Abstract
Long non-coding RNAs (lncRNAs) are transcripts with low protein-coding potential that represent a large proportion of the transcriptional output of the cell. Many lncRNAs exhibit features indicative of functionality including tissue-restricted expression, localization to distinct subcellular structures, regulated expression and evolutionary conservation. Some lncRNAs have been shown to associate with chromatin-modifying activities and transcription factors, suggesting that a common mode of action may be to guide protein complexes to target genomic loci. However, the functions (if any) of the vast majority of lncRNA transcripts are currently unknown, and the subject of investigation. Here, we consider the putative role(s) of lncRNAs in neurodevelopment and brain function with an emphasis on the epigenetic regulation of gene expression. Associations of lncRNAs with neurodevelopmental/neuropsychiatric disorders, neurodegeneration and brain cancers are also discussed.
Keywords: long non-coding RNA, epigenetics, neurodegeneration, neurodevelopment, brain
1. Introduction
It is now clear that the majority of the mammalian genome produces RNA transcripts despite only approximately 1% of the DNA sequence encoding proteins (a phenomenon known as pervasive transcription) [1]. The majority of loci produce a forest of interlaced [2] and overlapping [3] transcripts in both sense and antisense orientations [4,5]. Complementary results have been observed using multiple transcriptomics methodologies (i.e. RNA-seq [6], RNA tiling arrays [3,7–11], sequencing of full-length cDNA libraries [2,12], high-throughput rapid amplification of cDNA ends (RACE) [7] and sequencing of CAGE tags [2]) suggesting that the observed transcription is real and not a technical artefact or background genomic DNA/pre-mRNA.
Long non-coding RNAs (lncRNAs) are RNA transcripts more than 200 nucleotides in length that do not encode proteins. lncRNA transcripts are generally ‘mRNA-like’ [13,14] as they are frequently transcribed by RNA polymerase II, contain canonical splice sites (GU/AG), have similar intron/exon lengths to mRNAs, exhibit alternative splicing, may be polyadenylated or non-polyadenylated and associate with the same types of histone modification as protein-coding genes [14–16]. In contrast to mRNAs, a large fraction of lncRNAs (42% of lncRNAs in the GENCODE v7 catalogue) consist of only two exons [16]. lncRNAs generally exhibit low coding potential and are devoid of extended open reading frames (ORFs). Putative lncRNA ORFs have also been shown to be of similar quality to ORFs found in random genomic sequence [16], lack the pattern of cross-species mutation accumulation typical of protein-coding sequence [17] and show little similarity with ORFs of recently evolved proteins [18]. lncRNAs are associated with ribosomes (as are other non-coding RNAs and non-coding regions of mRNAs) but are distinct from coding transcripts in that they lack a characteristic ribosome drop-off signature found at the 3′-end of bona fide ORFs [18], suggesting that the majority are not translated into proteins. Nevertheless, some lncRNAs may encode short peptide sequences [19–21].
A major challenge in biology is to decode the genomic language that governs the architecture and function of the central nervous system (CNS). The mammalian CNS arguably represents the most complex system within all of biology. Not only does it comprise hundreds of billions of cells of neuronal and glial origin, but this complexity is amplified by the hundreds of trillions of synaptic interactions between these cells. Establishing this intricate cellular architecture during neurodevelopment and maintaining it effectively during adult life with appropriate adaptation and learning is a significant undertaking. It is highly likely that the cells of the CNS take advantage of all the subtleties of genomic evolution in order to achieve these complex cellular behaviours. Here, we explore the possible roles of lncRNAs as critical genomic regulators within the brain.
2. Are long non-coding RNAs functional?
The degree to which non-coding transcription is functional is currently a matter of debate, with some arguing that the majority is simply noise resulting from stochastic promoter firing or illegitimate transcripts arising from ‘promiscuous’ promoters. A study by van Bakel et al. [22] argued in favour of the transcriptional noise hypothesis and showed that the majority of non-coding RNA transcripts are associated with known genes. These conclusions have been vigorously opposed by others who have suggested that association of lncRNA transcripts with protein-coding loci is consistent with pervasive transcription, and point to insufficient sequencing depth in the van Bakel study [1]. Furthermore, a contradictory finding that the majority of lncRNAs are independent transcriptional units, was reported by the GENCODE consortium [16].
By contrast, many studies point to a functional role for non-coding transcription in the general case. Firstly, lncRNA genes are expressed in a tissue-specific manner. Investigation of the transcriptional landscape of multiple human cell lines found that 29% of lncRNAs were expressed specifically in a single cell type, while only 10% were expressed in all cell types (in stark contrast to protein-coding genes for which the numbers were 7% and 53%, respectively) [6]. Furthermore, among the most differentially expressed lncRNAs, approximately 40% are expressed specifically in the brain [16]. Using in situ hybridization data in mouse brain sections taken from the Allen Brain Atlas, Mercer et al. [23] found that most lncRNAs are associated with distinct neuroanatomical loci. For example, expression of the lncRNA AK037594 was found only in the dentate gyrus and CA1–3 regions of the hippocampus. Similarly, MIAT (Gomafu) a nuclear-localized lncRNA is expressed only in differentiating neural progenitors and a subset of postmitotic neurons [13].
A study by Ponjavic et al. [24] found that the genomic loci of lncRNAs expressed in the developing brain were preferentially located in the vicinity of protein-coding genes that are (i) highly expressed in brain, (ii) involved in transcriptional regulation or (iii) involved in CNS development. Furthermore, analysis of a subset of these lncRNA–protein-coding gene pairs revealed co-expression in the same specific brain regions consistent with cross-talk between coding and non-coding transcripts arising from the same loci [24]. Similarly, many overlapping sense mRNA–antisense lncRNA pairs are co-expressed and specifically localized to synaptoneurosomes (specialized structures enriched at the pinched-off dendritic spines of pyramidal neurons) in the adult mouse forebrain [25]. Some of these mRNAs have known roles in synaptogenesis (e.g. BC1, Camk2a, Dag1) or have been implicated in Alzheimer's disease (AD) pathophysiology (e.g. Bace1 and App). Additionally, many lncRNAs are localized to specific subcellular compartments or to subnuclear structures [13,26]. For example, the lncRNA Ntab was found to be expressed only in the developing and adult rat CNS and transported to processes distal from the cell soma [27].
Targeted sequencing of cDNAs eluted from tiling arrays has revealed a plethora of low abundance transcripts (some originating from so-called gene deserts) exhibiting well-defined exon–exon boundaries, indicative of high-fidelity lncRNA splicing [28]. Interestingly, a recent study by Tilgner et al. [29] showed that the efficiency of lncRNA splicing is significantly lower than for mRNAs, and that many lncRNA transcripts (including well-studied functional examples such as Airn and KCNQ1OT1) remain unspliced.
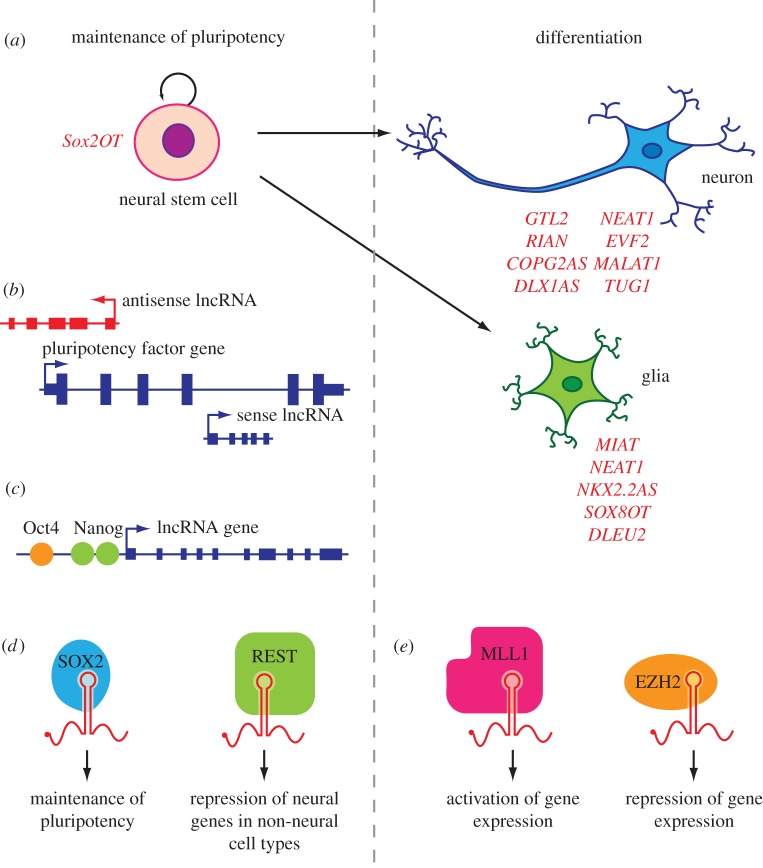
Secondly, lncRNAs exhibit signs of regulated expression [30]. For example, enhancer-derived lncRNAs are differentially expressed in an activity-dependent manner in neuronal cultures [31]. Similarly, 174 lncRNAs were differentially expressed during the 16-day differentiation of mouse embryonic stem (ES) cells to embryoid bodies [32]. Four lncRNAs (including Miat) also showed dynamic patterns of expression following retinoic acid-induced neuronal differentiation in a separate study in mouse ES cells [33]. The observation that pluripotency factors, such as Oct4 and Nanog, can bind to the promoters of lncRNA genes and modulate their transcription suggests that lncRNAs constitute an important component of the genetic circuitry that regulates the balance between maintenance of pluripotency and lineage commitment (figure 1). RNA interference (RNAi) knockdown and overexpression of two of these lncRNA transcripts led to alterations in Nanog and Oct4 expression, and promoted the adoption of lineage-specific differentiation programmes [33]. A separate study identified Sox2OT as a lncRNA gene that is dynamically expressed during neural cell differentiation [34]. Sox2OT encodes a sense-orientation transcript that overlaps with the pluripotency-associated transcription factor (TF) Sox2 (sex determining region Y-box 2). The genomic proximity of Sox2OT and Sox2 suggested a possible regulatory role for Sox2OT in the maintenance of pluripotency, which was recently confirmed experimentally [35].
