Abstract
The growing list of mutations implicated in monogenic disorders of the developing brain includes at least seven genes (ARX, CUL4B, KDM5A, KDM5C, KMT2A, KMT2C, KMT2D) with loss-of-function mutations affecting proper regulation of histone H3 lysine 4 methylation, a chromatin mark which on a genome-wide scale is broadly associated with active gene expression, with its mono-, di- and trimethylated forms differentially enriched at promoter and enhancer and other regulatory sequences. In addition to these rare genetic syndromes, dysregulated H3K4 methylation could also play a role in the pathophysiology of some cases diagnosed with autism or schizophrenia, two conditions which on a genome-wide scale are associated with H3K4 methylation changes at hundreds of loci in a subject-specific manner. Importantly, the reported alterations for some of the diseased brain specimens included a widespread broadening of H3K4 methylation profiles at gene promoters, a process that could be regulated by the UpSET(KMT2E/MLL5)-histone deacetylase complex. Furthermore, preclinical studies identified maternal immune activation, parental care and monoaminergic drugs as environmental determinants for brain-specific H3K4 methylation. These novel insights into the epigenetic risk architectures of neurodevelopmental disease will be highly relevant for efforts aimed at improved prevention and treatment of autism and psychosis spectrum disorders.
Keywords: chromatin, nucleosome, epigenetic, autism, schizophrenia, histone
1. Introduction
The ‘epi- (greek for ‘over’, ‘above’) genome’, with its rich cache of highly regulated, structural modifications—which include four different types of DNA methyl-cytosine derivatives and more than 100 different types of post-translational histone modifications and histone variants—defines the mouldings and three-dimensional structures of the genomic material inside the cell nucleus. Studies in human brain have indicated that the epigenetic landscape remains ‘plastic’ throughout all periods of brain development and ageing, with ongoing dynamic regulation occurring even in neurons and other postmitotic constituents of the brain [1–4]. Furthermore, mutations and structural variants in perhaps up to 50 genes, each encoding a different chromatin regulator, have been linked to a wide range of neurodevelopmental syndromes and rare, monogenic forms of autism and schizophrenia [5], and even adult-onset hereditary neurodegenerative disease [6–8]. Of note, on the list of mutations in chromatin-associated proteins linked to neurodevelopmental disease, some molecular systems stand out owing to a more prominent representation. These include, for example, at least seven members of the BAF family of molecules, which assemble into ATP-dependent chromatin remodelling complexes to regulate mobility of nucleosomes (the elementary units of chromatin, composed of 146 bp of DNA wrapped around an octamer of the core histones H2A/H2B/H3/H4) [5]. Here, we focus on methylation of the lysine (K) no. 4 residue of histone H3, a mark that on a genome-wide scale is broadly associated with transcriptional regulation and epigenetic tagging of promoter and enhancer sequences [9]. To date, mutations in five genes each encoding a H3K4-specific lysine methyltransferase or demethylase have been linked to developmental disability and impaired cognition, and several additional disease-associated genes encode proteins with a key regulatory role for H3K4 methylation [5]. Furthermore, there is evidence that H3K4 methylation landscapes in the human cerebral cortex are dynamically regulated during prenatal development and throughout early childhood years until adolescence [10], and potentially altered in some cases on the autism [11] or schizophrenia spectrum [12]. Therefore, the findings from clinical genetics and the conclusions drawn from the epigenome mappings in brain tissue coalesce, and when taken together, leave little doubt that fine-tuning of H3K4 methylation is of fundamental importance during an extended period of human brain development.
2. H3K4 methylation: an overview
The functions of post-translational histone modifications (histone PTM) are highly specific in a site- and residue-specific manner, and include methylation, acetylation and crotonylation, polyADP-ribosylation and small protein (ubiquitin, SUMO) modification of specific lysine residues, as well as arginine (R) methylation and citrullination, serine (S) phosphorylation, tyrosine (T) hydroxylation, among others [13–15]. Methylation of the various lysine residues, including H3K4, H3K9, H3K27, H3K36, H3K79 and H4K20, has been linked to transcriptional initiation and elongation, heterochromatic silencing and other chromatin functions [16]. The ammonium group of lysine side chains can carry up to three methyl groups, and for many of the aforementioned methyl-lysines, important differences in the distribution and regulation of the mono- (me1), di- (me2) and trimethylated (me3) forms emerged [16]. Powerful technologies, such as chromatin immunoprecipitation in conjunction with next-generation-sequencing-based quantification of the DNA (ChIP-seq), have been widely applied to various histone methylation markings in multiple tissues. In the case of H3K4, ChIP-seq studies have shown that the trimethylated form H3K4me3 is in human brain typically organized as approximately 30 000 sharp ‘peaks’ genome-wide [4,17]. These ‘peaks, which often are cell-type specific and limited to 1–2 kb in length, are mostly found around transcription start sites (TSSs) of proximal gene promoters and other regulatory sequences, many of which are CpG enriched [18,19]. On a genome-wide scale, the H3K4me3 mark broadly correlates with RNA polymerase II occupancy at sites of active gene expression [20], and is thought to provide an additional layer of transcriptional regulation [21,22]. In striking contrast, the di- and mono-methylated forms, H3K4me2 and H3K4me1, are organized as much broader peaks when compared with H3K4me3 and tag a much larger proportion of open chromatin, including promoters and enhancers that positively regulate expression of genes which could be positioned many thousands of basepairs further down- or upstream [9,23]. When occurring together with sharp peaks of histone H3 acetylated at lysine 27 (H3K27ac), enrichment for H3K4me1 and/or H3K4me2 and in some cases, me3, is likely to flag active enhancer sequences [9,24]. Conversely, inactive enhancer sequences are defined by high levels of H3K4me1 but in the presence of the repressive mark, histone H3 trimethylated at lysine 27 (H3K27me3) [25] and at the expense of acetyl-H3K27 [24]. It has been proposed that some of the inactive enhancers tagged with the H3K4me1 mark is a reflection of their developmental potential, because some of these sequences could be reset epigenetically into an active state under certain conditions, including differentiation, similar to the transiently occurring bivalent state of promoters dually tagged with open and repressive histone modifications in some stem cell types [24]. Interestingly, in brain chromatin, 70% of H3K4me1 sites are positioned within transcriptional units, which contrasts with many peripheral tissues that are defined by a somewhat higher representation of intergenic sequences at the expense of intragenic H3K4me1 [26]. The fact that brains show higher enrichments for H3K4me1 in intronic sequences in comparison with the peripheral tissues studied so far, could be explained by the fact that brain transcriptomes, in comparison with other tissues, show an overrepresentation of genes defined by high conservation of intronic sequence [26]. This finding could imply that transcriptional mechanisms in the brain rely more heavily on regulatory sequences within introns. In any case, proper H3K4 methylation is probably important to maintain open chromatin states in brain tissue. In this context, it is important to note that unmethylated H3K4 effectively serves as a DNA methylation signal which at promoters and enhancers is mostly associated with repression and inhibition of transcriptional activity [27,28].
