Abstract
Variations in maternal care in the rat influence the epigenetic state and transcriptional activity of glucocorticoid receptor (GR) gene in the hippocampus. The mechanisms underlying this maternal effect remained to be defined, including the nature of the relevant maternally regulated intracellular signalling pathways. We show here that increased maternal licking/grooming (LG), which stably enhances hippocampal GR expression, paradoxically increases hippocampal expression of the methyl-CpG binding domain protein-2 (MBD2) and MBD2 binding to the exon 17 GR promoter. Knockdown experiments of MBD2 in hippocampal primary cell culture show that MBD2 is required for activation of exon 17 GR promoter. Ectopic co-expression of nerve growth factor-inducible protein A (NGFI-A) with MBD2 in HEK 293 cells with site-directed mutagenesis of the NGFI-A response element within the methylated exon 17 GR promoter supports the hypothesis that MBD2 collaborates with NGFI-A in binding and activation of this promoter. These data suggest a possible mechanism linking signalling pathways, which are activated by behavioural stimuli and activation of target genes.
Keywords: maternal care, glucocorticoid receptor, methyl-CpG binding domain protein-2, serotonin, NGFI-A, hippocampal neurons
1. Introduction
Environmental conditions prevailing during perinatal development shape individual differences in defensive responses. Thus, in primates and rodents, there are maternal effects on the hypothalamic–pituitary–adrenal (HPA) response to stress [1–4] that are associated with stable differences in gene expression. In the rat, the adult offspring of mothers that exhibit increased levels of pup licking/grooming (i.e. high LG mothers) over the first week of life show increased hippocampal glucocorticoid receptor (GR) expression, enhanced glucocorticoid feedback sensitivity in comparison with those reared by low LG mothers and more modest HPA responses to stress [3,5]. Infusion of a GR antagonist directly into the hippocampus eliminates the maternal effect on HPA responses to stress. Cross-fostering studies [5,6] suggest direct effects of maternal care at the level of both gene expression and stress responses. These findings suggest that variations in maternal care ‘programme’ hippocampal GR gene expression, with resulting effects on HPA responses to stress.
An obvious question concerns the signalling pathways by which maternal care might stably alter hippocampal GR gene expression. In vivo and in vitro studies suggest that maternal LG increases GR gene expression in the offspring through a thyroid hormone-dependent increase in 5-hydroxytryptamine (serotonin, 5-HT) activity at 5-HT7 receptors, and the subsequent activation of cyclic adenosine 3′,5′ monophosphate (cAMP) and cAMP-dependent protein kinase A activity, and are accompanied by an increased hippocampal expression of a nerve growth factor-inducible protein A (NGFI-A) transcription factor [7–10]. NGFI-A overexpression increases GR expression at the level of both mRNA and protein in cultured hippocampal neurons [10].
The non-coding exon 1 region of the hippocampal GR includes a promoter region, exon 17, that contains an NGFI-A response element [11] and that is spliced onto the mature GR mRNA upon transcription, although without effect on the protein product. Co-transfection studies reveal that NGFI-A overexpression increases transcription through the exon 17 promoter. Moreover, GR mRNA transcripts bearing the exon 17 promoter sequence are significantly increased in the hippocampus of adult offspring of high compared with low LG mothers [12], suggesting increased in vivo transcription through the exon 17 promoter.
There is increased in vivo occupancy of the NGFI-A response element in the exon 17 promoter in hippocampal tissue from the adult offspring of high compared with low LG mothers [12,13]. This difference occurs despite the absence of any difference in hippocampal NGFI-A expression among the adult offspring as a function of maternal care [12,13]. However, increased maternal LG is associated with demethylation of the 5′ CpG dinucleotide within the NGFI-A response element and increased binding of NGFI-A to the exon 17 GR promoter in the hippocampus of the offspring [12]. The methylation status of the exon 17 region remains stable through adulthood and appears to mediate the differences in GR expression. Methylation of the exon 17 promoter disfavours NGFI-A binding [12,13] and GR transcription. However, the mechanisms that link maternal care and methylation status of the exon 17 promoter in the hippocampus of the offspring are unknown.
Methyl-CpG binding domain proteins (MBDs) mediate the biological effects of DNA methylation [14–16] through a capacity to ‘read’ the DNA methylation signal and attract chromatin modelling complexes that include histone deacetylases and result in transcriptional repression. Thus, both MBD2 and MeCP2 bind methylated genes and silence expression through recruitment of histone deacetylases [14,15]. In the following studies, we examined whether MeCP2 or MBD2 plays a role in epigenetic programming of the exon 17 GR promoter. MeCP2 did not bind to the exon 17 GR promoter in hippocampal tissue samples from the offspring of the high or low LG and therefore does not seem to be involved in the differential expression of GR associated with variations in maternal care. Conversely and surprisingly, MBD2 expression was increased by 5-HT treatment, and MBD2 occupancy of the exon 17 GR promoter was highly enriched in the hippocampus of offspring of high LG mothers: both conditions associate with increased GR expression. The results of subsequent co-transfection studies are consistent with previous reports concerning the potential role of MBD2 as a transcriptional cofactor [17]. Thus, we find that (i) increased MBD2 expression enhances the binding of NGFI-A to the exon 17 promoter and (ii) that a viral-mediated knockdown of MBD2 attenuates the effects of 5-HT on hippocampal GR expression.
2. Material and methods
(a). Animals and maternal behaviour
The animals used in all studies were derived from Long–Evans hooded rats born in our colony from animals originally obtained from Charles River Canada (St. Constant, Québec). All procedures were performed according to guidelines developed by the Canadian Council on Animal Care and protocol approved by the McGill University Animal Care Committee.
Maternal behaviour was scored as previously described [18]. The frequency of maternal LG behaviour was scored on postpartum days 1–4. Observers were trained to a high level of inter-rater reliability (greater than 0.90). Dams were observed in their home cage and undisturbed for the duration of the observation period. Daily observations occurred during five 75 min sessions: three occurring during the light phase (10.00, 13.00 and 17.00 h) and two during the dark phase (07.00 and 20.00 h) of the light cycle. Within each observation session, the behaviour of each mother was scored 25 times (one observation per 3 min) for pup LG (including both body and anogenital licking). Thus, the frequency score of pup LG was expressed as percentage occurrence (number of occurrences per number of observations × 100%). Mothers were designated as high or low LG dams on the basis of the pup LG frequency score relative to the mean ± 1 s.d. for the cohort (generally 60–80 mothers per cohort). High LG mothers were defined as females for which the LG frequency scores were greater than 1 s.d. above the cohort mean. Low LG mothers were defined as females for which the LG frequency scores were greater than 1 s.d. below the cohort mean.
(b). In situ hybridization
Whole brains were removed from day 6 and day 90 male offspring of high and low LG mothers (n = 5 animals per group) by rapid decapitation less than 1 min after their removal from the home cage. Coronal sections (16 μm) were taken corresponding to stereotaxic levels from −2.30 to −3.80 mm from bregma [19]. The rat MBD2 cDNA clone [20] was subcloned in pGem3Z and cut with HindIII to yield a 680 nt cRNA probe. The 35S-labelled cRNA riboprobes were transcribed using a T3 MAXIscript kit (Ambion, Austin, TX).
