Abstract
The pursuits of sustainable treatments for diseases and disorders that afflict the central nervous system (CNS) have proven challenging for the field of viral vector-based gene therapy. However, recent advances in viral vector technology coupled with efficient delivery methods have opened up new avenues that show promise at the preclinical testing stage. The development of the Herpes Simplex Virus/Sleeping Beauty (HSV/SB) hybrid vector represents such an advance for devising treatments targeting the CNS with its potential for stably integrating large transgenomic segments of DNA within the genomes of transduced cells. In utero administration of this hybrid vector into the embryonic mouse brain has revealed the capacity for widespread transgene dissemination due to the targeting of a neuronal precursor cell population. This unique feature has provided the means to stably express a transgene throughout the brain for prolonged periods, which is a prerequisite for the treatment of progressive CNS disorders. In this review we provide a comprehensive breakdown of the characteristics of the HSV/SB vector system and how it can be efficiently employed in the derivation of CNS-targeted gene therapeutic strategies.
Keywords: HSV-1, Sleeping Beauty, central nervous system, gene transfer, integration
1. Introduction
The development of gene delivery vehicles via the manipulation of pathogenic and nonpathogenic viruses has revolutionized the way certain diseases can be treated. Gene therapy attempts to correct a disease at the “grass-root” level specifically targeting the disease-causing genetic aberration itself to ensure long-lasting therapeutic benefit. Recently, a significant breakthrough in the field was achieved when an adeno-associated virus (AAV) vector was used to correct Type 2 Leber's congenital amaurosis (LCA) disease, a severe form of retinal degeneration, by safely delivering a correct copy of the human RPE65 gene to the retinal pigment epithelium of patients [1]. Such success stories are a synergistic result of advances made in understanding disease etiology, vector design and vector delivery precision. During the past two decades researchers have also made significant strides in the use of viral vectors to deliver genes directly to the CNS in a more efficient and safe way as a means to bypass the stringent control imposed by the blood-brain barrier to vectors themselves and traditional small molecule-based treatments [2].
Viruses such as adenovirus (Ad), AAV, lentivirus (LV), and Herpes simplex virus (HSV) provide an excellent means to deliver therapeutic genes for disease correction within the CNS due to their intrinsic ability to infect dividing and non-dividing cells that populate the human CNS. The genomes of these viruses have been meticulously characterized and researchers have successfully generated innocuous forms of the virus that retain their potency in infecting cells but are non-pathogenic and lack the capacity to replicate within mammals. As the etiologies of many CNS disorders such as Parkinson's disease, Huntingtons’ disease, and motor neuron disease, were being gradually unraveled at the molecular level in the early 1990's, researchers were enthused by the possibility of using viral vectors to deliver neuro-protective genes to the CNS with the hope of either curing the disease or at least slowing its progression in the patient. Thus, rigorous preclinical testing was set in motion using Ad, LV, and HSV-based vectors in animal models of neurodegenerative diseases. These studies have provided a wealth of information regarding the pros and cons of each vector system in their use as genetic vehicles for CNS-related diseases. While Ad vectors have proven to elicit potent inflammatory responses in patients [3], traditional HSV-derived vectors have been shown to provide only short-term transgene expression due to their entrance into latency (recombinant HSV vectors) or genome's episomal existence within the cell (amplicon type), qualities that do not suffice for the treatment of progressive neurodegenerative diseases [4, 5]. AAV and LV vectors too are limited in disease application due to their limited genetic cargo capacity [6, 7]. Furthermore, while many animal models of neurodegeneration attempt to mirror the human condition, translation of results from pre-clinical models to the human clinical setting has yielded divergent outcomes [8]. Such findings highlight the complexities of the human brain, epigenetics, and immune system.
In recent years AAV vectors have increased in prominence as the vectors of choice for the experimental treatment of numerous human diseases due to their minimal immunogenicity, long-term transgene expression profile, and versatility in targeting specific cell types by use of a vast array of naturally occurring serotypes [9]. Currently, AAV is being assessed at the preclinical and clinical stage for the treatment of Parkinson's disease (PD) via the delivery of the glial-derived neurotrophic factor (GDNF) and Neurturin to the putamen region of the brain in order to protect and restore dopaminergic neuron function during the progression of PD [10, 11].
However, with a limited size capacity of ~4.5 kb, which is dictated by its inherent genome packaging capacity, AAV utility is primarily restricted to diseases that can be corrected by the delivery of a relatively small transcription cassette. As an alternative, our laboratory and other groups have focused on the development of the HSV type-1 (HSV-1) amplicon vector, which is devoid of all viral genes except two non-coding sequences from the parental genome, to efficiently deliver cargo up to 130 kb efficiently to different cell types within the CNS. The versatility of the HSV-1 amplicon not only lies in its extensive cargo capacity, but also in its ability to tolerate components from other viral and non-viral systems. This characteristic has provided the means to generate hybrid forms such as the HSV/Sleeping Beauty platform: a vector system that is capable of stably integrating transgenes within the genomes of transduced cells for long-term therapeutic gene expression, which is ideal for the treatment of progressive CNS-related diseases.
2. Viral vector based gene transfer to the CNS
Systemic treatment of monogenic diseases that have severe CNS involvement with viral vector-based gene therapy is a challenging endeavor due to the potential for significant anti-vector immune responses and the blood brain barrier, which has been shown to be relatively impermeable to most vector particles. Hence, patients suffering from progressive CNS degeneration require alternate treatment strategies. Preclinical testing of viral vectors in animal models of neurodegenerative diseases has identified certain features that are essential for a particular viral vector to be an attractive candidate for CNS-targeted gene therapy.
2.1 Minimal immunogenicity
Initially viewed as an immune privileged site, the CNS has revealed a remarkable capacity to recognize and clear foreign antigens [12]. First-generation viral vectors contained several components from the parental wild-type virus in its structural make-up and non-virulent viral genes that triggered potent inflammatory and immune responses in vivo [13]. Further “gutting” of these delivery agents to remove accessory viral genes not required for viral replication or to delete all viral open reading frames was systematically performed to minimize vector-induced immunogenic responses. An extreme example of this strategy is the HSV-1 amplicon, which consists of only 2 non-coding sequences from the wild-type HSV-1 genome (the HSV origin of replication and the cleavage/packaging signal) within a standard plasmid-based design [14]. While packaging of the HSV amplicon into infectious viral particles was initially achieved through use of a replication-defective helper virus, the presence of the helper virus in packaged stocks was found to significantly participate in a prolonged CNS-resident inflammatory response following amplicon delivery [13]. Delivery of latest generation helper virus-free HSV amplicon stocks to the CNS incites only a mild inflammatory response that resolves in 5 days [15], thereby garnering a safer inflammatory profile in transduced brain.