3. H3K4-specific methyltransferases and demethylases
For a detailed review on lysine methyltransferases (KMTs) and demethylases (KDMs) as regulators of histone methylation, including the H3K4 residue, see Black et al. [29]. At least 11 genes in the human genome encode a protein with lysine methyltransferase activity specifically directed against the H3K4 residue. There are six H3K4-specific methyltransferases that share a highly conserved catalytic SET domain (KMT2A-D, KMT2F-G, which in the literature are better known under their older names SET1A-B, MLL1–4) [30] and additional genes (including MLL5 (KMT2E), ASH1 (KMT2H), SMYD3 (KMT3E), SET7/9 (KMT7), MEISETZ); each of these proteins methylates H3K4 with high specificity [31]. The one exception is MLL5 which is thought to harbour a SET domain that lacks catalytic activity [32,33]. The mixed-lineage leukaemia (MLL) group require additional subunits to attain maximal activity towards H3K4. A well-studied example would be the ‘WRAD’ (WDR5, RbBP5, ASH2L, DPY30) MLL-associated core complex [30,34]. Like the WRAD, the scaffolding protein MENIN has been shown to be important for MLL1-mediated H3K4me3/2/1 methylation [35–37]. Mechanistic studies in vivo are complicated by the frequent observation that dysregulation in one type of histone PTM causes secondary alterations in additional PTMs. For example, while MLL1 upregulates in vitro primarily mono- and di-methylated H3K4 [34], Mll1 deficiency in some brain regions results not only in decreased H3K4me2 (as predicted from the in vitro data) but also lower levels of the trimethylated form (H3K4me3) around a subset of TSSs [12,38]. In addition, Mll1 deficiency is associated with secondary effects on histone marks not directly regulated by the MLLs; for example, there is incomplete removal of a repressive histone mark, H3K27me3, at the H3K4me3-tagged promoters [39]. It is thought that the MLL1–4 enzymes each target a limited but very specific subset of genomic loci, including homeobox gene clusters as key regulators of morphogenesis and early development [29]. Thus, MLL1 regulates less than 5% of promoters that carry an H3K4 trimethylation signal, whereas SET1A/B are thought to function as general regulators for this histone mark [37]. The (gene) target selectivity that defines some of the H3K4 KMTs could be due to differential binding to various cofactors and chromatin-associated proteins [40,41].
The H3K4-specific KMTs are complemented by an equally complex system of KDMs, with to date at least eight genes encoding a KDM that primarily (albeit often not exclusively) targets the H3K4 residue [29,31]. Active histone demethylation has been linked to two different mechanisms. The first type, represented by LSD1/KDM1A, involves an amine oxidase domain and flavin adenine dinucleotide (FAD) as cofactor to demethylate mono- and dimethylated lysines, including H3K4me2 and H3K4me1 [42], albeit activity against the H3K9 residue, particularly in complex with oestrogen or androgen receptors, also has been described [43–45]. Monoamine oxidase inhibitors (MAOi) such as tranylcypromine or phenelzine—powerful antidepressants that exert their therapeutic effects mainly by elevating brain monoamine levels through inhibition of MAO-A/B—also block LSD1-type histone demethylases [44]. The second type of demethylase, which in contrast to LSD1/LSD2 is capable of demethylating trimethyl markings, involves Fe2+-dependent dioxygenation by Jumonji-C (JmJC) domain-mediated catalysis [44]. The KDMs, like the KMTs, each show a specific combinatorial set of functional domains and (protein) binding partners [44,46], suggesting at least partial non-redundancy in function. For example, LSD1/KDM1A is thought to regulate histone methylation at promoters, whereas LSD2/KDM2B is bound to transcriptional elongation complexes and removes H3K4 methyl markings in gene bodies, thereby facilitating gene expression by reducing spurious transcriptional initiation outside of promoters [47].
4. Neurological disease associated with mutations in H3K4 regulators
Normal gene dosage of both for MLL1 and MLL4 is essential for human health, and haploinsufficiency of either gene is associated with developmental delay [48] and disability [49–51]. However, whether genetic ablation of each of these two orthologues results in overlapping behavioural, electrophysiological and molecular phenotypes remains an open question. Mutations in MLL1 are thought to be responsible for the majority of cases with Wiedemann–Steiner syndrome (Mendelian inheritance in man (MIM) no. 605130), an extremely rare neurodevelopmental condition defined by intellectual disability, short stature, microcephaly and elbow hypertrichosis [51]. Furthermore, approximately 70% of patients with Kabuki syndrome (MIM no. 147920) carry deleterious mutations in MLL4 [48]. Note that MLL4 (a closer homologue to MLL3) in the literature is sometimes confused with MLL2 (a close homologue to MLL1) [30]. We refer to MLL4 (also known as ALR) as the KMT encoded on chromosome 12p13 that is mutated in Kabuki syndrome, and refer to MLL2 (also known as TRX2 or Wbp7) as the KMT encoded on chromosome 19q13. Kabuki syndrome, with more than 400 cases described worldwide, is defined by mild-to-moderate intellectual disability, failure to thrive in early childhood and short stature, and characteristic facial features, including elongated palpebral fissures [48]. The MLLs, including MLL1 and MLL4, are very large proteins 3972 (MLL1) and 5537 (MLL4) amino acids in length comprised 37 (MLL1) or 60 (MLL4) exons [30], with the majority of mutations thought to cause premature termination or frameshifts, and there is no apparent clustering of mutatons around a specific functional domain [51]. The majority of mutations in Wiedemann–Steiner and Kabuki patients are likely to occur de novo, albeit a significant number of familial cases have been observed [51]. Additional cases with intellectual disability have been linked to single-point mutations in MLL3 [52]. Strikingly, the list of monogenic neurodevelopmental disorders includes additional genes with an essential role for MLL-mediated H3K4 methylation. CUL4B, encoding a scaffold protein for the assembly of Cullin4B-ring ubiquitin ligase (CRL4B) complexes, is mutated in some cases with X-linked mental retardation (XLMR) [53–55]. Recently, it was discovered that CUL4B mediates ubiquitination and degradation of WDR5, a core subunit of the WRAD complex which is essential for MLL-mediated H3K4 methylation in vivo [56]. Consequently, downregulation of CUL4B resulted in elevated levels of WDR5 and H3K4 trimethylation on neuronal gene promoters [56].
