Abstract
In the mammalian brain, epigenetic mechanisms are clearly involved in the regulation of self-renewal of neural stem cells and the derivation of their descendants, i.e. neurons, astrocytes and oligodendrocytes, according to the developmental timing and the microenvironment, the ‘niche’. Interestingly, local epigenetic changes occur, concomitantly with genome-wide level changes, at a set of gene promoter regions for either down- or upregulation of the gene. In addition, intergenic regions also sensitize the availability of epigenetic modifiers, which affects gene expression through a relatively long-range chromatinic interaction with the transcription regulatory machineries including non-coding RNA (ncRNA) such as promoter-associated ncRNA and enhancer ncRNA. We show that such an epigenetic landscape in a neural cell is statically but flexibly formed together with a variable combination of generally and locally acting nuclear molecules including master transcription factors and cell-cycle regulators. We also discuss the possibility that revealing the epigenetic regulation by the local DNA–RNA–protein assemblies would promote methodological innovations, e.g. neural cell reprogramming, engineering and transplantation, to manipulate neuronal and glial cell fates for the purpose of medical use of these cells.
Keywords: DNA methylation, non-coding RNA, polycomb repressive complex 2, REST, histone methylation, histone acetylation
1. Introduction
Mouse neural stem cells (NSCs) can be induced to diverge from embryonic stem cells (ESCs) by the withdrawal of leukaemia inhibitory factor (LIF) [1]. Then, a single NSC can give rise to neuron, astrocyte or oligodendrocyte through balancing between symmetric and asymmetric cell divisions according to a given niche. For example, LIF can strongly stimulate NSCs to take the differentiation route to become astrocytes through activation of the janus kinase (JAK)/signal transducer and activator of transcription (STAT) signalling pathways in vitro [2]. Such a differentiation process can be reversed by the forced expression of defined factors, so-called ‘master regulators’, as exemplified by OCT4, SOX2, c-MYC and KLF4 in the technology of the efficient propagation of induced pluripotent stem cells (iPSCs), which are functionally comparable to ESCs [3]. It should be noted that, not only for iPSC/ESC generation but also for that of the NSC and its derivatives, a set of master regulators may influence the dynamic adaptation of core gene networks, by which cell-state-specific epigenome status is statically set along with gene-locus-level regulation (figure 1). However, considering that genes constituting core networks for the stabilization of a cell fate are different and sometimes very different from those functioning in the physiological output characteristic of a given fate, recapitulation of the cell status with the expression of master regulators is still an immature science and we must be prudent about using such reprogrammed cells, especially for therapeutic purposes. Meanwhile, the major effects of the core networks on their downstream gene expression through epigenetic mechanisms are now being analysed by many researchers, and non-coding RNAs (ncRNAs) are emerging as epigenetic players in embryogenesis and in developmental processes [4]. So far, most efforts aiming to understand ncRNA functions in pluripotency and neural differentiation have focused on the mouse as a model system [4–8]. Recent studies of human and mouse ESCs and iPSCs indicate that long ncRNAs (lncRNAs) are integral members of the ESC self-renewal regulatory circuit [7,8]. Here, we focus on the in vivo and in vitro epigenomic settings of the neural cells that are derived from the mouse cerebral cortex and those from human cell systems and discuss the associated information important for reconstituting the pattern of the epigenome that is usually specific to each neural cell.
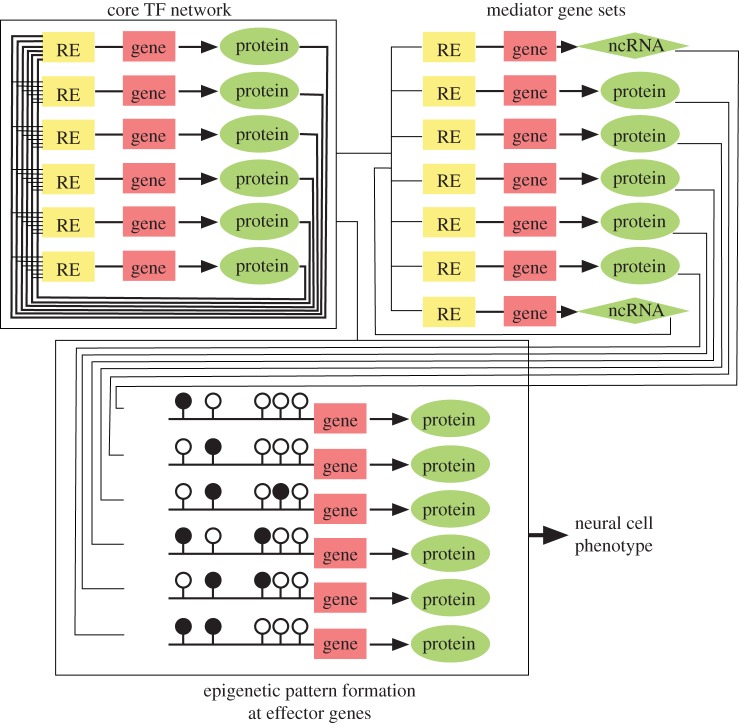
Figure 1.
Core networks and their predominant effects on effector genes in neural cells. Open and filled lollipops denote unmethylated and methylated CpG sites, respectively. In the central nervous system, TFs such as SOX2, NEUROG1 and ASCL1 direct formation of the robust network of neural cells. The TF network controls the expression of mediator and effector gene sets, thereby establishing the neural cell functions. Note that fluctuations in the core gene network can be amplified through these pathways, resulting in the generation of epigenetic variations such as those frequently seen after TF-based reprogramming.
2. Epigenetic overview of the neural cells constituting mouse cerebral cortex
Mammalian NSCs divide repeatedly in the ventricular zone (VZ) of the embryonic brain. After birth, NSCs are located in restricted areas such as the early postnatal and adult subventricular zones (SVZs) of the forebrain and subgranular zone (SGZ) of the hippocampal dentate gyrus. NSCs exhibit two defining characteristics: the capacities for self-renewal and for generating specialized cell types, i.e. neurons, astrocytes and oligodendrocytes. These capacities are controlled spatio-temporally to fully organize the morphology and function of the brain. For example, from embryonic day 11 (E11) to E18, NSCs preferentially produce neurons in the mouse developing brain. NSCs gradually acquire the capacity to generate astrocytes [9]. The majority of oligodendrocytes are generated after birth in the mouse cerebral cortex. These sequential steps enable the initial establishment of neuronal networks followed by integration of glial cells that support the functioning of the neuronal networks.
Extracellular signals can trigger the proliferation and differentiation of NSCs according to the variable levels of epigenetic modifiers. For example, in E8–E10 NSCs, histone H3 lysine 27 (H3K27) methyltransferase EZH2 is highly expressed and prevents Wnt-signal-mediated β-catenin action on neuronal genes and thus blocks neuronal differentiation. After E11, a decreasing level of EZH2 expression allows stabilized β-catenin to act in the nucleus, which causes neuronal differentiation of NSCs through upregulation of the proneural transcription factor (TF) neurogenin1 gene (NEUROG1) [10].
Many studies have shown that cytokine signalling pathways contribute to the regulation of astrocytic differentiation from NSCs. The interleukin-6 family members, including ciliary neurotrophic factor, cardiotrophin-1 and LIF, activate the JAK-STAT pathway through their interaction with a heterodimeric receptor complex of LIF receptor β and glycoprotein 130 (gp130), triggering the differentiation of NSCs into astrocytes [2]. Bone morphogenetic proteins (BMPs) also activate the expression of astrocytic genes via formation of a complex between BMP-downstream TF SMAD1 and STAT bridged by a transcriptional coactivator, cAMP response element-binding protein (CREB)-binding protein/p300 (CBP/p300), which has acetyltransferase activity [2]. In support of the notion of the common usage of JAK-STAT signalling for triggering the astrocytic fate determination of NSCs, knockout of either gp130 or Stat3 impairs astrocyte differentiation [11]. Acquisition of astrocyte differentiation potential of NSCs seems to be accomplished by cell-intrinsic DNA demethylation at astrocytic gene promoters, which is supported by the fact that NSCs exhibit extreme neurogenic characters before this DNA demethylation occurs both in vivo and in vitro [12,13]. Therefore, neuronal and astrocytic cell fate as well are clearly regulated by the niche and epigenetic mechanisms.