2.2 Ability to transduce non-dividing cells of the CNS and to efficiently disseminate
The adult human CNS is comprised of numerous cell types, including neurons and glia. Since many CNS conditions affect specific subsets of post-mitotic neurons, the initial strategy in CNS gene transfer was to identify viral vectors that were capable of transducing non-dividing cells. While Ad, AAV, and LV vectors have been shown to transduce non-dividing cells in the brain, HSV vectors became a primary choice given their inherent neuronal tropism [4]. This is mainly attributed to the efficiency of HSV-encoded envelope glycoprotein receptor interactions with ubiquitously expressed cellular receptors, such as Hve A, Hve C (Nectin) and proteoglycans (reviewed in [16]). Although the genomic organization of HSV-1 derived recombinant vectors and amplicons differ considerably, the packaged viral particles contain identical glycoprotein molecules embedded within the viral envelope for binding and entry into the target cell. Another important feature of some viral vectors, including HSV amplicons, is their ability to travel from the axonal termini to the soma of neurons, a process referred to as retrograde transport [17]. This characteristic can be important when attempting to deliver a gene therapeutic to a surgically intractable structure. Knowing the neuronal input into that region can allow for vector delivery to termini residing within a distal surgically tractable region. Another important feature of a CNS-targeted viral vector is its ability to spread from the point of delivery to proximally located target cells in an efficient manner to ensure optimal coverage for therapeutic gene expression. Since these viral vectors have been stripped of their replication abilities, the spread of the virus relies on its tropism and ability to navigate through the intraparenchymal space. More recently, to aid the spread of the viral vector in the brain, a convection enhanced delivery (CED)-based method has been honed and has proven to enhance AAV vector diffusion in mice and non-human primates [18], which precludes the requirement for multiple injections for vector delivery to targeted region(s).
2.3 Long-term expression of therapeutic gene for progressive disorders
As mentioned earlier, neurodegenerative diseases, including AD and PD, lead to the demise of specific neuronal cell populations over protracted periods of time. This has highlighted the need for viral vectors capable of expressing a given therapeutic gene stably over the course of the disease, which would circumvent the need for repeated vector administration and minimize invasive procedures to the patient. Some current viral vectors have limitations in their expression profile of vector-harbored transgenes due to the genomic status of the vector following transduction, specifically whether it remains in an extrachromosomal/episomal form or stably integrates within the cellular genome. The former scenario has been true for conventional HSV and Ad vectors in terminally differentiated and dividing cell types, which results in transient transgene expression due to forces exerted by the host cell to silence expression of exogenously derived DNA [19-21]. The notable exception has been AAV, which has been shown to express mainly in an episomal state (although 10% of transduced vectors are capable of stably integrating in the genome [22]) over a period of 8 years following vector transduction in vivo [23]. Naturally integrating retroviral and LV vectors display a longer expression profile due to integration within transcriptionally active regions of the host cell genome, but integration raises concerns relating to oncogene activation and disruption of host cell gene expression [24]. While researchers have shown that transgene expression from episomal vectors like HSV amplicons can be extended to up to 1 year using cellular promoters in combination with insulator elements [25], such vectors continue to fall short of the requirement for the lifelong treatment of progressive CNS diseases, due to progressive host cell-mediated silencing of their episomal vector genomes [20].
2.4 Large payload capacity
The payload capacity or transgene carrying capacity of a viral vector is dictated by the amount of genetic material it can package into a single viral particle. Each viral vector has a distinct payload capacity that can be delivered efficiently into the target cell. AAV vectors are capable of packaging 4.5-kb worth of genetic material, while LV and Ad vectors can carry up to 9 and 12-kb, respectively. HSV amplicon vectors are set apart from the competition due to their extended cargo capacity of nearly 130 kb. This feature has provided researchers an opportunity to deliver an entire genomic locus of a therapeutic gene [26, 27] and even multiple transcription units [28] via HSV amplicons to the CNS, which to date has not been possible using AAV, Ad, and LV vectors. Additionally, transcriptional regulatory elements can also be incorporated into the HSV amplicon to allow researchers to stringently control therapeutic gene expression [29] within the patient, which undoubtedly enhances the safety feature of this platform.
2.5 Minimal impact to CNS physiology following vector transduction
The adult CNS is the most complex organ of the human body. Comprised of a multitude of synaptic junctions and neuronal projections that connect different anatomical regions, the CNS coordinates intricate and highly integrated signaling processes. Thus, it is imperative that viral vector delivery to the CNS does not induce untoward alterations in cellular physiology. Evidence gleaned from pre-clinical studies and clinical trials have shown that due to their efficient receptor-mediated entry into cells, many viral vector classes have been engineered to minimize disruption of normal cellular physiology. However, it should be noted that careful assessment of the target location and the therapeutic gene to be expressed is essential to guarantee that its over-expression does not impart any detrimental side-effects on these processes.
3. The HSV amplicon and its hybrids
From its inception in 1982, the basic design of the HSV-1 amplicon has remained relatively unchanged, but apparent limitations in its application in vivo to settings requiring vector genome maintenance in mitotically active cells or lifelong therapeutic gene expression in post-mitotic cells have prompted the development of hybrid amplicon vector platforms via the incorporation of genetic elements from other viral and non-viral vector systems. This has been facilitated by the amenability of the HSV amplicon to genetic manipulation, and has expanded its utility as a vector for the treatment of conditions that afflict the CNS.