(c). Western blotting
Hippocampi were dissected immediately after sacrifice, pooled and flash-frozen in 2-methylbutane (Fisher, Pittsburgh, PA) on dry ice and stored at −80°C until processing. For experiments on whole-cell homogenates, roughly 60 mg tissue was homogenized by trituration with a pipette tip in lysis buffer consisting of 20 mM triethanolamine, 0.14 M NaCl and 0.05% each of sodium deoxycholate, sodium dodecyl sulfate and triton-X with pH adjusted to 7.5. For experiments on nuclear and cytosolic fractions, tissue was homogenized in a hypertonic buffer consisting of 10 mM HEPES, 10 mM KCl, 0.1 mM EDTA and 0.1 mM EGTA with pH 7.9 supplemented with a protease inhibitor cocktail (Sigma). After 15 min of incubation on ice, 6.3 μl of 1% NP-40 per 100 μl buffer was added, samples were vortexed briefly and centrifuged at 5000g for 1 min at 4°C. The resultant supernatant, enriched for the cytosolic fraction, was removed to a new tube for analysis by Western blot. The pellet was resuspended in a second buffer consisting of 20 mM HEPES, 0.4 M NaCl, 1 mM EDTA and 1 mM EGTA with pH adjusted to 7.9. The suspension was shaken for 15 min at 4°C and centrifuged at 13 000g for 5 min at 4°C. This supernatant, the nuclear fraction, was also removed to a new tube for further analysis.
Following homogenization, proteins were quantified using the bicinchoninic acid method using a commercially available kit (Pierce, Rockford, IL) in combination with a plate reader and associated software (SpectraMax Plus 384; Molecular Devices, Sunnyvale, CA). Equal amounts of protein (15–30 μg) were combined with sample buffer and dithiothreitol (5–10 mM final concentration) before incubation in boiling water for 3 min. Samples were loaded into pre-cast 4–12% bis–tris gels alongside a molecular weight ladder reactive against secondary antibodies (MagicMark) and run in a Mini-cell apparatus with commercial running buffer at 125 V until migration was complete (approx. 90–100 min; all products from Invitrogen). Proteins were transferred onto Amersham nitrocellulose membranes (GE Healthcare, Buckinghamshire, UK) according to the method of Towbin using a commercial transfer buffer (Invitrogen) in a Mini-cell apparatus for 90 min at 25 V. Non-specific binding was blocked by incubation for 1 h in 5% powdered skim milk in Tris-buffered saline (0.15 M Tris, 0.8% w/v NaCl; pH 7.5) with 0.1% Tween-20 (TBS-T) at room temperature (RT) with agitation. Membranes were rinsed briefly in TBS-T and incubated overnight with primary antibody at 4°C (except tubulin, which required only 1 h at RT).
Primary antibodies were used at empirically optimized concentrations (1 : 250–1 : 2500 dilutions in TBS-T) and included MBD2/3 (Upstate Biotechnology (subsidiary of Millipore, Billerica, MA) 07-199) and tubulin (Sigma T9026). Following three washes of 5 min, membranes were incubated with the appropriate horseradish peroxidase-linked secondary antibody (GE Healthcare) diluted 1 : 2500 in TBS-T. Protein was visualized by first exposing the membrane to enhanced chemiluminescent reagent (ECL; GE Healthcare) and then apposing to an appropriate high-sensitivity film (Hyperfilm-ECL; GE Healthcare) and developing manually using commercial developer and fixer (Kodak, Rochester, NY). Relative optical densities (RODs) of bands were quantified using a digital camera and computer-assisted densitometry software (MCID 4.0; Imaging Research). A ROD measure was also taken from the background and subtracted from the signal. Target bands were normalized to similarly quantified α-tubulin from the same lanes.
(d). Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays [21] were performed using the ChIP assay kit protocol (06-599, Upstate Biotechnology), as previously described [10,12,13,22]. Primary antibodies used at empirically optimized concentrations included CREB-binding protein (CBP; Cell Signalling 4772; Beverly, MA), MBD2/3 (Upstate Biotechnology 07-199), and NGFI-A (Rockland Immunochemicals 600-401-693, Gilbertsville, PA). Animals at postnatal day 4–6 were cryoanaesthetized and perfused with 4% paraformaldehyde to cross-link protein–DNA complexes and then stored at –80°C until dissection. Hippocampi were dissected, and chromatin was specific antibody immunoprecipitated or incubated with normal rabbit IgG non-immune antibody (from Santa Cruz sc-2027). One-tenth of the lysate was kept before immunoprecipitation and used to quantify DNA levels (input). The rat GR exon 17 of the un-cross-linked DNA was subjected to quantitative PCR amplification as shown in §2h (forward primer sequence: CTCCCGAGCGGTTCCAAG; reverse primer sequence: TTTAGTTTCTCTTCTCCCAGGCTCCC). Results are expressed as the amount of DNA detected in immunoprecipitated fraction minus the amount of DNA in negative control normalized to input DNA. For double ChIP experiments, the protein bound to the beads with the first antibody was incubated (30 min, 37°C) twice with DTT (20 mM), and the combined elutes were resuspended in ChIP dilution buffer, which was then immunoprecipitated (14 h, 4°C) with the second antibody.
(e). HEK 293 cell cultures and transient transfections
GR promoter : luciferase reporter vector containing the rat exon 17 GR promoter region (GenBank accession number AJ271870) was transiently cotransfected with a total amount of 1.5 μg of plasmid DNA (1.0 μg exon 17 GR promoter-pGL2 and/or 0.5 μg NGFI-A, MBD2b or MBD2b mutant expression plasmid) using the calcium phosphate method as described previously [13]. The cells were harvested 72 h after transfection, lysed, and luciferase activity was assayed using the luciferase assay system (Promega) according to the manufacturer's protocol. Transfections were repeated a minimum of three times using different cultures of HEK 293 cells.
(f). Hippocampal cell cultures
Hippocampal cell cultures from ED 19–20 Long–Evans rat fetuses were prepared as previously described [10,23,24]. Two days after seeding, uridine (20 mM) and 5-fluorodeoxyuridine (20 mM) were added to the medium to prevent proliferation of glial cells. Cell cultures were maintained at 37°C in a humid atmosphere with 5% CO2. Hippocampi were isolated by gross dissection under sterile conditions. Cells were mechanically dissociated by trituration after incubation for 15 min at 37°C in 0.25% trypsin (Invitrogen) and seeded onto poly-d-lysine coated 60 mm plates at a concentration of roughly 106 ml−1 medium. Medium consisted of minimal essential medium alpha (MEMα; Invitrogen, no. 12561-056) supplemented with 10% v/v heat-inactivated fetal bovine serum (Invitrogen, no. 10082-139), 0.5% w/v glucose, 15 mM HEPES and 20 mM KCl (Sigma-Aldrich). Medium was changed the day after seeding and every 2–3 days thereafter with medium containing a 1 : 1000 dilution of penicillin/streptomycin (Invitrogen) and 20 μM uridine/5-fluorodeoxyuridine to prevent glial proliferation. Previous characterizations of cultures generated with this protocol reveal more than 95% neuronal composition [24].
(g). Lentiviral vectors
Lentiviral vectors were obtained from a variety of commercial and academic sources. For overexpression vector, NGFI-A cDNA was ligated into the pLenti6/V5-Topo vector plasmid (Invitrogen). The resulting expression plasmid contains a CMV promoter-driving expression of NGFI-A, or a random sequence (empty vector, EV). For the siRNA-containing vector, NGFI-A siRNA was acquired commercially (clone ID TRCN00000081623; Open Biosystems, Huntsville, AL) and subcloned into a pLVTHM plasmid (TronoLab) along with a GFP expression tag. Control plasmids contained a random 21mer of cDNA.