Furthermore, missense mutations in the H3K4 demethylase KDM5A/JARID1A have been linked to an autosomal recessive form of intellectual disability [57] and, likewise, deleterious mutations in the X-linked KDM5C/JARID1C/SMCX could cause mental retardation and autism [58–60]. Furthermore, polyalanine (polyA)-tract-expansion-encoding mutations in the aristaless-related homeobox transcriptional regulator (ARX, MIM no. 300382), considered to rank among the most frequent causes of XLMR and epilepsy, are thought to lead to a dramatic downregulation of the ARX-driven transcription of KDM5C and, as a secondary effect, excessive H3K4 trimethylation [61]. To summarize then, at least seven genes—MLL1, MLL2, MLL3, KDM5A, KDM5C, ARX, CUL4B—each ascribed an essential role in the regulation of H3K4 methylation, are linked to rare monogenic forms of neurodevelopmental disease, including intellectual disability and autism (figure 1 and table 1).
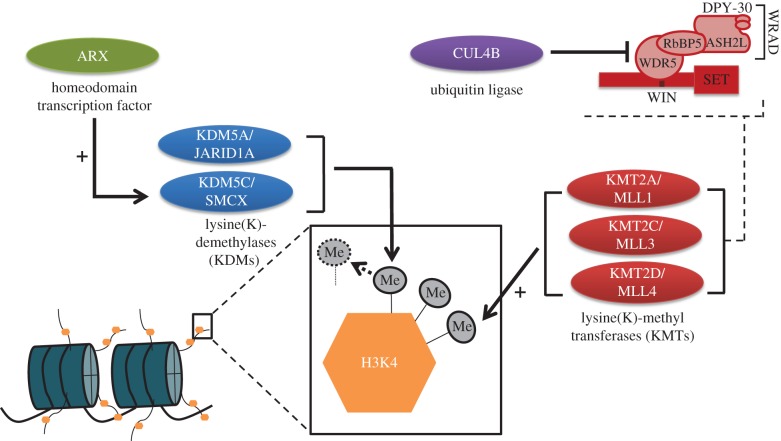
Figure 1.
Molecular mechanisms of the seven genes (ARX, CUL4B, KDM5A, KDM5C, KMT2A, KMT2C, KMT2D) that play an essential role in H3K4 methylation and for which loss-of-function mutations are implicated in monogenic disorders of the developing brain. ARX is a homeodomain transcription factor; CUL4B is a ubiquitin ligase; KDM5A and KDM5C are H3K4 demethylases; KMT2A, KMT2C and KMT2D are H3K4-specific methyl transferases. ARX is a key transcriptional regulator for KDM5C, and CUL4B plays a role in the degradation of WDR5, a key component of the WRAD (MLL cofactor) complex. See text for further details. (Online version in colour.)
Table 1.
List of monogenic neurodevelopmental diseases associated with the seven essential regulators of H3K4 methylation.
| function | gene | MIM ID | predicated alteration of H3K4 methylation owing to mutations or genetic variations | associated phenotype and MIM number | references |
|---|---|---|---|---|---|
| lysine(K)-methyl transferases (KMTs) | KMT2A/Mll1 | 159555 | decreased | Wiedemann–Steiner syndrome (605130); microcephaly; autism; aggression | [51] |
| KMT2C/Mll3 | 606833 | decreased | Kleefstra syndrome (610253); autism; microcephaly; intellectual disability; seizures; aphasia; anxiety; depression | [52,62,63] | |
| KMT2D/Mll4 (‘Mll2’) | 602113 | decreased | Kabuki syndrome 1 (147920); autism; seizures; microcephaly; cortical atrophy without hydrocephaly | [48–50,64,65] | |
| lysine(K)-specific demethylase (KDMs) | KDM5A/JARID1A | 180202 | increased | intellectual disability | [57] |
| KDM5C/SMCX | 314690 | increased | Claes-Jensen type X-linked mental retardation (300534); microcephaly; seizures | [57,66,67] | |
| homeodomain transcription factor | ARX | 300382 | increased | early infantile epileptic encephalopathy-1 (308350); hydranencephaly with abnormal genitalia (300215); X-linked lissencephaly (300215); X-linked mental retardation (300419); Partington syndrome (309510); Proud syndrome (300004); intellectual disability; seizures | [68–74] |
| ubiquitin ligase | CUL4B | 300304 | increased | Cabezas type X-linked mental retardation (300354); macrocephaly; intellectual disability; seizures | [53,55,75–78] |
5. Mouse models for genetic disorders associated with H3K4 methylation defects
The aforementioned subjects diagnosed with Wiedemann–Steiner or Kabuki syndrome are haploinsufficient for MLL1 or MLL4. Interestingly, like their human counterparts, brain function is abnormal in Mll1 mutant mice that carry only one intact copy of the gene. These Mll1−/+ heterozygous mice are affected by robust deficits in hippocampal learning and memory [79] and synaptic plasticity [80], whereas conditional deletion of both Mll1 alleles in stem cells residing in the subventricular zone results in severe defects in neurogenesis, whereas the glial lineage remains relatively unaffected [39]. Homozygous Mll1 null mutant mice die during early embryogenesis (around E10) but, interestingly, deletion of the catalytic SET domain alone is not lethal [81]. It is not known whether selective ablation of the SET domain would result in a brain phenotype in mice. Furthermore, conditional deletion of Mll2 (the closest homologue to Mll1 among all other Mlls and SET-domain containing H3K4 KMTs) also causes a deficit in hippocampus-dependent memory functions in mice [82], which would suggest that Mll1 and Mll2 are non-redundant in function. There is no information yet whether deficiency in Mll4 (the gene mutated in Kabuki syndrome) would result in abnormal brain function and behaviours. The molecular defects in the aforementioned Mll mutant mouse generally are in good agreement with current concepts on MLL function, including selective effects on a small subset of neuronal or glial transcripts, in the absence of global shifts in neuronal or glial transcriptomes. For example, conditional Mll2 deletion in hippocampal neurons affected transcript levels for as little as 22 genes in the CA pyramidal neurons, and 161 genes in the dentate gyrus, together with corresponding changes in H3K4me2 and H3K4me3 that were confined to promoters of only the genes that were subjected to altered expression [82].
To the best of our knowledge, there are no studies exploring behavioural alterations in H3K4-specific KDMs, including Kdm5a and Kdm5c. However, it has been reported that RNA-induced Kdm5c knockdown in rat cerebellar granule neurons reduces dendritic length, and furthermore, Kdm5c is essential for neuronal survival during zebrafish development [59]. Interestingly, in cultured Arx null mutant cells, Kdm5c levels are severely reduced in differentiating GABAergic neurons [61]. Furthermore, conditional Arx deletion in the ganglionic eminence which contributes the largest proportion of interneurons for the cerebral cortex, leads to seizure disorders and neurocognitive deficits in mice [83].