Figure 1.
Long non-coding RNAs regulate pluripotency and neuronal-glial differentiation. (a) Multipotent NSCs differentiate to form neurons and glia. lncRNAs are differentially expressed between the undifferentiated state and the neuronal-glia lineages. Lineage/state-specific upregulated lncRNAs are labelled in red. (b) Protein-coding genes involved in the maintenance of a pluripotent state may have associated sense or antisense lncRNAs which regulate their expression. (c) lncRNA genes are themselves transcriptionally regulated by pluripotency factors such as Oct4 and Nanog. (d) lncRNAs form ribonucleoprotein complexes with pluripotency factors such as SOX2 or the master regulator of neurogenesis REST. The lncRNA components act as guides to their respective complexes in order to direct them to specific chromatin loci. As a result lncRNAs directly contribute to the maintenance of pluripotency and the repression of neural genes in non-neural cell types. (e) Upon lineage commitment, lncRNAs act as guides to ribonucleoprotein complexes which epigenetically modulate gene expression. In so doing, lncRNAs regulate the patterns of differential gene expression required for differentiation. lncRNAs may have an activating or repressive effect on gene expression depending on their respective protein partners (e.g. the trithorax protein MLL1 is a H3K4 trimethylase which promotes gene activation, whereas the polycomb component EZH2 is a H3K27 trimethylase which has a repressive effect on gene expression).
While the precise processing, tissue-specificity, sub-cellular localization and differential expression of lncRNA transcripts have been used as arguments for functionality, it could be equally argued that many of these observations are also consistent with lncRNAs being the product of noisy transcription. In this case, lncRNA expression might be explained as a result of low-level TF binding and RNA polymerase engagement [36]. Given that the expression of TFs and other gene regulatory mechanisms are differentially active in specific tissues/cell types and during changes in cellular metabolism, this could give rise to patterns of tissue-specific or apparently regulated lncRNA noise. However, the observation that many lncRNA transcripts are localized to distinct subcellular compartments is more difficult to dismiss as noise [23]. In a recent study, Sauvageau et al. [37] developed 18 transgenic knockout mice strains in order to investigate possible lncRNA functions. These researchers focused on a subclass of lncRNAs called long intergenic non-coding RNAs (lincRNAs). lincRNAs are biochemically indistinct from other lncRNAs but differ in their genomic organization as they reside in the space between genes. As lincRNAs do not overlap with protein-coding genes, functions can unambiguously be ascribed to the non-coding transcript rather than as indirect effects on neighbouring protein-coding genes. lincRNAs targeted for knockout were replaced with a lacZ expression cassette such that transcription from each lincRNA loci was maintained. As a result, any phenotypes observed in the knockout mice can be attributed to the lincRNA sequence, rather than as sequence-independent effects mediated by the act of transcription itself. Of the 18 lincRNA knockout strains, three lncRNA knockout strains (Fendrr, Peril and Mdgt) had perinatal and postnatal lethal phenotypes indicating critical roles for these transcripts in development. Another strain knocked out for linc-Brn1b showed a reduction in the number of intermediate progenitor cells in the subventricular zone, suggesting that this lncRNA plays a key role in the developing cortex [23].
Importantly, linc-Brn1b showed many features consistent with the results of Mercer et al. [23,38]. linc-Brn1b expression is primarily restricted to specific brain regions (i.e. telencephalon, ventricular zone and subventricular zone), is predominantly nuclear localized in cultured neural progenitor cells derived from the cerebral cortex and shows spatio-temporally regulated patterns of expression during cortical development [37]. These findings lend support to the attribution of potential function on the basis of tissue-specific and regulated expression patterns. While the results presented by Sauvageau et al. are highly encouraging (especially given the inability to find essential functions for lncRNAs in other studies [39,40]) many more knockout studies are required to demonstrate further functions for lncRNAs in vivo.
Thirdly, lncRNA genes show evidence of being under evolutionary constraint (although generally to a lesser extent than for protein-coding genes). The exons of lncRNA genes show a tendency to have lower base substitution rates than their corresponding intronic regions, indicative of evolutionary conservation [17,41]. Similarly, lncRNA exons show other signs of conservation such as enrichment for phastCons elements and indel-purified sequence [42,43]. Additionally, the promoter regions and splice sites of lncRNA genes are conserved at rates higher than would be expected by chance [42,44]. A separate study found that while conservation of lncRNA genes was low when looking at the full-length transcript, the degree of conservation became much higher when transcripts were analysed in 50 nucleotide windows [45]. This is consistent with short conserved functional sequences residing within longer transcripts that are generally under less evolutionary constraint. Furthermore, conservation of RNA secondary structural motifs within lncRNA genes unambiguously points to functions for their RNA gene products [46–50]. RNA structure may be critical to the functionality of many lncRNAs, whereas the primary base sequence may be less important. As a result, conservation analyses which fail to take into account the preservation of RNA secondary structure motifs despite changes to the primary base sequence will tend to underestimate the degree of actual lncRNA conservation.
Although conventional metrics of evolutionary constraint suggest that lncRNAs are under selective pressure, these findings should be treated with a degree of caution. Annotation of a transcript as definitively non-coding is not trivial, and so it is possible that a substantial number of protein-coding transcripts have been misclassified as lncRNAs. Such an eventuality would ‘contaminate’ the pool of so-called lncRNA genes with conserved sequence and bias estimations of lncRNA conservation [14]. Similarly, estimates of lncRNA functionality based on conservation may be skewed as a result overlap with protein-coding genes or other conserved DNA elements (such as enhancers).
While primary sequence conservation of lncRNA genes may be limited, some show other signs of being under evolutionary constraint, such as positional conservation [5]. For example, the lncRNA MALAT1 is syntenically conserved across a wide variety of organisms [39]. Similarly, 68 lncRNAs derived from pseudogene loci [51] showed positional conservation between human and at least two other mammals [52].
Analysis of lncRNAs in the GENCODE v7 catalogue showed that approximately 30% of lncRNAs are specific to the primates and therefore lack evolutionary conservation outside that lineage [16]. Importantly, while evolutionary conservation is indicative of function, lack of conservation does not necessarily imply lack of function [45]. For example, several lncRNAs (i.e. Xist and Airn) with well-established epigenetic regulatory roles are poorly conserved between human and mouse at the primary sequence level [53,54]. Additionally, rapidly evolving lncRNAs that are lineage-specific likely represent recent evolutionary innovations. One such primate-specific lncRNA gene HAR1F (human accelerated region 1F) is expressed in Cajal–Retzius neurons of the neocortex [55]. Interestingly, despite considerable sequence changes, the expression pattern of HAR1F in developing cortex is highly conserved between humans and cynomolgus macaques, suggesting that HAR1F expression is functionally significant.
In some cases, lncRNAs may have sequence-independent functions, whereby the act of their transcription alone may regulate expression of neighbouring genes (a phenomenon called transcriptional interference [56] or promoter occlusion [57]). As a result, the nucleotide sequence of the lncRNA may be inconsequential with respect to its functionality and therefore not subject to evolutionary constraint [58–61]. In support of this, Derrien et al. showed that lncRNA promoters are generally more conserved (at a level similar to protein-coding exons) than lncRNA exons [16], suggesting that the transcription of many lncRNAs is more important than the lncRNA sequence itself.