Although NSCs are characterized by their multipotency to become not only neurons and astrocytes but also oligodendrocytes, we do not yet know whether all NSCs can function as ancestors of oligodendrocyte precursors (OPCs) in the early postnatal cerebral cortex. OPC markers were first detected at E9 in the ventricular germinal layer of the laterobasal plate of the diencephalon during mouse brain development. By E14, OPC marker-positive cells had largely disappeared from the original location and had colonized at the ventral mantle layer in the posterior part of the basal diencephalon [14]. In parallel, the activation of sonic hedgehog signalling promotes derivation of OPCs from NSCs after birth [15]. An appropriate level of histone acetylation is important for maintaining the NSC capacity to commit to OPCs [16,17]. Histone deacetylase (HDAC) inhibition in OPCs by treatment with pharmacological inhibitors caused these committed progenitors to revert to multipotent cells characterized by expression of an important TF in NSCs, SRY-box containing gene 2 (SOX2) [17]. After cells are committed to OPCs, mitogen withdrawal can induce oligodendrocytic differentiation and myelination in vitro [18]. It should be noted that, during oligodendrocytic differentiation of OPCs, histone acetylation levels are globally decreased [18], suggesting that histone deacetylation is important not only for NSC commitment to OPCs but also for OPC differentiation into mature oligodendrocytes. HDAC1 and 2 contribute to this genome-wide histone deacetylation and compete with the Wnt signalling pathway at the promoters of the inhibitor of differentiation 2 and 4 genes which inhibit myelin gene expression [17,19]. In parallel, increasing the expression level of HDAC11 results in its recruitment to the myelin basic protein and proteolipid protein genes, both of which are important for oligodendrocyte maturation.
In adult mice, multipotent NSCs exist persistently in restricted brain areas such as the SVZ and SGZ. Maintenance and differentiation of these particular NSCs also seems to be regulated physiologically by epigenetic factors. For example, Methyl-CpG-binding domain protein 1 (MBD1) is relatively highly expressed in adult brain neurons and downregulates the expression of basic fibroblast growth factor 2 (FGF2), which usually functions to expand NSCs. Conversely, NSCs from adult Mbd1 knockout mice showed hypomethylation of the promoter region of Fgf2, leading to its increased expression and a great reduction of neuronal differentiation capacity [20]. DNA methylation inhibitor treatment of NSCs also caused similar effects.
3. Gene-specific effects of epigenetic modifiers with local chromatin regulators
In ESCs, many gene promoters are in a poised state. Subsequently, epigenetic changes that facilitate closed chromatin formation at a global level are usually associated with the stemness of somatic stem cells with restricted potential, including that of NSCs. Conversely, global level epigenetic changes that facilitate open chromatin formation are frequently associated with NSC differentiation to the neuronal cell fate. For example, H3K27 methyltransferase EZH2, H3K9 methyltransferase ESET (also called SETDB1), H3K4 demethylase LSH1 and HDACs (HDAC1 and 2), all of which are involved in the formation of closed chromatin structures, are significantly expressed in NSCs, and their inhibition by gene targeting or pharmacological drugs attenuates the cell-cycle progression. Conversely, decreasing the level of closed chromatin-associated epigenetic modifiers allows neuronal differentiation at the midgestational stage in the mouse [21]. In addition to these general effects of epigenetic alterations, we need to consider the local effect that enables the epigenetic changes on a particular set of genes. For example, treatment of mouse E14 NSCs with valproic acid, an HDAC inhibitor, enhances the LIF-mediated astrocytic differentiation partly via the facilitation of histone acetylation around the STAT3 binding site of the glial fibrillary acidic protein gene promoter [2], suggesting that histone hypoacetylation has a role in preventing NSCs from astrocytic differentiation to produce more neurons. Paradoxically, the global level of histone acetylation was found to be higher in neurons than in astrocytes, when these cells were generated from adult rat hippocampus-derived NSCs in vitro [16]. Therefore, global level changes are most likely different from the changes at the gene level. Regarding the DNA methylation pattern in neurons, differential DNA methylation is overrepresented in CpG island shores and enriched within gene bodies but not in intergenic regions, indicating that DNA methylation is unevenly distributed across a cell's genome. Furthermore, non-CpG methylation is also unevenly distributed and substantially more prevalent in neurons than in non-neuronal cells, reinforcing the idea that DNA methylation machineries act in cell- and gene-dependent manners [22,23]. Currently, one key issue is identification of the molecules that establish/maintain the epigenetic modification in a sequence-specific manner. In this context, ncRNAs are emerging as an additional layer of epigenetic regulation to attain the long-lasting stabilization of cell identity exhibited especially by neurons, which usually exhibit an extremely low capacity for regeneration if damaged. We describe below the recent understandings of ncRNA-mediated mechanisms in neural cells. As little is known about the exact epigenetic mechanisms that are mediated by ncRNAs in neural cells, we also refer to the findings in non-neural cells that can be extrapolated to neural cells.
(a). Long non-coding RNAs
Thousands of ncRNAs have been found, and more than 60% of the genomic DNA contributes to the transcriptome [24]. For a fraction of the transcriptionally competent genomic regions, both DNA strands are used for RNA generation [25]. These facts imply that the total number of functional ncRNAs will not be negligible. Indeed, ncRNAs have been shown to act to regulate gene expression negatively at the post-transcriptional level in animals via processes such as RNA editing, RNA degradation, RNA interference, splicing and translation, by forming RNA duplexes [26]. However, as only 1–2% of the genome provides templates for protein-coding gene expression [27,28], bulk RNA from most of the genome theoretically would not form perfectly matched duplexes with mRNA. Rather, extensive RNA-mediated gene regulation would be possible if double-stranded structures were formed between RNA and DNA [29–32]. In addition to regulation at the post-transcriptional level, double-stranded RNAs (dsRNAs), including siRNA, microRNA and Piwi-interacting RNA, also seem to be essential for chromatin-level regulation, especially for achieving a transcriptionally inert status [32,33]. For example, dsRNA derived from transposon-like inverted repeats stabilizes the heterochromatin structure by inducing histone H3K9 methylation [34,35].
Increasing evidence has shown that single-stranded RNA (ssRNA) functions in a chromatin context. Examples include thousands of large intergenic ncRNAs, named lincRNAs, about 20% of which are physically associated with polycomb repressive complex 2 (PRC2) [36–38]. However, we do not yet know if ssRNA functions only to set up a closed chromatin structure. Previous reports have shown that DNA demethylation is directed by antisense promoter-associated ncRNAs (pancRNAs) in the Sphk1/Khps1 locus [39,40]. Therefore, additional information about the functional properties of ssRNAs will be necessary to understand how such RNAs direct gene activation as well as gene repression. As many more lncRNAs have been found thus far, lincRNAs have been now integrated into a category of long ssRNA, i.e. lncRNAs, which is now widely accepted as a generic category.
Ng et al. [41] screened the expression of lncRNAs before and after induction of human ESC differentiation into dopaminergic neurons and found that lncRNA_N1, _N2 and _N3 were weakly but significantly expressed after induction of neuronal differentiation. lncRNA_N1 and lncRNA_N3 might function as scaffolds for the interaction with transcription repressors, repressor element 1 (RE1; also called NRSE) silencing TF (REST; also called NRSF) and a PRC2 protein, suppressor of zeste 12 homologue (SUZ12), respectively, resulting in inhibition of their suppressor activity [41]. On the other hand, small modulatory dsRNAs encoding the RE1 sequence were found in the nucleus, where they interacted with the REST complex to promote transcription of RE1-associated genes [42]. These observations of the modulation of REST activity by ncRNA highlight the intricate relationships that link REST function with the expression of small dsRNA and lncRNA in the nervous system.
As noted above, a growing body of evidence suggests that lncRNA is frequently associated with upregulation of the associated genes. Rhabdomyosarcoma 2-associated transcript (RMST) is an activation-related RNA component in the SOX2-triggered neural regulatory network. In human NSCs, where RMST is not expressed, SOX2 binds to and activates its target genes in order to maintain NSC identity. During neuronal differentiation, the downregulation of REST leads to increased expression of RMST, which binds to SOX2 as well as chromatin with the help of an RNA-binding protein, hnRNPA2/B1. In this way, RMST activates the transcription of neurogenic genes such as ASCL1 and DLX1, which drives the neuronal differentiation pathway [43].