6. Disordered H3K4 methylation in brain of subjects diagnosed with autism and schizophrenia, and in the animal model
As discussed above, multiple monogenic forms of neurodevelopmental disease have been linked to dysregulated H3K4 methylation. It is very likely that proper regulation and fine-tuning of the H3K4-methyl marks is pivotal for healthy brain development, because both mutations associated with loss (MLL1, MLL4) and gain of H3K4 methylation (ARX, CUL4B, KDM5A, KDM5C) potentially result in intellectual disability and autism, microcephaly, seizure disorder and other neurological disease in early childhood. Furthermore, there is increasing evidence that H3K4 methylation landscapes undergo genome-wide reorganization during early development. For example, in the human cerebral cortex, neuronal and glial H3K4-trimethyl profiles are subjected to significant remodelling at the site of 1000–1500 genes during the transitions from the late gestational period to early and late childhood [10]. Such developmentally programmed remodelling of neuronal and glial H3K4 methylation marks most certainly overlaps with the time period for vulnerability of common psychiatric disorders with a neurodevelopmental aetiology, including autism and schizophrenia. Autism spectrum disorders (ASD) are a group of conditions bound together by broad syndromic overlap of three key behavioural deficits in social interaction, communication and restricted, stereotypical or repetitive behaviour. ASDs also present with intellectual disability and neurological manifestations, including seizure disorder, and other movement disorders. The age of onset of ASD falls within early childhood, mostly within the first 36 months after birth. It is currently estimated that one of every 88 children born in the USA will be diagnosed with ASD [84]. Schizophrenia is a disorder affecting approximately 1% of the general population. Its core symptoms include defects of motivation, thought and cognition, and delusions and hallucinations [85]. The disease typically becomes manifest during young–adult years, but shows, in terms of genetic [86,87] and neurodevelopmental risk architectures [88,89], substantial overlap with ASD and related psychiatric disorders.
Therefore, given that multiple regulators of H3K4 methylation are associated with neurodevelopmental disease, and given that the H3K4 methylation is subject to dynamic changes during the extended period of development and maturation of the human brain, it remains possible that this epigenetic mark is more broadly involved in the pathophysiology of the aforementioned common psychiatric conditions. While it is unlikely that mutations or genetic variations in H3K4-specific KMTs and KDMs, or their regulatory networks, play a role outside of affecting a very small subset of subjects diagnosed with autism, schizophrenia and other common psychiatric disease such as depression, dysregulated H3K4 methylation could play an important role in the pathophysiology, by providing a molecular bridge that links the internal and external environment of the neuronal and glial genomes in the brain. As illustrated by the following findings from independent animal models of relevance for the neurobiology of autism and psychosis spectrum disorders, there can be little doubt that H3K4 methylation landscapes of brain cells are sensitive to a wide range of environmental perturbations. First, the H3K4me3 mark undergoes both global and gene-specific alterations in hippocampus of fear-conditioned animals [79]. Second, different levels of maternal care are associated with changes in H3K4me3 at the metabotropic glutamate receptor gene (mGluR1) in hippocampus of adult offspring [90]. Third, studies in mice have shown that activation of the maternal immune system by the cytokine activator and viral RNA mimic polyriboinosinic–polyribocytidilic acid (poly IC), which leads to behavioural deficits in the adult offspring reminiscent of autism and schizophrenia [91,92], could result in robust but transient changes in H3K4 methylation at cytokine signalling and other genes in the fetal brain, together with more subtle changes in adult offspring brain [93]. Fourth, prenatal exposure to the alkylating and antimitotic agent methylazoxymethanol, such as the aforementioned immune activation which is a popular paradigm to model the neurodevelopmental origins of psychosis [94], results in decreased H3K4 methylation in the adult prefrontal cortex [95]. Fifth, some drugs, including the atypical antipsychotic clozapine and the stimulant metamphetamine, alter H3K4 methylation signatures at specific gene promoters in cerebral cortex and striatum [12,96]. While there is no evidence to date that overall levels of H3K4 methylation are altered in brain tissue of subjects diagnosed with autism or schizophrenia [11,97], changes at specific loci have been reported. In a recent genome-wide study, H3K4me3 was quantified in neuronal chromatin from the prefrontal cortex of 16 subjects on the autism spectrum [11]. Remarkably, not a single locus reached statistical significance on the cohort level. However, more than 700 sequences genome-wide showed a significant methylation change in at least one of the autism brains, in comparison with each of the controls, without exception. Interestingly, there was a two- to threefold, significant enrichment for genes and loci conferring genetic risk for neurodevelopmental disease. Furthermore, for many loci with an abnormal epigenetic signal in one (or several) autistic individuals, changes in gene expression could be documented for at least some of the very same cases with altered histone methylation. Based on these findings [11], one could conclude that each of the diseased brains showed a subject-specific H3K4 methylation signature, with each individual affected by a unique combinatorial set of abnormal histone methylation levels at select TSSs, together with altered expression of the associated gene transcripts. These individual-specific epigenetic alterations in the autism postmortem cohort included AUTS2, PARK2, RIA1, RIMS3, SHANK3, VGEL and many other susceptibility loci with high penetrance for disease risk. This finding suggests that in autism, epigenetic and genetic risk maps overlap significantly. Whether the observed H3K4 methylation changes are linked to a DNA sequence alteration in cis or elsewhere in the genome (trans); reflect an adaptive or maladaptive response to environmental effects; are the outcome of some secondary or tertiary process in the neurobiology of disease; or are simply an epiphenomenon, remains to be determined in future studies.
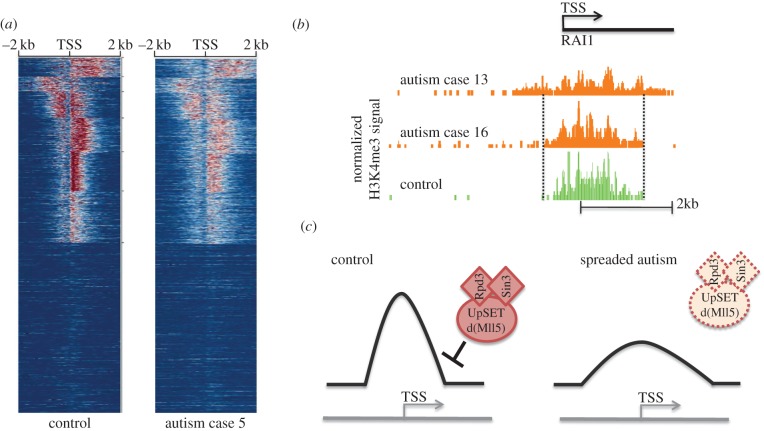
One interesting finding that emerged from the genome-wide, next-generation-sequencing-based mapping of H3K4me3 in prefrontal neurons of subjects with autism spectrum was the ‘spreading’ of this histone mark away from the TSSs and encroaching into gene bodies and upstream sequences [11] (figure 2a,b). This alteration, which affected 4/16 cases but none of the controls, resulted in a significant broadening of the neuronal H3K4me3 peaks [11], which in normal brain are overwhelmingly organized into sharp peaks extending over 1–2 kb or less [4]. Interestingly, UpSET, the MLL5 homologue of Drosophila, assembles with Rpd/Sin3-repressor complexes and thereby regulates promoter-associated histone acetylation and H3K4 tri- and di-methylation. Strikingly, loss or decrease of UpSET alters chromatin architectures of promoters, including broadening of H3K4me3 peaks [32]. Whether or not MLL5 plays a role in the observed H3K4me3 peak broadening that affects neuronal chromatin of some subjects with autism [11] will await further investigations (figure 2c). In either case, these findings serve as important reminders that epigenetic alterations in neuropsychiatric disease should not solely be viewed in quantitative terms as ‘increase’ or ‘decrease’, and should include shape, spread and spatial profiles of epigenetic markings at specific sequences.