3. What are the functions of long non-coding RNAs?
To date, lncRNAs have been implicated in a wide variety of processes including modulation of splicing [62,63], organelle formation [26,64], telomere function [65], post-transcriptional gene regulation [66–69], sequestration of signalling proteins [70], generation of small RNAs (e.g. nucleolar RNAs (snoRNAs), microRNAs (miRNAs) and endogenous small interfering RNAs) [71–73], competition for miRNA binding [74–76] and regulation of protein localization [77]. A major function of lncRNAs appears to be in the epigenetic regulation of gene transcription and, as such, lncRNAs have been implicated in practically every epigenetic process: X-chromosome dosage compensation [78,79], mono-allelic expression of imprinted genes [80–82], control of chromatin macro structure [83], direction of genomic loci to distinct nuclear sub-substructures [84] and lineage commitment/cell fate determination [33,85,86].
Epigenetics is the study of heritable traits that are not encoded in the primary DNA sequence itself, but rather in the patterns of covalent alteration of DNA nucleobases (e.g. cytosine methylation) and histone protein post-translational modification (e.g. the histone code) [87–91]. Epigenetic modifications regulate the accessibility of the genome to the transcriptional machinery [92] and are thus important controllers of gene expression. As such, it has been proposed that lncRNAs act as ‘analogue–digital convertors’ [93] capable of facilitating the flow of information between proteins and nucleic acids. The structural plasticity of RNA enables the simultaneous binding of lncRNAs to proteins by forming secondary structure motifs (i.e. analogue interactions), and to nucleic acids through Watson–Crick, Hoogsteen and reverse Hoogsteen base pairings (i.e. digital interactions). lncRNAs may consist of multiple binding modules and are therefore, in theory, capable of bringing together any cellular component [94–96]. Specifically, lncRNAs act to direct epigenetic modifying complexes and TFs to specific chromatin loci. The observation that lncRNAs tend to be enriched in nuclear extracts [6,15], and more specifically in the chromatin fraction [15,16] (whereas coding transcripts are primarily cytoplasmic), is consistent with specific interactions of lncRNAs with genomic DNA (figure 2).
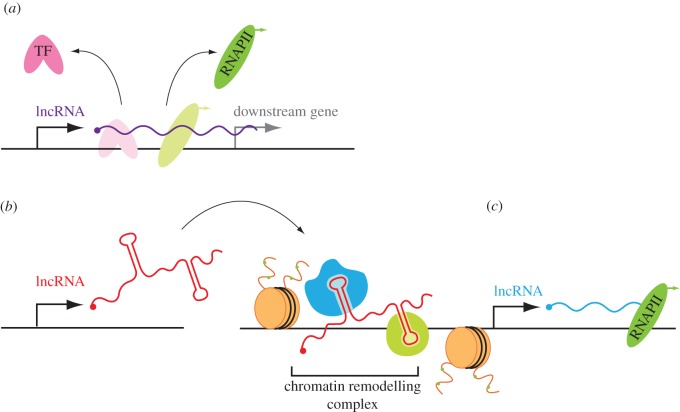
Figure 2.
Mechanisms of gene regulation by long non-coding RNAs. (a) Transcriptional interference by an adjacent lncRNA gene. lncRNAs can regulate neighbouring genes in cis in a sequence-independent manner by inhibiting the assembly of the transcriptional machinery (i.e. RNA polymerase II, RNAPII and TFs) at the promoter of a downstream gene. (b) lncRNAs can act as guides for chromatin remodelling activities and transcription factors in both trans (depicted) and cis. The lncRNA forms a ribonucleoprotein complex with one or more transcriptional regulators and guides them to specific chromatin loci in order to induce local changes in chromatin structure (active chromatin marks indicated by green circles). (c) lncRNA genes themselves are targets of epigenetic regulation, thereby facilitating a feed-forward cascade of gene expression states.
Two landmark studies used RNA-immunoprecipitation methodologies in order to systematically identify lncRNAs which bind to chromatin-modifying proteins. Khalil et al. [97] performed RIP-chip using antibodies against PRC2, SMCX and CoREST (a general transcriptional co-repressor which acts to regulate neural-specific genes) in order to precipitate and analyse bound lncRNA transcripts. These epigenetic modifier complexes were found to associate with 38% of the approximately 1100 lncRNA genes featured on the microarray chips. Furthermore, there was little overlap between the lncRNA-binding partners for each protein complex, suggesting that each complex binds a distinct repertoire of lncRNAs [97]. By contrast, very few mRNAs (approx. 2% of those featured on the arrays) associated with PRC2, suggesting that PRC2 binding is a lncRNA-specific phenomenon. Similar results were obtained by Zhao et al. [98] who immunoprecipitated Ezh2 (the component of PRC2 which trimethylates H3K27 in order to induced transcriptional silencing) and identified approximately 9000 bound transcripts by RNA sequencing. Subsequently, a plethora of other epigenetic modifier complexes were shown to associate with lncRNAs including PRC1, Cbx1, Cbx3, Tip60/P400, Setd8, ESET, and Suv39h1, Jarid1b, Jarid1c, HDAC1 and YY1 [85]. While the majority of studies have identified lncRNAs that bind to repressive epigenetic modifying complexes, associations with activating complexes have also been observed [32,97,99]. For example, the lncRNAs Evx1as and Hoxb5/6as (which show concordant expression with their overlapping sense-orientation protein-coding genes during mouse ES cell differentiation) immunoprecipitated with the H3K4 trimethylase Mll1, suggesting that they may be cis positive regulators [32].
In several cases, lncRNAs have been shown to be composed of distinct protein or nucleic-acid-binding modules, and this has been proposed as a general mode of lncRNA function [94,100]. Modular binding of proteins allows for the activities of multiple epigenetic modifier complexes to be directed to specific genomic loci in a coordinated manner. The best described example of this is the lncRNA HOTAIR which is a trans negative regulator of the HOXC cluster [101,102] and other loci [103]. The HOTAIR transcript acts as a scaffold for PRC2 and a complex of LSD1/CoREST/REST (Repressor Element 1-Silencing TF) at its 5′ and 3′ termini, respectively [101,104]. As a result, HOTAIR coordinates the H3K27 trimethylase and H3K4 demethylase activities of these protein complexes in order to facilitate gene silencing at specific target loci.
RNAi screening loss-of-function studies targeting lncRNAs in mouse ES cells have shown that many non-coding transcripts act to control pluripotency and differentiation [33,85,105]. Interestingly, knockdown of lncRNA generally resulted in comparable numbers of up- and downregulated transcripts. Given that the majority of studies have focused on lncRNAs with gene silencing functions, this observation suggests that gene-activating lncRNAs may be of equal importance and that many positive regulators of gene expression remain to be discovered. Knockdown of many lncRNAs produced gene expression changes associated with a loss of pluripotency and the adoption of early differentiation lineages (including neuroectoderm). lncRNA knockdown did not, in general, affect neighbouring genomic loci, suggesting that the primary mode of gene regulation is in trans rather than in cis [85]. Similar results were obtained by Khalil et al. [97], who showed that RNAi knockdown of PRC2-associated lncRNAs resulted in activation of polycomb targets while not significantly affecting lncRNA-neighbouring genes, again indicative of trans regulation.
An alternative and complementary approach, termed ‘guilt-by-association’ has been used to infer lncRNA functions. Firstly, lncRNAs and protein-coding genes are clustered according to the degree of correlation between their expression patterns. The degree of association of each lncRNA with each gene ontology term is determined and biclustering used to identify groups of lncRNAs associated with specific functions [17]. Similarly, Liao et al. [106] identified probable functions (including neuronal development) for 340 lncRNAs based on coding/non-coding gene co-expression networks.
4. Long non-coding RNAs are involved in neural development and brain function
Multiple studies have implicated non-coding RNAs in brain development and function. Here, we focus only on lncRNAs, although small non-coding RNAs, such as miRNAs, are also important and have been discussed elsewhere [107,108]. Dynamic expression of lncRNAs has been observed in human-induced-pluripotent stem cells (iPSCs) [109] and human ES cells [105] during neuronal differentiation using RNA-seq and custom microarray, respectively. Neurogenesis-associated lncRNAs were found to associate directly with SUZ12 (a component of the polycomb repressive complex 2, PRC2), REST (discussed below) and SOX2 (a pluripotency-associated TF) suggesting that lncRNAs may act as guides for these proteins. Importantly, knockdown of these lncRNAs by RNAi resulted in impaired neuronal differentiation, suggesting that lncRNAs are critical regulators of neurogenesis [105]. A landmark study by Lipovich et al. [110] measured lncRNA expression in surgically resected in vivo human neocortical samples. Analysis of a range of samples from patients of different ages identified eight lncRNAs which showed strong statistical associations with aging and, by extension, brain development [110]. The majority of these lncRNAs were antisense to neighbouring protein-coding genes, suggesting possible gene regulatory functions. Interestingly, these lncRNAs also exhibited features consistent with recent evolutionary origins, including anthropoid-specific exons and mRNA processing sites which reside within primate-specific sequence [110]. Taken together, these findings implicate lncRNAs in the development of the human brain.