3.1 Conventional amplicons
The conventional HSV-1 amplicon consists of (1) a plasmid backbone harboring a bacterial origin of DNA replication (e.g., ColE1) and an antibiotic resistance gene (e.g., Ampr) for propagation in bacteria; and (2) two non-coding sequences from the wild-type HSV-1 genome required for replication (ori) and packaging (pac) of the amplicon into infectious particles [30]. A transgene expression cassette can be cloned into the amplicon plasmid, and subsequently replicated and packaged into defective viral particles in the presence of helper function, which can be provided in the form of a helper virus or helper virus-free system [31]. Coordinated expression of viral genes from these helper functions within the packaging cell results in the mono-directional replication of the transgene-harbored amplicon plasmid via a rolling circle mechanism [32]. This leads to the generation of a concatemer consisting of several copies of the amplicon unit arranged in a head-to-tail configuration, which is subsequently packaged into viral particles as 150-kb units. The number of copies contained within a concatemeric genome is determined by the original size of the transgene-harbored amplicon plasmid. For example, if the size of the amplicon plasmid is 10 kb, the packaged viral particle will contain a concatemeric amplicon genome consisting of approximately 15 copies of the amplicon. Once packaged into viral particles the amplicon retains the ability to infect numerous cell types, and its genome maintains an episomal state within the nucleus of the transduced cell. Due to the absence of viral gene expression, the amplicon is completely replication-defective, and its episomal existence results in stable maintenance in post-mitotic cells, but leads to unequal segregation in mitotically active cells. As a gene delivery vector, it is capable of delivering transgene unit(s) up to 150-kb in size and provides transient gene expression in dividing cells and relatively extended gene expression in post-mitotic cells. Since it does not integrate into the host cell genome, the conventional amplicon does not lead to insertional mutagenesis, thus increasing its safety profile as a gene therapy vector. While helper virus-based and helper virus-free systems are currently used to package HSV amplicons, the latter is considered a safer option due to the minimal immunogenic profile induced by its resultant stocks in vivo [15].
3.2 Integration-competent amplicons
One of the main features of the conventional HSV amplicon is the episomal existence of its genome within the nucleus of the transduced cell, which is similar to wild-type HSV-1. While this characteristic may appear to be a potential safety advantage of the platform, Suzuki and colleagues demonstrated that HSV amplicon transduction induces host cell STAT1 and interferon signaling responses that result in progressive silencing of the episomal vector genome [20], thereby limiting transgene expression to less than 1 year. It has been long believed that stable integration of the transgene unit within the transduced cell could provide the means to extend expression duration in post-mitotic cells by linking it with host cell gene expression and to ensure retention of the transgene within the chromosomes of mitotically active cells. This concept has led to the development of hybrid amplicon vector platforms engineered to contain elements from viral and non-viral systems that facilitate the integration of the amplicon-ferried transgene unit into host cell chromosomes (reviewed in [33]). The first hybrid amplicon vector to be generated was the HSV-1/adeno-associated virus (AAV) hybrid amplicon in 1997, which incorporated the inverted terminal repeat (ITR) elements and the Rep gene from AAV into the HSV amplicon backbone ([34] and reviewed in [35]). These elements are directly involved in the integration of the AAV genome into the AAVS1 site located on human chromosome 19q13.3-qter during the latent phase of wild-type AAV infection. Cell culture-based testing and in vivo testing has established that the HSV/AAV amplicon platform is competent in stably integrating the ITR-flanked transgene cassette within the genome resulting in sustained transgene expression [36, 37]. Analysis of stably transduced clonal lines has revealed that the ITR-flanked transgene cassette integrates at a significant efficiency into the AAVS1 site located on hChr19. The advantage of this hybrid over AAV vectors is its ability to integrate transgenes up to 100 kb in size (versus 4.5 kb for AAV vectors) with some specificity of integration occurring at the AAVS1 site. One of the noted drawbacks of the HSV/AAV amplicon design has been the cytotoxic and inhibitory effect of AAV-derived Rep protein expressed during HSV/AAV amplicon production, which has made it challenging to produce high-titer hybrid amplicon vector stocks [34, 38]. Furthermore, stable integration mediated by HSV/AAV amplicon vectors does not exclusively occur at the AAVS1 site, indicating there is room for optimization regarding this promising hybrid vector system platform.
An alternative integration-competent HSV amplicon hybrid vector was developed in 2006 using the elements of the non-viral Sleeping Beauty (SB) transposon system [39]. The HSV/SB hybrid vector has proven to be a versatile vector platform for stable integration of large transgene units, the details of which are presented below.
4. The HSV/SB amplicon vector for integration of large transgenes
4.1 HSV/SB vector design
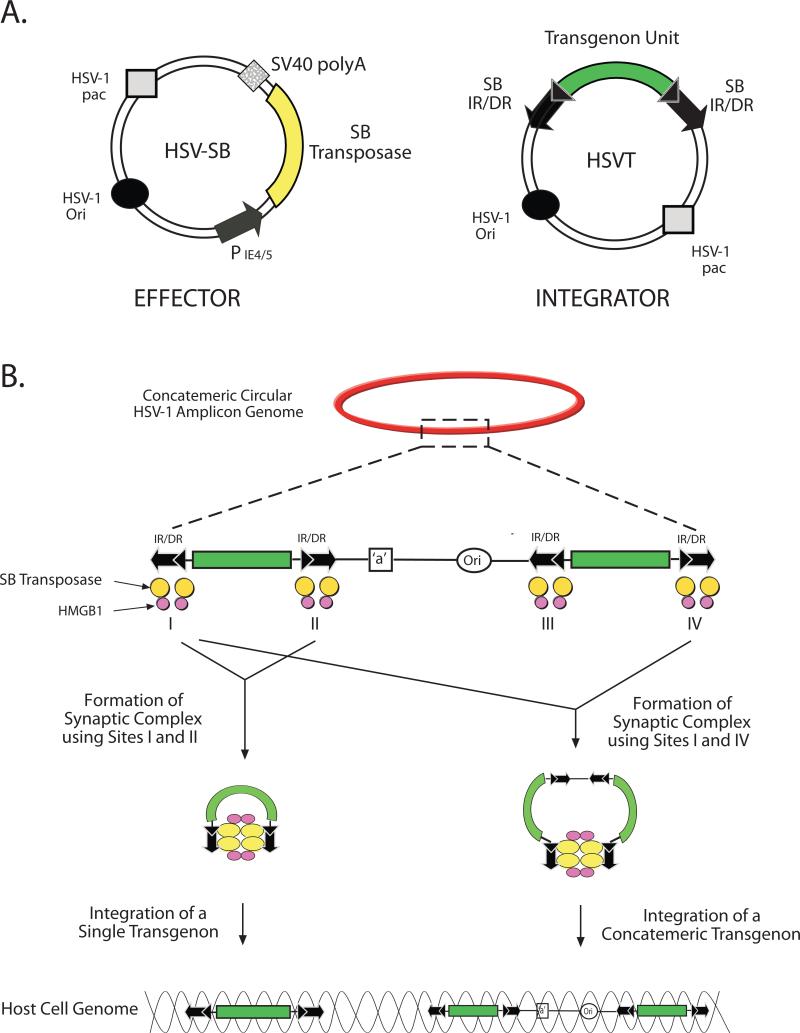
