Abstract
Consumption of dietary flavonoids has been associated with reduced mortality and risk of cardiovascular disease, partially by reducing triglyceridemia. We have previously reported that a grape seed procyanidin extract (GSPE) reduces postprandial triglyceridemia in normolipidemic animals signaling through the orphan nuclear receptor Small Heterodimer Partner (SHP) a target of the bile acid receptor Farnesoid X Receptor (FXR). Our aim was to elucidate whether FXR mediates the hypotriglyceridemic effect of procyanidins. In FXR-driven luciferase expression assays GSPE dose-dependently enhanced FXR activity in the presence of chenodeoxycholic acid. GSPE gavage reduced triglyceridemia in wild type mice but not in FXR-null mice, revealing FXR as an essential mediator of the hypotriglyceridemic actions of procyanidins in vivo. In the liver, GSPE down-regulated, in a FXR-dependent manner, the expression of the transcription factor Steroid Response Element Binding Protein 1 (SREBP1) and several SREBP1 target genes involved in lipogenesis, and upregulated ApoA5 expression. Altogether, our results indicate that procyanidins lower triglyceridemia following the same pathway as bile acids: activation of FXR, transient upregulation of SHP expression and subsequent downregulation of SREBP1 expression. This study adds dietary procyanidins to the arsenal of FXR ligands with potential therapeutic use to combat hypertriglyceridemia, type 2 diabetes and metabolic syndrome.
Keywords: Bile Acids, FXR, liver, procyanidins, SHP, SBARM, SREBP1, triglycerides
INTRODUCTION
Procyanidins, which are oligomers and polymers of polyhydroxyflavan-3-ol units, are the most abundant polyphenols in grapes, apples, red grape juice, red wine and chocolate [1, 2]. These flavonoids have been shown to prevent and ameliorate atherosclerosis and other factors of cardiovascular disease, a fact that was primarily ascribed to their antioxidant activity and the modulation of diverse signaling pathways in vascular system [2, 3]. However, the antiatherogenic properties of procyanidins are also attributable to a reduction of plasma levels of ApoB-containing TG-rich proatherogenic lipoproteins, i.e. intestinal chylomicrons and hepatic VLDL and LDL, as well as to an improved serum cholesterol profile. Thus, in hamster models of diet-induced atherosclerosis, chronic administration of procyanidins inhibit aortic fatty streak area and progression of atherosclerosis, and lowers plasma TG, ApoB, and non-HDL-cholesterol [4, 5]. In ovariectomized Guinea pigs and in postmenopausal women, grape-seed procyanidin-rich extracts diminish plasma TG and VLDL-cholesterol, as well as cholesterol accumulation in the aorta [6]. In rats fed hypercholesterolemic diets, chronic consumption of a grape seed procyanidin extract (GSPE) reduced plasma TG, LDL-cholesterol and VLDL concentrations, while increasing plasma HDL-cholesterol in the fasted state [7, 8]. The hypolipidemic effect of procyanidins may be exerted in part through the inhibition of the absorption of dietary lipids, and diminished chylomicron secretion by enterocytes [9, 10]. But also hepatocytes respond to red wine and red grape juice polyphenols diminishing the secretion of ApoB100 and increasing LDL-receptor activity [11–13]. Nevertheless, the molecular mechanisms that underlie the improvement of plasma lipid profile by procyanidins are largely unknown.
We have previously shown that an acute dose of GSPE reduces postprandial triglyceridemia and plasma ApoB levels in normolipidemic rats while concomitantly increasing hepatic mRNA levels of SHP [14], an orphan nuclear receptor that regulates bile acids, cholesterol, TG and glucose homeostasis in the liver (recently reviewed in [15]). In HepG2 cells, grape procyanidins require the activity of SHP to reduce secretion of TG whereas, in contrast, they reduce the secretion of ApoB in a SHP-independent way [16]. Also, SHP mediates the hypotriglyceridemic effect of grape procyanidins in wild-type mouse in the postprandial state, which is accompanied by the downregulation of hepatic expression of transcription factor SREBP1 [16], a master mediator for insulin/glucose signaling to lipogenesis (reviewed in [17, 18]).
It is well established that bile acids (BA) are potent hypotriglyceridemic agents. These effects of BA are mediated by their binding to the bile acid receptor FXR, a transcription factor that, like SHP, exerts metabolic control on bile acids, lipid and glucose homeostasis (reviewed in [19–22]). FXR activation by bile acids lowers plasma TG levels by repressing hepatic lipogenesis and TG secretion, and by increasing the clearance of TG-rich lipoproteins from the blood (reviewed in [22, 23]). In the liver, BA-activated FXR upregulates the expression of SHP, which in turn represses the expression of SREBP1, which is translated into a diminished hepatic FA synthesis and an increased plasma TG catabolism [24].
Besides procyanidins, two other phytochemicals, guggulsterone from the guggul tree [25, 26] and xanthohumol from beer hops [27] have been described to lower triglyceridemia in vivo regulating the hepatic expression of a subset of FXR target genes, including SHP and SREBP1. Both xanthohumol and guggulsterone behave as selective bile acid receptor modulators (SBARM) that, like bile acids, enhance transcriptional activity of FXR in in vitro assays [25–27].
The partial similarity between activators of FXR and GSPE regarding changes elicited in liver gene expression profile and in plasma lipid parameters prompted us to hypothesize that procyanidins might enhance the transcriptional activity of FXR and, consequently, that FXR could mediate the hypotriglyceridemic effects of GSPE. To test these hypotheses we have here evaluated the effect of GSPE on plasma TG levels in wild-type and FXR−/− mice, and have assessed the effects of GSPE on a cell-based FXR-responsive luciferase expression assay. The results show that procyanidins enhance the activity of CDCA-activated FXR, and that, like bile acids, signal through FXR to lower triglyceridemia, concomitantly inhibiting hepatic expression of SREBP1 and several SREBP1 target genes involved in lipogenesis in a FXR-dependent manner.
MATERIALS AND METHODS
Chemicals
A grape seed procyanidin extract (GSPE) was kindly provided by Les Dérives Résiniques et Térpeniques (Dax, France). This extract contains monomeric catechins (polyhydroxyflavan-3-ol) (16.55%), dimeric (18.77%), trimeric (16%), tetrameric (9.3%) and oligomeric (5–13 units) (35.7%) procyanidins, as well as phenolic acids (4.22 %). Chenodeoxycholic acid (CDCA) was from Sigma and GW4064 was a kind gift from Tim Willson (GlaxoSmithKline).