Similar results have also been observed in mouse cells where lncRNAs have been shown to control neuronal-glial cell fate decisions. Using custom microarray analysis of both coding and non-coding transcripts, Mercer et al. [38] identified lncRNAs that were differentially expressed between mouse embryonic forebrain-derived neural stem cells (NSCs), bipotent GABAergic neuron/oligodendrocyte cells and the various stages of terminally differentiated neurons and glia [38]. For example, the lncRNAs Neat1 and Neat2 (Malat1) were downregulated in the bipotent precursor cells but upregulated in differentiated neuronal and glial cells. Treatment of oligodendrocyte progenitor cells with the histone deacetylase (HDAC) inhibitor trichostatin A (known to suppress the maturation of oligodrocyte precursors and induce a more neuronal-like pattern of gene expression) also affected expression of lncRNAs, suggesting that their expression is under HDAC control [38]. A separate study identified the lncRNA Nkx2.2AS, which is a natural antisense transcript overlapping the TF gene Nkx2.2, as a further regulator of oligodendrocyte differentiation [111]. Overexpression of Nkx2.2AS induced differentiation and resulted in an increase in Nkx2.2 mRNA expression.
lncRNAs have also been implicated in the differentiation of other types of CNS tissue. Photoreceptors are specialized neurons in the retina which facilitate vision through the process of phototransduction [112]. The lncRNA TUG1 (which is highly expressed in brain) has been shown to be required for photoreceptor differentiation, although the mechanism of action has not yet been identified [86].
REST is a TF that represses expression of genes involved in neurogenesis and neuronal function in non-neural and immature neural cell types [113]. REST is therefore a key player in maintaining pluripotency and regulating neurogenesis. Johnson et al. showed that two brain-restricted lncRNAs are repressed by REST in NSCs [114] and the HAR1F/R lncRNA locus (discussed in §2) in a separate study [115]. Similarly, RCOR1 (also known as CoREST) is another protein that acts to repress expression of neural genes [116]. RIP-chip analysis using antibodies against RCOR1 identified 63 associated lncRNAs, many of which were also found to bind PRC2, suggesting that non-coding transcripts may play a key role in neural cell differentiation [97].
Imprinting is an epigenetic process by which certain genes are expressed in a parent-allele specific manner. A common theme in epigenetic imprinting is the reciprocal allelic expression of an imprinted gene and an imprinted non-coding RNA cis regulator. One of the most well-understood examples of this is the lncRNA Airn (also known as Air) which mediates epigenetic silencing of the Igf2r/Slc22a2/Slc22a3 locus on chromosome 17 [82]. Airn encodes an antisense transcript which overlaps with the Igf2r gene but not Slc22a2 or Slc22a3. Airn is expressed only from the parental allele, leading to epigenetic silencing of the parental Igf2r/Slc22a2/Slc22a3 locus in cis. Conversely, on the maternal allele, Airn is itself silenced by a reciprocal imprinting process and expression of the maternal Igf2r/Slc22a2/Slc22a3 locus is unhindered. Airn-mediated silencing occurs by at least two different mechanisms. In the case of Igf2r, transcription alone is sufficient to induce silencing [117]. Conversely, Slc22a3 silencing is dependent upon Airn-dependent recruitment of EHMT2 (a H3K9 histone methylase also known as G9a) [118]. In the majority of tissues, Igf2r is expressed only from the maternal allele, whereas Airn is expressed only from the paternal allele. However, this pattern of reciprocal allelic expression is not observed in brain where Igf2r is expressed in a biallelic manner as a result of neuron-specific relaxation of Airn-mediate imprinting [119].
Dlx genes encode homeodomain proteins that play key roles in the regulation of neuronal differentiation and migration [120,121]. The lncRNA Evf2 is transcribed from an ultraconserved region between the Dlx5 and Dlx6 protein-coding genes and is a direct target of SHH (Sonic hedgehog), a master regulator of vertebrate CNS development. Evf2 RNA forms a stable complex with Dlx4 protein and enhances its transcriptional activation functionality in C17 NSCs [122]. In a follow-up study, Evf2 was shown to act via both cis- and trans-acting mechanisms to recruit both Dlx and Mecp2 (methyl CpG-binding protein 2) to the Dlx5/6 ultraconserved region in the ventral forebrain [123]. Transgenic mice deficient in Evf2 transcription exhibited an imbalance in gene expression that led to a decrease in the number of GABAergic interneurons in the postnatal hippocampus, thereby illustrating the importance of this lncRNA in the patterning of the brain [123].
Malat1 is one of the most well-studied lncRNAs. It is well conserved, highly abundant and expressed in a wide range of tissues [39]. In the brain, Malat1 is expressed at high levels in neurons and low levels in glia and astrocytes, suggesting an important neuronal function [124]. Genes affected by antisense oligonucleotide-mediated Malat1 depletion were enriched for gene ontology terms associated with synaptic function and dendrite development. Knockdown of Malat1 in primary hippocampal neuron cultures resulted in reduced synaptic density, whereas Malat1 overexpression showed the opposite effect. Changes in the expression of Nlgn1 and SynCAM1 were observed upon Malat1 knockdown, suggesting that Malat1 regulates synaptogenesis by modulating the expression of genes in synapse formation [124].
5. Long non-coding RNAs and neurodegeneration
The human genome overwhelmingly (approx. 99%) consists of non-protein-coding sequence and it is therefore not surprising that the majority of mutations identified by genome-wide association studies (GWAS) occur in non-coding regions [125,126]. As such, a number of neurodegenerative disorders are known to be caused by mutations in lncRNA genes.
Perhaps the clearest example is spinocerebellar ataxia type 8 (SCA8) which is caused by a CTG triplet expansion in the brain-expressed ATXN8OS gene which is an antisense lncRNA transcript that partially overlaps with its neighbouring protein-coding gene, KLHL1 [127]. Although the aetiology of the disease is not well understood, the microsatellite expansion in the antisense transcript is believed to interfere with its endogenous role in regulating KLHL1 expression [128]. Microsatellite expansions in non-coding regions are also known to cause toxic RNA gain-of-function pathologies (such as in myotonic dystrophy) by sequestering factors involved in alternative splicing such as MBNL and CELF [129].
In 2011, a hexanucleotide (GGGGCC) repeat expansion in a protein-coding gene, C9ORF72 (chromosome 9 ORF 72) was identified as the first causative mutation for both amyotrophic lateral sclerosis (ALS) and frontotemporal dementia [130,131]. Since this landmark discovery, non-coding transcripts have now also been identified at the C9ORF72 locus. The C9ORF72 repeat expansion region undergoes bidirectional transcription [132]. Antisense C9ORF72 transcripts are elevated in the brains of ALS patients with both sense and antisense transcripts forming nuclear RNA foci [132,133]. The importance of the antisense C9ORF72 transcript is exemplified by the observation that targeted degradation of the corresponding sense transcript using antisense oligonucleotides is insufficient to correct the disease-associated gene expression signature in patient-derived fibroblasts [134]. These findings would be consistent with a toxic RNA-type cellular pathology, although the reality may be more complex as both sense and antisense transcripts produce dipeptide repeat proteins by repeat-associated non-ATG translation [132,135,136].
By interrogating published microarray gene expression data from Huntington's disease (HD) patient caudate nucleus [137], Johnson [138] was able to identify lncRNAs with a HD-specific pattern of differential expression. Three novel lncRNAs were elevated in HD brains in addition to TUG1 and NEAT1 (which were upregulated in HD) and MEG3 and DGCR5 (which were downregulated). The role of these lncRNAs in HD pathophysiology is currently unknown, although the observation that MEG3 and TUG1 associate with PRC2 suggests that they may act as epigenetic regulators which induce disease-specific gene expression signatures [97]. Similarly, a separate study found that the expression of lncRNAs originating from the HAR1F/R locus was repressed in the striatum of post-mortem HD brains [115]. Using a similar data mining approach, Michelhaugh et al. [139] found that the MIAT, MEG3, NEAT1 and NEAT2 lncRNAs were all upregulated in the post-mortem dissected nucleus accumbens of heroin users, suggesting a possible role for lncRNAs in addictive behaviours.
BACE1 (β-site amyloid precursor protein cleaving enzyme 1, also known as β-secretase) is an enzyme central to the pathology of AD. BACE1 catalyses the cleavage of amyloid precursor protein to generate β-amyloid peptides which aggregate to form plaques [140]. Studies by Faghihi and co-workers identified a conserved antisense transcript overlapping (BACE1-AS) at the BACE1 locus [66,141]. BACE1-AS is concordantly expressed with BACE1 sense mRNA and acts as a feed-forward positive regulator of BACE1 expression. Expression of BACE1-AS was also found to be elevated in the hippocampus, superior frontal gyrus and entorhinal cortex in post-mortem AD brain tissue [66]. The mechanism of BACE1 regulation by BACE1-AS was subsequently shown to be via the formation of an RNA duplex between the overlapping transcripts which masks the binding site for miR-485–5p, thereby relieving miRNA-mediated gene silencing [67].