Another example of gene activation-related lncRNA is mouse utNgn1, whose expression seems to be regulated by EZH2. This lncRNA is transcribed upstream of mouse Neurog1 in the E11 cortex. The expression of utNgn1 is highly correlated with that of Neurog1 during neuronal differentiation of NSCs. Knockdown of utNgn1 caused repression of Neurog1. Furthermore, the amounts of utNgn1 and Neurog1 transcription were concomitantly increased upon β-catenin activation and were downregulated by binding of EZH2-containing PRC2 to H3K27 at the genomic utNgn1 region [44]. These results suggest that decreasing EZH2 converts the epigenetic ability of the Neurog1 locus in NSCs towards neuronal differentiation through utNgn1 upregulation. Neurog1 is not the sole target of EZH2, because chromatin immunoprecipitation sequencing analysis showed that, in E14 proliferating NSCs, EZH2 preferentially binds to not only the genomic utNgn1 region but also the promoters of a set of neural differentiation-associated genes, such as a neurogenic differentiation factor Neurod2 and a T-cell leukaemia homeobox protein gene [45].
lncRNAs are also dynamically expressed during neuronal–glial fate specification and appear to regulate the expression of protein-coding genes within the same genomic locus, further suggesting locus-specific functions of lncRNAs [46]. Although little information is available regarding lncRNAs in the regulation of astrocyte differentiation, forced expression of either NKX2.2 or Nkx2.2AS, an antisense ncRNA to Nkx2.2, can enhance induction of differentiation along the oligodendrocytic lineage [5]. NKX2.2 is known to be one of the TFs that direct NSCs into the oligodendrocytic lineage. Further analyses showed that overexpression of Nkx2.2AS induced a modest increase in Nkx2.2 level, suggesting that the upregulation of Nkx2.2 level can be a minor cause of enhanced induction of oligodendrocytic differentiation as a result of increasing Nkx2.2AS expression. A set of lncRNAs is located in a different layer that is still related to a nuclear function. For instance, MALAT1 was reported to function as a nuclear structural component to specify neuronal or glial cell fate and function [47].
A recent study indicated that pancRNAs shows concordant expression with that of the associated mRNAs at about 80% of protein-coding gene promoters [48]. Although many pancRNAs are cleaved and polyadenylated at polyA sites shortly after initiation, such rapid decay of lncRNA may be modulated by depletion of such sites during evolutionary processes. In contrast to a large fraction of pancRNA, mRNA shows biased existence of U1 small nuclear ribonucleoprotein (snRNP) recognition sites, and mRNA–U1 snRNP interaction is probably involved in efficient RNA splicing and maturation followed by RNA stabilization. Nonetheless, a certain set of pancRNAs seem to have gained functionality in neuronal genes. We previously investigated neuronal cytoskeletal genes for microtubule-associated protein 2B (MAP2B) and a neurofilament protein, NEFL [49]. MAP2B is a neuron-specific protein that is relatively abundant in the central nervous system and is localized mainly in dendrites of mature neurons. NEFL is an abundant cytoskeletal component in mature neurons. More than 200-nt polyA+ pancRNAs were endogenously generated from the sense strand at Map2b and antisense strand at Nefl. Forced expression of the fragments expressing the antisense pancRNA caused sequence-specific DNA demethylation, whereas a decrease of the expression induced methylation of the same sequences. By contrast, perturbing the expression of the sense pancRNA did not change the DNA methylation status, indicating that pancRNA for Nefl is functional, but the pancRNA for Map2b is not. Therefore, a fraction of naturally occurring ncRNAs act in cis as a single-stranded form and the transcriptional orientation of pancRNA is important for the establishment of sequence-specific epigenetic modifications consistent with open chromatin structure [49].
Notably, RNA polymerase II (RNAPII) transcribes a novel class of enhancer RNAs (eRNAs) within enhancer domains defined by the presence of H3K4 monomethylation. The level of eRNA expression at neuronal enhancers positively correlates with the level of mRNA synthesis at nearby genes, suggesting that eRNA synthesis occurs specifically at enhancers that are actively engaged in promoting mRNA synthesis [50]. For example, genetic deletion of DLX genes in mice demonstrated their critical role in neuronal differentiation and migration, as well as craniofacial and limb patterning during development. The Evf-2 ncRNA, an alternatively spliced form of Evf-1, is expressed in immature neurons. It is transcribed from the ultraconserved region located between Dlx5 and Dlx6 and functions as an enhancer for production of these transcripts [51]. It has been proposed that Evf-2 ncRNA prevents the inhibitory actions or binding of MSX, which is also known to inhibit activation of the Wnt1 enhancer and allows DLX2 to target the Dlx5/6 enhancer region.
Although we do not yet know the details of the association among eRNA, pancRNA and mRNA, long-range chromatin conformations can facilitate the RNA-based epigenetic setting that locally memorizes the gene expression status, potentially leading to establishment of an ideal situation for the phenotypic output. Figure 2 shows three possible steps triggered by the association between lncRNAs (eRNA and pancRNA) and mRNAs. This model may be of interest to not only neuroscientists but also evolutionary scientists because ncRNA genes can be expanded more easily in the genome and diverged much more during the evolutionary process than protein-coding genes due to the way that ncRNA functions, which is not restricted by the necessity for protein-coding potential. Evolutionary expansion of functional ncRNAs may explain in part why brain structures are different among species, and even among mammals, in spite of the fact that their protein-coding genes are well conserved. Thus, further expansion of the array of known ncRNA functions should definitely help to understand the complexity of neural cell networks.
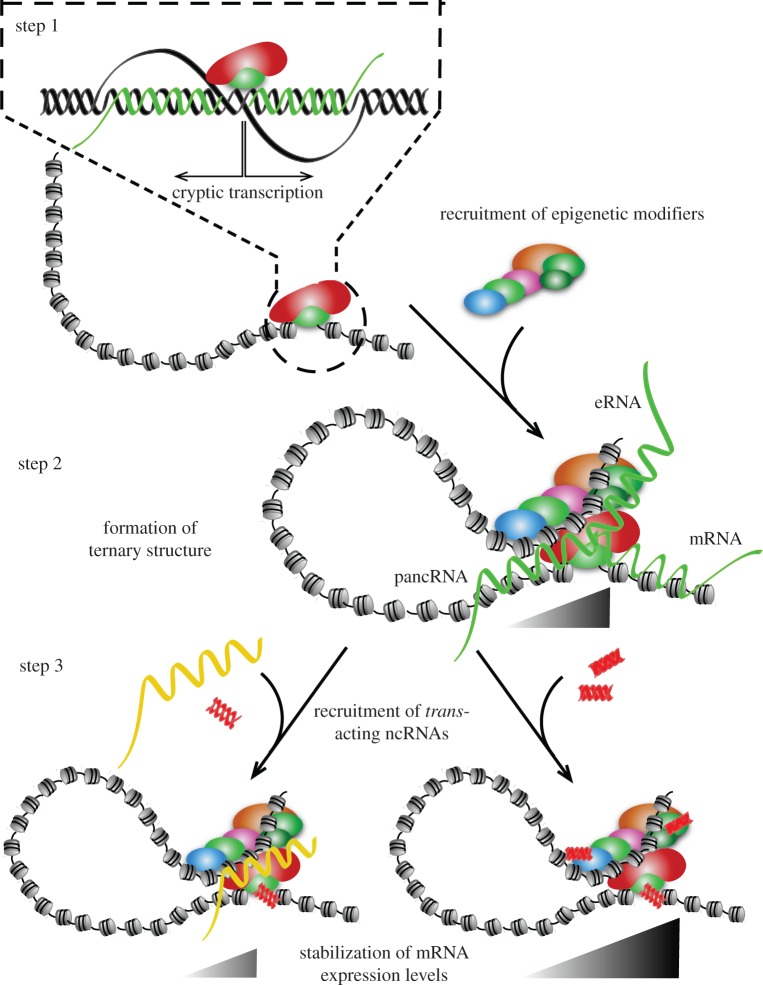
Figure 2.