Figure 2.
Broadening and ‘spreading’ of H3K4me3 peaks observed in some autism cases and the possible molecular explanation. (a) Clustered H3K4me3 profiles for entire Refseq transcription start sites (TSSs), within a −2 kilobase (kb) to +2 kb window around TSSs for prefrontal neurons from control and autism case no. 5. H3K4me3 signal intensity is illustrated using colour scaling with red representing a strong signal, white representing an intermediate signal and blue representing a weak signal. Note the extensive spreading of H3K4me3 profiles in the diseased case, compared with control. (b) Genome Browser tracks (from the University of California, Santa Cruz, CA) illustrate altered H3K4me3 profiles of the autism-susceptible gene RAI1 in cortical neurons of autism cases no. 13 and 16 compared with the control subject. Note abnormally broad and flattened (‘spreaded’) peak shape in case no. 13. (c) Hypothesized role of UpSET (Mll5) repressive chromatin complex to maintain normal promoter H3K4 methylation profiles. See text and Shulha et al. [11] for further details. (Online version in colour.)
In addition to the aforementioned work in autism brains, genome-wide histone methylation surveys have been conducted in schizophrenia. One study explored H3K4me3 changes in cultured cells, including immature neurons, which were derived from a biopsy of the olfactory epithelium of four subjects with schizophrenia and four matched controls [98]. Again, hundreds of loci showed H3K4 methylation changes in the disease cases, and at least 72 genes, some of which were of relevance for oxidative stress, metabolism and cellular (incl. synaptic) signalling, were affected by altered H3K4me3 in conjunction with altered expression. In addition to these genome-wide surveys, a number of studies reported changes in H3K4 methylation at neuronal signalling genes in brains of subjects diagnosed with schizophrenia or depression [12,99,100].
7. Outlook
As discussed above, multiple genes encoding regulators of H3K4 methylation are linked to monogenic forms of neurodevelopmental disease, thus accounting for some of the strongest genetic risk factors for intellectual disability and ASD. Changes in H3K4me1/2/3 have also been observed at numerous loci in cerebral cortex of subjects diagnosed with autism, schizophrenia and even depression, though it should be noted that these studies cannot clarify whether the observed H3K4 methylation changes were upstream of the disease process as opposed to a secondary effect, or epiphenomemon of the disease process. Interestingly, in the animal model, H3K4 methylation markings are sensitive to drugs interfering with dopamine and other monoamine signalling, and to various environmental perturbations. Thus, the question arises whether regulators of H3K4 methylation could emerge as promising drug targets for the treatment of brain disorders. To the best of our knowledge, preclinical or clinical studies targeting regulators of H3K4 methylation for drug-based treatment of neurological or psychiatric disease have not yet been reported. Given that other types of ‘epigenetic’ drug treatments, including histone deacetylase inhibitors, exhibit a surprisingly broad therapeutic profile in the preclinical model, including depression and other psychiatric illnesses [101–103] in addition to various neurological disorders [8], it will be extremely interesting to target H3K4 methylation in the treatment of neurodevelopmental and neuropsychiatric disease. Furthermore, the rationale for such an approach is provided by recent studies reporting that RNA-mediated knockdown of H3K4-specific methyltransferases and demethylases, including Mll1 and Kdm5c, affects reward- and addiction-related behaviours in the adult animal [96]. Finally, while the focus of this review was on H3K4 methylation and neurodevelopment, it should be mentioned that regulatory mechanisms associated with several additional types of histone methylation, including H3K9, H3K27 and other repressive marks, have also been implicated in various disorders of the developing and adult brain [31,104–106].
Note added in proof
While this paper was in print, Takata et al. [107] reported that loss-of-function variants of the H3K4-specific methyltransferase SETD1 (SET domain-containing 1A/SET1A) are associated with genetic susceptibility for schizophrenia.
Funding statement
Work in the authors' laboratory is supported by grants from the National Institutes of Health and the Brain Behavior Research Foundation.
References
- 1.Numata S, et al. 2012. DNA methylation signatures in development and aging of the human prefrontal cortex. Am. J. Hum. Genet. 90, 260–272. ( 10.1016/j.ajhg.2011.12.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. 2007. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE 2, e895 ( 10.1371/journal.pone.0000895) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hernandez DG, et al. 2011. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum. Mol. Genet. 20, 1164–1172. ( 10.1093/hmg/ddq561) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung I, Shulha HP, Jiang Y, Matevossian A, Wang J, Weng Z, Akbarian S. 2010. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc. Natl Acad. Sci. USA 107, 8824–8829. ( 10.1073/pnas.1001702107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ronan JL, Wu W, Crabtree GR. 2013. From neural development to cognition: unexpected roles for chromatin. Nat. Rev. Genet. 14, 347–359. ( 10.1038/nrg3413) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein CJ, et al. 2011. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat. Genet. 43, 595–600. ( 10.1038/ng.830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winkelmann J, et al. 2012. Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum. Mol. Genet. 21, 2205–2210. ( 10.1093/hmg/dds035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakovcevski M, Akbarian S. 2012. Epigenetic mechanisms in neurological disease. Nat. Med. 18, 1194–1204. ( 10.1038/nm.2828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. ( 10.1016/j.cell.2007.05.009) [DOI] [PubMed] [Google Scholar]
- 10.Shulha HP, Cheung I, Guo Y, Akbarian S, Weng Z. 2013. Coordinated cell type-specific epigenetic remodeling in prefrontal cortex begins before birth and continues into early adulthood. PLoS Genet. 9, e1003433 ( 10.1371/journal.pgen.1003433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shulha HP, et al. 2012. Epigenetic signatures of autism: trimethylated H3K4 landscapes in prefrontal neurons. Arch. Gen. Psychiatry 69, 314–324. ( 10.1001/archgenpsychiatry.2011.151) [DOI] [PubMed] [Google Scholar]