Following the derivation of the highly active SB transposon by Ivics and co-workers [40], it became evident that its components could be used to efficiently mediate the integration of vector-delivered episomal transgenes into the host chromosome for genomic retention during mitosis and long-term transgene expression [41]. Consequently, the key elements comprising the SB system were adapted to the HSV amplicon in a bi-partite manner to develop the HSV/SB hybrid vector [39]. This two-vector platform consists of an “effector” amplicon, which expresses the SB transposase, and an “integrator” amplicon that contains a transgene unit flanked by the IR/DR elements of SB (Figure 1A). Upon co-transduction of the two amplicons, the SB transposase catalyzes the precise excision and subsequent integration of the IR/DR-flanked transposon into a TA dinucleotide site within the host cell genome via a cut-and-paste mechanism similar to transposition from plasmid-based SB vectors [42]. The bi-partite design was opted over a single amplicon design as a practical measure to avert likely transposition events that would occur in the packaging cell line during amplicon production. In addition, transient expression of the SB transposase from the non-integrating “effector” amplicon reduces the risk of remobilization of integrated transgenes, hence providing an additional level of safety to the vector design. Initial cell line-based studies using the HSV/SB vector revealed that transposition via SB from the packaged amplicon was permissive in multiple cell types including baby hamster kidney cells (BHK), human embryonic kidney cells (HEK 293), HeLa cells, and mouse primary neurons at varying efficiencies [39]. One could envision the creation of a single-component HSV/SB platform if SB expression could be stringently suppressed during packaging and controlled following transduction, as this more optimal configuration could enhance the efficiency of transposition akin to what was proposed by Yant et al. with their gutless Ad/SB hybrid system [41].
Figure 1. A model of transposition-based gene transfer by the HSV-1 amplicon/Sleeping Beauty (SB) hybrid platform.
A) The two-amplicon HSV/SB vector design includes a non-integrating “effector” amplicon that expresses the SB transposase and an “integrator” amplicon that contains a transposon unit flanked by the inverted direct repeat elements (IR/DR) for SB-mediated transposition from the HSV amplicon. B) Following vector packaging, the concatemeric organization of the transgene-harbored HSV amplicon provides an optimal substrate for SB-mediated transposition of either single transgene units via the use of IR/DR sites I and II or a concatemer of transgene units via the use of non-consecutive binding sites, I and IV. The interaction of cellular and virion-associated high mobility group DNA-binding protein 1 (HMGB1) with the SB transposase can facilitate the formation of the synaptic complex, and enhance transposition from the HSV amplicon.
4.2 Transposition capacity and efficiency
One of the limitations of SB transposition from plasmid-based vectors is the inefficient transposition of transgenes >10-kb [43-45]. This has been mainly attributed to the large intervening segments of DNA that is thought to hinder the formation of a stable transposon/transposase complex (termed the ‘synaptic complex’), a pre-requisite of SB-mediated transposition. The formation of this complex requires the juxtaposition of the IR/DR elements and looping out of the intervening DNA as predicted by the model of transposition, which was originally presented by Ivics and colleagues [40]. In an effort to overcome this limitation, Zayed and colleagues designed a sandwich transposon vector, wherein the transgene was flanked by two complete SB elements in an inverted orientation [46]. This novel design with additional SB binding sites surrounding the transgene supported the transposition of a 12.2-kb fragment albeit at a low efficiency.
Given its large packaging capacity and its rolling circle mechanism of replication, the HSV amplicon contains multiple IR/DR-flanked transgene units arranged in a head-to-tail fashion (Figure 1B). We recently provided evidence showing that this unique multi-transposon configuration within a single substrate allows for transposition of transgenes at least 12-kb in length, which surpasses the integration capacity of current lentiviral vectors [47]. An interesting phenomenon observed during HSV amplicon-based transposition is that SB transposase is able to utilize non-consecutive IR/DR sites during transposition, which leads to the integration of multiple copies of the transgene unit including intervening amplicon sequences. It should also be noted that the variability in the sequence that is excised from the concatemeric amplicon genome could be viewed as a potential weakness that will need to be further studied and likely minimized in occurrence. We envision such events could be minimized by extending the size of the transposon using ”filler” DNA, which would limit the number of IR/DR-flanked transgene units packaged into a single HSV amplicon.
While the upper bounds of transposition from the HSV amplicon still remain to be determined in vivo, it is evident that the HSV amplicon is well suited for transposition of large transgenes. There may be two non-mutually exclusive reasons for why HSV/SB vectors can integrate larger sized transposons. First, the enclosure of the transposon-encoded HSV amplicon genome within the HSV nucleocapsid leads to efficient delivery of its genetic cargo in pristine condition into the transduced cell nucleus. Second, certain viral tegument proteins as well as packaging cell line-derived proteins co-packaged within the HSV virion [48] may enhance the transposition process of large transposable units from the HSV amplicon. This latter point is discussed in more detail in the next section.
4.3 HMGB1 involvement during amplicon-based transposition
High mobility group DNA-box protein 1 (HMGB1), was the first cellular co-factor to be identified as a key facilitator of SB transposition from plasmid-based vectors [49]. HMGB1 plays a pivotal role in altering DNA structure during replication, recombination, etc., and facilitates the recruitment of transcriptional co-activators. Zayed and colleagues discovered that HMGB1 directly interacted with SB transposase and allowed tighter binding of the latter to the inner IR/DR element for efficient bending of the transposon. In a similar line of investigation we found that host cell HMGB1 was not essential for SB transposition from the HSV amplicon vector due to the fact that the HSV particle incorporates HMGB1 within its tegument, which is derived from the packaging cell line during amplicon production [50]. While this was the first time it was reported that HSV amplicons co-package HMGB1, it was also shown that virion-associated HMGB1 could facilitate SB-transposition. Thus, we hypothesize that HSV/SB vectors provide the target cell with a level of HMGB1 sufficient to enhance SB-mediated transposition, whereas plasmid-delivered SB is solely dependent upon host cell-expressed HMGB1. The potential exists to regulate HMGB1 in the packaging cell line during amplicon production in order to enhance levels of virion-associated HMGB1, which could facilitate SB-mediated transposition in cell types where HMGB1 is a limiting factor.