Possible steps of local chromatin formation triggered by associations with lncRNAs. Step 1: basal transcription of mRNA and cis-acting lncRNA. Cryptic RNA transcription occurs in conjunction with mRNA expression [48]. Many CpG island-bearing genes show bidirectional transcription. It has been reported that G-skew in a CpG island leads to a directionality of the transcription, generating mRNAs that start from G-rich sequences. Associated cryptic transcripts with G-rich structures are involved in cis in a local chromatin set-up that is reminiscent of the transcriptionally competent structure, called the R-loop [52]. The R-loop structure is favoured by H3K4 methylation, but not by DNMT activity, which may explain the unmethylated characteristics of CpG island type promoters. However, it should be noted that R-loop structure constitutes a repressed state in a different context in plants [53]. The mechanisms of switching between opening and closing of chromatin structure via lncRNA might be coupled with association with different components in steps 2 and 3. Step 2: upregulation of epigenetic modifiers to strengthen transcription through ternary structure formation. Transcriptionally competent structure can spread beyond the promoter regions. Such expanded open chromatin status can be recognized by a set of distally located enhancer-associated proteins to form a ternary structure for effective transcription of the target genes. Recent studies have suggested that, for example, SOX2 can be located not only on the target gene promoter regions but also on a fraction of enhancers together with BRN2 [54]. Although we do not yet know how polyA+ eRNA is generated, association of eRNA transcription may be coupled with such a structure involving RNAPII that is originally associated with the promoter sequences. Step 3: stabilization or antagonization of gene activation by trans-acting lncRNA and dsRNA. In addition, if interaction of chromatinic lncRNA with small dsRNA occurs, it would add further complexity to the transcription regulatory dimensions. In fact, association between lncRNAs (eRNA and pancRNA) and mRNAs have been reported to be modulated further by the association of small dsRNAs with the lncRNAs and/or genomic DNA, as described above. In addition, small dsRNA can associate with trans-acting proteins including DNMT and MBD proteins, to allosterically modulate their functions or to mask their catalytic domains, as described in the text.
(b). Chromatin-associated proteins that target local components in the neural cell genome
There is increasing evidence that RNA plays a role in directing DNA methylation-related machineries to specific genomic loci within mammalian cells. It is also possible that DNA methyltransferase (DNMT) and MBD proteins enable RNA molecules to participate in DNA methylation-mediated chromatin control. For example, a subset of the DNMT and MBD proteins can form RNA–protein complexes. The RNA-binding activity of MBD proteins is encoded distinctly from the MBD domain and mediates a high-affinity interaction with RNA [55]. DNMT3A, one of the two de novo DNMTs, is essential for transcriptional regulation during cellular development and differentiation [56]. Two modes of RNA regulation of DNMT3A have been discovered in vitro: ssRNA that is antisense to the E-cadherin promoter binds tightly to the catalytic domain, resulting in the inhibition of DNMT3A activity, whereas two other RNA molecules bind to DNMT3A at an allosteric site outside the catalytic domain [57]. More recently, it has been found that not only DNMT3A but also the maintenance DNMT1 activity is modulated by lncRNAs to set the DNA methylation patterns at specific gene loci. One such novel RNA is produced relatively far from the transcription start site of the CEBPA mRNA and completely encompasses the corresponding protein-coding gene body for regulating its own DNA methylation profile. Indeed, this lncRNA binds to DNMT1 to prevent methylation at the CEBPA locus. Deep sequencing of transcripts associated with DNMT1 combined with genome scale methylation and expression profiling has extended the generality of this phenomenon of maintaining a hypomethylated status at a specific locus via DNMT1–ncRNA interaction to numerous gene loci [58]. DNA methylation-related machineries including ncRNAs also involved RNA-binding proteins to counteract the epigenetic stability. A molecular complex including p68/p72, also known as DEAD-box RNA helicase 5/17, and DNMT1, 3A and 3B are implicated in the rapid DNA demethylation of the TFF1 promoter [59], suggesting the involvement of various RNA-binding proteins in DNA demethylation in genomic context-dependent manners.
A recently discovered form of modified base in DNA, 5′-hydroxymethylcytosine (5hmC), which can be a target of base excision repair, is associated with an active DNA demethylation process. Ten-eleven translocation (TET) 1, 2 and 3 convert 5′-methylcytosine (5mC) to 5hmC in an Fe (II)- and α-ketoglutarate-dependent manner [60]. During embryonic brain development, high levels of 5hmC are detected in the genome. Tet1 or 2 single knockout mice appear to undergo normal embryonic and early postnatal development, although Tet1 and 2 are normally highly expressed in adult brain and in neuronal layers of the embryonic cortex, respectively [60]. Recently, more detailed analysis has revealed that TET1-deficiency impairs self-renewal of NSCs and causes aberrant DNA methylation, resulting in a significant decrease of adult neurogenesis [61]. In the adult mouse dentate gyrus, TET1 was shown to be involved in neuronal activity-induced DNA demethylation of the promoter regions of proliferation-related Fgf1b and of the brain-derived neurotrophic factor gene, followed by their transcriptional upregulation [60]. However, little is known about how TET proteins target these specific gene loci.
In addition to DNA methylation-related machineries, other chromosomal proteins also modulate the epigenetic status in instructive and context-dependent manners by acting in concert with many different components for neural cell development. As described above, REST is an important repressor that limits neuronal differentiation. At its N-terminus, REST recruits mSin3, a scaffold for HDACs 1, 2, 4 and 5. The C-terminal repression domain of REST interacts with CoREST [62], which additionally recruits HDACs (HDAC1 and HDAC2), methyl-CpG-binding protein 2, histone H3K4 lysine demethylase, LSD1, histone H3K9 methyltransferases, G9a and SUV39H1, and a component of the SWI/SNF chromatin remodelling complex, BRG1. In addition, REST also associates with a number of other epigenetic and regulatory cofactors that include DNMTs, MBDs and chromatin remodelling enzymes [63]. In this way, REST acts as an adaptable molecular platform to which these factors may all be recruited and promotes dynamic modifications of DNA, histones, nucleosomes and higher order chromatin codes and helps maintain genomic stability. These locus-specific epigenetic changes promote context-dependent gene repression and long-term gene silencing. Also, REST potentially modulates genes critical for microRNA biogenesis and function, such as Dicer1, Ago1, Ago3, Ago4 and Xpo5, which are all RE1-associated [64]. REST also regulates the expression of certain microRNAs, including the nervous system-specific miR-124, which suppresses hundreds of non-neuronal genes. In non-neuronal cells and in neural progenitors, REST represses miR-124 [65]. However, when progenitors differentiate into mature neurons, REST is downregulated and consequently miR-124 is de-repressed, leading to the degradation of non-neuronal transcripts. Furthermore, REST and potential members of the REST complex are also targets of multiple microRNAs, including miR-124, miR-9 and miR-132 [65]. Many of these microRNA genes also contain cAMP response elements in their regulatory regions, suggesting that the transcriptional regulators CREB and CBP/p300 are also integrated into REST–microRNA regulatory networks that mediate neural gene expression programmes [65].
PRC2 is also a famous platform that uses lncRNAs as chromosomal scaffolds in neurons [66]. Recent studies of HOX genes and X inactivation have provided evidence for RNA cofactors in PRC2. PRC2 is the Ezh2 histone methyltransferase-containing complex required for epigenetic silencing during neural development. A fraction of lncRNAs recruit PRC2 to chromatin, but the general role of RNA in maintaining repressed chromatin is yet to be determined. PRC2-binding affinity for lncRNAs is size dependent, with lower affinity for shorter RNAs. In vivo, PRC2 predominantly occupies repressed genes, and RNA binding leads to maintenance of the repressed state in some cases. Importantly, PRC2 is also associated with active genes, but most of them are not regulated by PRC2. Rather, RNAs may also act as decoys for PRC2 [67]. A genome-wide capture of the PRC2 transcriptome identified a pool of more than 9000 PRC2-interacting RNAs in ESCs [68]. This transcriptome includes antisense, intergenic and promoter-associated transcripts, as well as many unannotated RNAs. In this case, direct RNA–protein interactions most likely occur via the EZH2 subunit. Although repressed, PRC2 targets are also generally associated with the transcriptional initiation marker H3K4 trimethylation. A class of short RNAs, 50–200 nt in length, are transcribed from the 5′ end of PRC2 target genes in ESCs [69]. Transcription of such short RNA is associated with RNAPII and H3K4 trimethylation and is independent of PRC activity. Although it has not been demonstrated during neural cell development, such short RNAs may play a role in the association of PRC2 to keep the NSC identity by repressing differentiation-associated genes.