6. Long non-coding RNAs and neurodevelopmental/neuropsychiatric disorders
The non-coding RNA BC200 is restricted to brain tissue (specifically to the neurite outgrowths of neurons) and its expression gradually declines with aging. However, BC200 expression is elevated in the brains of AD patients and mislocalized to the neuronal cell bodies rather than at dendritic spines [142]. The molecular function of BC200 appears to be in the regulation of neuronal protein translation and so it may contribute to amyloid plaque formation and subsequent AD [143,144]. The murine homologue of BC200, BC1, was targeted in a transgenic knockout model. Interestingly, BC1 knockout mice showed no obvious phenotype in a laboratory cage environment. However, when introduced into a controlled ‘natural outdoor’ environment, these mice showed signs of increased anxiety and reduced survival [145]. This study demonstrates that non-coding transcripts may exert subtle effects on complex behaviour and raises the intriguing possibility that lncRNAs may be involved in the pathogenesis of neurodevelopmental and neuropsychiatric diseases with poorly understood aetiologies.
Autism spectrum disorder (ASD) refers to a heterogeneous group of neurodevelopmental disorders that are characterized by defects in social interactions, communication and repetitive stereotyped behaviours. Although ASD is known to have a strong genetic basis, its pathophysiology is poorly understood [146]. Microarray analysis of human post-mortem brain tissue (prefrontal cortex and cerebellum) from ASD patients and unaffected controls identified 222 differentially expressed lncRNAs which were enriched at protein-coding gene loci associated with brain development. Interestingly, the ASD brains were more transcriptionally homogeneous than the controls, both in terms of mRNA and lncRNA expression [147]. Similarly, interrogation of publicly available RNA-seq data identified overlapping antisense lncRNAs at 38 protein-coding loci associated with ASD. Furthermore, one of these antisense transcripts, SYNGAP1-AS, was found to be upregulated in the ASD post-mortem prefrontal cortex and superior temporal gyrus [148].
A study by Kerin et al. [149] identified a single nucleotide polymorphism (SNP) associated with ASD at a non-coding locus in a GWAS. This locus was found to encode an lncRNA (MSNP1AS) antisense to a processed pseudogene of moesin (MSNP1) which shows no evidence of being transcribed in the sense orientation. The SNP-containing MSNP1AS transcript was shown to be elevated in post-mortem brain tissue (temporal cerebral cortex) of ASD patients and regulated expression of Moesin protein (a known regulator of nuclear architecture [150]) in human cells [149], suggesting a possible role in ASD pathophysiology.
Fragile X syndrome (FXS) and fragile X tremor ataxia syndrome (FXTAS) are intellectual disabilities caused by expansions of a CGG repeat in the 5′-UTR of the FMR1 protein-coding gene [151]. Normal individuals typically carry 5–54 repeats, whereas 55–200 repeats (so-called premutation alleles) lead to FXTAS, and more than 200 repeats lead to FXS. As in the case of SCA8, FMR1 has an upstream partially overlapping antisense transcript, FMR4 (also known as FMR1-AS1), which is presumably driven by a bidirectional promoter. In FXS, the repeat expansion region becomes hypermethylated and transcription of the gene products in both orientations is diminished. siRNA-mediated knockdown of either FMR1 or FMR4 did not affect the expression of each transcript's antisense partner, suggesting that FMR4 is not a regulator of FMR1. Instead, FMR4 knockdown was shown to promote apoptosis, suggesting that its endogenous function is as an RNA anti-apoptotic signal [152]. By contrast, FMR1 and FMR4/FMR1-AS are upregulated in carriers of premutation alleles (i.e. FXTAS) [153]. The application of a high-throughput sequencing RACE methodology to the FMR1 locus identified a further two lncRNA transcripts, FMR5 and FMR6 (in sense and antisense orientations, respectively). Analysis of post-mortem brain tissues from carriers of both full and premutation alleles showed that expression of FMR6 was suppressed in both cases relative to wild-type controls [154]. Furthermore, a recent study identified a role for CTCF in regulating bidirectional transcription of FMR1 through chromatin structure [155]. Consequently, the variable patterns of antisense RNA expression at the FMR1 gene as a result of different repeat region lengths have been proposed as an explanation for the different clinical features of FXS and FXTAS, despite both syndromes being caused by CGG expansions [153].
Several lncRNAs have been implicated in the pathogenesis of schizophrenia (SZ). The lncRNA MIAT is downregulated upon neuronal activation [156]. Investigation of SZ patient post-mortem brain tissue (superior temporal gyrus) found that MIAT was downregulated. The MIAT transcripts directly interact with the splicing factors QKI and SRSF1 and loss of MIAT expression results in global changes in alternative splicing similar to those observed for other SZ-associated genes (i.e. DISC1) [156,157].
7. Long non-coding RNAs and brain cancers
Multiple studies have identified lncRNAs involved in cancer [102,158]. For example, lncRNAs have been shown to be direct targets of p53, including linc-p21, PANDA, TUG1 and Pint [159,160]. The lncRNA ANRIL is implicated in melanoma-neural system tumour [161] and has been shown to interact with PRC2 in order to epigenetically silence the p15 tumour suppressor [162].
The lncRNA CRNDE is highly upregulated in gliomas [163] and in iPSCs undergoing neuronal differentiation [109]. CRNDE shares a bidirectional promoter with the IRX5 gene (which is involved in neurogenesis) and the two genes exhibit concordant expression patterns. CRNDE binds to CoREST [97] and in the human adult brain CRNDE is predominantly expressed in the basal ganglia, thalamus, cerebellum and surrounding structures [164]. Conversely, the imprinted lncRNA gene, MEG3, is a brain-specific tumour suppressor that suppresses cell growth, promotes p53-mediated apoptosis and is lost in pituitary tumours [165,166], and in meningiomas [167].
8. Conclusion
In summary, the proposition that lncRNAs are functional is supported by the following: (i) specific spatial and temporal expression patterns, (ii) high-fidelity transcript processing, (iii) differential expression during cellular processes, (iv) evolutionary conservation, (v) knockout mouse models, (vi) RNAi loss-of-function screens, (vii) guilt-by-association co-expression studies, (viii) interactions with chromatin-modifying proteins and TFs, (ix) implication in disease pathophysiology and (x) focused studies demonstrating function in specific cases. The relatively low abundance and tissue-restricted expression of lncRNA transcripts suggest that they function as subtle regulators in the determination of cell fate and identity, rather than in the execution of housekeeping functions. The preponderance of evidence suggests that lncRNAs constitute a previously under-appreciated component of cellular metabolism that together with TFs, chromatin remodelling complexes and miRNAs, regulates differential gene expression. Given that the degree of organismal complexity scales with the amount of non-coding DNA sequences [168], it is tempting to speculate that the increase in regulatory complexity afforded by the interplay of lncRNAs and protein-coding genes may be responsible for the difference in cognitive abilities between humans and other animals [169].
The importance of lncRNAs in the brain is exemplified by their involvement in the maintenance of pluripotency, neuroectodermal differentiation, neuronal-glial cell fate determination, neuron-specific relaxation of epigenetic imprinting, repression of neural genes in non-neural cells, brain tissue patterning and synaptogenesis. Given that epigenetic mechanisms underlie memory formation, it is likely that lncRNAs may also be involved in this process [170]. The involvement of lncRNAs in neurodegenerative, neurodevelopmental and neuropsychiatric disorders, and in brain cancers further underlines their importance in CNS development and function. lncRNAs may themselves drive or mediate the disease pathophysiology (as in the case of ATXN8OS and FMR4), or they may regulate the expression of disease-associated genes (as in the case of BACE1-AS). As a result, lncRNAs are promising novel targets for therapeutic intervention [171,172].