4.4 Integration profile
SB-mediated integration exclusively occurs at TA dinucleotide sites within the host cell genome. Extensive mapping of SB integration sites in human cell lines and mouse tissue following plasmid-based transposition has revealed a random integration profile with less likelihood of targeting transcriptionally active regions [51, 52], unlike retrovirus and lentivirus vectors [53]. Integration from the HSV/SB vector follows a similar trend and occurs at TA sites both in human cell lines and in vivo. In a recent study we mapped transposon insertion sites in mouse brain tissue (n=5 mice) following in utero vector delivery to the developing mouse brain, and found that integration did not occur in transcriptionally active regions [54]. However, more integration sites need to be analyzed to draw a definite conclusion regarding the integration site preference of SB in the brain. More recently, the SB transposon has been used in the context of non-integrating lentiviral vectors to shift integration preference towards non-actively transcribed chromosomal regions to enhance safety [51, 55, 56]. The fact that integration does not seem to favor transcriptionally active regions is expected to be advantageous for SB-based vector platforms, although random genomic insertion may still pose a risk for insertional mutagenesis and oncogene activation in a clinical setting. Thus, efforts are ongoing to target SB integration into “safe” genomic loci within the human genome [57].
4.5 Targeting neuronal progenitor cells in the embryonic mouse brain
A serendipitous finding of the HSV/SB vector is its ability to target neuronal-restricted precursor cells in the developing mouse brain, and execute transposition for stable transgene integration [39]. The rationale for testing the vector in the embryonic mouse brain was to 1) exploit the proliferating cell environment for efficient vector/transgene dissemination, 2) circumvent possible immune responses that could arise against the transgene product, and 3) target neurological diseases for which genetic correction would be imperative at the prenatal stage. Based on these rationales the HSV/SB vector was delivered in utero into the intraventricular space of E14.5 mouse embryos and integration was assessed at various postnatal time points. While expression from the integration-competent reporter amplicon, which contained an IR/DR-flanked LacZ/neo unit (termed T-βgeo), in the absence SB transposase was undetectable at 3 weeks post-birth due to dilution of the episomal genomes as a result of cell division, the mice receiving the HSV/SB vector combination revealed widely disseminated β-galactosidase expression that was restricted to neuronal cells at 90 days post-birth. This finding was a result of chromosomal integration of the T-βgeo by the SB transposase. The neuron-restricted expression profile of the T-βgeo reporter was a direct consequence of HSV/SB vector transduction of a specific neuronal precursor cell population that line the ventricle of the E14.5 mouse embryo. These cells have been shown to be positive for the canonical HSV receptors and highly express the SB co-factor HMGB1, and thus, serve as an optimal target cell type for HSV/SB transduction [58]. We speculate that the portion of neuronal precursor cells that underwent SB transposition subsequently proliferated and migrated throughout the brain, which resulted in widespread reporter transgene expression. However, further in vivo testing is required to confirm this hypothesis.
5. Exploiting the developing CNS for widespread transgene dissemination
Unlike post-mitotic neurons, embryonic neuronal precursor cells offer an effective mode to widely disseminate an SB-integrated transgene, given their capacity to proliferate and migrate to various regions of the developing brain. To determine the primary target cell type(s) of HSV/SB in the fetal brain, Peterson et al. intracerebroventricularly infused E14.5 embryos with an HSVlac reporter amplicon vector and performed immunohistochemical analyses 24-h post-infusion [58]. β-galactosidase expression from the HSVlac amplicon clearly overlapped with the expression of the neuronal competence factor Sox-1 in the ependymal and subventricular regions proximal to the site of vector infusion, hence providing correlative evidence that Sox-1 positive neuronal progenitor cells are a major target cell type of the HSV/SB amplicon vector platform in utero. These neuronal progenitor cells also express HveA and Nectin-1 receptors for HSV-1 entry (Peterson and Bowers, unpublished data) and HMGB1 [58], which render them viable targets for HSV/SB vector transduction and SB transposition in this setting.
Interestingly, β-galactosidase expression from HSVlac was barely detected in Doublecortin (DCX)-positive migratory neuroblasts lining the ventricular space at E14.5, although robust β-galactosidase expression from the integrated T-βgeo transposon was detected in the same cell type at 90 days following in utero HSV/SB vector delivery [39]. This finding not only indicated that the βgeo transgene-expressing neuroblasts in the adult brain arose from in utero transduced Sox-1 positive neuroprogenitor cells, but also revealed a self-renewing population of DCX-positive cells containing the integrated T-βgeo transposon that line the adult ventricle and can act as a reservoir for the dissemination of the transgene to various regions of the brain.
The widespread β-galactosidase expression detected in neuronal cells in the subventricular zone, septofimbrial region, dentate gyrus, hippocampus, and primary and secondary cortices from the integrated T-βgeo transposon at 90 days post-vector delivery is remarkable given the number of HSV/SB vector particles (2×104 total transducing units) that was delivered in a single dose to the ventricle at E14.5. This underscores the potency of the HSV/SB vector in exploiting the existing neuronal precursor cells for transgene conveyance and also circumvents the need for repeated vector administration. Currently, efforts are ongoing to track the migratory pattern and fate of the HSV/SB transduced neuronal precursors to identify which regions of the adult brain are ultimately populated by them. This would aid in developing therapies for specific neurological diseases that affect specific subsets of neuronal cell populations. Cell-type specific promoters could also be employed to transcriptionally regulate the expression of the IR/DR-flanked therapeutic gene within the affected cell population.
Noticeably, β-galactosidase expression was not detected in glial-fibrillary acidic protein (GFAP)-positive glial cells at the above time points, since precursors of this cell type were extremely rare in regions encompassing the E14.5 ventricles as evidenced by the minimal S100B and NG2 immunostaining, which are progenitor markers for astrocyte and oligodendrocyte cell types, respectively. However, it is known that Sox-1 positive progenitor cells are capable of differentiating into cells of the neuronal and glial lineages [59]. Thus, the question remains whether HSV/SB vector transduction and/or SB transposition within these Sox-1 positive progenitor cells inadvertently predisposes them towards a neuronal lineage.