Infectious viral particles were generated by transient cotransfection of the expression plasmid (15 µg), envelope plasmid pMD2.G (5 µg; TronoLab), and the packaging plasmids (for overexpression, 10 µg of third-generation packaging plasmids pRSVrev and pMDLg pRRE (TronoLab) were used; for siRNA, 10 µg of second-generation packaging plasmid psPAX2 (TronoLab)) into a 150 mm plate of 90% confluent HEK 293 T cells by calcium phosphate precipitation. Medium was collected 48 and 72 h after transfection, cleared of debris by low-speed centrifugation and filtered through 0.45 µm filters. High-titre stocks were prepared by an initial ultracentrifugation for 1 h at 138 000g. Viral pellet was resuspended in sterile PBS and stored at −80°C. After concentration, typical titres ranged from 107 to 108 TU ml−1. Sufficient amount of virus was added to culture medium to provide multiplicity of infection of at least 20 in all experiments.
Experiments with NGFI-A overexpression and co-immunoprecipitation were performed with viral particles obtained through commercial sources. NGFI-A overexpression vectors were prepared by Systems Bioscience (Mountain View, CA) and contained NGFI-A cDNA coupled with cDNA coding for a haemoagglutinin (HA) protein tag along with a distinct GFP expression tag, separated by a T2A site driven by a CMV promoter. Control viral constructs expressed GFP only. This vector also contained a puromycin resistance cassette. Titres were reported in the range 108–109 TU ml−1.
(h). Quantitative real-time analysis
Samples were analysed at least in triplicate (n = 3 independent samples per group). Primers for MBD2 were as follows: forward sequence: GGAGAAGAGGCTACAAGGACTTAG; reverse sequence: CAAACAGCAGGGTTCTTTTCCAC. Primers for β-2M (forward sequence: CCGTGAT CTTTCTGGTGCTT; reverse sequence: AAGTTGGGCTTCCCATTCTC) were used for normalization. PCR mixtures (20 µl) were loaded into LightCycler capillaries (Roche Molecular Biochemicals). The temperature cycles were as follows: 10 min at 95°C (denatures DNA and activates HotStart polymerase) followed by 45 cycles of 95°C for 20 s (denaturation), 55°C for 15 s (primer annealing) and 72°C for 15 s (cDNA elongation). SYBR green fluoresces in the presence of double-stranded DNA, and the intensity of fluorescence was measured after each elongation step. The resultant amplification curve was analysed to provide a cycle number where the amplification is at a linear maximum. DNA concentration was determined by comparing this cycle number with those from the standard curve. The mean value for each duplicate or triplicate measure was counted as a single independent measure for statistical analysis. Samples from the standard curve were also used to calculate efficiency and error for each reaction; reactions were rejected if the efficiency was more than ±0.2 from the ideal 2.0, and if error values exceeded 0.1.
Specificity of amplification was confirmed by melting peak analysis. The temperature of the PCR products was raised from 65 to 97°C, whereas fluorescence measures were taken at a rate of five for every rise of 1°C in temperature. Specific products denature (‘melt’) at the same temperature and are easily identified. Samples showing non-specific amplification were excluded from analysis.
3. Results
(a). Maternal care alters hippocampal MBD2 expression and association with the exon 17 GR promoter
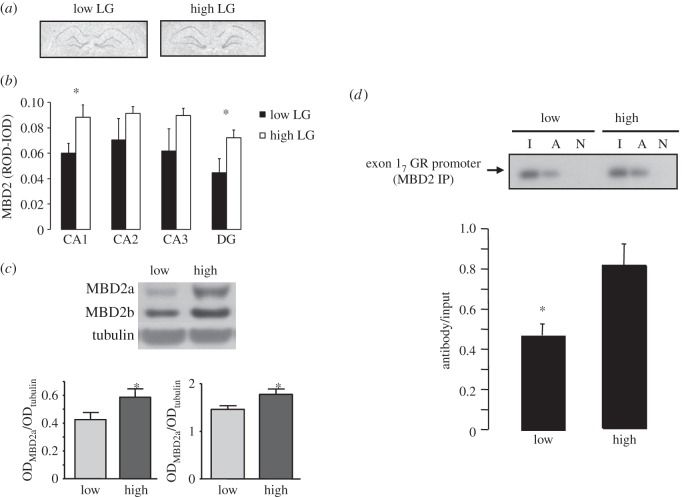
We first examined hippocampal MBD2/3 expression in the neonatal and adult offspring of high and low LG mothers (figure 1a,b). The results revealed a significant (p < 0.05) effect of maternal care on MBD2 mRNA levels in the hippocampus in day 6 animals. Post hoc analysis revealed increased levels of MBD2 mRNA in the CA1 region of Ammon's horn and dentate gyrus of the hippocampus. By contrast, we found no effect of maternal care on hippocampal MBD2 expression in adult animals. Additionally, we found no differences in MBD3 mRNA expression as a function of maternal care at any age (data not shown).
Figure 1.
Maternal effect on hippocampal MBD2 function and exon 17 GR expression. (a) Representative photomicrographs indicating MBD2 expression on hippocampal coronal sections from postnatal day 6 animals (n = 8 animals per group). (b) Relative optical density (ROD; mean ± s.e.m.) of hippocampal MBD2 expression (n = 8 animals per group, *p < 0.01). (c) Western blot analysis of MBD2a and MBD2b from hippocampal nuclear extracts. Data are presented as the mean ± s.e.m. optical densities normalized for tubulin. (d) (top) Representative Southern blots of the amplified exon 17 GR promoter region of MBD2 chromatin immunoprecipitated hippocampal tissue (194 bp band) from postnatal day 6 high and low LG offspring (n = 4 animals per group). Lanes were loaded with non-immunoprecipitated input (I); MBD2 primary antibody immunoprecipitated (A); or non-immune IgG antibody immunoprecipitated (N) hippocampal extracts. (Bottom) Mean ± s.e.m. antibody bound of exon 17 sequence amplified from MBD2-immunoprecipitated hippocampal tissue from high and low LG offspring (n = 4 animals per group) expressed as a function of input and determined using qRT-PCR (*p < 0.05).
We used Western blotting to examine protein levels of MBD2 isoforms in nuclear fractions of postnatal day 4 pups. We found significantly (p < 0.05) higher levels of both MBD2a and MBD2b in hippocampal samples derived from the offspring of high compared with low LG mothers (figure 1c).
We then used ChIP assays to examine MBD2 occupancy of the exon 17 GR promoter (figure 1d). ChIP assays with hippocampal tissue obtained from postnatal day 6 pups showed increased MBD2 binding at the exon 17 GR promoter in the offspring of high compared with low LG mothers. In contrast with MBD2, binding of MECP2 to the exon 17 GR promoter was not observed in the day 6 offspring of high and low LG mothers (data not shown). Binding of MeCP2 to the exon III brain-derived neurotrophic factor (BDNF) promoter had been previously reported [25] and thus served as a control for our MeCP2 ChIP. Interestingly, elevated MeCP2 was detected at the exon III BDNF promoter in low LG offspring (data not shown). This finding is consistent with previous reports of increased hippocampal BDNF expression in the neonatal offspring of high compared with low LG mothers [26]. The finding of both increased MBD2 expression and increased MBD2 occupancy of the exon 17 GR promoter was surprising, because GR expression is increased in the neonatal offspring of high compared with low LG mothers.