- 12.Huang HS, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP, Akbarian S. 2007. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J. Neurosci. 27, 11 254–11 262. ( 10.1523/JNEUROSCI.3272-07.2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128, 693–705. ( 10.1016/j.cell.2007.02.005) [DOI] [PubMed] [Google Scholar]
- 14.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–40. ( 10.1038/nsmb1338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan M, et al. 2011. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028. ( 10.1016/j.cell.2011.08.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou VW, Goren A, Bernstein BE. 2011. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 12, 7–18. ( 10.1038/nrg2905) [DOI] [PubMed] [Google Scholar]
- 17.Shulha HP, et al. 2012. Human-specific histone methylation signatures at transcription start sites in prefrontal neurons. PLoS Biol. 10, e1001427 ( 10.1371/journal.pbio.1001427) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrera LO, Li Z, Smith AD, Arden KC, Cavenee WK, Zhang MQ, Green RD, Ren B. 2008. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 18, 46–59. ( 10.1101/gr.6654808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88. ( 10.1016/j.cell.2007.05.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. 2005. Global and Hox-specific roles for the MLL1 methyltransferase. Proc. Natl Acad. Sci. USA 102, 8603–8608. ( 10.1073/pnas.0503072102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shilatifard A. 2008. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 20, 341–348. ( 10.1016/j.ceb.2008.03.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81, 65–95. ( 10.1146/annurev-biochem-051710-134100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maston GA, Landt SG, Snyder M, Green MR. 2012. Characterization of enhancer function from genome-wide analyses. Annu. Rev. Genom. Hum. Genet. 13, 29–57. ( 10.1146/annurev-genom-090711-163723) [DOI] [PubMed] [Google Scholar]
- 24.Creyghton MP, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21 931–21 936. ( 10.1073/pnas.1016071107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283. ( 10.1038/nature09692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J, et al. 2013. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell 152, 642–654. ( 10.1016/j.cell.2012.12.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu JL, Zhou BO, Zhang RR, Zhang KL, Zhou JQ, Xu GL. 2009. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc. Natl Acad. Sci. USA 106, 22 187–22 192. ( 10.1073/pnas.0905767106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ooi SK, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717. ( 10.1038/nature05987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Black JC, Van Rechem C, Whetstine JR. 2012. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell 48, 491–507. ( 10.1016/j.molcel.2012.11.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ernst P, Vakoc CR. 2012. WRAD: enabler of the SET1-family of H3K4 methyltransferases. Brief. Funct. Genomics 11, 217–226. ( 10.1093/bfgp/els017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peter CJ, Akbarian S. 2011. Balancing histone methylation activities in psychiatric disorders. Trends Mol. Med. 17, 372–379. ( 10.1016/j.molmed.2011.02.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rincon-Arano H, Halow J, Delrow JJ, Parkhurst SM, Groudine M. 2012. UpSET recruits HDAC complexes and restricts chromatin accessibility and acetylation at promoter regions. Cell 151, 1214–1228. ( 10.1016/j.cell.2012.11.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sebastian S, Sreenivas P, Sambasivan R, Cheedipudi S, Kandalla P, Pavlath GK, Dhawan J. 2009. MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc. Natl Acad. Sci. USA 106, 4719–4724. ( 10.1073/pnas.0807136106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Del Rizzo PA, Trievel RC. 2011. Substrate and product specificities of SET domain methyltransferases. Epigenetics 6, 1059–1067. ( 10.4161/epi.6.9.16069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal SK, Jothi R. 2012. Genome-wide characterization of menin-dependent H3K4me3 reveals a specific role for menin in the regulation of genes implicated in MEN1-like tumors. PLoS ONE 7, e37952 ( 10.1371/journal.pone.0037952) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J, Gurung B, Wan B, Matkar S, Veniaminova NA, Wan K, Merchant JL, Hua X, Lei M. 2012. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature 482, 542–546. ( 10.1038/nature10806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P, et al. 2009. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell. Biol. 29, 6074–6085. ( 10.1128/MCB.00924-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katada S, Sassone-Corsi P. 2010. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat. Struct. Mol. Biol. 17, 1414–1421. ( 10.1038/nsmb.1961) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. 2009. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458, 529–533. ( 10.1038/nature07726) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. 2008. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol. Cell. Biol. 28, 7337–7344. ( 10.1128/MCB.00976-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Nuland R, Smits AH, Pallaki P, Jansen PW, Vermeulen M, Timmers HT. 2013. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol. Cell. Biol. 33, 2067–2077. ( 10.1128/MCB.01742-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. 2009. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461, 415–418. ( 10.1038/nature08315) [DOI] [PubMed] [Google Scholar]
- 43.Ruthenburg AJ, Allis CD, Wysocka J. 2007. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 25, 15–30. ( 10.1016/j.molcel.2006.12.014) [DOI] [PubMed] [Google Scholar]
- 44.Mosammaparast N, Shi Y. 2010. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79, 155–179. ( 10.1146/annurev.biochem.78.070907.103946) [DOI] [PubMed] [Google Scholar]
- 45.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439. [DOI] [PubMed] [Google Scholar]
- 46.Selvi BR, Mohankrishna DV, Ostwal YB, Kundu TK. 2010. Small molecule modulators of histone acetylation and methylation: a disease perspective. Biochim. Biophys. Acta 1799, 810–828. ( 10.1016/j.bbagrm.2010.09.005) [DOI] [PubMed] [Google Scholar]
- 47.Fang R, et al. 2010. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol. Cell 39, 222–233. ( 10.1016/j.molcel.2010.07.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hannibal MC, et al. 2011. Spectrum of MLL2 (ALR) mutations in 110 cases of Kabuki syndrome. Am. J. Med. Genet. A 155, 1511–1516. ( 10.1002/ajmg.a.34074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ng SB, et al. 2010. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 42, 790–793. ( 10.1038/ng.646) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banka S, et al. 2013. MLL2 mosaic mutations and intragenic deletion-duplications in patients with Kabuki syndrome. Clin. Genet. 83, 467–471. ( 10.1111/j.1399-0004.2012.01955.x) [DOI] [PubMed] [Google Scholar]