6. Considerations for in utero CNS-targeted gene transfer
Although still at an “embryonic” stage, in utero gene therapy is being vetted as a potential preventive therapy for early-onset pediatric diseases with known genetic and molecular basis (e.g., lysosomal storage disorders) that do not provide a sufficient window of opportunity for postnatal treatment. The hope is that it would provide a therapeutic option to families faced with prenatal diagnosis of a severe genetic disease. When proposals for in utero gene therapy were first presented in 1999, several ethical concerns and questions were raised regarding this controversial treatment option. These involved the risks associated with fetal intervention, to both fetus and mother, and the possibility of inadvertent germline transmission of the therapeutic gene and its consequences (reviewed in [60, 61].
Targeting the developing human CNS for viral vector-based gene therapy for the treatment of early onset neurological diseases is a delicate undertaking. While much is know about CNS formation during human fetal development, the precise timeframe during which therapeutic intervention can be efficacious for a particular disease would have to be carefully determined. Thus, thorough evaluation of safety and efficacy need to be conducted in animal models of human disease to address these concerns before contemplating human application. A recent in vivo study by our group revealed that intracerebroventricular HSV/SB vector infusion at E14.5 did not trigger a long-lasting inflammatory response [54] which, as stated previously, is a serious concern with respect to viral vector-based gene transfer. Structural abnormalities in the brain were undetectable following HSV/SB vector transduction and SB transposition, thus revealing the innocuous nature of this vector platform. Furthermore, a survival study of the mice that received the HSV/SB vector showed that SB transposition in the developing mouse brain did not result in fatalities besides the ones that were a direct result of the invasive surgical procedure itself.
Viral vector-based in utero gene therapy has significant advantages, which include: 1) beneficial vector-to-cell ratio; 2) the ability to harness the potency of expanding stem cells for widespread transgene dissemination; and 3) induction of tolerance to vector and transgene protein product due to the naïve nature of the immune system (reviewed in [61]). Furthermore, advanced imaging techniques and minimally invasive ultra-sound guided vector delivery have enabled the testing of in utero gene therapy in large animals that serve as models of human genetic conditions [62]. Thus, the next step in assessing the true potential of the HSV/SB vector would be its evaluation in large animal models of neurological disease.
7. Potential CNS diseases that can be targeted for hybrid amplicon-based therapy
Certain lysosomal storage diseases (LSDs) such as Niemann-Pick disease and mucopolysaccharidosis disease (MPS) are viable candidates for HSV/SB-mediated gene therapy. LSDs are mainly autosomal recessive diseases that arise due to a deficiency in one or more lysosomal enzymes, which causes the aberrant accumulation of a certain macromolecules such as glycoproteins, lipids, etc., in the lysosomal compartment of the cell [63]. The gradual accumulation of these macromolecules that begin during fetal development in all cell types not only affects the function of visceral organs, but also the CNS, which leads to progressive neurodegeneration and mental retardation (reviewed in [64]). While it remains unclear whether correction of the neurological component of the disease would also lead to overall improvement in the visceral system, current therapies have mainly focused on enzyme replacement therapy for the latter. Genetic correction of these diseases at the prenatal stage via gene therapy has the potential to be more efficacious in improving prognosis and even curing the disease.
Summary
Gene therapy trials and preclinical studies have revealed a wealth of information regarding ways by which current viral vectors can be improved for more effective therapeutic outcome devoid of detrimental side effects to the patient. Initially viewed as a promising viral vector for CNS targeted gene therapy, the HSV-1 amplicon has encountered quite a few challenges, among which the inability to sustain long-term transgene expression has stymied its application for the treatment of progressive neurological diseases. Derivation of the HSV/Sleeping Beauty amplicon vector platform has opened up the possibility of treating early-onset neurological disorders with its capacity to integrate a therapeutic gene within a neuronal precursor cell population within the fetal brain to mediate widespread dissemination of the therapeutic gene and long-term expression. Rigorous preclinical testing of this vector platform in animal models of early-onset neurological disorders is warranted to evaluate its true potential and enable further optimization, which would facilitate its transition to a clinical setting.
Acknowledgements
We would like to thank Dr. Howard Federoff (Georgetown University) for helpful discussions. NIH R01-AG023593 (WJB) supported this work.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
Bibliography
- 1.Simonelli F, Maguire AM, Testa F, et al. Gene therapy for Leber's congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol Ther. 2010;18:643–50. doi: 10.1038/mt.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowers WJ, de Silva S, Federoff HJ. Gene Therapy for Neurological Diseases. In: Templeton NS, editor. Gene and Cell Therapy: Therapeutic Mechanisms and Strategies. 3rd ed. CRC Press; Boca Raton: 2009. pp. 833–68. [Google Scholar]
- 3.Somia N, Verma IM. Gene therapy: trials and tribulations. Nat Rev Genet. 2000;1:91–9. doi: 10.1038/35038533. [DOI] [PubMed] [Google Scholar]
- 4.Geller AI, Breakefield XO. A defective HSV-1 vector expresses Escherichia coli beta-galactosidase in cultured peripheral neurons. Science. 1988;241:1667–9. doi: 10.1126/science.241.4873.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki M, Kasai K, Saeki Y. Plasmid DNA sequences present in conventional herpes simplex virus amplicon vectors cause rapid transgene silencing by forming inactive chromatin. J Virol. 2006;80:3293–300. doi: 10.1128/JVI.80.7.3293-3300.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grieger JC, Samulski RJ. Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J Virol. 2005;79:9933–44. doi: 10.1128/JVI.79.15.9933-9944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar M, Keller B, Makalou N, Sutton RE. Systematic determination of the packaging limit of lentiviral vectors. Hum Gene Ther. 2001;12:1893–905. doi: 10.1089/104303401753153947. [DOI] [PubMed] [Google Scholar]