Cell transfections and luciferase reporter assays
Human epithelial cells (HeLa) and African Green Monkey Fibroblasts (CV-1) were obtained from the American Type Tissue Culture Collection. HeLa and CV-1 cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% FBS at 37°C and 5% C02. For transfections, cells were plated into 24-well plates (Becton Dickinson) (1.3 × 105 (HeLa) or 1 × 105 (CV-1)) in DMEM plus 10% charcoal stripped serum to 80% confluence, then cotransfected the next day using the calcium phosphate precipitation method. Plasmids for expression of full-length mouse FXR, for expression of Gal4 DBD fused to FXR LBD, the FXR-responsive luciferase reporter plasmid ((PLTP)2 TKluc), the Gal4-driven luciferase reporter plasmid, and the plasmid expressing β-galactosidase (CMX-β-gal) have been previously described [25, 28]. The CDM-RXRα plasmid (expressing full-length human RXRα) has also been described [29]. Cells were plated in 24-well dishes with DMEM supplemented with 10% charcoal-stripped serum. Transfections included 100 ng of the plasmid encoding full-length FXR or the Gal4 DBD-FXR LBD fusion, 200 ng of the luciferase reporter plasmid, 10 ng of CDM-RXRα, 200 ng of CMX-β-gal (used as internal control for transfection efficiency) and 490 ng of pGEM4 (Promega), used as carrier DNA, to make a total of 1μg of plasmid DNA per well. The next morning, cells were washed with phosphate buffered saline and FXR ligands (CDCA, GW4064) or GSPE were added as indicated. Ligands were dissolved in DMSO whereas GSPE was dissolved in ethanol. Cells were assayed for luciferase activities (Promega luciferase assay kit) 24 hours after addition of vehicle, ligands or GSPE, and reporter expression was normalized to β-galactosidase activity (β-gal Assay kit, Applied Biosystems), measured with a MLX luminometer (DYNEX Technologies). Results were obtained from at least three independent experiments, each performed in triplicate. For luciferase-based studies, T-test analyses were performed using SPSS software.
In vivo feeding studies
Mice were housed under standard conditions. Experimental procedures were approved by the local Committee for Care and Use of Laboratory Animals at Baylor College of Medicine. The FXR-deficient mice have been previously described [30], and were backcrossed with C57BL6 mice to the 10th generation. The correct genotype was verified using previously reported primers and PCR conditions [30]. Age-matched groups of 8–10 week-old male mice were used in all experiments (n=5 per experimental group). Mice were fed a standard rodent chow and water ad libitum. On experimental day, mice were fed either vehicle (water), or procyanidins (250 mg/kg) via oral gavage. A first dose was administered at 9:00 pm and a second dose 12 hours later; food was then retired and 2 hours later mice were anesthetized with isoflurane. Blood was collected from the orbital plexus and livers were snap-frozen and stored at −80°C until use. Plasma cholesterol and TG levels were assessed with enzymatic kits as described [14].
Gene expression analysis
Total RNA was obtained using Trizol reagent (Invitrogen). and further purified using NucleoSpin RNA2 kit (Macherie-Naegel). For microarray hybridizations, the 5 RNA samples from each of the 4 treatment groups were pooled and its integrity was assessed using the Agilent 2100 Bioanalyzer. Cy3- or Cy5-labeled cRNA was obtained from each RNA pool using the Agilent Low RNA Input Fluorescent Linear Amplification Kit (Agilent manual 5185–5818). Labeled cRNAs were hybridized against Agilent Mouse 60-mer Oligo Microarrays (Part Number G4122A) following Agilent’s instruction. Duplicate hybridizations with dye-swap labeling were performed with each pair of RNA samples being compared. Images of hybridized microarrays were acquired with the Agilent G2565BA scanner, and data from the microarray images were obtained and analyzed with the Agilent Feature Extraction software. For validation of microarray data, relative mRNA levels of SREBP1, CYP7A1, ApoA5, and SHP genes were analyzed by real-time PCR, using GAPDH as the endogenous control. RNA was retrotranscribed using TaqMan Reverse Transcription Reagents kit (Applied Biosystems) and gene expression was evaluated in the ABIPrism 7300 SDS Real Time PCR system (Applied Biosystems) using SYBR Green PCR Master Mix (Applied Biosystems) and gene-specific primers (sequences of the primers used in real-time PCR reactions are available upon request).
Microarray data processing
A whole array of data was constructed matching each gene symbol or Genbank ID with its fold-change value from the microarray analysis. Genes were clustered into different biological processes using Panther software [31]. The gene expression profile deviation of each biological process group from the whole array expression pattern was calculated using the Mann-Whitney U Test (Wilcoxon Rank-Sum test) as described [32]. The resulting p-values were considered significant when smaller than 0.05.
RESULTS
GSPE requires FXR activity to reduce triglyceridemia in mice
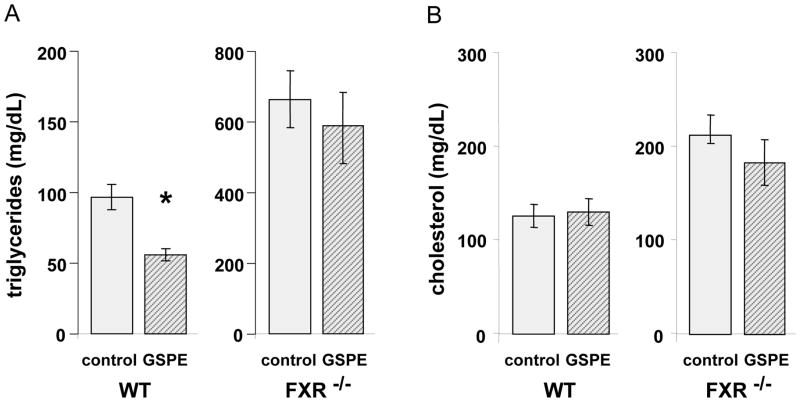
In order to assess the relevance of FXR as a mediator of GSPE hypotriglyceridemic actions in an in vivo model, we compared the effects of GSPE administration in FXR−/−versus wild type mice. As previously described [30], FXR−/− mice present elevated basal levels of plasma TG and cholesterol when compared to wild type mice (Figure 1). Oral GSPE gavage triggered a 40% reduction in plasma TG levels in wild type mice in the postprandial state whereas it did not modify plasma total cholesterol levels. This response to GSPE administration is identical to that previously found in rats [14]. In contrast, GSPE gavage did not cause any statistically significant reduction in plasma TG levels in FXR−/− animals. Therefore, FXR is a key mediator of the hypotriglyceridemic activity of procyanidins in mice.