- 51.Jones WD, et al. 2012. De novo mutations in MLL cause Wiedemann–Steiner syndrome. Am. J. Hum. Genet. 91, 358–364. ( 10.1016/j.ajhg.2012.06.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kleefstra T, et al. 2012. Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am. J. Hum. Genet. 91, 73–82. ( 10.1016/j.ajhg.2012.05.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Isidor B, Pichon O, Baron S, David A, Le Caignec C. 2010. Deletion of the CUL4B gene in a boy with mental retardation, minor facial anomalies, short stature, hypogonadism, and ataxia. Am. J. Med. Genet. A 152, 175–180. ( 10.1002/ajmg.a.33152) [DOI] [PubMed] [Google Scholar]
- 54.Badura-Stronka M, Jamsheer A, Materna-Kiryluk A, Sowinska A, Kiryluk K, Budny B, Latos-Bieleńska A. 2010. A novel nonsense mutation in CUL4B gene in three brothers with X-linked mental retardation syndrome. Clin. Genet. 77, 141–144. ( 10.1111/j.1399-0004.2009.01331.x) [DOI] [PubMed] [Google Scholar]
- 55.Tarpey PS, et al. 2007. Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am. J. Hum. Genet. 80, 345–352. ( 10.1086/511134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakagawa T, Xiong Y. 2011. X-linked mental retardation gene CUL4B targets ubiquitylation of H3K4 methyltransferase component WDR5 and regulates neuronal gene expression. Molecular Cell 43, 381–391. ( 10.1016/j.molcel.2011.05.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Najmabadi H, et al. 2011. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478, 57–63. ( 10.1038/nature10423) [DOI] [PubMed] [Google Scholar]
- 58.Adegbola A, Gao H, Sommer S, Browning M. 2008. A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD). Am. J. Med. Genet. A 146, 505–511. ( 10.1002/ajmg.a.32142) [DOI] [PubMed] [Google Scholar]
- 59.Iwase S, et al. 2007. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128, 1077–1088. ( 10.1016/j.cell.2007.02.017) [DOI] [PubMed] [Google Scholar]
- 60.Abidi FE, Holloway L, Moore CA, Weaver DD, Simensen RJ, Stevenson RE, Rogers RC, Schwartz CE. 2008. Mutations in JARID1C are associated with X-linked mental retardation, short stature and hyperreflexia. J. Med. Genet. 45, 787–793. ( 10.1136/jmg.2008.058990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poeta L, et al. 2013. A regulatory path associated with X-linked intellectual disability and epilepsy links KDM5C to the polyalanine expansions in ARX. Am. J. Hum. Genet. 92, 114–125. ( 10.1016/j.ajhg.2012.11.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neale BM, et al. 2012. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245. ( 10.1038/nature11011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Roak BJ, et al. 2012. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250. ( 10.1038/nature10989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Banka S, et al. 2012. How genetically heterogeneous is Kabuki syndrome?: MLL2 testing in 116 patients, review and analyses of mutation and phenotypic spectrum. Eur. J. Hum. Genet. 20, 381–388. ( 10.1038/ejhg.2011.220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Y, et al. 2011. A mutation screen in patients with Kabuki syndrome. Hum. Genet. 130, 715–724. ( 10.1007/s00439-011-1004-y) [DOI] [PubMed] [Google Scholar]
- 66.Jensen LR, et al. 2005. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 76, 227–236. ( 10.1086/427563) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. 2007. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 447, 601–605. ( 10.1038/nature05823) [DOI] [PubMed] [Google Scholar]
- 68.Bienvenu T, et al. 2002. ARX, a novel Prd-class-homeobox gene highly expressed in the telencephalon, is mutated in X-linked mental retardation. Hum. Mol. Genet. 11, 981–991. ( 10.1093/hmg/11.8.981) [DOI] [PubMed] [Google Scholar]
- 69.Grønskov K, Hjalgrim H, Nielsen IM, Brøndum-Nielsen K. 2004. Screening of the ARX gene in 682 retarded males. Eur. J. Hum. Genet. 12, 701–705. ( 10.1038/sj.ejhg.5201222) [DOI] [PubMed] [Google Scholar]
- 70.Kato M, et al. 2004. Mutations of ARX are associated with striking pleiotropy and consistent genotype–phenotype correlation. Hum. Mutat. 23, 147–59. ( 10.1002/humu.10310) [DOI] [PubMed] [Google Scholar]
- 71.Kato M, Saitoh S, Kamei A, Shiraishi H, Ueda Y, Akasaka M, Hayasaka K. 2007. A longer polyalanine expansion mutation in the ARX gene causes early infantile epileptic encephalopathy with suppression-burst pattern (Ohtahara syndrome). Am. J. Hum. Genet. 81, 361–366. ( 10.1086/518903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kitamura K, et al. 2002. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat. Genet. 32, 359–369. ( 10.1038/ng1009) [DOI] [PubMed] [Google Scholar]
- 73.Proud VK, Levine C, Carpenter NJ. 1992. New X-linked syndrome with seizures, acquired micrencephaly, and agenesis of the corpus callosum. Am. J. Med. Genet. 43, 458–466. ( 10.1002/ajmg.1320430169) [DOI] [PubMed] [Google Scholar]
- 74.Strømme P, et al. 2002. Mutations in the human ortholog of Aristaless cause X linked mental retardation and epilepsy. Nat. Genet. 30, 441–445. ( 10.1038/ng862) [DOI] [PubMed] [Google Scholar]
- 75.Cabezas DA, Slaugh R, Abidi F, Arena JF, Stevenson RE, Schwartz CE, Lubs HA. 2000. A new X linked mental retardation (XLMR) syndrome with short stature, small testes, muscle wasting, and tremor localises to Xq24–q25. J. Med. Genet. 37, 663–668. ( 10.1136/jmg.37.9.663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ravn K, Lindquist SG, Nielsen K, Dahm TL, Tümer Z. 2012. Deletion of CUL4B leads to concordant phenotype in a monozygotic twin pair. Clin. Genet. 82, 292–294. ( 10.1111/j.1399-0004.2011.01839.x) [DOI] [PubMed] [Google Scholar]
- 77.Tarpey PS, et al. 2009. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat. Genet. 41, 535–543. ( 10.1038/ng.367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zou Y, et al. 2007. Mutation in CUL4B, which encodes a member of cullin-RING ubiquitin ligase complex, causes X-linked mental retardation. Am. J. Hum. Genet. 80, 561–566. ( 10.1086/512489) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. 2010. Histone methylation regulates memory formation. J. Neurosci. 30, 3589–3599. ( 10.1523/JNEUROSCI.3732-09.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim SY, Levenson JM, Korsmeyer S, Sweatt JD, Schumacher A. 2007. Developmental regulation of Eed complex composition governs a switch in global histone modification in brain. J. Biol. Chem. 282, 9962–9972. ( 10.1074/jbc.M608722200) [DOI] [PubMed] [Google Scholar]