- 8.Hartung T. Thoughts on limitations of animal models. Parkinsonism Relat Disord. 2008;14(Suppl 2):S81–3. doi: 10.1016/j.parkreldis.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Gao G, Vandenberghe LH, Wilson JM. New recombinant serotypes of AAV vectors. Curr Gene Ther. 2005;5:285–97. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- 10.Su X, Kells AP, Huang EJ, et al. Safety evaluation of AAV2-GDNF gene transfer into the dopaminergic nigrostriatal pathway in aged and parkinsonian rhesus monkeys. Hum Gene Ther. 2009;20:1627–40. doi: 10.1089/hum.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herzog CD, Brown L, Gammon D, et al. Expression, bioactivity, and safety 1 year after adeno-associated viral vector type 2-mediated delivery of neurturin to the monkey nigrostriatal system support cere-120 for Parkinson's disease. Neurosurgery. 2009;64:602–12. doi: 10.1227/01.NEU.0000340682.06068.01. discussion 12-3. [DOI] [PubMed] [Google Scholar]
- 12.Kreutzberg GW. Microglia, the first line of defence in brain pathologies. Arzneimittelforschung. 1995;45:357–60. [PubMed] [Google Scholar]
- 13.Wood MJ, Byrnes AP, Pfaff DW, Rabkin SD, Charlton HM. Inflammatory effects of gene transfer into the CNS with defective HSV-1 vectors. Gene Ther. 1994;1:283–91. [PubMed] [Google Scholar]
- 14.Spaete RR, Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982;30:295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- 15.Olschowka JA, Bowers WJ, Hurley SD, Mastrangelo MA, Federoff HJ. Helper-free HSV-1 amplicons elicit a markedly less robust innate immune response in the CNS. Mol Ther. 2003;7:218–27. doi: 10.1016/s1525-0016(02)00036-9. [DOI] [PubMed] [Google Scholar]
- 16.Burton EA, Fink DJ, Glorioso JC. Gene delivery using herpes simplex virus vectors. DNA Cell Biol. 2002;21:915–36. doi: 10.1089/104454902762053864. [DOI] [PubMed] [Google Scholar]
- 17.Agudo M, Trejo JL, Lim F, et al. Highly efficient and specific gene transfer to Purkinje cells in vivo using a herpes simplex virus I amplicon. Hum Gene Ther. 2002;13:665–74. doi: 10.1089/10430340252837251. [DOI] [PubMed] [Google Scholar]
- 18.Hadaczek P, Kohutnicka M, Krauze MT, et al. Convection-enhanced delivery of adeno-associated virus type 2 (AAV2) into the striatum and transport of AAV2 within monkey brain. Hum Gene Ther. 2006;17:291–302. doi: 10.1089/hum.2006.17.291. [DOI] [PubMed] [Google Scholar]
- 19.Michou AI, Santoro L, Christ M, Julliard V, Pavirani A, Mehtali M. Adenovirus-mediated gene transfer: influence of transgene, mouse strain and type of immune response on persistence of transgene expression. Gene Ther. 1997;4:473–82. doi: 10.1038/sj.gt.3300412. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki M, Chiocca EA, Saeki Y. Early STAT1 activation after systemic delivery of HSV amplicon vectors suppresses transcription of the vector-encoded transgene. Mol Ther. 2007;15:2017–26. doi: 10.1038/sj.mt.6300273. [DOI] [PubMed] [Google Scholar]
- 21.Santos K, Simon DA, Conway E, et al. Spatial and temporal expression of herpes simplex virus type 1 amplicon-encoded genes: implications for their use as immunization vectors. Hum Gene Ther. 2007;18:93–105. doi: 10.1089/hum.2006.082. [DOI] [PubMed] [Google Scholar]
- 22.Nakai H, Iwaki Y, Kay MA, Couto LB. Isolation of recombinant adeno-associated virus vector-cellular DNA junctions from mouse liver. J Virol. 1999;73:5438–47. doi: 10.1128/jvi.73.7.5438-5447.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadaczek P, Eberling JL, Pivirotto P, Bringas J, Forsayeth J, Bankiewicz KS. Eight years of clinical improvement in MPTP-lesioned primates after gene therapy with AAV2-hAADC. Mol Ther. 2010;18:1458–61. doi: 10.1038/mt.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–6. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Kong L, Zhang GR, Sun M, Geller AI. A preproenkephalin-neurofilament chimeric promoter in a helper virus-free herpes simplex virus vector enhances long-term expression in the rat striatum. Neurobiol Dis. 2004;16:596–603. doi: 10.1016/j.nbd.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 26.Gomez-Sebastian S, Gimenez-Cassina A, Diaz-Nido J, Lim F, Wade-Martins R. Infectious delivery and expression of a 135 kb human FRDA genomic DNA locus complements Friedreich's ataxia deficiency in human cells. Mol Ther. 2007;15:248–54. doi: 10.1038/sj.mt.6300021. [DOI] [PubMed] [Google Scholar]
- 27.Senior SL, Wade-Martins R. Herpes simplex virus type 1 amplicon vectors for the infectious delivery and expression of genomic DNA loci. Curr Opin Mol Ther. 2005;7:337–45. [PubMed] [Google Scholar]
- 28.Sun M, Kong L, Wang X, et al. Coexpression of tyrosine hydroxylase, GTP cyclohydrolase I, aromatic amino acid decarboxylase, and vesicular monoamine transporter 2 from a helper virus-free herpes simplex virus type 1 vector supports high-level, long-term biochemical and behavioral correction of a rat model of Parkinson's disease. Hum Gene Ther. 2004;15:1177–96. doi: 10.1089/hum.2004.15.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Q, Sun M, Wang X, Zhang GR, Geller AI. Long-term inducible expression in striatal neurons from helper virus-free HSV-1 vectors that contain the tetracycline-inducible promoter system. Brain Res. 2006;1083:1–13. doi: 10.1016/j.brainres.2006.01.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spaete RR, Frenkel N. The herpes simplex virus amplicon: analyses of cis-acting replication functions. Proc Natl Acad Sci U S A. 1985;82:694–8. doi: 10.1073/pnas.82.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeki Y, Breakefield XO, Chiocca EA. Improved HSV-1 amplicon packaging system using ICP27-deleted, oversized HSV-1 BAC DNA. Methods Mol Med. 2003;76:51–60. doi: 10.1385/1-59259-304-6:51. [DOI] [PubMed] [Google Scholar]
- 32.Vlazny DA, Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci U S A. 1981;78:742–6. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oehmig A, Fraefel C, Breakefield XO. Update on herpesvirus amplicon vectors. Mol Ther. 2004;10:630–43. doi: 10.1016/j.ymthe.2004.06.641. [DOI] [PubMed] [Google Scholar]