References
- 1.Clark MB, et al. 2011. The reality of pervasive transcription. PLoS Biol. 9, e1000625 (discussion e1001102). ( 10.1371/journal.pbio.1000625) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carninci P, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309, 1559–1563. ( 10.1126/science.1112014) [DOI] [PubMed] [Google Scholar]
- 3.ENCODE Project Consortium et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. ( 10.1038/nature05874) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katayama S, et al. 2005. Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566. ( 10.1126/science.1112009) [DOI] [PubMed] [Google Scholar]
- 5.Engström PG, et al. 2006. Complex loci in human and mouse genomes. PLoS Genet. 2, e47 ( 10.1371/journal.pgen.0020047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djebali S, et al. 2012. Landscape of transcription in human cells. Nature 489, 101–108. ( 10.1038/nature11233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapranov P, Drenkow J, Cheng J, Long J, Helt G, Dike S, Gingeras TR. 2005. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res 15, 987–997. ( 10.1101/gr.3455305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SPA, Gingeras TR. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296, 916–919. ( 10.1126/science.1068597) [DOI] [PubMed] [Google Scholar]
- 9.Rinn JL, et al. 2003. The transcriptional activity of human chromosome 22. Genes Dev. 17, 529–540. ( 10.1101/gad.1055203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertone P, et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306, 2242–2246. ( 10.1126/science.1103388) [DOI] [PubMed] [Google Scholar]
- 11.Cheng J, et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308, 1149–1154. ( 10.1126/science.1108625) [DOI] [PubMed] [Google Scholar]
- 12.Okazaki Y, et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420, 563–573. ( 10.1038/nature01266) [DOI] [PubMed] [Google Scholar]
- 13.Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. 2007. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell. Sci. 120, 2498–2506. ( 10.1242/jcs.009357) [DOI] [PubMed] [Google Scholar]
- 14.Dinger ME, Pang KC, Mercer TR, Mattick JS. 2008. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 4, e1000176 ( 10.1371/journal.pcbi.1000176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiyosawa H, Mise N, Iwase S, Hayashizaki Y, Abe K. 2005. Disclosing hidden transcripts: mouse natural sense-antisense transcripts tend to be poly(A) negative and nuclear localized. Genome Res. 15, 463–474. ( 10.1101/gr.3155905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derrien T, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. ( 10.1101/gr.132159.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guttman M, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227. ( 10.1038/nature07672) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. 2013. Ribosome profiling provides evidence that large non-coding RNAs do not encode proteins. Cell 154, 240–251. ( 10.1016/j.cell.2013.06.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802. ( 10.1016/j.cell.2011.10.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. 2007. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 5, e106 ( 10.1371/journal.pbio.0050106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bánfai B, et al. 2012. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 22, 1646–1657. ( 10.1101/gr.134767.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Bakel H, Nislow C, Blencowe BJ, Hughes TR. 2010. Most ‘dark matter’ transcripts are associated with known genes. PLoS Biol. 8, e1000371 ( 10.1371/journal.pbio.1000371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. 2008. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA 105, 716–721. ( 10.1073/pnas.0706729105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponjavic J, Oliver PL, Lunter G, Ponting CP. 2009. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 5, e1000617 ( 10.1371/journal.pgen.1000617) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smalheiser NR, Lugli G, Torvik VI, Mise N, Ikeda R, Abe K. 2008. Natural antisense transcripts are co-expressed with sense mRNAs in synaptoneurosomes of adult mouse forebrain. Neurosci. Res. 62, 236–239. ( 10.1016/j.neures.2008.08.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. 2009. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 19, 347–359. ( 10.1101/gr.087775.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.French PJ, Bliss TV, O'Connor V. 2001. Ntab, a novel non-coding RNA abundantly expressed in rat brain. Neuroscience 108, 207–215. ( 10.1016/S0306-4522(01)00408-0) [DOI] [PubMed] [Google Scholar]
- 28.Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. 2012. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotech. 30, 99–104. ( 10.1038/nbt.2024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tilgner H, et al. 2012. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 22, 1616–1625. ( 10.1101/gr.134445.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravasi T, et al. 2006. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 16, 11–19. ( 10.1101/gr.4200206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim T-K, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187. ( 10.1038/nature09033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinger ME, et al. 2008. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 18, 1433–1445. ( 10.1101/gr.078378.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohamed JS, Gaughwin PM, Lim B, Robson P, Lipovich L. 2010. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 16, 324–337. ( 10.1261/rna.1441510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC, Mattick JS. 2009. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA 15, 2013–2027. ( 10.1261/rna.1705309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shahryari A, Rafiee MR, Fouani Y, Oliae NA, Samaei NM, Shafiee M, Semnani S, Vasei M, Mowla SJ. 2014. Two novel splice variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma. Stem Cells 32, 126–134. ( 10.1002/stem.1542) [DOI] [PubMed] [Google Scholar]
- 36.Struhl K. 2007. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 14, 103–105. ( 10.1038/nsmb0207-103) [DOI] [PubMed] [Google Scholar]
- 37.Sauvageau M, et al. 2013. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2, e01749 ( 10.7554/eLife.01749) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. 2010. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 11, 14 ( 10.1186/1471-2202-11-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang B, et al. 2012. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2, 111–123. ( 10.1016/j.celrep.2012.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakagawa S, Naganuma T, Shioi G, Hirose T. 2011. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 193, 31–39. ( 10.1083/jcb.201011110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guttman M, et al. 2010. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 28, 503–510. ( 10.1038/nbt.1633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marques AC, Ponting CP. 2009. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 10, R124 ( 10.1186/gb-2009-10-11-r124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. 2009. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics 25, i54–i62. ( 10.1093/bioinformatics/btp190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ponjavic J, Ponting CP, Lunter G. 2007. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 17, 556–565. ( 10.1101/gr.6036807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pang KC, Frith MC, Mattick JS. 2006. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 22, 1–5. ( 10.1016/j.tig.2005.10.003) [DOI] [PubMed] [Google Scholar]
- 46.Novikova IV, Hennelly SP, Sanbonmatsu KY. 2012. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 40, 5034–5051. ( 10.1093/nar/gks071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Havgaard J, Kaur S, Gorodkin J. 2012. Comparative ncRNA gene and structure prediction using Foldalign and FoldalignM. Curr. Protoc. Bioinform. 39, 12.11.1–12.11.15 ( 10.1002/0471250953.bi1211s39) [DOI] [PubMed] [Google Scholar]
- 48.Torarinsson E, Sawera M, Havgaard JH, Fredholm M, Gorodkin J. 2006. Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res. 16, 885–889. ( 10.1101/gr.5226606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. 2011. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–1550. ( 10.1016/j.cell.2011.11.055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Washietl S, Hofacker IL, Lukasser M, Hüttenhofer A, Stadler PF. 2005. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 23, 1383–1390. ( 10.1038/nbt1144) [DOI] [PubMed] [Google Scholar]
- 51.Roberts TC, Morris KV. 2013. Not so pseudo anymore: pseudogenes as therapeutic targets. Pharmacogenomics 14, 2023–2034. ( 10.2217/pgs.13.172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khachane AN, Harrison PM. 2009. Assessing the genomic evidence for conserved transcribed pseudogenes under selection. BMC Genomics 10, 435 ( 10.1186/1471-2164-10-435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nesterova TB, et al. 2001. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res. 11, 833–849. ( 10.1101/gr.174901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oudejans CB, Westerman B, Wouters D, Gooyer S, Leegwater PA, van Wijk IJ, Sleutels F. 2001. Allelic IGF2R repression does not correlate with expression of antisense RNA in human extraembryonic tissues. Genomics 73, 331–337. ( 10.1006/geno.2001.6522) [DOI] [PubMed] [Google Scholar]
- 55.Pollard KS, et al. 2006. An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443, 167–172. ( 10.1038/nature05113) [DOI] [PubMed] [Google Scholar]
- 56.Shearwin KE, Callen BP, Egan JB. 2005. Transcriptional interference: a crash course. Trends Genet. 21, 339–345. ( 10.1016/j.tig.2005.04.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adhya S, Gottesman M. 1982. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell 29, 939–944. ( 10.1016/0092-8674(82)90456-1) [DOI] [PubMed] [Google Scholar]
- 58.Schmitt S, Paro R. 2004. Gene regulation: a reason for reading nonsense. Nature 429, 510–511. ( 10.1038/429510a) [DOI] [PubMed] [Google Scholar]
- 59.Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol. Cell 5, 377–386. ( 10.1016/S1097-2765(00)80432-3) [DOI] [PubMed] [Google Scholar]