Recent studies have indicated the importance of unique epigenetic profiles that keep key developmental genes ‘poised’ in a repressed but activatable state. In this context, in addition to PRC1 and 2, TrxG members are required for neurogenesis in the mouse postnatal brain. Mixed-lineage leukaemia 1 (MLL1) gene-deficient SVZ NSCs survive, proliferate and efficiently differentiate into glial lineages; however, neuronal differentiation is severely impaired. In Mll1-deficient cells, Dlx2, a key downstream regulator gene of neurogenesis in the SVZ, is not expressed. Dlx2 is a direct target of MLL1 in the SVZ, and overexpression of DLX2 can rescue neurogenesis in Mll1-deficient cells. In Mll1-deficient NSCs, chromatin at Dlx2 is bivalently marked by both H3K4 and H3K27 trimethylation, and Dlx2 fails to be properly expressed [70]. The MLL1 system might be a trigger of the coordinated expression of DLX family genes in neurons as described above.
4. Cell-intrinsic epigenetic mechanisms targeted by cell-cycle regulators
Epigenetic status can change dramatically at the point of DNA synthesis during the cell-cycle progression [71,72]. For example, methylated CpGs are passed on to one of the two daughter DNAs, allowing a difference between the original DNA and daughter DNA in terms of DNA methylation status. In this way, the epigenetic memory derived from the original cell type is attenuated through continuous passaging of iPSCs [73], although the epigenetic modification profiles of the iPSCs retain a fraction of the epigenetic marks of the original cell [74]. It is thus possible that cell-cycle progression can induce global epigenetic changes. There are many reports showing that cell-cycle exit is coupled with neural cell differentiation. From the viewpoint of local epigenetic regulation, it is also valuable to highlight the molecules that constitute core networks up- or downregulating the cell cycle for maintaining NSC properties and for acquiring differentiation capacities via epigenetic processes (figure 3).
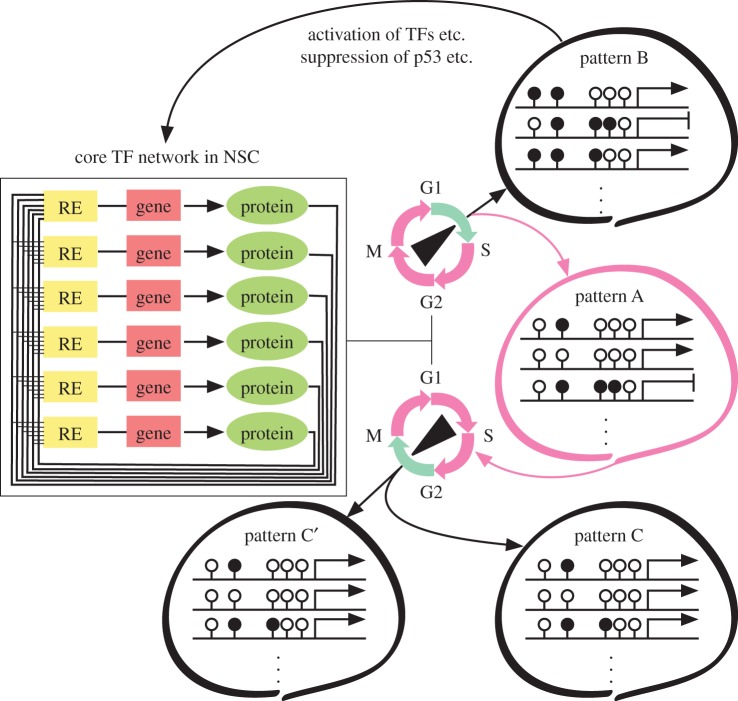
Figure 3.
A model for cell-cycle-dependent epigenetic choice driven by master regulator networks. Pink and blue curved arrows indicate hypothetical cell-cycle phases at which cellular genomes are affected by stable and fluctuating epigenetic modifiers, respectively. Neural differentiation occurs in a restricted time window at the transition from G1 to S phase [75]. If the asymmetric division occurs to produce a differentiating cell (pattern A) and a proliferative cell (pattern B), the respective epigenetic patterns become different from each other. If epigenetic choice occurs after S phase, such cells may produce two daughter cells in which the epigenetic patterns are similar to each other (patterns C and C′).
(a). Association between epigenetic and cell-cycle regulators
To clarify neurological development, a wealth of genetic studies have been performed and established essential roles of the RTK-PI3K-PTEN, ARF-MDM2-p53 and p16/INK4A-RB pathways in gliomagenesis [76]. Inactivation of p53 and PTEN promotes an undifferentiated state with high renewal potential and drives increased MYC protein levels and their associated signatures [77]. These molecular complexes seem to be associated with epigenetic machineries. For example, LSD1 inhibition in embryonic mouse NSCs results in reduced proliferation through upregulation of cell-cycle inhibitors PTEN and p21/WAF1 [78]. PTEN controls NSCs by negatively regulating their cell-cycle progression rather than preventing post-mitotic neurons from re-entering the cell cycle. As HDAC3 and 5 also form repressive complexes with LSD1, it seems more likely that multiple epigenetic mechanisms collaborate on the basis of cell-cycle regulators in normal neural development.
Regarding p53 machineries, the RNA helicase p68 is involved as an established co-activator that itself has a pivotal role in orchestrating the cellular response to DNA damage. Several factors influence the biological outcome of p53 activation. For example, p68 is critical for p53-mediated activation of p21. p68 depletion results in a striking inhibition of recruitment of p53 and RNAPII to the p21 promoter but not to the other promoters related to apoptosis, which highlights a function of p68 as a modulator of the decision between p53-mediated growth arrest and apoptosis in vitro and in vivo [79]. As described above, p68 is known to be associated with DNMTs. Therefore, not only PTEN but also p53 provides an interface that is accessed by various epigenetic modifiers. In vitro experiments as models of neuronal maturation, and in vivo analyses of axonal injury and regeneration suggest that atypical p53-dependent cellular functions could depend on specific patterns of p53 modification such as acetylation in its C-terminus [80]. These modifications directly affect the transcriptional activity of p53 and regulate its affinity for diverse cofactors, which in turn regulate the occupancy of p53 in specific promoters [81]. In adult NSCs in the SVZ of p53-deficient mice, loss of p53 was not sufficient for tumour formation, but led to increased cell proliferation and altered differentiation under physiological conditions [82]. p53 was not essential to promote cell death in any of these studies, which also supports the idea that p53 is required to maintain the physiological proliferation rate in NSCs.
A p53–CBP/p300 transcription module seems to be required for axon outgrowth and regeneration. It is possible that p53 and CBP/p300 function as scaffolds for epigenetic settings. Studies of neuronal precursor-like PC12 and neuroblastoma cells have shown that p53 gene expression is induced and required during neuronal differentiation and maturation [83]. p53 was also reported to bind to the nerve growth factor (NGF) receptor trkA, which is known to induce PC12 neuronal differentiation and to activate trkA expression [84]. p53 associates with histone acetylases CBP/p300, PCAF and hGCN5 at three distinct regions, respectively. CBP/p300 acetylates Lys 370, 372, 373 and 382 of p53. On the other hand, nuclear translocation of PCAF and hGCN5 upon the phosphorylation of Ser and Thr residues within their histone acetyltransferase domains is rapidly induced by NGF, and increases Lys 320 acetylation of p53, leading to the activation of the p21 promoter, which triggers G1 arrest and promotes neuronal differentiation in PC12 cells [85]. This observation clearly shows the presence of crosstalk between the phosphorylation and acetylation axes. Moreover, Wnt7b was found to be one of the new putative p53 target genes during NGF-mediated PC12 neuronal differentiation [86]. Studies in neuronal cells have suggested that the interaction of p53 with the neuron-specific and pro-differentiation TF BRN3A facilitates a shift of p53 transcriptional activity from cell death to neuronal differentiation [87]. As there are approximately 12 000 neuronal activity-regulated enhancers that are bound by the general transcriptional co-activator CBP/p300 in a neuronal activity-dependent manner, the function of CBP/p300 at enhancers may be to recruit RNAPII together with p53, as activity-regulated RNAPII binding to thousands of enhancers has been observed [50]. These facts taken together reinforce the idea that ncRNA is involved in the formation of ternary structures between enhancer and promoter sequences as described in figure 2.