- 34.Johnston KM, Jacoby D, Pechan PA, et al. HSV/AAV hybrid amplicon vectors extend transgene expression in human glioma cells. Hum Gene Ther. 1997;8:359–70. doi: 10.1089/hum.1997.8.3-359. [DOI] [PubMed] [Google Scholar]
- 35.Glauser DL, Ackermann M, Saydam O, Fraefel C. Chimeric herpes simplex virus/adeno-associated virus amplicon vectors. Curr Gene Ther. 2006;6:315–24. doi: 10.2174/156652306777592090. [DOI] [PubMed] [Google Scholar]
- 36.Costantini LC, Jacoby DR, Wang S, Fraefel C, Breakefield XO, Isacson O. Gene transfer to the nigrostriatal system by hybrid herpes simplex virus/adeno-associated virus amplicon vectors. Hum Gene Ther. 1999;10:2481–94. doi: 10.1089/10430349950016825. [DOI] [PubMed] [Google Scholar]
- 37.Fraefel C, Jacoby DR, Lage C, et al. Gene transfer into hepatocytes mediated by helper virus-free HSV/AAV hybrid vectors. Mol Med. 1997;3:813–25. [PMC free article] [PubMed] [Google Scholar]
- 38.Heilbronn R, Burkle A, Stephan S, zur Hausen H. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J Virol. 1990;64:3012–8. doi: 10.1128/jvi.64.6.3012-3018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowers WJ, Mastrangelo MA, Howard DF, Southerland HA, Maguire-Zeiss KA, Federoff HJ. Neuronal precursor-restricted transduction via in utero CNS gene delivery of a novel bipartite HSV amplicon/transposase hybrid vector. Mol Ther. 2006;13:580–8. doi: 10.1016/j.ymthe.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 40.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–10. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 41.Yant SR, Ehrhardt A, Mikkelsen JG, Meuse L, Pham T, Kay MA. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat Biotechnol. 2002;20:999–1005. doi: 10.1038/nbt738. [DOI] [PubMed] [Google Scholar]
- 42.Luo G, Ivics Z, Izsvak Z, Bradley A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 1998;95:10769–73. doi: 10.1073/pnas.95.18.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Izsvak Z, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J Mol Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 44.Karsi A, Moav B, Hackett P, Liu Z. Effects of insert size on transposition efficiency of the sleeping beauty transposon in mouse cells. Mar Biotechnol (NY) 2001;3:241–5. doi: 10.1007/s101260000072. [DOI] [PubMed] [Google Scholar]
- 45.Geurts AM, Yang Y, Clark KJ, et al. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8:108–17. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 46.Zayed H, Izsvak Z, Walisko O, Ivics Z. Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 47.de Silva S, Mastrangelo MA, Lotta LT, Jr., Burris CA, Federoff HJ, Bowers WJ. Extending the transposable payload limit of Sleeping Beauty (SB) using the Herpes Simplex Virus (HSV)/SB amplicon-vector platform. Gene Ther. 2010;17:424–31. doi: 10.1038/gt.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowers WJ, Howard DF, Brooks AI, Halterman MW, Federoff HJ. Expression of vhs and VP16 during HSV-1 helper virus-free amplicon packaging enhances titers. Gene Ther. 2001;8:111–20. doi: 10.1038/sj.gt.3301340. [DOI] [PubMed] [Google Scholar]
- 49.Zayed H, Izsvak Z, Khare D, Heinemann U, Ivics Z. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 2003;31:2313–22. doi: 10.1093/nar/gkg341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Silva S, Lotta LT, Burris CA, Bowers WJ. Virion-Associated Cofactor High-Mobility Group DNA-Binding Protein-1 Facilitates Transposition from the Herpes Simplex Virus/Sleeping Beauty Amplicon Vector Platform. Hum Gene Ther. 2010;21:1615–22. doi: 10.1089/hum.2010.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moldt B, Miskey C, Staunstrup NH, et al. Comparative Genomic Integration Profiling of Sleeping Beauty Transposons Mobilized With High Efficacy From Integrase-defective Lentiviral Vectors in Primary Human Cells. Mol Ther. 2011 doi: 10.1038/mt.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–94. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bushman F, Lewinski M, Ciuffi A, et al. Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol. 2005;3:848–58. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 54.de Silva S, Mastrangelo MA, Lotta LT, et al. Herpes Simplex Virus/Sleeping Beauty Vector-Based Embryonic Gene Transfer Using the HSB5 Mutant: Loss of Apparent Transposition Hyperactivity In Vivo. Hum Gene Ther. 2010;21:1603–13. doi: 10.1089/hum.2010.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staunstrup NH, Moldt B, Mates L, et al. Hybrid lentivirus-transposon vectors with a random integration profile in human cells. Mol Ther. 2009;17:1205–14. doi: 10.1038/mt.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vink CA, Gaspar HB, Gabriel R, et al. Sleeping beauty transposition from nonintegrating lentivirus. Mol Ther. 2009;17:1197–204. doi: 10.1038/mt.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivics Z, Katzer A, Stuwe EE, Fiedler D, Knespel S, Izsvak Z. Targeted Sleeping Beauty transposition in human cells. Mol Ther. 2007;15:1137–44. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- 58.Peterson EB, Mastrangelo MA, Federoff HJ, Bowers WJ. Neuronal specificity of HSV/sleeping beauty amplicon transduction in utero is driven primarily by tropism and cell type composition. Mol Ther. 2007;15:1848–55. doi: 10.1038/sj.mt.6300267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wegner M, Stolt CC. From stem cells to neurons and glia: a Soxist's view of neural development. Trends Neurosci. 2005;28:583–8. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 60.Caplan AL, Wilson JM. The ethical challenges of in utero gene therapy. Nat Genet. 2000;24:107. doi: 10.1038/72747. [DOI] [PubMed] [Google Scholar]
- 61.Coutelle C, Rodeck C. On the scientific and ethical issues of fetal somatic gene therapy. Gene Ther. 2002;9:670–3. doi: 10.1038/sj.gt.3301761. [DOI] [PubMed] [Google Scholar]
- 62.Punzo C, Cepko CL. Ultrasound-guided in utero injections allow studies of the development and function of the eye. Dev Dyn. 2008;237:1034–42. doi: 10.1002/dvdy.21500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winchester B, Vellodi A, Young E. The molecular basis of lysosomal storage diseases and their treatment. Biochem Soc Trans. 2000;28:150–4. doi: 10.1042/bst0280150. [DOI] [PubMed] [Google Scholar]
- 64.Paul CA, Boegle AK, Maue RA. Before the loss: neuronal dysfunction in Niemann-Pick Type C disease. Biochim Biophys Acta. 2004;1685:63–76. doi: 10.1016/j.bbalip.2004.08.012. [DOI] [PubMed] [Google Scholar]