Figure 1. Effect of GSPE gavage on plasma TG and cholesterol levels in wild-type and FXR-null mice.
Wild type (WT) and FXR−/− mice were fed with vehicle (control) or 250 mg/kg GSPE and plasma TG (panel A) and total cholesterol (panel B) were determined as described in Materials and Methods. * denotes significant differences versus control (p< 0.05).
GSPE represses the expression of SREBP1 and SREBP1-target genes in wild-type but not FXR−/− mice
To gain further insight into the FXR-dependent actions of procyanidins, we next analyzed the differential response in gene expression changes induced by GSPE gavage in the livers of wild type and FXR−/− mice using oligonucleotide microarray hybridization. The changes induced by GSPE treatment in the expression level of all genes, clustered by biological process, were subjected to unbiased analysis using Panther software [31], evaluating the changes in each cluster of genes by means of the Mann-Whitney U Test (Wilcoxon Rank-Sum test) [32]. In wild type mice, changes in genes clustered in the biological process “Lipid, fatty acid and steroid metabolism”, including 747 genes, showed a significant overall repression (p-value 0.018). In contrast, in FXR−/− mice, GSPE treatment did not significantly affect this gene cluster (p-value 0.5). These results could indicate that lipid metabolism is repressed to some extent by GSPE in wild type but not in FXR−/− mice, thus placing FXR as a mediator of the repression of lipid related genes by procyanidins in liver.
Next, in order to identify FXR-dependent changes in the expression of genes putatively involved in the hypotriglyceridemic effect of GSPE in wild type mice, we selected those genes clustered into the “Lipid, fatty acid and steroid metabolism” by PANTHER method [31] whose expression was altered by GSPE treatment in wild-type mice but remained unaltered in FXR−/− mice, setting a fold-change threshold of 1.5 for up-regulated and 0.7 for down-regulated genes (Table 1). In total, 31 lipid-related genes were identified that showed FXR-dependent repression by GSPE, including transcription factor SREBP1, a key regulator of fatty acid and TG synthesis and lipoprotein metabolism [17, 18], acyl-CoA synthetase Acss2/Acsl1 (involved in fatty acid synthesis) [33], the fatty acid desaturases Scd1 and Scd2 [34], and several genes encoding cholesterol biosynthetic enzymes. Two other genes involved in lipid and lipoprotein metabolism, although not classified by Panther software in this cluster, also changed in a FXR-dependent manner, and were included in Table 1: ApoA5 (involved in VLDL catabolism) [35, 36] and the transcription factor C/EBP-β (a regulator of glucose and lipid homeostasis [37]. Remarkably, several of the genes which showed an FXR-dependent response to GSPE, have been previously described as targets of SREBP1. Also, many of the genes which have been identified in this screening, including SREBP1, have been previously characterized as SHP-dependent GSPE-responsive genes (marked with an asterisk in Table 1) [16]. Therefore, SREBP1 and SREBP1 target genes emerge as putative FXR-and SHP-dependent effectors of the hypotriglyceridemic response triggered by procyanidins in vivo.
Table 1.
FXR-dependent changes induced by GSPE in the hepatic expression of lipid and lipoprotein related genes.
| Genebank ID | Gene name; symbol | fold change upon GSPE treatment | SREBP1 target | |
|---|---|---|---|---|
| FA and TG synthesis and metabolism | WT | FXR−/− | ||
| NM_011480 | sterol regulatory element binding factor 1;Srebf1 (SREBP1) * | 0.7 | 1.0 | [53] |
| 0.6 | 1.0 | |||
| NM_019811 | acyl-CoA synthetase short-chain family member 2; Acss2/Acsl1 * | 0.6 | 0.9 | [33] |
| NM_146197 | acyl-CoA synthetase medium-chain family member 2; Acsm2 * | 0.6 | 0.8 | |
| NM_009127 | stearoyl-Coenzyme A desaturase 1; Scd1 | 0.6 | 1.1 | [34] |
| NM_009128 | stearoyl-Coenzyme A desaturase 2; Scd2 * | 0.7 | 1.0 | |
| NM_028089 | cytochrome P450, family 2, subfamily c, polypeptide 55; Cyp2c55 * | 0.6 | 1.0 | |
| NM_175443 | ethanolamine kinase 2; Etnk2 * | 0.7 | 0.9 | |
| NM_008903 | phosphatidic acid phosphatase 2a; Ppap2a * | 0.6 | 0.8 | |
| NM_008846 | phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha; Pip5k1a | 0.7 | 0.9 | |
| NM_008845 | phosphatidylinositol-4-phosphate 5-kinase, type II, alpha; Pip5k2a * | 0.7 | 0.9 | |
| NM_207683 | phosphatidylinositol 3-kinase, C2 domain containing, gamma polypeptide; Pik3c2g | 0.7 | 1.0 | |
| NM_013490 | choline kinase alpha; Chka * | 0.7 | 0.8 | |
| NM_019677 | phospholipase C, beta 1; Plcb1 | 0.7 | 1.0 | |
| NM_080434 | Apolipoprotein A5; ApoA5 * | 1.4 | 1.0 | [36] |
| 1.7 | 1.0 | |||
| Cholesterol Biosynthesis | ||||
| NM_145942 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1; Hmgcs1 * | 0.7 | 0.9 | [39, 42] |