Although it has not been found in neural cells, there is an inverse relation between p15/INK4B antisense (p15AS) and p15 sense expression in leukaemia. A p15AS expression construct induced p15 silencing in cis and in trans through heterochromatin formation but not DNA methylation, and the silencing persisted after p15AS expression was turned off. The p15AS-induced silencing was DICER-independent. Expression of exogenous p15AS in mouse ESCs caused p15 silencing and increased growth through closed chromatin formation as well as DNA methylation after differentiation of the ESCs [88]. Similarly, at steady state, endogenous p21 transcripts consist of comparable levels of both sense and antisense transcripts in human MCF7 [89]. When a reduction in p21 antisense transcription occurs, there is a loss of the low-level antisense-directed H3K27 trimethylation at the p21 sense promoter and an increase in p21 sense/mRNA expression. Conversely, a decrease in p21 sense/mRNA expression results in p21AS-mediated AGO1 recruitment to the p21 sense/mRNA promoter, followed shortly thereafter by an enrichment of H3K27 trimethylation, similar to the observed mechanism whereby siRNAs direct transcriptional gene silencing.
NSCs expand their population during mid-to-late embryogenesis by dividing symmetrically, while also increasing their cell-cycle length [90], raising the possibility that the timing of the decision for neural differentiation depends not only on cell-intrinsic epigenetic mechanisms but also on the phase of the cell cycle. Recently, it has been shown that human ESCs in early G1 phase can only initiate differentiation into endoderm, whereas the ESCs in late G1 are restricted to neuroectoderm differentiation [75]. This is supported by cyclin D overexpression experiments showing that, in ESCs, neuroectoderm differentiation is specifically induced. Functional experiments reveal that the activity of Activin/Nodal signalling during cell-cycle progression is controlled by cyclin D proteins that activate CDK4/6 and lead to the phosphorylation of SMAD2 and SMAD3 in their linker region. This mechanism blocks SMAD2/3 translocation into the nucleus in late G1, thereby preventing endoderm specification and allowing neuroectoderm specification. Together with epigenetic regulation, intrinsic mechanisms for cell-cycle progression can thus be used for the stochastic neural cell fate choices directed by extracellular differentiation signals according to the timing of their signal input coming from the niche.
(b). Towards reprogramming to or from neurons
Mature neurons, which have exited from the cell cycle, have low efficiency of reprogramming to iPSCs by simultaneous expression of OCT4, SOX2, C-MYC and KLF4 [91]. In this context, it is interesting to note that p53 suppression is crucial for efficient reprogramming of neurons to iPSCs. Although neurons and glial cells are generated from common NSCs, it is much easier to reprogramme glial cells and NSCs into iPSCs than to similarly reprogramme neurons [92,93]. The efficiency of transdifferentiation, like that of reprogramming to iPSCs, also depends on the cell type. These facts would lead to the expectation that neurons may exhibit lower potency of transdifferentiation into different lineages compared with the transdifferentiation potency of glial cells, because neurons are post-mitotic cells.
Transdifferentiation is the conversion of a differentiated cell to alternative lineage(s), such as conversion from fibroblasts to neurons [94–97]. Regarding glial cells, we have previously shown that activation of the JAK/STAT signalling pathway can induce transdifferentiation from oligodendrocytes into astrocytes [98]. Pericytes, a cell type implicated in the establishment and maintenance of the blood–brain barrier, isolated from human cerebral cortex were also reported to be transdifferentiated into neurons by the ectopic coexpression of SOX2 and ASCL1 [99]. In addition, ectopic expression of PAX6 or NEUROG2 in astrocytes could induce their conversion to immature neurons in mice [100]. Moreover, the plasticity of astrocytes was shown by their transdifferentiation into fully differentiated glutamatergic and GABAergic neurons by ectopic expression of NEUROG2 or DLX2, respectively [100]. In contrast to glial cell transdifferentiation, there are few reports showing the transdifferentiation of neurons into different cell lineages. However, a recent study showed that overexpression of the FEZ family zinc finger 2 gene could induce the direct conversion of post-mitotic callosal neurons into corticofugal neurons [101]. Nonetheless, there are no reports in which neurons have been converted to glial cells. Collectively, these facts support the notion that the robust epigenetic profile of neurons may underlie their extremely low capacity for cellular reprogramming. What molecular mechanisms contribute to the robustness of the differentiated state, or the low differentiation plasticity of neurons? One important clue is the fact that p53 inhibition efficiently reprogrammes neurons to iPSCs as described above [91]. Also in fibroblasts, an increase in the cell proliferation rate by either inhibition of the p53/p21 pathway or overexpression of LIN28 could accelerate conversion to iPSCs [102]. Considering that knockdown of p53 helps re-entry of post-mitotic neurons into the cell cycle during the reprogramming to iPSCs [91], it is conceivable that the cell-cycle-regulating machineries play a critical role in the reprogramming of the epigenetically stable neurons with extremely low conversion capacity to the highly dynamic epigenetic state represented by the poised chromatinic structure in iPSCs.
5. Concluding remarks
Spatio-temporal gain and/or loss of the differentiation capacities of NSCs are defined by both a set of epigenetic modifiers and a given epigenomic profile. In this way, neurons acquire robust or refractory cell identities that underlie the complex neuronal networks. Perhaps as a consequence of these identities, neurons have low ability to be reprogrammed into other lineages. It would be ideal for the therapeutic use of reprogrammed cells if, even in such terminally differentiated cells, the epigenomic profile could be re-written by a set of epigenetic modifiers. Key elements of the sequence-specific alterations of epigenomic status are now becoming known to be components of a molecular axis comprising trans-acting factors, ncRNA and genomic DNA, that constitute cell-specific chromatin. Manipulations of these three types of factors and cell-cycle progression will lead to the ability to generate a particular neural network and even to jailbreak from the neuronal cell lineage, which could then be applied for medical purposes. Especially, clarifying the plasticity and robustness of lncRNA-dependent epigenetic regulations should be further pursued together with elucidating the mechanisms of the brain functions that underlie animal behaviours.
Acknowledgements
We thank Elizabeth Nakajima for proofreading of this manuscript.
Funding statements
This work was supported in part by grants from MEXT (N°24380158 to T.I., N°25123714 to K.N. and the Excellent Graduate Schools Program of Kyoto University). M.U. was supported by a JSPS Research Fellowship for Young Scientists.