(b). Role of MBD2 in 5-HT-induced glucocorticoid receptor expression
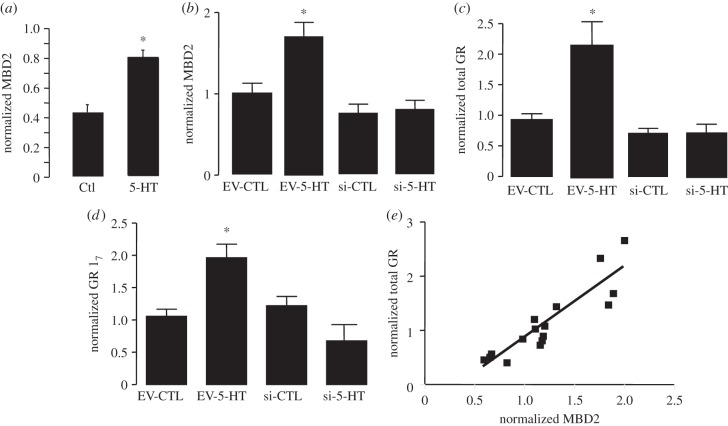
5-HT binds to 5-HT7 receptors to activate NGFI-A expression and enhance GR transcription in hippocampal neurons [7,10,13,24,27,28]. We first examined the effect of 5-HT treatment on hippocampal MBD2 expression using primary hippocampal neuronal cultures. Cultures were incubated with medium alone or medium containing 100 nM 5-HT. This concentration was based on the previous studies examining the effect of 5-HT on neuronal GR expression (see references [7,24]). Cultures were harvested after 4 days of 5-HT treatment and samples prepared for qRT-PCR analysis of MBD2 mRNA levels. The results (figure 2a) revealed a highly significant (p < 0.01) increase in MBD2 expression in 5-HT-treated cultures compared with cultures treated with medium alone.
Figure 2.
Mean ± s.e.m. of target gene expression in primary hippocampal neuron cultures normalized to β2M as determined by qRT-PCR (n = 4–6 independent samples per group). (a) Effect of 100 nM 5-HT or media alone treatment on MBD2 expression. (b–d) Effect of 100 nM 5-HT or media alone ± MBD2 siRNA-bearing or empty lentiviral vector on expression of MBD2 (b), total GR mRNA (c) or exon 17 GR transcripts (d). (e) Scatterplot of the relation between total GR mRNA and MBD2 across all samples used in studies described in panels (b–d). *p < 0.05 by comparison to all other groups.
To examine the potential contribution of MBD2 to the 5-HT-induced induction of GR expression, primary dissociated hippocampal neuronal cultures were transduced 4 days after plating with lentiviral vectors expressing either cDNA coding an siRNA (si) against MBD2 or a non-silencing siRNA-like sequence (EV) and a GFP tag. After 3 days, medium was changed, and cells from both conditions were treated with control medium or medium supplemented with 100 nM 5-HT. Cultures were harvested after 4 days of 5-HT treatment. Statistical analysis revealed a significant (p = 0.02) interaction between the effects of 5-HT and the siRNA vector (figure 2b) such that there was an elevation of MBD2 expression in the 5-HT-treated neurons expressing the non-silencing siRNA sequence that was not present in the siRNA-expressing cells. These results indicate that 5-HT induces a significant increase in MBD2 expression that is reduced in the presence of the siRNA, thus verifying the efficacy of the MBD2 siRNA. Analysis of total GR mRNA expression (figure 2c) in these cells revealed a similar pattern, with a significant (p < 0.01) interaction between the effects of 5-HT and the MBD2 siRNA vector, indicating a significant induction of total GR expression by 5-HT in the cells treated with the EV that was blocked by MBD2 knockdown. Virtually the same pattern of results was obtained for the alternative mRNA leader sequences of exon 17 (figure 2d). Thus, we found a significant (p = 0.03) interaction between the effects of 5-HT and the MBD2 siRNA vector, indicating a significant induction of GR mRNA transcripts bearing the exon 17 sequence in 5-HT-treated cells treated with the empty lentiviral vector and that this effect was not apparent following MBD2 knockdown. Taken together, these findings suggest that 5-HT increases MBD2 expression, which then mediates the effect of 5-HT on GR transcription.
Overall, we found a highly significant correlation between MBD2 expression in hippocampal neurons and levels of GR mRNA (Pearson's r = 0.81; p < 0.01; figure 2e). We then performed an in silico analysis of the relation between MBD2 expression and that of the GR gene using data hippocampal expression analyses with recombinant mouse strains available through the NIH-supported project (http://www.genenetwork.org) [29]. The GeneNetwork database contains data from a microarray analysis of whole brain in which mRNA levels were treated as quantitative traits. Correlational analysis was determined by Pearson's r using the BXD published phenotypes database that includes hippocampal mRNA expression within WebQTL database (http://www.genenetwork.org). The results (figure 3) show a significant Pearson's correlation (r = 0.50; p < 0.0001) across 71 recombinant B×D mouse strains between hippocampal expression of MBD2 and that of the NR3C1 gene that codes for the GR protein.
Figure 3.
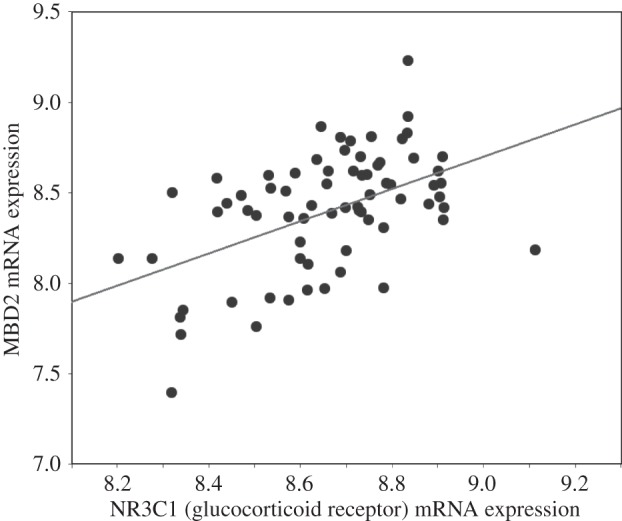
Scatterplot of hippocampal MBD2 (probeset on Chr 18; methyl-CpG binding domain protein 2, first two exons) and NR3C1 (probeset on Chr 18; nuclear receptor subfamily 3, group C, member 1; last three exons and 3′-UTR) mRNA expression in B × D recombinant mouse strains (n = 71).
(c). MBD2 interacts with NGFI-A
NGFI-A directly activates GR transcription through binding to the exon 17 GR gene promoter [7,10,13,24]. To explore the potential contribution of MBD2 to NGFI-A-dependent induction of hippocampal GR expression, we determined whether the two proteins interact in HEK cells. HEK cell cultures transduced with the HA-tagged NGFI-A overexpression construct were harvested in PBS 7 days after transduction. Co-immunoprecipitation (co-IP) was performed with protein G-conjugated magnetic beads linked to anti-HA antibody, and Western blot was performed on the co-IP product for MBD2 protein (figure 4). MBD2 protein was present in the co-IP fraction, but absent from non-transducing cells or IgG-precipitated product. This finding demonstrates that NGFI-A and MBD2 may physically interact as a complex.
Figure 4.
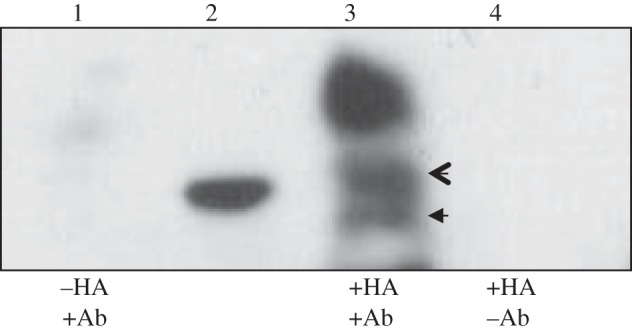
MBD2 binds NGFI-A in hippocampal cultures and co-transfection to HEK 293 cells synergistically drives luciferase expression. Panel (a) shows results from co-immunoprecipitation experiments with primary hippocampal cultures transducing an HA-tagged NGFI-A construct. Samples were immunoprecipitated with an anti-HA antibody, subjected to Western blot, and probed with anti-MBD2/3. Lane 1 is co-IP product from non-transduced cells, lane 2 shows 30 kD mark of ladder, lane 3 shows MBD3 (open arrow) and MBD2b (closed arrow) and an unidentified protein, lane 4 is co-IP product from IgG-precipitated samples.