| NM_008255 | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase; Hmgcr * | 0.7 | 0.8 | |
| NM_023556 | mevalonate kinase; Mvk * | 0.7 | 0.9 | |
| NM_026784 | phosphomevalonate kinase; Pmvk * | 0.7 | 0.9 | |
| NM_138656 | mevalonate (diphospho) decarboxylase; Mvd | 0.7 | 0.9 | |
| NM_134469 | farnesyl diphosphate synthetase; Fdps * | 0.7 | 0.9 | |
| NM_010941 | NAD(P) dependent steroid dehydrogenase-like; Nsdhl * | 0.7 | 0.9 | |
| NM_172769 | sterol-C5-desaturase ; Sc5d * | 0.7 | 0.9 | |
| NM_007856 | 7-dehydrocholesterol reductase; Dhcr7 * | 0.7 | 0.9 | |
| Other lipid-related | ||||
| NM_009883 | CCAAT/enhancer binding protein (C/EBP), beta; Cebpb * | 0.7 | 0.9 | [37] |
| NM_013634 | peroxisome proliferator activated receptor binding protein; Pparbp * | 0.7 | 1.0 | |
| NM_011374 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1; St8sia1 | 0.7 | 0.9 | |
| NM_018784 | ST3 beta-galactoside alpha-2,3-sialyltransferase 6; St3gal6 * | 0.7 | 1.3 | |
| NM_011372 | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3; St6galnac3 | 0.7 | 0.9 | |
| NM_018830 | N-acylsphingosine amidohydrolase 2; Asah2 | 0.5 | 1.2 | |
| NM_153389 | ATPase, Class V, type 10D; Atp10d | 0.7 | 1.0 | |
| NM_028057 | cytochrome b5 reductase 1; Cyb5r1 * | 0.7 | 1.5 | |
| NM_012054 | acyloxyacyl hydrolase; Aoah | 0.6 | 1.0 | |
Wild type and FXR−/− mice (n=5 in each treatment group, 8–10 weeks old) were fed two doses of either vehicle or GSPE (250 mg/Kg), separated by a 12 h time lapse, as described in Materials and Methods. Two hours after the second gavage, liver total RNA from the 4 groups was obtained and pooled. Microarray data was obtained by comparing gene expression of WT control versus WT GSPE-treated mice and FXR−/− control versus FXR−/− GSPE-treated mice. The whole microarray fold-changes were processed using Panther software in order to identify FXR-dependent changes induced by GSPE in genes clustered in the “Lipid, fatty acid and steroid metabolism” metabolic pathway. ApoA5 and CEBP/β are not included in this cluster by Panther software and were added separately. Fold-change thresholds were fixed as 0.7 and 1.5 for down-regulation and up-regulation respectively. Real time quantitative PCR was performed with selected genes to confirm the microarray data (shown in bold characters). Selected references are given for known SREBP1 target genes. Asterisks highlight those genes previously found to be responsive to GSPE in a SHP-dependent way [16]. WT column: fold-change in expression induced by GSPE in wild-type mice relative to the expression in wild type mice treated with vehicle. FXR−/− column: fold-change in expression induced by GSPE in FXR−/− mice relative to the expression in FXR−/− mice treated with vehicle only.
GSPE enhances the transcriptional activity of CDCA-activated FXR in CV-1 and HeLa cells
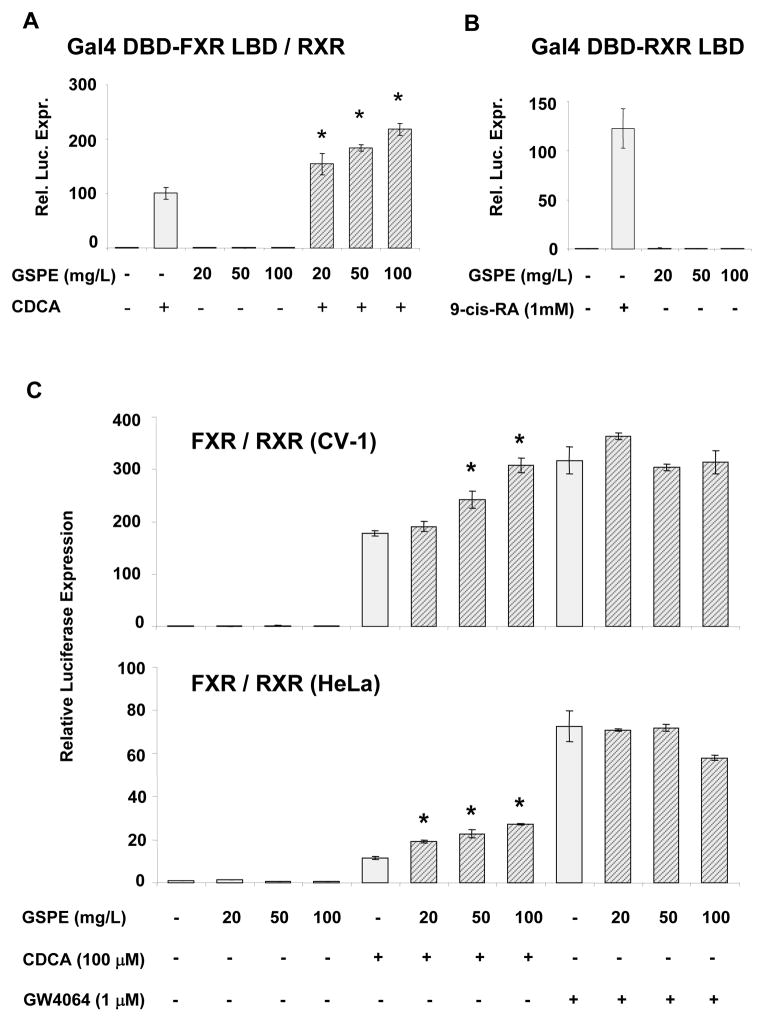
The lack of hypotriglyceridemic effect of GSPE in FXR-null mice prompted us to test whether procyanidins can modulate the transcriptional activity of the FXR/RXR heterodimer in an in vitro system, co-transfecting CV-1 cells with different constructs expressing full-length RXR and either a Gal4 DBD-FXR LBD chimera or full-length FXR together with a FXR-responsive luciferase reporter plasmid (Figure 2). A vector expressing full-length Gal4 was co-transfected as a control, in order to discard interactions of GSPE with the DNA-binding domain of this protein (data not shown). GSPE displayed no significant effects on RXR activity as assayed using a Gal4 DBD-RXR LBD chimera (Figure 2B). In order to assess the interactions of GSPE with FXR, a Gal4 DBD-FXR LBD chimera (Figure 2A) or full length FXR (Figure 2C) was cotransfected with an RXR expression plasmid. In both cases, GSPE alone did not cause transactivation of FXR/RXR. In contrast, when GSPE was added to transfected cells together with the bile acid CDCA, a natural FXR ligand, it enhanced the transactivation of FXR in a dose-dependent manner, reaching a 2-fold increase when cells where incubated with 100 mg/L of GSPE and 100 μM CDCA compared with the CDCA treatment alone. On the contrary, GSPE did not increase the transactivation of FXR induced by GW4064, a synthetic non-steroidal FXR agonist (Figure 2C). In order to discard cell-specific actions of GSPE on FXR transactivation, the vector expressing full-length FXR was transfected, along with the vector expressing full-length RXR and the FXR-responsive luciferase reporter plasmid, (PLTP)2 TKluc, in HeLa cells, and equivalent transactivation to those found in CV-1 was observed (Figure 2C). These results show that procyanidins enhance the transcriptional activity of CDCA-activated FXR, but not that of GW4064-activated FXR, and thus they behave as activator-dependent FXR co-agonists in a cell-based FXR-driven luciferase expression assay.
