20050 (previously published glow-discharged2), 32751 and 9819 for am-C.
Orientation distribution analysis
We assessed the orientational distribution of the ribosomes on the graphene substrate and compared it to am-C using the same number of particles randomly chosen from a previously published dataset on the same ribosome sample2. We plotted the result using a Mollweide equal area projection (Supplementary Fig. 5). While proteins generally exhibit preferential orientations relative to any interacting surface, we found the orientational distribution for 70S on hydrogen plasma-treated graphene was better (more optimal coverage of Fourier space) than on glow-discharged am-C, whereas for 80S, it was somewhat worse compared to the previously published result2 (Supplementary Fig. 5). Interestingly, even though the preparation of the graphene surface and the resultant particle surface density are nearly the same for 70S and 80S samples, the orientational distribution is different. While every surface, including an air-water interface, is expected to influence the orientational distribution of proteins in thin layers of water, it will be specific to each protein-surface interaction and may be influenced by solution conditions and the presence of other amphiphilic species like detergents. Further work will be required to understand, control and improve the orientational distributions of molecules on different substrates.
Speed plots
To measure the motion of the particles on the various grids, we first determined their position at each movie frame in the micrograph using cross-correlation as implemented in Relion. To improve positional accuracy, five-frame running averages were used for the calculation. At the two ends of the movie, the averages for the first and last frames and the second and penultimate frames were averaged over the adjacent three and four frames respectively. The coordinates assigned to each particle for each frame were used to calculate the RMS particle displacements for the entire particle ensemble vs. time/dose. For speed plot calculations, an ensemble comprised of a set of particles from an individual grid. Each grid ensemble is plotted separately in Fig. 3 and Supplementary Fig. 8 where a dotted line connects each data point. We calculated the mean value for the various ensemble trajectories on a particular type of grid and plot those (large markers) as well as the standard error of the mean (error bars). Using these data points, we calculated linear fits using regression analysis including a weighting function to account for the errors. The results are tabulated in Supplementary Table 1.
Since the initial position of the particle before irradiation is unknown, we must make some assumption to set the zero of the displacement curve. Here we took the y-intercept from the fit to the first phase of motion as the best available estimate of the initial position of the particle. This includes as much information as possible about where the particle was located at the initiation of irradiation, but is only approximate as the equation of motion for the particles during electron irradiation is unknown. In addition, to set the zero exposure time, we had to account for the speed of the blanking relative to the timing of frame acquisition. Since the detector is continuously acquiring frames, the first collected frame in the exposure is triggered by the unblanking of the beam, which occurs somewhat randomly, but on average is in the middle of the frame. This frame is discarded but we add time equivalent to half a frame in the ensemble calculations to set the absolute zero of exposure time and fluence shown in Fig. 3. Based on the accuracy of the clocking of the detector frame acquisition times, the accuracy of the beam current measurement and stability, and the beam blanking speeds, we estimate the error in the exposure time per frame and the variation in electron fluence per frame to be <1% for all frames, and the error in the absolute electron flux to be 20-30%, and the relative variation in electron flux between the different experiments to be <5%.
Finally, we also used newly developed motion correction algorithms2 to correct for particle motion during 3D reconstruction. We found that this gave very little improvement of the reconstructed map for graphene data, whereas the same algorithms applied to the same sample with larger particle motions on am-C did result in improvement of the reconstruction (Supplementary Fig. 7b). This agrees with the fact that we have reduced the motion of the particles, so motion correction is less effective.
Atomic force microscopy
To accurately measure the thickness of carbon films, a portion of the carbon layer as evaporated on mica was cleaved off using adhesive tape (3M Scotch Crystal) leaving behind a step edge. This was imaged in constant force, DC mode using an oxide sharpened, contact mode silicon tip/cantilever (Mikromasch CSC37) in air at ambient conditions in an atomic force microscope (MFP3D, Asylum Research). After obtaining a 2D topographic image, the height was flattened using a two-dimensional, 2nd order polynomial fit to the masked mica surface and rendered using a grayscale as indicated in the images (Supplementary Fig. 2d). A histogram of the height values was created, and the two peaks representing the substrate and carbon surfaces were fit with Gaussian functions (Supplementary Fig. 2e). The parameters of these fits were then used to measure the height and calculate the error in the measurement (std. dev.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. A. Golovchenko for the use of a chemical vapor deposition instrument at Harvard for graphene synthesis during the initial phases of this work; S. Scotcher for fabrication of custom sample holders and copper punch machines; I.S. Fernandez, A. Kelley, and V. Ramakrishnan of the MRC-LMB for the gift of ribosomes; J. Grimmett, T. Darling, G. McMullan and S. Chen for technical assistance; E. Rajendra, R.A. Crowther, D. Neuhaus, X.C. Bai, S. Scheres, N. Unwin, A.R. Faruqi and R. Henderson for helpful discussions and comments. This work was supported by European Research Council grant no. 261151 to L.A.P., a MRC (UK) Centenary award to C.J.R. and MRC (UK) grant U105192715 (L.A.P.).