(d). MBD2 binding to the exon 17 GR promoter is NGFI-A-dependent
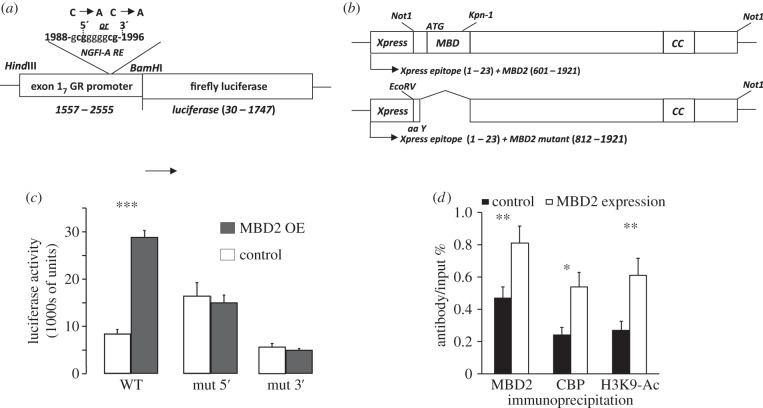
The previous data suggest that MBD2 binds to NGFI-A and participates in activation of exon 17 GR promoter. We used site-directed mutagenesis of the 5′ or 3′ CpG dinucleotides in the NGFI-A response element (figure 5a; also see reference [13]) to examine whether MBD2 overexpression (figure 5b) activates transcription through the exon 17 GR promoter and whether such transactivation by MBD2 requires the presence of NGFI-A response element within this promoter. We [13] previously determined that the 3′ CpG dinucleotide is crucial for NGFI-A binding to the exon 17 GR promoter: conversion of the 3′ CpG sites to ApG dinucleotides eliminates NGFI-A binding. Here, we showed that luciferase activity was significantly (p < 0.0001) increased in cells co-transfected with the non-mutated (wild-type, WT) exon 17 GR promoter in the presence of ectopically expressed MBD2 (figure 5c). These findings reveal that MBD2 overexpression within this context enhances transcription through the exon 17 GR promoter. Conversely, MBD2 did not significantly alter luciferase activity from the mutant 5′ CpG exon 17 GR promoter. Nor did MBD2 activate the mutant 3′ CpG exon 17 GR promoter. These findings suggest that MBD2 induces transcriptional activity from the methylated exon 17 GR promoter region and that this effect requires an intact NGFI-A response element, implying that NGFI-A binding to the promoter is for MBD2 action.
Figure 5.
(a) Physical map of the exon 17 GR promoter–luciferase reporter construct. In vitro mutation of the exon 17 GR promoter was performed using primers designed for cytosine to adenine conversion within the 5′ CpG and 3′ CpG dinucleotide in the NGFI-A response element. (b) Physical map of the Xpress-MBD2 (top) and Xpress-MBD2 mutant (bottom) expression vector. (c) Mean ± s.e.m. of luciferase expression (***p < 0.0001) in HEK 293 cells from the non-mutated and mutated exon 17 GR promoter–luciferase reporter plasmid co-transfected without or with an MBD2 expression plasmid (n = 4 samples per treatment). (d) MBD2, CBP or acetylated histone H3-K9 association with the non-mutated exon 17 GR promoter–luciferase reporter plasmid co-transfected without or with an MBD2 expression plasmid in HEK 293 cells was determined by ChIP analysis (n = 4 samples per treatment). Mean ± s.e.m. ROD from Southern blots of the exon 17 sequence amplified from CBP, acetylated histone H3-K9 or MBD2-immunoprecipitated cell extract normalized to the input values (**p < 0.001; *p < 0.05). For all experiments, the signal from non-immune IgG antibody immunoprecipitated samples was not different from background.
Exon 17 GR promoter activation is accompanied by histone acetylation and binding of a histone acetyltransferase, termed CREB-binding protein (CBP). The results from an accompanying ChIP analysis (figure 5d) shows that MBD2 overexpression significantly (p < 0.05) increased the association of both MBD2 and the CBP with the exon 17 GR promoter. Moreover, we found histone H3-K9 acetylation in cells co-transfected with the MBD2 overexpression vector. These findings further suggest that MBD2 binding induces transcriptional activation of the exon 17 GR promoter region through interaction with the CBP and histone acetylation. It has been previously reported that MBD2 induces gene expression through recruitment of CBP [30].
(e). NGFI-A binding enhances the interaction of MBD2 with the exon 17 GR promoter
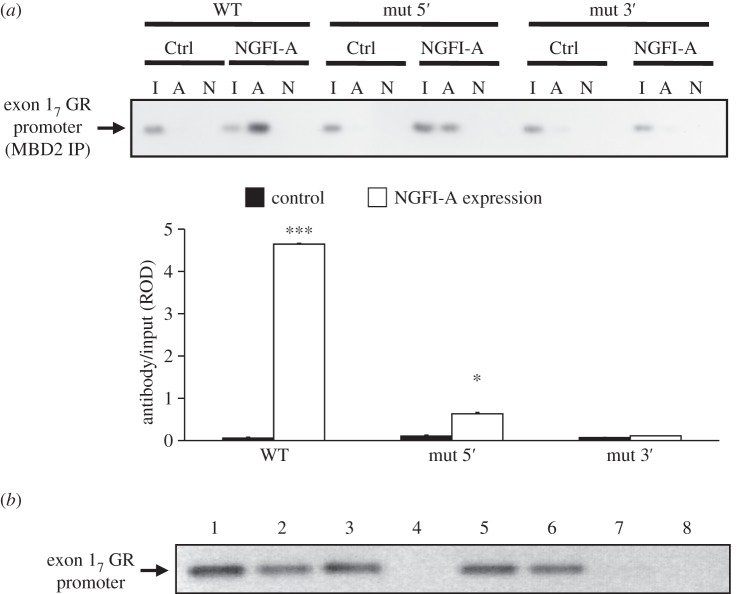
The results of the site-specific mutagenesis of NGFI-A response element are consistent with the hypothesis that MBD2 action requires the interaction of NGFI-A with the promoter. We therefore determined whether ectopic expression of NGFI-A would increase MBD2 association with the exon 17 GR promoter. Mutation of the 5′ CpG dinucleotide within the NGFI-A response element reduces, whereas mutation of the 3′ CpG dinucleotide precludes, NGFI-A binding to the exon 17 GR promoter sequence [13]. Here, we showed that NGFI-A interaction with the exon 17 GR promoter significantly increased MBD2 association in cells co-transfected with WT (p < 0.00001) or, to a lesser extent, the 5′ CpG dinucleotide mutated exon 17 GR promoter sequences (p < 0.05; figure 6a). There was, however, no effect of NGFI-A overexpression on MBD2 binding to the 3′ CpG dinucleotide mutated exon 17 GR promoter, which is consistent with the previous results.
Figure 6.
(a) MBD2 binding of the exon 17 GR promoter–luciferase reporter plasmid containing either a non-mutated (WT), 5′ CpG mutated or 3′ CpG mutated NGFI-A response element, co-transfected into HEK 293 cells without or with an NGFI-A expression plasmid (n = 4 samples per treatment). Mean ± s.e.m. ROD of exon 17 sequences amplified from MBD2-immunoprecipitated cell extract (***p < 0.001; *p < 0.02). (b) Double ChIP assay showing the association of MBD2 and NGFI-A with the same exon 17 GR promoter molecules. Lanes were loaded with: 1, non-immunoprecipitated input; 2, anti-MBD2; 3, anti-NGFI-A; 4, non-immune IgG; 5, anti-MBD2 followed by anti-NGFI-A; 6, anti-NGFI-A followed by anti-MBD2; 7, non-immune IgG followed by non-immune IgG and 8, immunoprecipitated HEK 293 cell extracts. The results are representative of two independent experiments.