- 81.Ansari KI, Mandal SS. 2010. Mixed lineage leukemia: roles in gene expression, hormone signaling and mRNA processing. FEBS J. 277, 1790–1804. ( 10.1111/j.1742-4658.2010.07606.x) [DOI] [PubMed] [Google Scholar]
- 82.Kerimoglu C, et al. 2013. Histone-methyltransferase MLL2 (KMT2B) is required for memory formation in mice. J. Neurosci. 33, 3452–3464. ( 10.1523/JNEUROSCI.3356-12.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marsh E, et al. 2009. Targeted loss of Arx results in a developmental epilepsy mouse model and recapitulates the human phenotype in heterozygous females. Brain 132, 1563–1576. ( 10.1093/brain/awp107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Christensen D, et al. 2013. Prevalence of cerebral palsy, co-occurring autism spectrum disorders, and motor functioning: autism and Developmental Disabilities Monitoring Network, USA, 2008. Dev. Med. Child Neurol. 56, 59–65. ( 10.1111/dmcn.12268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ibrahim HM, Tamminga CA. 2010. Schizophrenia: treatment targets beyond monoamine systems. Annu. Rev. Pharmacol. Toxicol. 51, 189–209. ( 10.1146/annurev.pharmtox.010909.105851) [DOI] [PubMed] [Google Scholar]
- 86.Kenny EM, et al. 2013. Excess of rare novel loss-of-function variants in synaptic genes in schizophrenia and autism spectrum disorders. Mol. Psychiatry epub ahead of print. ( 10.1038/mp.2013.127) [DOI] [PubMed] [Google Scholar]
- 87.Murdoch JD, State MW. 2013. Recent developments in the genetics of autism spectrum disorders. Curr. Opin. Genet. Dev. 23, 310–315. ( 10.1016/j.gde.2013.02.003) [DOI] [PubMed] [Google Scholar]
- 88.Akbarian S, Huang HS. 2006. Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res. Rev. 52, 293–304. ( 10.1016/j.brainresrev.2006.04.001) [DOI] [PubMed] [Google Scholar]
- 89.Connor CM, Crawford BC, Akbarian S. 2011. White matter neuron alterations in schizophrenia and related disorders. Int. J. Dev. Neurosci. 29, 325–334. ( 10.1016/j.ijdevneu.2010.07.236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bagot RC, Zhang TY, Wen X, Nguyen TT, Nguyen HB, Diorio J, Wong TP, Meaney MJ. 2012. Variations in postnatal maternal care and the epigenetic regulation of metabotropic glutamate receptor 1 expression and hippocampal function in the rat. Proc. Natl Acad. Sci. USA 109(Suppl. 2), 17 200–17 207. ( 10.1073/pnas.1204599109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meyer U. 2013. Prenatal poly(I:C) exposure and other developmental immune activation models in rodent systems. Biol. Psychiatry 75, 307–315. ( 10.1016/j.biopsych.2013.07.011) [DOI] [PubMed] [Google Scholar]
- 92.Bauman MD, Iosif AM, Smith SE, Bregere C, Amaral DG, Patterson PH. 2013. Activation of the maternal immune system during pregnancy alters behavioral development of rhesus monkey offspring. Biol. Psychiatry 75, 332–341. ( 10.1016/j.biopsych.2013.06.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Connor CM, Dincer A, Straubhaar J, Galler JR, Houston IB, Akbarian S. 2012. Maternal immune activation alters behavior in adult offspring, with subtle changes in the cortical transcriptome and epigenome. Schizophr. Res. 140, 175–184. ( 10.1016/j.schres.2012.06.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moore H, Jentsch JD, Ghajarnia M, Geyer MA, Grace AA. 2006. A neurobehavioral systems analysis of adult rats exposed to methylazoxymethanol acetate on E17: implications for the neuropathology of schizophrenia. Biol. Psychiatry 60, 253–264. ( 10.1016/j.biopsych.2006.01.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mackowiak M, Bator E, Latusz J, Mordalska P, Wedzony K. 2013. Prenatal MAM administration affects histone H3 methylation in postnatal life in the rat medial prefrontal cortex. Eur. Neuropsychopharmacol. 24, 271–289. ( 10.1016/j.euroneuro.2013.05.013) [DOI] [PubMed] [Google Scholar]
- 96.Aguilar-Valles A, Vaissiere T, Griggs EM, Mikaelsson MA, Takacs IF, Young EJ, Rumbaugh G, Miller CA. 2013. Methamphetamine-associated memory is regulated by a writer and an eraser of permissive histone methylation. Biol. Psychiatry 76, 57–65. ( 10.1016/j.biopsych.2013.09.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Akbarian S, et al. 2005. Chromatin alterations associated with down-regulated metabolic gene expression in the prefrontal cortex of subjects with schizophrenia. Arch. Gen. Psychiatry 62, 829–840. ( 10.1001/archpsyc.62.8.829) [DOI] [PubMed] [Google Scholar]
- 98.Kano S, et al. 2013. Genome-wide profiling of multiple histone methylations in olfactory cells: further implications for cellular susceptibility to oxidative stress in schizophrenia. Mol. Psychiatry 18, 740–742. ( 10.1038/mp.2012.120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cruceanu C, Alda M, Nagy C, Freemantle E, Rouleau GA, Turecki G. 2013. H3K4 tri-methylation in synapsin genes leads to different expression patterns in bipolar disorder and major depression. Int. J. Neuropsychopharmacol. 16, 289–299. ( 10.1017/S1461145712000363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ernst C, Chen ES, Turecki G. 2009. Histone methylation and decreased expression of TrkB.T1 in orbital frontal cortex of suicide completers. Mol. Psychiatry 14, 830–832. ( 10.1038/mp.2009.35) [DOI] [PubMed] [Google Scholar]
- 101.Morris MJ, Karra AS, Monteggia LM. 2010. Histone deacetylases govern cellular mechanisms underlying behavioral and synaptic plasticity in the developing and adult brain. Behav. Pharmacol. 21, 409–419. ( 10.1097/FBP.0b013e32833c20c0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Covington HE, 3rd, et al. 2009. Antidepressant actions of histone deacetylase inhibitors. J. Neurosci. 29, 11 451–11 460. ( 10.1523/JNEUROSCI.1758-09.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schroeder FA, Lin CL, Crusio WE, Akbarian S. 2007. Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. Biol. Psychiatry 62, 55–64. ( 10.1016/j.biopsych.2006.06.036) [DOI] [PubMed] [Google Scholar]
- 104.Parkel S, Lopez-Atalaya JP, Barco A. 2013. Histone H3 lysine methylation in cognition and intellectual disability disorders. Learn. Mem. 20, 570–579. ( 10.1101/lm.029363.112) [DOI] [PubMed] [Google Scholar]
- 105.Jarome TJ, Lubin FD. 2013. Histone lysine methylation: critical regulator of memory and behavior. Rev. Neurosci. 24, 375–387. ( 10.1515/revneuro-2013-0008) [DOI] [PubMed] [Google Scholar]
- 106.Gilman SR, Chang J, Xu B, Bawa TS, Gogos JA, Karayiorgou M, Vitkup D. 2012. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat. Neurosci. 15, 1723–1728. ( 10.1038/nn.3261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Takata A, Xu B, Ionita-Laza I, Roos JL, Gogos JA, Karayiorgou M. 2014. Loss-of-function variants in schizophrenia risk and SETD1A as a candidate susceptibility gene. Neuron 82, 773–780. ( 10.1016/j.neuron.2014.04.043) [DOI] [PMC free article] [PubMed] [Google Scholar]