Figure 2. Cell-based assays of GSPE effects on FXR-dependent luciferase expression.
(A) To study the effect of GSPE on transcriptional activity of FXR/RXR, the Gal4 DBD-FXR LBD expression vector and the Gal4 luciferase reporter plasmid were cotransfected in CV-1 cells, and these were treated with vehicle, GSPE and/or 100 μM CDCA. (B) RXR interactions with GSPE were assayed using the Gal4:DBD-RXR:LBD expression vector, along with the Gal4 luciferase reporter plasmid. Transfected cells were treated with vehicle (−) or 1 mM 9-cis-retinoic acid (+) and GSPE at the indicated concentrations. (C) To assess activation of full-length FXR by GSPE, a full length FXR expression plasmid along with a reporter construct containing a (PLTP)2 TKluc were co-transfected in CV-1 cells (upper panel) or in HeLa cells (lower panel). Ligands for FXR were CDCA (100μM) or GW4064 (1μM). GSPE was added in the indicated concentrations. All controls (−) were treated with the respective vehicles in a final concentration lower than 0.1%. All transfections included the expression vector for RXR to allow the formation of FXR/RXR heterodimers, and CMX-β-Gal as internal control. All DNA constructs have been previously described [25, 28, 29]. Values represent the mean fold-change with respect to control values, obtained from three independent experiments. * denotes significant differences at the p<0.05 level respect the CDCA treatment.
DISCUSSION
The essential role of FXR in mediating the hypotriglyceridemic actions of procyanidins in vivo has been revealed by administering GSPE to wild-type and FXR-null mice. In accordance with our previous results in wild type rats and mice [14, 16], oral gavage of GSPE elicited an hypotriglyceridemic effect in wild type mice in the postprandial state, without affecting total plasma cholesterol levels. In contrast, GSPE was ineffective in reducing plasma TG levels in FXR-null mice, which, as previously reported [30] displayed hypertriglyceridemia and hypercholesterolemia. Therefore, procyanidins need the presence and activity of FXR, i.e. they act through an FXR-dependent pathway, to exert hypotriglyceridemic actions in vivo. Genome-wide analysis of liver gene expression profile has identified a group of genes whose expression is responsive to GSPE treatment in wild-type mouse but not in the FXR-null genotype, i.e., FXR-dependent GSPE target genes, that provide some clues to understand the FXR-dependent mechanisms used by procyanidins to lower plasma TG levels. Remarkably, most of these FXR-dependent GSPE targets have been already identified as SHP-dependent targets of GSPE [16], including the transcription factor SREBP1 and several genes known to be regulated by it. SREBP1 is a key mediator for insulin/glucose signaling to lipogenesis; overexpression of the mature form of SREBP1a or SREBP1c leads to increased hepatic FA biosynthesis and TG levels and its inhibition has been proposed as a method for lowering triglyceridemia [17, 38, 39]. Known targets of SREBP1 found here to be regulated by GSPE include genes involved in the synthesis of monounsaturated and polyunsaturated FA: Acetyl-CoA synthetase Acss2/Acsl1 [33], and two stearoyl-coenzyme A desaturases (SCD), Scd1 and Scd2 [34]. Deficiency in SCD activity, the rate-limiting enzyme for the biosynthesis of monounsaturated FA, greatly reduces hepatic TG synthesis and protects mice against hypertriglyceridemia induced by LXR activation [40]. Since the liver lipid pool is a limiting factor in the synthesis and secretion of VLDLs by the liver [41], a decrease in hepatic lipogenesis should reduce the number of VLDLs or the TG content of these lipoproteins. Also the genes encoding cholesterol biosynthetic enzymes that are downregulated by GSPE in a FXR-dependent and a SHP-dependent manner are targets of SREBP1a, which regulates their expression in concert with SREBP2 [39, 42]. Downregulation of these genes in WT mice suggests that GSPE could potentially inhibit cholesterol biosynthesis in the liver, although this was not translated into total plasma cholesterol levels in the postprandial phase. In this regard, our previous study in rats [14] showed no effects of GSPE on plasma total cholesterol, although significantly lowered the cholesterol associated with TG-rich lipoproteins. However, chronic ingestion of monomeric catechins or oligomeric procyanidins is effective in lowering not only triglyceridemia, but also cholesterolemia, VLD- and LDL-cholesterol, while increasing HDL-cholesterol [7] suggesting that, on the long term, downregulation of cholesterol biosynthetic enzymes by GSPE may be translated into diminished plasma cholesterol levels. In addition, GSPE upregulated ApoA5 expression in mouse liver in a FXR- and SHP-dependent manner. It is known that ApoA5 gene expression is downregulated by LXR ligands through upregulation of SREBP1c and that overexpression of ApoA5 reduces plasma TG in hypertriglyceridemic mice; the hypotriglyceridemic action of ApoA5 is attributed to the inhibition of lipidation of ApoB and the activation of lipase-mediated VLDL-TG hydrolysis and consequent acceleration of VLDL catabolism [35, 36]. Taken together, this pattern of FXR-dependent changes elicited by GSPE strongly suggest that the hypotriglyceridemic effect elicited by dietary procyanidins is brought about, at least in part, through inhibition of hepatic lipogenesis and acceleration of TG-rich lipoproteins catabolism.