We then tested whether MBD2 and NGFI-A co-occupy the same exon 17 GR promoter molecules as predicted by our hypothesis using a double ChIP with both NGFI-A and MBD2 antibodies. Only DNA sequences that bind both proteins concurrently are detected by this assay. The results from this assay (figure 6b) indicate that both MBD2 and NGFI-A co-occupy the exon 17 GR promoter region.
(f). Ectopic MBD2 enhances transcriptional activation of exon 17 glucocorticoid receptor promoter by NGFI-A
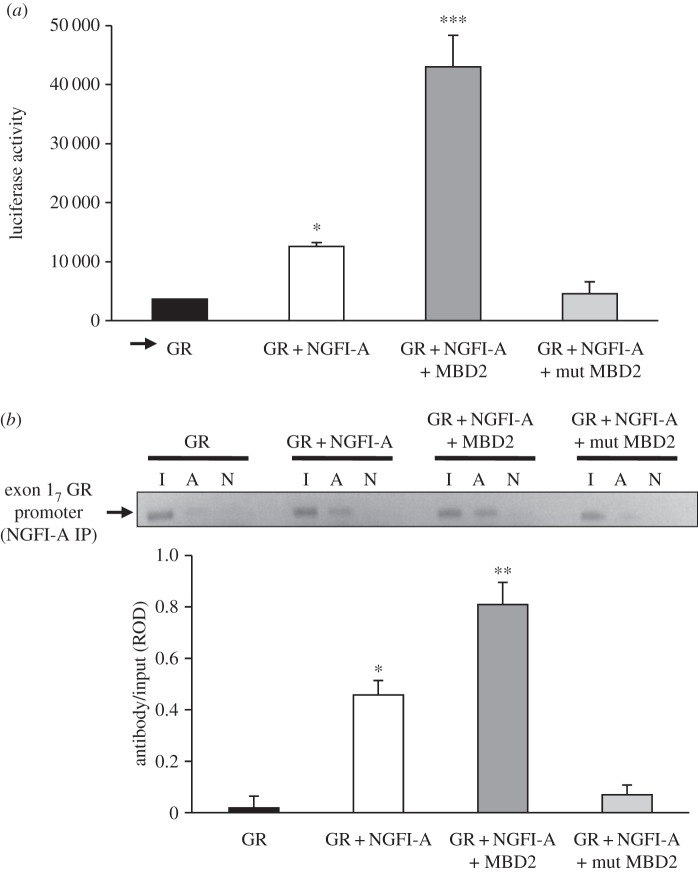
The above-described results are consistent with the hypothesis that NGFI-A facilitates MBD2 interaction with the methylated exon 17 GR promoter. If endogenous MBD2 levels are not saturating, then MBD2 overexpression could further enhance transactivation of the exon 17 GR promoter by NGFI-A overexpression. We first measured the effect of ectopic MBD2 expression on NGFI-A binding to the exon 17 GR promoter. The ChIP analysis (figure 7a) shows that in cells overexpressing NGFI-A, ectopic MBD2 expression enhanced the association of NGFI-A with the GR exon 17 promoter, whereas an MBD2 mutant (see lower panel of figure 5b for construct) lacking the MBD domain seems to have a dominant-negative effect on NGFI-A binding to the promoter (compare the second and last bars).
Figure 7.
Mean ± s.e.m. of (a) luciferase expression (**p < 0.001; ***p < 0.0002) or (b) NGFI-A binding (*p < 0.05; **p < 0.001) to the exon 17 GR promoter–luciferase reporter plasmid cotransfected into HEK 293 cells without or with an NGFI-A alone, NGFI-A with MBD2 or NGFI-A with MBD2 mutant (see lower panel of figure 5b for construct) expression plasmid (n = 4 samples per treatment). (b) Mean ± s.e.m. antibody bound of exon 17 sequence amplified from NGFI-A immunoprecipitated cell extract as a function of input, determined using qRT-PCR, using the same conditions as described in panel (a) (*p < 0.05; **p < 0.001).
We next measured the effect of ectopic MBD2 and NGFI-A expression on exon 17 GR promoter activity (figure 7b). Although NGFI-A overexpression alone enhanced exon 17 promoter activity, luciferase expression was enhanced further in cells co-expressing the MBD2. There was no effect of NGFI-A overexpression on transcriptional activation in cells co-expressing the mutant MBD2, suggesting that the MBD domain of MBD2 is critical for transcriptional activation from the exon 17 GR promoter.
4. Discussion
Previous findings suggested that the increased GR transcription from the exon 17 promoter as a function of maternal care is programmed by epigenetic effects that include differential demethylation of the exon 17 promoter in the hippocampus of the offspring of high and low LG mothers [12]. However, the molecular signalling at the level that the exon 17 GR promoter triggers the epigenetic remodelling is not fully understood. Maternal licking increases NGFI-A expression in hippocampal neurons and the binding of NGFI-A to the exon 17 GR promoter [10,12]. Likewise, maternal licking enhances hippocampal NGFI-A binding to other NGFI-A-regulated genes, such as GAD1 [31]. Interestingly, the GAD1 promoter shows the same hypomethylated pattern in hippocampal tissue from the adult offspring of high compared with low licking mothers [31]. Binding of the CBP to the exon 17 GR promoter shows a similar pattern, and is dependent upon an intact NGFI-A response element [12,13]. In the studies reported here, we examined whether methylated-DNA binding proteins, notably MBD3 and MBD2, might mediate the differential expression of the GR gene as a function of postnatal maternal care.
Maternal licking increases 5-HT signalling at the level of the hippocampus and, in vitro, 5-HT increases GR expression in hippocampal neurons. The results of studies described here somewhat unexpectedly suggest that 5-HT treatment of hippocampal neurons increases the expression of MBD2. Likewise, maternal licking associates with increased in vivo MBD2 binding to the exon 17 GR promoter, and viral-mediated knockdown of MBD2 strongly suppressed the effect of 5-HT on hippocampal GR expression. These findings were consistent with the results of subsequent experiments showing that (i) NGFI-A co-precipitates and appears to interact with NGFI-A and (ii) enhances NGFI-A binding to the exon 17 GR promoter. Additional support for the synergistic effects of NGFI-A and MBD2 derives from site-directed mutagenesis studies showing that alterations of the NGFI-A response element that eliminate NGFI-A binding likewise abolish MBD2 binding.