References
- 1.Yoshida K, Chambers I, Nichols J, Smith A, Saito M, Yasukawa K, Shoyab M, Taga T, Kishimoto T. 1994. Maintenance of the pluripotential phenotype of embryonic stem cells through direct activation of gp130 signalling pathways. Mech. Dev. 45, 163–171. ( 10.1016/0925-4773(94)90030-2) [DOI] [PubMed] [Google Scholar]
- 2.Namihira M, Nakashima K. 2013. Mechanisms of astrocytogenesis in the mammalian brain. Curr. Opin. Neurobiol. 23, 921–927. ( 10.1016/j.conb.2013.06.002) [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. ( 10.1016/j.cell.2006.07.024) [DOI] [PubMed] [Google Scholar]
- 4.Dinger ME, Pang KC, Mercer TR, Mattick JS. 2008. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 4, e1000176 ( 10.1371/journal.pcbi.1000176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tochitani S, Hayashizaki Y. 2008. Nkx2.2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem. Biophys. Res. Commun. 372, 691–696. ( 10.1016/j.bbrc.2008.05.127) [DOI] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. 2008. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA 105, 716–721. ( 10.1073/pnas.0706729105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. 2010. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 16, 324–337. ( 10.1261/rna.1441510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttman M, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300. ( 10.1038/nature10398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, Temple S. 2000. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron 28, 69–80. ( 10.1016/S0896-6273(00)00086-6) [DOI] [PubMed] [Google Scholar]
- 10.Hirabayashi Y, Gotoh Y. 2010. Epigenetic control of neural precursor cell fate during development. Nat. Rev. Neurosci. 11, 377–388. ( 10.1038/nrn2810) [DOI] [PubMed] [Google Scholar]
- 11.Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. 2004. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat. Cell Biol. 6, 547–554. ( 10.1038/ncb1138) [DOI] [PubMed] [Google Scholar]
- 12.Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. 2001. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell 1, 749–758. ( 10.1016/S1534-5807(01)00101-0) [DOI] [PubMed] [Google Scholar]
- 13.Hatada I, Namihira M, Morita S, Kimura M, Horii T, Nakashima K. 2008. Astrocyte-specific genes are generally demethylated in neural precursor cells prior to astrocytic differentiation. PLoS ONE 3, e3189 ( 10.1371/journal.pone.0003189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Timsit S, Martinez S, Allinquant B, Peyron F, Puelles L, Zalc B. 1995. Oligodendrocytes originate in a restricted zone of the embryonic ventral neural tube defined by DM-20 mRNA expression. J. Neurosci. 15, 1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orentas DM, Hayes JE, Dyer KL, Miller RH. 1999. Sonic hedgehog signaling is required during the appearance of spinal cord oligodendrocyte precursors. Development 126, 2419–2429. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. 2004. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc. Natl Acad. Sci. USA 101, 16 659–16 664. ( 10.1073/pnas.0407643101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Casaccia P. 2010. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 33, 193–201. ( 10.1016/j.tins.2010.01.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. 2002. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J. Neurosci. 22, 10 333–10 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye F, et al. 2009. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin–TCF interaction. Nat. Neurosci. 12, 829–838. ( 10.1038/nn.2333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Barkho BZ, Luo Y, Smrt RD, Santistevan NJ, Liu C, Kuwabara T, Gage FH, Zhao X. 2008. Epigenetic regulation of the stem cell mitogen Fgf-2 by Mbd1 in adult neural stem/progenitor cells. J. Biol. Chem. 283, 27 644–27 652. ( 10.1074/jbc.M804899200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pereira JD, Sansom SN, Smith J, Dobenecker M-W, Tarakhovsky A, Livesey FJ. 2010. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl Acad. Sci. USA 107, 15 957–15 962. ( 10.1073/pnas.1002530107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwamoto K, Bundo M, Ueda J, Oldham MC, Ukai W, Hashimoto E, Saito T, Geschwind DH, Kato T. 2011. Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome Res. 21, 688–696. ( 10.1101/gr.112755.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozlenkov A, et al. 2013. Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic Acids Res. 42, 109–127. ( 10.1093/nar/gkt838) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carninci P, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309, 1559–1563. ( 10.1126/science.1112014) [DOI] [PubMed] [Google Scholar]
- 25.Katayama S, et al. 2005. Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566. ( 10.1126/science.1112009) [DOI] [PubMed] [Google Scholar]
- 26.Wagner RW, Smith JE, Cooperman BS, Nishikura K. 1989. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl Acad. Sci. USA 86, 2647–2651. ( 10.1073/pnas.86.8.2647) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. ( 10.1038/nature05874) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattick JS, Gagen MJ. 2001. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol. Biol. Evol. 18, 1611–1630. ( 10.1093/oxfordjournals.molbev.a003951) [DOI] [PubMed] [Google Scholar]
- 29.Li L-C, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. 2006. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl Acad. Sci. USA 103, 17 337–17 342. ( 10.1073/pnas.0607015103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JT, Lu N. 1999. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99, 47–57. ( 10.1016/S0092-8674(00)80061-6) [DOI] [PubMed] [Google Scholar]
- 31.Sleutels F, Zwart R, Barlow DP. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813. ( 10.1038/415810a) [DOI] [PubMed] [Google Scholar]
- 32.Ting AH, Schuebel KE, Herman JG, Baylin SB. 2005. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat. Genet. 37, 906–910. ( 10.1038/ng1611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SCR, Lin H. 2007. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 21, 2300–2311. ( 10.1101/gad.1564307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837. ( 10.1126/science.1074973) [DOI] [PubMed] [Google Scholar]
- 35.Maison C, Bailly D, Peters AHFM, Quivy J-P, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30, 329–334. ( 10.1038/ng843) [DOI] [PubMed] [Google Scholar]
- 36.Khalil AM, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA 106, 11 667–11 672. ( 10.1073/pnas.0904715106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guttman M, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227. ( 10.1038/nature07672) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. ( 10.1126/science.1192002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imamura T, Yamamoto S, Ohgane J, Hattori N, Tanaka S, Shiota K. 2004. Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem. Biophys. Res. Commun. 322, 593–600. ( 10.1016/j.bbrc.2004.07.159) [DOI] [PubMed] [Google Scholar]
- 40.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322, 1851–1854. ( 10.1126/science.1164096) [DOI] [PubMed] [Google Scholar]
- 41.Ng S-Y, Johnson R, Stanton LW. 2012. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31, 522–533. ( 10.1038/emboj.2011.459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. 2004. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 116, 779–793. ( 10.1016/S0092-8674(04)00248-X) [DOI] [PubMed] [Google Scholar]
- 43.Ng S-Y, Bogu GK, Soh BS, Stanton LW. 2013. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51, 349–359. ( 10.1016/j.molcel.2013.07.017) [DOI] [PubMed] [Google Scholar]
- 44.Onoguchi M, Hirabayashi Y, Koseki H, Gotoh Y. 2012. A noncoding RNA regulates the neurogenin1 gene locus during mouse neocortical development. Proc. Natl Acad. Sci. USA 109, 16 939–16 944. ( 10.1073/pnas.1202956109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sher F, Boddeke E, Olah M, Copray S. 2012. Dynamic changes in Ezh2 gene occupancy underlie its involvement in neural stem cell self-renewal and differentiation towards oligodendrocytes. PLoS ONE 7, e40399 ( 10.1371/journal.pone.0040399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. 2010. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 11, 14 ( 10.1186/1471-2202-11-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernard D, et al. 2010. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082–3093. ( 10.1038/emboj.2010.199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. 2013. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499, 360–363. ( 10.1038/nature12349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomikawa J, Shimokawa H, Uesaka M, Yamamoto N, Mori Y, Tsukamura H, Maeda K-I, Imamura T. 2011. Single-stranded noncoding RNAs mediate local epigenetic alterations at gene promoters in rat cell lines. J. Biol. Chem. 286, 34 788–34 799. ( 10.1074/jbc.M111.275750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim T-K, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187. ( 10.1038/nature09033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. 2006. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20, 1470–1484. ( 10.1101/gad.1416106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. 2012. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 45, 814–825. ( 10.1016/j.molcel.2012.01.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. 2013. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340, 619–621. ( 10.1126/science.1234848) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lodato MA, Ng CW, Wamstad JA, Cheng AW, Thai KK, Fraenkel E, Jaenisch R, Boyer LA. 2013. SOX2 co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specify cell state. PLoS Genet. 9, e1003288 ( 10.1371/journal.pgen.1003288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeffery L, Nakielny S. 2004. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J. Biol. Chem. 279, 49 479–49 487. ( 10.1074/jbc.M409070200) [DOI] [PubMed] [Google Scholar]