It is noteworthy that the changes in liver gene expression observed here in GSPE-treated mice differ from that previously observed in the liver of rats treated with GSPE [14]. Thus, SHP and CYP7A1 were found to be upregulated by GSPE in rats livers, but none of these changes have been observed here in the liver of GSPE-treated mice. However, since microarray analysis was performed at one time point in each case, it is feasible that the observed differences in the expression of known FXR targets just reflect time-dependent variations in gene expression. In support of this view, it is known that bile acids and GW4064 enhance hepatic expression of SHP only transiently, and that transient inductions of SHP are sufficient to exert an hypotriglyceridemic effect [24, 43]. Upregulation of SHP expression by GSPE is also transient in HepG2 cells, were GSPE inhibits TG secretion [16]. Another possible factor to explain the observed variations between hepatic gene expression induced by GSPE in rats and mice is the fact that the strength and specificity of FXR activity is ligand and promoter-dependent, as has been shown comparing the transcriptional activity of FXR bound to the different bile acids (i.e., cholate, lithocholate, deoxycholate, chenodeoxycholate and ursodeoxycholate) [44] and to synthetic FXR agonists such as GW4064 and fexaramine [45, 46]. The binding of each ligand results in a different FXR conformation, which in turn differentially regulates expression of a subset of FXR targets, and with different potency. This is related to the binding of different transcriptional coactivators, such as TRRAP [47] or PGC-1α [48], to the ligand binding domain of FXR, which occurs in a ligand and promoter-specific fashion [45]. According to the results of the FXR-responsive luciferase expression assays, procyanidins from the grape seed extract cannot by themselves enhance the transcriptional activity of the FXR/RXR heterodimer, but instead behave as ligand-dependent co-agonists, enhancing the transcriptional strength of CDCA-activated FXR, but not that of GW4064-activated FXR. Although this result strongly suggest that procyanidin species present in GSPE, or their metabolites, directly bind to CDCA-bound FXR to enhance its transcriptional activity, it does not exclude the possibility that procyanidins might enhance FXR activity by binding through a FXR cofactor which should be commonly present in HeLa and CV-1 cells. In any case, different procyanidin/bile acid combinations, ultimately due to species specific differences in bile acids metabolism and in the absorption and metabolization of procyanidins, should result in activation of different subsets of FXR-target genes, different strengths of activation and/or different temporal patterns of expression. The enhancement of bile acid-activated FXR activity by procyanidins is expected to occur in vivo, both in hepatic cells and enterocytes, where FXR is highly expressed and hepatic synthesis and enterohepatic circulation guarantee the presence of bile acids. In this regard, monomeric catechins and dimeric to trimeric procyanidins have been detected in urine after oral gavage of GSPE to rats [49]. Likewise, catechins and dimeric to pentameric procyanidins have been detected in plasma after administration of apple procyanidin extracts to rats [50].
In summary, our results indicate that procyanidins act through an FXR-dependent pathway to exert hypotriglyceridemic actions in vivo. This effect is also dependent on SHP-activity [16] and is concomitant with downregulation of hepatic expression of SREBP1. FXR-responsive luciferase expression assays indicate that procyanidins act as bile acid dependent coactivators of FXR activity. Taken together, these results suggest that, in vivo, procyanidins exert hypotriglyceridemic effect following a pathway that goes with, and is dependent on, that followed by bile acids, i.e., activation of FXR, transient upregulation of SHP expression and subsequent downregulation of SREBP1 expression, which is translated into a diminished hepatic FA synthesis and an increased plasma TG catabolism.
FXR activity plays a key role in controlling not only triglyceridemia but also cholesterol, bile acid and glucose homeostasis and modulation of FXR has been proposed as a therapeutic target in the treatment of hyperlipidemia, hyperglycemia and metabolic syndrome [21, 22, 51, 52]. Consequently, dietary procyanidins, acting as activators of FXR, emerge as promising natural agents for the treatment of these metabolic disorders. Further research is required to identify the individual procyanidins that enhance FXR activity and to clear the mechanisms and metabolic consequences of this activation.
Acknowledgments
This study was supported by grant number AGL2005-04889 from the Spanish Comisión Interministerial de Ciencia y Tecnología (CICYT) and by grant number CO3/O8 from the Spanish Fondo de Investigaciones Sanitarias (FIS). In the US this work was supported by the USDA (ARS, CRIS 6250-51000-034) (D.D.M.) and the National Institutes of Health (RO1-DK53366) (D.D.M.) J.M. del Bas was the recipient of a fellowship from the Spanish Ministry of Science and Technology. M. Vaqué was the recipient of a fellowship from grant number CO3/O8. The authors thanks Tim Willson (GlaxoSmithKline) for the gift of GW4064. We gratefully acknowledge the expert technical assistance of the Microarray Core Facility at Centro Nacional de Investigaciones Oncológicas (Madrid, Spain) in performing microarray hybridizations and data acquisition.