Footnotes
COMPETING FINANCIAL INTERESTS: C.J.R. and L.A.P. are inventors on a patent application related to technology described in this manuscript (US 61/865,359).
REFERENCES
- 1.Grigorieff N, Harrison SC. Curr. Opin. Struct. Biol. 2011;21:265–273. doi: 10.1016/j.sbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai X-C, Fernandez IS, McMullan G, Scheres SH. Elife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubochet J, et al. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 4.Robertson J. Advances in Physics. 1986;35:317–374. [Google Scholar]
- 5.Miyazawa A, et al. J Mol Biol. 1999;288:765–786. doi: 10.1006/jmbi.1999.2721. [DOI] [PubMed] [Google Scholar]
- 6.Brilot AF, et al. J Struct Biol. 2012;177:630–637. doi: 10.1016/j.jsb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henderson R, McMullan G. Microscopy. 2013;62:43–50. doi: 10.1093/jmicro/dfs094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geim AK. Science. 2009;324:1530–1534. doi: 10.1126/science.1158877. [DOI] [PubMed] [Google Scholar]
- 9.Lee C, Wei X, Kysar JW, Hone J. Science. 2008;321:385–388. doi: 10.1126/science.1157996. [DOI] [PubMed] [Google Scholar]
- 10.Russo CJ. PhD. Thesis. Harvard; 2010. [Google Scholar]
- 11.Pantelic RS, Meyer JC, Kaiser U, Baumeister W, Plitzko JM. J Struct Biol. 2010;170:152–156. doi: 10.1016/j.jsb.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 12.Pantelic RS, et al. J Struct Biol. 2011;174:234–238. doi: 10.1016/j.jsb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Sader K, et al. J Struct Biol. 2013 doi: 10.1016/j.jsb.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Navarro C, et al. Nano Lett. 2007;7:3499–3503. doi: 10.1021/nl072090c. [DOI] [PubMed] [Google Scholar]
- 15.Elias DC, et al. Science. 2009;323:610–613. doi: 10.1126/science.1167130. [DOI] [PubMed] [Google Scholar]
- 16.Méndez I, et al. J. Phys. Chem. A. 2006;110:6060–6066. doi: 10.1021/jp057182+. [DOI] [PubMed] [Google Scholar]
- 17.Russo CJ, Golovchenko JA. P Natl Acad Sci Usa. 2012;109:5953–5957. doi: 10.1073/pnas.1119827109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Israelachvili J, Pashley R. Nature. 1982;300:341–342. doi: 10.1038/300341a0. [DOI] [PubMed] [Google Scholar]
- 19.Curtis GH, Ferrier RP. Journal of Physics D: Applied Physics. 1969;2:1035. [Google Scholar]
- 20.Downing KH, McCartney MR, Glaeser RM. MAM. 2004;10 doi: 10.1017/s143192760404067x. [DOI] [PubMed] [Google Scholar]
- 21.Reina A, et al. Nano Lett. 2009;9:30–35. doi: 10.1021/nl801827v. [DOI] [PubMed] [Google Scholar]
- 22.Li X, et al. Science. 2009;324:1312–1314. doi: 10.1126/science.1171245. [DOI] [PubMed] [Google Scholar]
- 23.Lieberman MA, Lichtenberg AJ. Principles of Plasma Discharges and Materials Processing: Second Edition. (Second Edition) 2005 [Google Scholar]
- 24.Taherian F, Marcon V, van der Vegt NFA, Leroy F. Langmuir. 2013;29:1457–1465. doi: 10.1021/la304645w. [DOI] [PubMed] [Google Scholar]
- 25.Dubochet J, Groom M, Mueller-Neuteboom S. Advances in Optical and Electron Microscopy. 1982;8:107–135. [Google Scholar]
- 26.Regan W, et al. Appl. Phys. Lett. 2010;96:113102. [Google Scholar]
- 27.Selmer M, et al. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 28.Wasilewski S, Karelina D, Berriman JA, Rosenthal PB. J Struct Biol. 2012;180:243–248. doi: 10.1016/j.jsb.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Tang G, et al. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Scheres SHW. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mindell J, Grigorieff N. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 32.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen EF, et al. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Armache J-P, et al. P Natl Acad Sci Usa. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheres SHW, Chen S. Nature Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenthal P, Henderson R. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.