- 56.Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. ( 10.1016/S0092-8674(00)81656-6) [DOI] [PubMed] [Google Scholar]
- 57.Holz-Schietinger C, Reich NO. 2012. RNA modulation of the human DNA methyltransferase 3A. Nucleic Acids Res. 40, 8550–8557. ( 10.1093/nar/gks537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Di Ruscio A, et al. 2013. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 503, 371–376. ( 10.1038/nature12598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Métivier R, et al. 2008. Cyclical DNA methylation of a transcriptionally active promoter. Nature 452, 45–50. ( 10.1038/nature06544) [DOI] [PubMed] [Google Scholar]
- 60.Pastor WA, Aravind L, Rao A. 2013. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14, 341–356. ( 10.1038/nrm3589) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang R-R, et al. 2013. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 13, 237–245. ( 10.1016/j.stem.2013.05.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. 1999. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl Acad. Sci. USA 96, 9873–9878. ( 10.1073/pnas.96.17.9873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ding N, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Boyer TG. 2009. MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression. J. Biol. Chem. 284, 2648–2656. ( 10.1074/jbc.M806514200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nishihara S, Tsuda L, Ogura T. 2003. The canonical Wnt pathway directly regulates NRSF/REST expression in chick spinal cord. Biochem. Biophys. Res. Commun. 311, 55–63. ( 10.1016/j.bbrc.2003.09.158) [DOI] [PubMed] [Google Scholar]
- 65.Wu J, Xie X. 2006. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 7, R85 ( 10.1186/gb-2006-7-9-r85) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gupta RA, et al. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076. ( 10.1038/nature08975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davidovich C, Zheng L, Goodrich KJ, Cech TR. 2013. Promiscuous RNA binding by polycomb repressive complex 2. Nat. Struct. Mol. Biol. 20, 1250–1257. ( 10.1038/nsmb.2679) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao J, et al. 2010. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 40, 939–953. ( 10.1016/j.molcel.2010.12.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanhere A, et al. 2010. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell 38, 675–688. ( 10.1016/j.molcel.2010.03.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lim DA, Huang Y-C, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. 2009. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458, 529–533. ( 10.1038/nature07726) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petruk S, et al. 2012. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell 150, 922–933. ( 10.1016/j.cell.2012.06.046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alabert C, Groth A. 2012. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 13, 153–167. ( 10.1038/nrm3288) [DOI] [PubMed] [Google Scholar]
- 73.Polo JM, et al. 2010. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 28, 848–855. ( 10.1038/nbt.1667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim K, et al. 2010. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290. ( 10.1038/nature09342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pauklin S, Vallier L. 2013. The cell-cycle state of stem cells determines cell fate propensity. Cell 155, 135–147. ( 10.1016/j.cell.2013.08.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wiedemeyer R, et al. 2008. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell 13, 355–364. ( 10.1016/j.ccr.2008.02.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng H, et al. 2008. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 455, 1129–1133. ( 10.1038/nature07443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun G, Alzayady K, Stewart R, Ye P, Yang S, Li W, Shi Y. 2010. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol. Cell. Biol. 30, 1997–2005. ( 10.1128/MCB.01116-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nicol SM, Bray SE, Black HD, Lorimore SA, Wright EG, Lane DP, Meek DW, Coates PJ, Fuller-Pace FV. 2013. The RNA helicase p68 (DDX5) is selectively required for the induction of p53-dependent p21 expression and cell-cycle arrest after DNA damage. Oncogene 32, 3461–3469. ( 10.1038/onc.2012.426) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tedeschi A, Di Giovanni S. 2009. The non-apoptotic role of p53 in neuronal biology: enlightening the dark side of the moon. EMBO Rep. 10, 576–583. ( 10.1038/embor.2009.89) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sims RJ, Reinberg D. 2008. Is there a code embedded in proteins that is based on post-translational modifications? Nat. Rev. Mol. Cell Biol. 9, 815–820. ( 10.1038/nrm2502) [DOI] [PubMed] [Google Scholar]
- 82.Gil-Perotin S, Marin-Husstege M, Li J, Soriano-Navarro M, Zindy F, Roussel MF, Garcia-Verdugo JM, Casaccia-Bonnefil P. 2006. Loss of p53 induces changes in the behavior of subventricular zone cells: implication for the genesis of glial tumors. J. Neurosci. 26, 1107–1116. ( 10.1523/JNEUROSCI.3970-05.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hughes AL, Gollapudi L, Sladek TL, Neet KE. 2000. Mediation of nerve growth factor-driven cell cycle arrest in PC12 cells by p53. Simultaneous differentiation and proliferation subsequent to p53 functional inactivation. J. Biol. Chem. 275, 37 829–37 837. ( 10.1074/jbc.M003146200) [DOI] [PubMed] [Google Scholar]
- 84.Browes C, Rowe J, Brown A, Montano X. 2001. Analysis of trk A and p53 association. FEBS Lett. 497, 20–25. ( 10.1016/S0014-5793(01)02429-2) [DOI] [PubMed] [Google Scholar]
- 85.Wong K, et al. 2004. Nerve growth factor receptor signaling induces histone acetyltransferase domain-dependent nuclear translocation of p300/CREB-binding protein-associated factor and hGCN5 acetyltransferases. J. Biol. Chem. 279, 55 667–55 674. ( 10.1074/jbc.M408174200) [DOI] [PubMed] [Google Scholar]
- 86.Brynczka C, Labhart P, Merrick BA. 2007. NGF-mediated transcriptional targets of p53 in PC12 neuronal differentiation. BMC Genomics 8, 139 ( 10.1186/1471-2164-8-139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hudson CD, Podesta J, Henderson D, Latchman DS, Budhram-Mahadeo V. 2004. Coexpression of Brn-3a POU protein with p53 in a population of neuronal progenitor cells is associated with differentiation and protection against apoptosis. J. Neurosci. Res. 78, 803–814. ( 10.1002/jnr.20299) [DOI] [PubMed] [Google Scholar]
- 88.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. 2008. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451, 202–206. ( 10.1038/nature06468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morris KV, Santoso S, Turner A-M, Pastori C, Hawkins PG. 2008. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 4, e1000258 ( 10.1371/journal.pgen.1000258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hitoshi S, et al. 2002. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 16, 846–858. ( 10.1101/gad.975202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim J, et al. 2011. Reprogramming of postnatal neurons into induced pluripotent stem cells by defined factors. Stem Cells 29, 992–1000. ( 10.1002/stem.641) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ruiz S, Brennand K, Panopoulos AD, Herrerías A, Gage FH, Izpisua Belmonte JC. 2010. High-efficient generation of induced pluripotent stem cells from human astrocytes. PLoS ONE 5, e15526 ( 10.1371/journal.pone.0015526) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim JB, et al. 2009. Oct4-induced pluripotency in adult neural stem cells. Cell 136, 411–419. ( 10.1016/j.cell.2009.01.023) [DOI] [PubMed] [Google Scholar]
- 94.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. 2010. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041. ( 10.1038/nature08797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guo Z, Zhang L, Wu Z, Chen Y, Wang F, Chen G. 2013. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer's disease model. Cell Stem Cell 14, 188–202. ( 10.1016/j.stem.2013.12.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Niu W, Zang T, Zou Y, Fang S, Smith DK, Bachoo R, Zhang C-L. 2013. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat. Cell Biol. 15, 1164–1175. ( 10.1038/ncb2843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Torper O, Pfisterer U, Wolf DA, Pereira M, Lau S, Jakobsson J, Björklund A, Grealish S, Parmar M. 2013. Generation of induced neurons via direct conversion in vivo. Proc. Natl Acad. Sci. USA 110, 7038–7043. ( 10.1073/pnas.1303829110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kohyama J, Sanosaka T, Tokunaga A, Takatsuka E, Tsujimura K, Okano H, Nakashima K. 2010. BMP-induced REST regulates the establishment and maintenance of astrocytic identity. J. Cell Biol. 189, 159–170. ( 10.1083/jcb.200908048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karow M, et al. 2012. Reprogramming of pericyte-derived cells of the adult human brain into induced neuronal cells. Cell Stem Cell 11, 471–476. ( 10.1016/j.stem.2012.07.007) [DOI] [PubMed] [Google Scholar]
- 100.Rouaux C, Bhai S, Arlotta P. 2012. Programming and reprogramming neuronal subtypes in the central nervous system. Dev. Neurobiol. 72, 1085–1098. ( 10.1002/dneu.22018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rouaux C, Arlotta P. 2013. Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nat. Cell Biol. 15, 214–221. ( 10.1038/ncb2660) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. 2009. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462, 595–601. ( 10.1038/nature08592) [DOI] [PMC free article] [PubMed] [Google Scholar]