List of abbreviations
- ApoA5
apolipoprotein A5
- ApoB
apolipoprotein B
- BA
bile acids
- CDCA
chenodeoxycholic acid
- CYP7A1
cholesterol-7α-hydroxylase
- FA
fatty acid/s
- FXR
farnesoid X receptor (NR1H4)
- GSPE
grape seed procyanidin extract
- HDL
high density lipoproteins
- LBD
ligand binding domain
- LDL
low density lipoproteins
- RXR
retinoid X receptor (NR2B1)
- SBARM
selective bile acid receptor modulator
- SHP
small heterodimer partner (NR0B2)
- SREBP1
steroid response element binding protein 1
- TG
triglycerides
- VLDL
very low density lipoproteins
References
- 1.Gu L, Kelm MA, Hammerstone JF, Beecher G, et al. Concentrations of proanthocyanidins in common foods and estimations of normal consumption. J Nutr. 2004;134:613–617. doi: 10.1093/jn/134.3.613. [DOI] [PubMed] [Google Scholar]
- 2.Rasmussen SE, Frederiksen H, Struntze Krogholm K, Poulsen L. Dietary proanthocyanidins: occurrence, dietary intake, bioavailability, and protection against cardiovascular disease. Mol Nutr Food Res. 2005;49:159–174. doi: 10.1002/mnfr.200400082. [DOI] [PubMed] [Google Scholar]
- 3.Dell’Agli M, Busciala A, Bosisio E. Vascular effects of wine polyphenols. Cardiovasc Res. 2004;63:593–602. doi: 10.1016/j.cardiores.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Auger C, Gerain P, Laurent-Bichon F, Portet K, et al. Phenolics from commercialized grape extracts prevent early atherosclerotic lesions in hamsters by mechanisms other than antioxidant effect. J Agric Food Chem. 2004;52:5297–5302. doi: 10.1021/jf040125d. [DOI] [PubMed] [Google Scholar]
- 5.Decorde K, Teissedre PL, Auger C, Cristol JP, Rouanet JM. Phenolics from purple grape, apple, purple grape juice and apple juice prevent early atherosclerosis induced by an atherogenic diet in hamsters. Mol Nutr Food Res. 2008;52:400–407. doi: 10.1002/mnfr.200700141. [DOI] [PubMed] [Google Scholar]
- 6.Zern TL, Wood RJ, Greene C, West KL, et al. Grape polyphenols exert a cardioprotective effect in pre- and postmenopausal women by lowering plasma lipids and reducing oxidative stress. J Nutr. 2005;135:1911–1917. doi: 10.1093/jn/135.8.1911. [DOI] [PubMed] [Google Scholar]
- 7.Tebib K, Besancon P, Rouanet JM. Dietary grape seed tannins affect lipoproteins, lipoprotein lipases and tissue lipids in rats fed hypercholesterolemic diets. J Nutr. 1994;124:2451–2457. doi: 10.1093/jn/124.12.451. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura Y, Tonogai Y. Effects of grape seed polyphenols on serum and hepatic lipid contents and fecal steroid excretion in normal and hypercholesterolemic rats. Journal of Health Science. 2002;48:570–578. [Google Scholar]
- 9.Vidal R, Hernandez-Vallejo S, Pauquai T, Texier O, et al. Apple procyanidins decrease cholesterol esterification and lipoprotein secretion in Caco-2/TC7 enterocytes. J Lipid Res. 2005;46:258–268. doi: 10.1194/jlr.M400209-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Sugiyama H, Akazome Y, Shoji T, Yamaguchi A, et al. Oligomeric procyanidins in apple polyphenol are main active components for inhibition of pancreatic lipase and triglyceride absorption. J Agric Food Chem. 2007;55:4604–4609. doi: 10.1021/jf070569k. [DOI] [PubMed] [Google Scholar]
- 11.Pal S, Ho N, Santos C, Dubois P, et al. Red wine polyphenolics increase LDL receptor expression and activity and suppress the secretion of ApoB100 from human HepG2 cells. J Nutr. 2003;133:700–706. doi: 10.1093/jn/133.3.700. [DOI] [PubMed] [Google Scholar]
- 12.Davalos A, Fernandez-Hernando C, Cerrato F, Martinez-Botas J, et al. Red grape juice polyphenols alter cholesterol homeostasis and increase LDL-receptor activity in human cells in vitro. J Nutr. 2006;136:1766–1773. doi: 10.1093/jn/136.7.1766. [DOI] [PubMed] [Google Scholar]
- 13.Castilla P, Echarri R, Davalos A, Cerrato F, et al. Concentrated red grape juice exerts antioxidant, hypolipidemic, and antiinflammatory effects in both hemodialysis patients and healthy subjects. Am J Clin Nutr. 2006;84:252–262. doi: 10.1093/ajcn/84.1.252. [DOI] [PubMed] [Google Scholar]
- 14.Del Bas JM, Fernandez-Larrea J, Blay M, Ardevol A, et al. Grape seed procyanidins improve atherosclerotic risk index and induce liver CYP7A1 and SHP expression in healthy rats. Faseb J. 2005;19:479–481. doi: 10.1096/fj.04-3095fje. [DOI] [PubMed] [Google Scholar]
- 15.Lee YS, Chanda D, Sim J, Park YY, Choi HS. Structure and function of the atypical orphan nuclear receptor small heterodimer partner. Int Rev Cytol. 2007;261:117–158. doi: 10.1016/S0074-7696(07)61003-1. [DOI] [PubMed] [Google Scholar]
- 16.Del Bas JM, Ricketts M-L, Baiges I, Quesada H, et al. Dietary procyanidins lower triglyceride levels signaling through the nuclear receptor small heterodimer partner. Mol Nutr Food Res. doi: 10.1002/mnfr.200800054. in press. [DOI] [PubMed] [Google Scholar]
- 17.Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 18.Raghow R, Yellaturu C, Deng X, Park EA, Elam MB. SREBPs: the crossroads of physiological and pathological lipid homeostasis. Trends Endocrinol Metab. 2008;19:65–73. doi: 10.1016/j.tem.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Lee FY, Lee H, Hubbert ML, Edwards PA, Zhang Y. FXR, a multipurpose nuclear receptor. Trends Biochem Sci. 2006;31:572–580. doi: 10.1016/j.tibs.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. Embo J. 2006;25:1419–1425. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuipers F, Stroeve JH, Caron S, Staels B. Bile acids, farnesoid X receptor, atherosclerosis and metabolic control. Curr Opin Lipidol. 2007;18:289–297. doi: 10.1097/MOL.0b013e3281338d08. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Edwards PA. FXR signaling in metabolic disease. FEBS Lett. 2008;582:10–18. doi: 10.1016/j.febslet.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Claudel T, Staels B, Kuipers F. The Farnesoid X receptor: a molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol. 2005;25:2020–2030. doi: 10.1161/01.ATV.0000178994.21828.a7. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe M, Houten SM, Wang L, Moschetta A, et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urizar NL, Liverman AB, Dodds DT, Silva FV, et al. A natural product that lowers cholesterol as an antagonist ligand for FXR. Science. 2002;296:1703–1706. doi: 10.1126/science.1072891. [DOI] [PubMed] [Google Scholar]