It is of interest to note that MBD2 is primarily known also for silencing of methylated genes [14–16]. For example, MBD2 links methylated DNA with the nucleosome remodelling and histone deacetylation complex (also known as Mi-2), an essential complex for chromatin remodelling. However, our apparently paradoxical findings are not without precedent [17]. There are multiple models where MBD2 can mediate transcriptional activation, even when bound to methylated CpG sites. For example, MBD2 complexes with transforming-acid-coiled-coil 3 (TACC3) and the pCAF histone acetyltransferase (p300/CBP associated factor) to enhance transcriptional activity [32]. The presence of MBD2 within the TACC3–pCAF complex increases histone acetyltransferase activity. Likewise, we found that MBD2 overexpression increased both CBP association with the exon 17 GR promoter and histone 3 K9 acetylation of the promoter. CBP acts as a histone acetyltransferase [33]. Similarly, an MBD2–RNase helicaseA complex enhances gene expression and the effect is CBP-dependent [30]. Studies of focal adhesion kinase (FAK)-induced muscle differentiation show that MBD2 mediates both transcriptional repression and activation from the same site depending on the presence or absence of FAK [34]. FAK translocates into nuclei in response to oxidative stress of muscle cells where it forms a complex with MBD2, attenuates MBD2-mediated transcriptional repression, activates myogenin expression, which then promotes muscle-terminal differentiation. Under these conditions, MBD2 interacts with FAK and enhances FAK nuclear translocation and myogenin expression, effectively operating as a transcriptional cofactor. These examples dovetail with array-based studies with neuronal tissues showing that binding of the methylated-DNA binding protein MeCP2 is associated with both active and repressed transcriptional states depending on the presence of CREB [35]. Increased CREB expression is associated with MeCP2-associated transcriptional activation. Interestingly, both maternal licking in vivo and 5-HT in vitro enhance phosphorylation of CREB, increase CBP levels and CBP binding to the exon 17 GR promoter [10,12]. The CBP acts as a histone acetyltransferase, which likely explains the increased H3K9ac that was paradoxically associated with increased MBD2 binding to the exon 17 GR promoter. Moreover, the same mutagenesis analysis applied in the experiments described here reveals that the binding of CBP to the exon 17 GR promoter, such as that of MBD2, is NGFI-A-dependent [10]. Moreover, we previously [12] showed evidence for the physical interaction of NGFI-A and CBP. Taken together, these findings suggest that maternal licking and the accompanying increase in hippocampal 5-HT signalling increases NGFI-A expression and the formation of NGFI-A complexes with CBP and/or MBD2. The results of the current studies suggest that MBD2 enhances NGFI-A association with the exon 17 GR promoter, although the mechanism for this effect is unknown.
Overexpression of either NGFI-A or MBD2 increases CBP association with the exon 17 GR promoter and histone acetylation of the promoter region. Earlier studies reveal that increased histone acetylation through the inhibition of histone deacetylase activity is associated with DNA demethylation [36–38]. Likewise, the binding of SP1 is associated with alterations of existing methylation states [39,40], and maternal licking increases SP1 binding to the exon 17 GR promoter [12]. The Sp1 site lies adjacent to the NGFI-A response element, within which increased maternal licking is associated with a demethylation of the 5′ CpG within the NGFI-A response element [13]. Previous studies have associated MBD2 with DNA demethylation [41], although the precise physiochemical mechanisms underlying the relation between MBD2 expression and alterations in DNA methylation remain to be fully elucidated. The results of these studies do suggest that variations in maternal care are associated with both chromatin remodelling of and increased MBD2 binding to the exon 17 GR promoter. Future studies will determine how these events are related to alterations in DNA methylation.
A critical question here concerns the mechanisms that target MBD2, which is a general methylated DNA binding protein with very limited sequence selectivity, to the exon 17 GR promoter. We have hypothesized here that MBD2 was targeted to specific promoters by sequence selective transcription factors; three lines of evidence support this. First, ectopic expression of NGFI-A increases MBD2 association with exon 17 GR promoter. Second, site-specific mutagenesis of the NGFI-A recognition element in the exon 17 GR promoter, which abolishes NGFI-A binding, also eliminates MBD2 binding to the promoter and transcriptional activation (figure 5c,d). Third, using double ChIP analysis, we show that NGFI-A and MBD2 co-occupy the same exon 17 GR promoter molecule (figure 6b). Moreover, we provide evidence for a physical interaction between NGFI-A and MBD2 (figure 4). We favour a model whereby recruitment of NGFI-A to the exon 17 GR promoter facilitates the accessibility of MBD2 to the promoter through changes in histone acetylation caused by the recruitment of CBP by NGFI-A. Binding of MBD2 to the promoter ensues. Recruitment of transcription factors to a gene might be a general mechanism for facilitating site-specific alterations in DNA methylation. One example is the requirement for NF-κB and the cis-acting Igκ enhancer sequences in B-cell-specific demethylation of the Igκ locus [42]. Interestingly, both maternal licking in vivo and 5-HT treatment in vitro enhance NGFI-A binding to the GAD1 promoter in hippocampal neurons [10,42]. The GAD1 promoter, such as the exon 17 GR promoter, shows decreased methylation of the NGFI-A response element [42]. These findings are consistent the hypothesis that environmental events may alter stable epigenetic states through effects on specific intracellular signalling pathways.
Funding statement
These studies were supported by grants from the Canadian Institutes for Health Research (CIHR) and the National Institute for Child Health and Development to M.J.M. and M.S. The authors further acknowledge continuing support from the Sackler Foundation and the Ludmer Family Foundation.
References
- 1.Agrawal AA. 2001. Phenotypic plasticity in the interactions and evolution of species. Science 294, 321–326. ( 10.1126/science.1060701) [DOI] [PubMed] [Google Scholar]
- 2.Higley JD, Hasert MF, Suomi SJ, Linnoila M. 1991. Nonhuman primate model of alcohol abuse: effects of early experience, personality, and stress on alcohol consumption. Proc. Natl Acad. Sci. USA 88, 7261–7265. ( 10.1073/pnas.88.16.7261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu D, et al. 1997. Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science 277, 1659–1662. ( 10.1126/science.277.5332.1659) [DOI] [PubMed] [Google Scholar]
- 4.Meaney MJ. 2001. Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annu. Rev. Neurosci. 24, 1161–1192. ( 10.1146/annurev.neuro.24.1.1161) [DOI] [PubMed] [Google Scholar]
- 5.Francis D, Diorio J, Liu D, Meaney MJ. 1999. Nongenomic transmission across generations of maternal behavior and stress responses in the rat. Science 286, 1155–1158. ( 10.1126/science.286.5442.1155) [DOI] [PubMed] [Google Scholar]
- 6.Caldji C, Diorio J, Meaney MJ. 2003. Variations in maternal care alter GABA(A) receptor subunit expression in brain regions associated with fear. Neuropsychopharmacology 28, 1950–1959. ( 10.1038/sj.npp.1300237) [DOI] [PubMed] [Google Scholar]
- 7.Laplante P, Diorio J, Meaney MJ. 2002. Serotonin regulates hippocampal glucocorticoid receptor expression via a 5-HT7 receptor. Brain Res. Dev. Brain Res. 139, 199–203. ( 10.1016/S0165-3806(02)00550-3) [DOI] [PubMed] [Google Scholar]