- 60.Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. 2006. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell 127, 1209–1221. ( 10.1016/j.cell.2006.10.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Osato N, Suzuki Y, Ikeo K, Gojobori T. 2007. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics 176, 1299–1306. ( 10.1534/genetics.106.069484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan M-D, Hong C-C, Lai G-M, Cheng A-L, Lin Y-W, Chuang S-E. 2005. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum. Mol. Genet. 14, 1465–1474. ( 10.1093/hmg/ddi156) [DOI] [PubMed] [Google Scholar]
- 63.Tripathi V, et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938. ( 10.1016/j.molcel.2010.08.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726. ( 10.1016/j.molcel.2009.01.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blackburn EH, Greider CW, Szostak JW. 2006. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 12, 1133–1138. ( 10.1038/nm1006-1133) [DOI] [PubMed] [Google Scholar]
- 66.Faghihi MA, et al. 2008. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14, 723–730. ( 10.1038/nm1784) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G, 3rd, Wahlestedt C. 2010. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 11, R56 ( 10.1186/gb-2010-11-5-r56) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kretz M, et al. 2013. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493, 231–235. ( 10.1038/nature11661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Annilo T, Kepp K, Laan M. 2009. Natural antisense transcript of natriuretic peptide precursor A (NPPA): structural organization and modulation of NPPA expression. BMC Mol. Biol. 10, 81 ( 10.1186/1471-2199-10-81) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. 2010. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3, ra8 ( 10.1126/scisignal.2000568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. 2006. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 20, 1732–1743. ( 10.1101/gad.1425706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yin Q-F, Yang L, Zhang Y, Xiang J-F, Wu Y-W, Carmichael GG, Chen L-L. 2012. Long noncoding RNAs with snoRNA ends. Mol. Cell 48, 219–230. ( 10.1016/j.molcel.2012.07.033) [DOI] [PubMed] [Google Scholar]
- 73.Augoff K, McCue B, Plow EF, Sossey-Alaoui K. 2012. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol. Cancer 11, 5 ( 10.1186/1476-4598-11-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038. ( 10.1038/nature09144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358. ( 10.1016/j.cell.2011.07.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369. ( 10.1016/j.cell.2011.09.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. 2005. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309, 1570–1573. ( 10.1126/science.1115901) [DOI] [PubMed] [Google Scholar]
- 78.Panning B, Jaenisch R. 1998. RNA and the epigenetic regulation of X chromosome inactivation. Cell 93, 305–308. ( 10.1016/S0092-8674(00)81155-1) [DOI] [PubMed] [Google Scholar]
- 79.Sun BK, Deaton AM, Lee JT. 2006. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol. Cell 21, 617–628. ( 10.1016/j.molcel.2006.01.028) [DOI] [PubMed] [Google Scholar]
- 80.Rougeulle C, Heard E. 2002. Antisense RNA in imprinting: spreading silence through Air. Trends Genet. 18, 434–437. ( 10.1016/S0168-9525(02)02749-X) [DOI] [PubMed] [Google Scholar]
- 81.Thakur N, Tiwari VK, Thomassin H, Pandey RR, Kanduri M, Göndör A, Grange T, Ohlsson R, Kanduri C. 2004. An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol. Cell. Biol. 24, 7855–7862. ( 10.1128/MCB.24.18.7855-7862.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sleutels F, Zwart R, Barlow DP. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813. ( 10.1038/415810a) [DOI] [PubMed] [Google Scholar]
- 83.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. 2010. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 24, 2543–2555. ( 10.1101/gad.1967810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. 2011. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147, 773–788. ( 10.1016/j.cell.2011.08.054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guttman M, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300. ( 10.1038/nature10398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Young TL, Matsuda T, Cepko CL. 2005. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr. Biol. 15, 501–512. ( 10.1016/j.cub.2005.02.027) [DOI] [PubMed] [Google Scholar]
- 87.Egger G, Liang G, Aparicio A, Jones PA. 2004. Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463. ( 10.1038/nature02625) [DOI] [PubMed] [Google Scholar]
- 88.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. 2009. An operational definition of epigenetics. Genes Dev. 23, 781–783. ( 10.1101/gad.1787609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403, 41–45. ( 10.1038/47412) [DOI] [PubMed] [Google Scholar]
- 90.Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. ( 10.1016/j.cell.2007.05.009) [DOI] [PubMed] [Google Scholar]
- 91.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128, 693–705. ( 10.1016/j.cell.2007.02.005) [DOI] [PubMed] [Google Scholar]
- 92.Teif VB, Rippe K. 2009. Predicting nucleosome positions on the DNA: combining intrinsic sequence preferences and remodeler activities. Nucleic Acids Res. 37, 5641–5655. ( 10.1093/nar/gkp610) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.St Laurent G, 3rd, Wahlestedt C. 2007. Noncoding RNAs: couplers of analog and digital information in nervous system function? Trends Neurosci. 30, 612–621. ( 10.1016/j.tins.2007.10.002) [DOI] [PubMed] [Google Scholar]
- 94.Guttman M, Rinn JL. 2012. Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346. ( 10.1038/nature10887) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roberts TC, Morris KV, Weinberg MS. 2013. Perspectives on the mechanism of transcriptional regulation by long non-coding RNAs. Epigenetics 9, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang KC, Chang HY. 2011. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914. ( 10.1016/j.molcel.2011.08.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khalil AM, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA 106, 11 667–11 672. ( 10.1073/pnas.0904715106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao J, et al. 2010. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 40, 939–953. ( 10.1016/j.molcel.2010.12.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang KC, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124. ( 10.1038/nature09819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wutz A, Rasmussen TP, Jaenisch R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167–174. ( 10.1038/ng820) [DOI] [PubMed] [Google Scholar]
- 101.Rinn JL, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. ( 10.1016/j.cell.2007.05.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gupta RA, et al. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076. ( 10.1038/nature08975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA–chromatin interactions. Mol. Cell 44, 667–678. ( 10.1016/j.molcel.2011.08.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. ( 10.1126/science.1192002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ng S-Y, Johnson R, Stanton LW. 2012. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31, 522–533. ( 10.1038/emboj.2011.459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liao Q, et al. 2011. Large-scale prediction of long non-coding RNA functions in a coding–non-coding gene co-expression network. Nucleic Acids Res. 39, 3864–3878. ( 10.1093/nar/gkq1348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RHA. 2006. Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 38, 1375–1377. ( 10.1038/ng1914) [DOI] [PubMed] [Google Scholar]
- 108.Varela MA, Roberts TC, Wood MJA. 2013. Epigenetics and ncRNAs in brain function and disease: mechanisms and prospects for therapy. Neurotherapeutics 10, 621–631. ( 10.1007/s13311-013-0212-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D, Lachman HM. 2011. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS ONE 6, e23356 ( 10.1371/journal.pone.0023356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lipovich L, et al. 2013. Developmental changes in the transcriptome of human cerebral cortex tissue: long noncoding RNA transcripts. Cereb. Cortex 24, 1451–1459. ( 10.1093/cercor/bhs414) [DOI] [PubMed] [Google Scholar]
- 111.Tochitani S, Hayashizaki Y. 2008. Nkx2.2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem. Biophys. Res. Commun. 372, 691–696. ( 10.1016/j.bbrc.2008.05.127) [DOI] [PubMed] [Google Scholar]
- 112.Ebrey T, Koutalos Y. 2001. Vertebrate photoreceptors. Prog. Retin. Eye Res. 20, 49–94. ( 10.1016/S1350-9462(00)00014-8) [DOI] [PubMed] [Google Scholar]
- 113.Chong JA, et al. 1995. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80, 949–957. ( 10.1016/0092-8674(95)90298-8) [DOI] [PubMed] [Google Scholar]
- 114.Johnson R, Teh CH-L, Jia H, Vanisri RR, Pandey T, Lu Z-H, Buckley NJ, Stanton LW, Lipovich L. 2009. Regulation of neural macroRNAs by the transcriptional repressor REST. RNA 15, 85–96. ( 10.1261/rna.1127009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Johnson R, Richter N, Jauch R, Gaughwin PM, Zuccato C, Cattaneo E, Stanton LW. 2010. The human accelerated region 1 noncoding RNA is repressed by REST in Huntington's disease. Physiol. Genomics 41, 269–274. ( 10.1152/physiolgenomics.00019.2010) [DOI] [PubMed] [Google Scholar]
- 116.Andrés ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. 1999. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl Acad. Sci. USA 96, 9873–9878. ( 10.1073/pnas.96.17.9873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Latos PA, et al. 2012. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472. ( 10.1126/science.1228110) [DOI] [PubMed] [Google Scholar]
- 118.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720. ( 10.1126/science.1163802) [DOI] [PubMed] [Google Scholar]
- 119.Yamasaki Y, et al. 2005. Neuron-specific relaxation of Igf2r imprinting is associated with neuron-specific histone modifications and lack of its antisense transcript Air. Hum. Mol. Genet. 14, 2511–2520. ( 10.1093/hmg/ddi255) [DOI] [PubMed] [Google Scholar]
- 120.Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. 1997. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278, 474–476. ( 10.1126/science.278.5337.474) [DOI] [PubMed] [Google Scholar]