- 26.Cui J, Huang L, Zhao A, Lew JL, et al. Guggulsterone is a farnesoid X receptor antagonist in coactivator association assays but acts to enhance transcription of bile salt export pump. J Biol Chem. 2003;278:10214–10220. doi: 10.1074/jbc.M209323200. [DOI] [PubMed] [Google Scholar]
- 27.Nozawa H. Xanthohumol, the chalcone from beer hops (Humulus lupulus L.), is the ligand for farnesoid X receptor and ameliorates lipid and glucose metabolism in KK-A(y) mice. Biochem Biophys Res Commun. 2005;336:754–761. doi: 10.1016/j.bbrc.2005.08.159. [DOI] [PubMed] [Google Scholar]
- 28.Urizar NL, Dowhan DH, Moore DD. The farnesoid X-activated receptor mediates bile acid activation of phospholipid transfer protein gene expression. J Biol Chem. 2000;275:39313–39317. doi: 10.1074/jbc.M007998200. [DOI] [PubMed] [Google Scholar]
- 29.Zavacki AM, Lehmann JM, Seol W, Willson TM, et al. Activation of the orphan receptor RIP14 by retinoids. Proc Natl Acad Sci U S A. 1997;94:7909–7914. doi: 10.1073/pnas.94.15.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinal CJ, Tohkin M, Miyata M, Ward JM, et al. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 31.Thomas PD, Campbell MJ, Kejariwal A, Mi H, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas PD, Kejariwal A, Guo N, Mi H, et al. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sone H, Shimano H, Sakakura Y, Inoue N, et al. Acetyl-coenzyme A synthetase is a lipogenic enzyme controlled by SREBP-1 and energy status. Am J Physiol Endocrinol Metab. 2002;282:E222–230. doi: 10.1152/ajpendo.00189.2001. [DOI] [PubMed] [Google Scholar]
- 34.Tabor DE, Kim JB, Spiegelman BM, Edwards PA. Identification of conserved cis-elements and transcription factors required for sterol-regulated transcription of stearoyl-CoA desaturase 1 and 2. J Biol Chem. 1999;274:20603–20610. doi: 10.1074/jbc.274.29.20603. [DOI] [PubMed] [Google Scholar]
- 35.Schaap FG, Rensen PC, Voshol PJ, Vrins C, et al. ApoAV reduces plasma triglycerides by inhibiting very low density lipoprotein-triglyceride (VLDL-TG) production and stimulating lipoprotein lipase-mediated VLDL-TG hydrolysis. J Biol Chem. 2004;279:27941–27947. doi: 10.1074/jbc.M403240200. [DOI] [PubMed] [Google Scholar]
- 36.Jakel H, Nowak M, Helleboid-Chapman A, Fruchart-Najib J, Fruchart JC. Is apolipoprotein A5 a novel regulator of triglyceride-rich lipoproteins? Ann Med. 2006;38:2–10. doi: 10.1080/07853890500407488. [DOI] [PubMed] [Google Scholar]
- 37.Le Lay S, Lefrere I, Trautwein C, Dugail I, Krief S. Insulin and sterol-regulatory element-binding protein-1c (SREBP-1C) regulation of gene expression in 3T3-L1 adipocytes, Identification of CCAAT/enhancer-binding protein beta as an SREBP-1C target. J Biol Chem. 2002;277:35625–35634. doi: 10.1074/jbc.M203913200. [DOI] [PubMed] [Google Scholar]
- 38.Shimano H. Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog Lipid Res. 2001;40:439–452. doi: 10.1016/s0163-7827(01)00010-8. [DOI] [PubMed] [Google Scholar]
- 39.Horton JD, Shah NA, Warrington JA, Anderson NN, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu K, Miyazaki M, Man WC, Ntambi JM. Stearoyl-coenzyme A desaturase 1 deficiency protects against hypertriglyceridemia and increases plasma high-density lipoprotein cholesterol induced by liver X receptor activation. Mol Cell Biol. 2006;26:6786–6798. doi: 10.1128/MCB.00077-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olofsson SO, Boren J. Apolipoprotein B: a clinically important apolipoprotein which assembles atherogenic lipoproteins and promotes the development of atherosclerosis. J Intern Med. 2005;258:395–410. doi: 10.1111/j.1365-2796.2005.01556.x. [DOI] [PubMed] [Google Scholar]
- 42.Sakakura Y, Shimano H, Sone H, Takahashi A, et al. Sterol regulatory element-binding proteins induce an entire pathway of cholesterol synthesis. Biochem Biophys Res Commun. 2001;286:176–183. doi: 10.1006/bbrc.2001.5375. [DOI] [PubMed] [Google Scholar]
- 43.Boulias K, Katrakili N, Bamberg K, Underhill P, et al. Regulation of hepatic metabolic pathways by the orphan nuclear receptor SHP. Embo J. 2005;24:2624–2633. doi: 10.1038/sj.emboj.7600728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lew JL, Zhao A, Yu J, Huang L, et al. The farnesoid X receptor controls gene expression in a ligand- and promoter-selective fashion. J Biol Chem. 2004;279:8856–8861. doi: 10.1074/jbc.M306422200. [DOI] [PubMed] [Google Scholar]
- 45.Nettles KW, Greene GL. Nuclear receptor ligands and cofactor recruitment: is there a coactivator “on deck”? Mol Cell. 2003;11:850–851. doi: 10.1016/s1097-2765(03)00133-3. [DOI] [PubMed] [Google Scholar]
- 46.Downes M, Verdecia MA, Roecker AJ, Hughes R, et al. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol Cell. 2003;11:1079–1092. doi: 10.1016/s1097-2765(03)00104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unno A, Takada I, Takezawa S, Oishi H, et al. TRRAP as a hepatic coactivator of LXR and FXR function. Biochem Biophys Res Commun. 2005;327:933–938. doi: 10.1016/j.bbrc.2004.12.095. [DOI] [PubMed] [Google Scholar]
- 48.Kanaya E, Shiraki T, Jingami H. The nuclear bile acid receptor FXR is activated by PGC-1alpha in a ligand-dependent manner. Biochem J. 2004;382:913–921. doi: 10.1042/BJ20040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsang C, Auger C, Mullen W, Bornet A, et al. The absorption, metabolism and excretion of flavan-3-ols and procyanidins following the ingestion of a grape seed extract by rats. Br J Nutr. 2005;94:170–181. doi: 10.1079/bjn20051480. [DOI] [PubMed] [Google Scholar]
- 50.Shoji T, Masumoto S, Moriichi N, Akiyama H, et al. Apple procyanidin oligomers absorption in rats after oral administration: analysis of procyanidins in plasma using the porter method and high-performance liquid chromatography/tandem mass spectrometry. J Agric Food Chem. 2006;54:884–892. doi: 10.1021/jf052260b. [DOI] [PubMed] [Google Scholar]
- 51.Cariou B, Staels B. FXR: a promising target for the metabolic syndrome? Trends Pharmacol Sci. 2007;28:236–243. doi: 10.1016/j.tips.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Wang YD, Chen WD, Huang W. FXR, a target for different diseases. Histol Histopathol. 2008;23:621–627. doi: 10.14670/HH-23.621. [DOI] [PubMed] [Google Scholar]
- 53.Amemiya-Kudo M, Shimano H, Yoshikawa T, Yahagi N, et al. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J Biol Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]