- 8.Meaney MJ, Aitken DH, Sapolsky RM. 1987. Thyroid hormones influence the development of hippocampal glucocorticoid receptors in the rat: a mechanism for the effects of postnatal handling on the development of the adrenocortical stress response. Neuroendocrinology 45, 278–283. ( 10.1159/000124741) [DOI] [PubMed] [Google Scholar]
- 9.Meaney MJ, Diorio J, Francis D, Weaver S, Yau J, Chapman K, Seckl JR. 2000. Postnatal handling increases the expression of cAMP-inducible transcription factors in the rat hippocampus: the effects of thyroid hormones and serotonin. J. Neurosci. 20, 3926–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellstrom IC, Dhir KS, Diorio JC, Meaney MJ. 2012. Maternal licking regulates hippocampal glucocorticoid receptor transcription through a thyroid hormone–serotonin–NGFI-A signalling cascade. Phil. Trans. R. Soc. B 367, 2495–2510. ( 10.1098/rstb.2012.0223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCormick JA, et al. 2000. 5′-Heterogeneity of glucocorticoid receptor messenger RNA is tissue specific: differential regulation of variant transcripts by early-life events. Mol. Endocrinol. 14, 506–517. [DOI] [PubMed] [Google Scholar]
- 12.Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. 2004. Epigenetic programming by maternal behavior. Nat. Neurosci. 7, 847–854. ( 10.1038/nn1276) [DOI] [PubMed] [Google Scholar]
- 13.Weaver IC, D'Alessio AC, Brown SE, Hellstrom IC, Dymov S, Sharma S, Szyf M, Meaney MJ. 2007. The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: altering epigenetic marks by immediate-early genes. J. Neurosci. 27, 1756–1768. ( 10.1523/JNEUROSCI.4164-06.2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389. ( 10.1038/30764) [DOI] [PubMed] [Google Scholar]
- 15.Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. 1999. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 23, 58–61. ( 10.1038/12659) [DOI] [PubMed] [Google Scholar]
- 16.Klose RJ, Bird AP. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, 89–97. ( 10.1016/j.tibs.2005.12.008) [DOI] [PubMed] [Google Scholar]
- 17.Meaney MJ, Ferguson-Smith AC. 2010. Epigenomic regulation of the neural transcriptome: the meaning of the marks. Nat. Neurosci. 13, 1313–1318. ( 10.1038/nn1110-1313) [DOI] [PubMed] [Google Scholar]
- 18.Champagne FA, Francis DD, Mar A, Meaney MJ. 2003. Variations in maternal care in the rat as a mediating influence for the effects of environment on development. Physiol. Behav. 79, 359–371. ( 10.1016/S0031-9384(03)00149-5) [DOI] [PubMed] [Google Scholar]
- 19.Paxinos G, Watson C. 1996. The rat brain in stereotaxic coordinates. San Diego, CA: Academic Press. [Google Scholar]
- 20.Campbell PM, Bovenzi V, Szyf M. 2004. Methylated DNA-binding protein 2 antisense inhibitors suppress tumourigenesis of human cancer cell lines in vitro and in vivo. Carcinogenesis 25, 499–507. ( 10.1093/carcin/bgh045) [DOI] [PubMed] [Google Scholar]
- 21.Crane-Robinson C, Myers FA, Hebbes TR, Clayton AL, Thorne AW. 1999. Chromatin immunoprecipitation assays in acetylation mapping of higher eukaryotes. Methods Enzymol. 304, 533–547. ( 10.1016/S0076-6879(99)04031-8) [DOI] [PubMed] [Google Scholar]
- 22.Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. 2005. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J. Neurosci. 25, 110 45–110 54. ( 10.1523/JNEUROSCI.3652-05.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banker GA, Cowan WM. 1977. Rat hippocampal neurons in dispersed cell culture. Brain Res. 126, 397–325. ( 10.1016/0006-8993(77)90594-7) [DOI] [PubMed] [Google Scholar]
- 24.Mitchell JB, Rowe W, Boksa P, Meaney MJ. 1990. Serotonin regulates type II corticosteroid receptor binding in hippocampal cell cultures. J. Neurosci. 10, 1745–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. 2003. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302, 885–889. ( 10.1126/science.1086446) [DOI] [PubMed] [Google Scholar]
- 26.Liu D, Diorio J, Day JC, Francis DD, Meaney MJ. 2000. Maternal care, hippocampal synaptogenesis and cognitive development in rats. Nat. Neurosci. 3, 799–806. ( 10.1038/77702) [DOI] [PubMed] [Google Scholar]
- 27.Hery M, Semont A, Fache MP, Faudon M, Hery F. 2000. The effects of serotonin on glucocorticoid receptor binding in rat raphe nuclei and hippocampal cells in culture. J. Neurochem. 74, 406–413. ( 10.1046/j.1471-4159.2000.0740406.x) [DOI] [PubMed] [Google Scholar]
- 28.Lai M, McCormick JA, Chapman KE, Kelly PA, Seckl JR, Yau JL. 2003. Differential regulation of corticosteroid receptors by monoamine neurotransmitters and antidepressant drugs in primary hippocampal culture. Neuroscience 118, 975–984. ( 10.1016/S0306-4522(03)00038-1) [DOI] [PubMed] [Google Scholar]
- 29.Chesler EJ, Wang JT, Lu L, Qu YH, Manly KF, Williams RW. 2003. Genetic correlates of gene expression in recombinant inbred strains: a relational model system to explore neurobehavioral phenotypes. Neuroinformatics 1, 343–357. ( 10.1385/NI:1:4:343) [DOI] [PubMed] [Google Scholar]
- 30.Fujita H, Fujii R, Aratani S, Amano T, Fukamizu A, Nakajima T. 2003. Antithetic effects of MBD2a on gene regulation. Mol. Cell Biol. 23, 2645–2657. ( 10.1128/MCB.23.8.2645-2657.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang TY, Hellstrom IC, Bagot RC, Wen X, Diorio J, Meaney MJ. 2010. Maternal care and DNA methylation of a glutamic acid decarboxylase 1 promoter in rat hippocampus. J. Neurosci. 30, 13130–13137. ( 10.1523/JNEUROSCI.1039-10.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angrisano T, Lembo F, Pero R, Natale F, Fusco A, Avvedimento VE, Bruni CB, Chiariotti L. 2006. TACC3 mediates the association of MBD2 with histone acetyltransferases and relieves transcriptional repression of methylated promoters. Nucleic Acids Res. 34, 364–372. ( 10.1093/nar/gkj400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grunstein M. 1997. Histone acetylation in chromatin structure and trasncription. Nature 389, 349–352. ( 10.1038/38664) [DOI] [PubMed] [Google Scholar]
- 34.Luo SW, Zhang C, Zhang B, Kim C-H, Qiu Y-Z, Du Q-S, Mei L, Xiong WC. 2009. Regulation of heterochromatin remodelling and myogenin expression during muscle differentiation by FAK interaction with MBD2. EMBO J. 28, 2568–2582. ( 10.1038/emboj.2009.178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chahrour M, Jung SY, Shaw C, Zhou X, Wong STC, Qin J, Zoghbi HY. 2008. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320, 1224–1229. ( 10.1126/science.1153252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cervoni N, Detich N, Seo SB, Chakravarti D, Szyf M. 2002. The oncoprotein Set/TAF-1beta, an inhibitor of histone acetyltransferase, inhibits active demethylation of DNA, integrating DNA methylation and transcriptional silencing. J. Biol. Chem. 277, 250 26–250 31. ( 10.1074/jbc.M202256200) [DOI] [PubMed] [Google Scholar]
- 37.Cervoni N, Szyf M. 2001. Demethylase activity is directed by histone acetylation. J. Biol. Chem. 27, 27. [DOI] [PubMed] [Google Scholar]
- 38.Detich N, Bovenzi V, Szyf M. 2003. Valproate induces replication-independent active DNA demethylation. J. Biol. Chem. 278, 27 586–27 592. ( 10.1074/jbc.M303740200) [DOI] [PubMed] [Google Scholar]
- 39.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. 1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371, 435–438. ( 10.1038/371435a0) [DOI] [PubMed] [Google Scholar]
- 40.Macleod D, Charlton J, Mullins J, Bird AP. 1994. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 8, 2282–2292. ( 10.1101/gad.8.19.2282) [DOI] [PubMed] [Google Scholar]
- 41.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. 1999. A mammalian protein with specific demethylase activity for mCpG DNA. Nature 397, 579–583. ( 10.1038/17533) [DOI] [PubMed] [Google Scholar]
- 42.Kirillov A, Kistler B, Mostoslavsky R, Cedar H, Wirth T, Bergman Y. 1996. A role for nuclear NF-kappaB in B-cell-specific demethylation of the Igkappa locus. Nat. Genet. 13, 435–441. ( 10.1038/ng0895-435) [DOI] [PubMed] [Google Scholar]