- 121.Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL. 1997. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron 19, 27–37. ( 10.1016/S0896-6273(00)80345-1) [DOI] [PubMed] [Google Scholar]
- 122.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. 2006. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20, 1470–1484. ( 10.1101/gad.1416106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bond AM, Vangompel MJW, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. 2009. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020–1027. ( 10.1038/nn.2371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bernard D, et al. 2010. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082–3093. ( 10.1038/emboj.2010.199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Halvorsen M, Martin JS, Broadaway S, Laederach A. 2010. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet. 6, e1001074 ( 10.1371/journal.pgen.1001074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA 106, 9362–9367. ( 10.1073/pnas.0903103106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nemes JP, Benzow KA, Moseley ML, Ranum LP, Koob MD. 2000. The SCA8 transcript is an antisense RNA to a brain-specific transcript encoding a novel actin-binding protein (KLHL1). Hum. Mol. Genet. 9, 1543–1551. ( 10.1093/hmg/9.10.1543) [DOI] [PubMed] [Google Scholar]
- 128.Chen W-L, Lin J-W, Huang H-J, Wang S-M, Su M-T, Lee-Chen G-J, Chen C-M, Hsieh-Li HM. 2008. SCA8 mRNA expression suggests an antisense regulation of KLHL1 and correlates to SCA8 pathology. Brain Res. 1233, 176–184. ( 10.1016/j.brainres.2008.07.096) [DOI] [PubMed] [Google Scholar]
- 129.Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LPW. 2009. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 5, e1000600 ( 10.1371/journal.pgen.1000600) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.DeJesus-Hernandez M, et al. 2011. Expanded GGGGCC hexanucleotide repeat in non-coding region of C9ORF72 causes chromosome 9p-linked frontotemporal dementia and amyotrophic lateral sclerosis. Neuron 72, 245–256. ( 10.1016/j.neuron.2011.09.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Renton AE, et al. 2011. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268. ( 10.1016/j.neuron.2011.09.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zu T, et al. 2013. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl Acad. Sci. USA 110, E4968–E4977. ( 10.1073/pnas.1315438110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mizielinska S, Lashley T, Norona FE, Clayton EL, Ridler CE, Fratta P, Isaacs AM. 2013. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 126, 845–857. ( 10.1007/s00401-013-1200-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lagier-Tourenne C, et al. 2013. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl Acad. Sci. USA 110, E4530–E4539. ( 10.1073/pnas.1318835110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gendron TF, et al. 2013. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126, 829–844. ( 10.1007/s00401-013-1192-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mori K, et al. 2013. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 126, 881–893. ( 10.1007/s00401-013-1189-3) [DOI] [PubMed] [Google Scholar]
- 137.Hodges A, et al. 2006. Regional and cellular gene expression changes in human Huntington's disease brain. Hum. Mol. Genet. 15, 965–977. ( 10.1093/hmg/ddl013) [DOI] [PubMed] [Google Scholar]
- 138.Johnson R. 2012. Long non-coding RNAs in Huntington's disease neurodegeneration. Neurobiol. Dis. 46, 245–254. ( 10.1016/j.nbd.2011.12.006) [DOI] [PubMed] [Google Scholar]
- 139.Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ. 2011. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J. Neurochem. 116, 459–466. ( 10.1111/j.1471-4159.2010.07126.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Querfurth HW, LaFerla FM. 2010. Alzheimer's disease. N. Engl. J. Med. 362, 329–344. ( 10.1056/NEJMra0909142) [DOI] [PubMed] [Google Scholar]
- 141.Modarresi F, Faghihi MA, Patel NS, Sahagan BG, Wahlestedt C, Lopez-Toledano MA. 2011. Knockdown of BACE1-AS nonprotein-coding transcript modulates beta-amyloid-related hippocampal neurogenesis. Int. J. Alzheimers Dis. 2011, 929042 ( 10.4061/2011/929042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mus E, Hof PR, Tiedge H. 2007. Dendritic BC200 RNA in aging and in Alzheimer's disease. Proc. Natl Acad. Sci. USA 104, 10 679–10 684. ( 10.1073/pnas.0701532104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tiedge H, Chen W, Brosius J. 1993. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J. Neurosci. 13, 2382–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Muddashetty R, et al. 2002. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. 321, 433–445. ( 10.1016/S0022-2836(02)00655-1) [DOI] [PubMed] [Google Scholar]
- 145.Lewejohann L, et al. 2004. Role of a neuronal small non-messenger RNA: behavioural alterations in BC1 RNA-deleted mice. Behav. Brain Res. 154, 273–289. ( 10.1016/j.bbr.2004.02.015) [DOI] [PubMed] [Google Scholar]
- 146.Geschwind DH. 2008. Autism: many genes, common pathways? Cell 135, 391–395. ( 10.1016/j.cell.2008.10.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ziats MN, Rennert OM. 2013. Aberrant expression of long noncoding RNAs in autistic brain. J. Mol. Neurosci. 49, 589–593. ( 10.1007/s12031-012-9880-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Velmeshev D, Magistri M, Faghihi MA. 2013. Expression of non-protein-coding antisense RNAs in genomic regions related to autism spectrum disorders. Mol. Autism 4, 32 ( 10.1186/2040-2392-4-32) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kerin T, Ramanathan A, Rivas K, Grepo N, Coetzee GA, Campbell DB. 2012. A noncoding RNA antisense to moesin at 5p14.1 in autism. Sci. Transl. Med. 4, 128ra40 ( 10.1126/scitranslmed.3003479) [DOI] [PubMed] [Google Scholar]
- 150.Paglini G, Kunda P, Quiroga S, Kosik K, Caceres A. 1998. Suppression of radixin and moesin alters growth cone morphology, motility, and process formation in primary cultured neurons. J. Cell Biol. 143, 443–455. ( 10.1083/jcb.143.2.443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Willemsen R, Levenga J, Oostra BA. 2011. CGG repeat in the FMR1 gene: size matters. Clin. Genet. 80, 214–225. ( 10.1111/j.1399-0004.2011.01723.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. 2008. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS ONE 3, e1486 ( 10.1371/journal.pone.0001486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ladd PD, et al. 2007. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 16, 3174–3187. ( 10.1093/hmg/ddm293) [DOI] [PubMed] [Google Scholar]
- 154.Pastori C, Peschansky VJ, Barbouth D, Mehta A, Silva JP, Wahlestedt C. 2014. Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in fragile X syndrome and fragile X-associated tremor/ataxia syndrome. Hum. Genet. 133, 59–67. ( 10.1007/s00439-013-1356-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lanni S, et al. 2013. Role of CTCF protein in regulating FMR1 locus transcription. PLoS Genet. 9, e1003601 ( 10.1371/journal.pgen.1003601) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Barry G, et al. 2013. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 19, 486–494. ( 10.1038/mp.2013.45) [DOI] [PubMed] [Google Scholar]
- 157.Millar JK, et al. 2000. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9, 1415–1423. ( 10.1093/hmg/9.9.1415) [DOI] [PubMed] [Google Scholar]
- 158.Ji P, et al. 2003. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041. ( 10.1038/sj.onc.1206928) [DOI] [PubMed] [Google Scholar]
- 159.Huarte M, et al. 2010. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419. ( 10.1016/j.cell.2010.06.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Marín-Béjar O, et al. 2013. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 14, R104 ( 10.1186/gb-2013-14-9-r104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pasmant E, Laurendeau I, Héron D, Vidaud M, Vidaud D, Bièche I. 2007. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 67, 3963–3969. ( 10.1158/0008-5472.CAN-06-2004) [DOI] [PubMed] [Google Scholar]
- 162.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. 2011. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30, 1956–1962. ( 10.1038/onc.2010.568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang X, Sun S, Pu JKS, Tsang ACO, Lee D, Man VOY, Lui WM, Wong STS, Leung GKK. 2012. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 48, 1–8. ( 10.1016/j.nbd.2012.06.004) [DOI] [PubMed] [Google Scholar]
- 164.Ellis BC, Molloy PL, Graham LD. 2012. CRNDE: a long non-coding RNA involved in cancer, neurobiology, and development. Front. Genet. 3, 270 ( 10.3389/fgene.2012.00270) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gordon FE, Nutt CL, Cheunsuchon P, Nakayama Y, Provencher KA, Rice KA, Zhou Y, Zhang X, Klibanski A. 2010. Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology 151, 2443–2452. ( 10.1210/en.2009-1151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, Klibanski A. 2003. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J. Clin. Endocrinol. Metab. 88, 5119–5126. ( 10.1210/jc.2003-030222) [DOI] [PubMed] [Google Scholar]
- 167.Balik V, Srovnal J, Sulla I, Kalita O, Foltanova T, Vaverka M, Hrabalek L, Hajduch M. 2013. MEG3: a novel long noncoding potentially tumour-suppressing RNA in meningiomas. J. Neurooncol. 112, 1–8. ( 10.1007/s11060-012-1038-6) [DOI] [PubMed] [Google Scholar]
- 168.Taft RJ, Mattick JS. 2003. Increasing biological complexity is positively correlated with the relative genome-wide expansion of non-protein-coding DNA sequences. Genome Biol. 5, P1 ( 10.1186/gb-2003-5-1-p1) [DOI] [Google Scholar]
- 169.Mehler MF, Mattick JS. 2006. Non-coding RNAs in the nervous system. J. Physiol. 575, 333–341. ( 10.1113/jphysiol.2006.113191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mercer TR, Dinger ME, Mariani J, Kosik KS, Mehler MF, Mattick JS. 2008. Noncoding RNAs in long-term memory formation. Neuroscientist 14, 434–445. ( 10.1177/1073858408319187) [DOI] [PubMed] [Google Scholar]
- 171.Roberts TC, Wood MJA. 2013. Therapeutic targeting of non-coding RNAs. Essays Biochem. 54, 127–145. ( 10.1042/bse0540127) [DOI] [PubMed] [Google Scholar]
- 172.Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. 2012. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30, 453–459. ( 10.1038/nbt.2158) [DOI] [PMC free article] [PubMed] [Google Scholar]