Abstract
The coiled-coil is one of the most common protein structural motifs. Amino acid sequences of regions that participate in coiled-coils contain a heptad repeat in which every third then forth residue is occupied by a hydrophobic residue. Here we examine the consequences of a “stutter,” a deviation of the idealized heptad repeat that is found in the central coiled-coil of influenza hemagluttinin HA2. Characterization of a peptide containing the native stutter-containing HA2 sequence, as well as several variants in which the stutter was engineered out to restore an idealized heptad repeat pattern, revealed that the stutter is important for allowing coiled-coil formation in the WT HA2 at both neutral and low pH (7.1 and 4.5). By contrast, all variants that contained idealized heptad repeats exhibited marked pH-dependent coiled-coil formation with structures forming much more stably at low pH. A crystal structure of one variant containing an idealized heptad repeat, and comparison to the WT HA2 structure, suggest that the stutter distorts the optimal interhelical core packing arrangement, resulting in unwinding of the coiled-coil superhelix. Interactions between acidic side chains, in particular E69 and E74 (present in all peptides studied), are suggested to play a role in mediating these pH-dependent conformational effects. This conclusion is partially supported by studies on HA2 variant peptides in which these positions were altered to aspartic acid. These results provide new insight into the structural role of the heptad repeat stutter in HA2.
Keywords: Protein structure, protein design, viral membrane fusion, influenza, pH-dependent structure
INTRODUCTION
It is estimated that 3–10% of all proteins contain coiled-coil segments.1 These domains, which consist of two or more associating α-helices, play important functional roles in transcription, motility, membrane fusion, and other biological processes.2–4 An expansive body of work has provided guidelines for design of coiled-coil proteins and various applications ranging from materials synthesis to therapeutic development. Simple design rules, based on spacing of hydrophobic and hydrophilic residues, can yield stable coiled-coils with well-defined topology.5 Incorporation of more subtle design features expand the range of potential structures and, in some cases, endow environmentally sensitive behavior.6–9
Protein sequences of coiled-coil segments contain a regular hydrophobic repeat pattern known as the heptad repeat.2–4 Positions are denoted abcdefg, with a and d occupied by a hydrophobic residue (typically one with an aliphatic side chain) that create a stripe that wraps around the α-helix. At the core, the a and d side chains pack in “knob-into-holes” fashion where each side chain “knob” fits into a “hole” generated by four side chains from an apposing α-helix.13 The identity of residues at the core a and d positions as well as flanking e and g positions can specify number of α-helices, relative α-helix orientation (parallel or antiparallel), pairing preferences among different α-helical segments.2–4,6,8
Some coiled-coil proteins contain slight deviations in the periodicity of the heptad repeat pattern.14,15 One such example is the heptad repeat stutter, which is caused the insertion of four residues into the heptad (abcdefgdefg), generating a 3-4-4-3 hydrophobic pattern instead of the canonical 3-4-3-4 pattern.14 A stutter alters the left-handed nature of the hydrophobic stripe, resuting in local unwinding of the coiled-coil and an increase in super-helical pitch. Knobs-into-holes packing is disrupted at the stutter site, caused by orientation of core side chains on the α-helices directly toward one other. This arrangement creates “knobs-into-knobs” packing in what is referred to as x layer geometry. The presence of a stutter therefore locally alters ideal interhelical interactions within the coiled-coil and is predicted to be destabilizing for short segments.
A conserved stutter can be found in the “postfusion” coiled-coils of influenza hemagglutinin HA2 as well most other class I (α-helical) and many class III (mixed α/β structure) fusion proteins.10,16,17 For class I viral glycoproteins, the location of the stutter can be used as a register to align the coiled-coils in relation to the membrane.10 That a structural feature would exhibit such a high level of conservation across disparate phylogenetic families suggests functional significance. The stutter positions in Ebola virus GP2 and influenza virus HA2, both endosomal viruses, are found in regions that have drastically different conformations in the prefusion and postfusion structures.18,19 In both cases, pH has a dramatic effect on conformational preferences of the glycoprotein, with the postfusion conformation preferred at low pH. This may implicate a specific role for the stutter in the fusion events that require pH-dependent structural transitions.
Here we explore the effect of a heptad repeat stutter on the pH-dependent conformational behavior of the central coiled-coil from influenza hemagglutinin HA2. Although stutters have been noted in many virus glycoproteins, their functional effects have not previously been investigated.10–12
MATERIALS AND METHODS
Peptide synthesis and purification
All peptides were synthesized by solid-phase peptide synthesis using standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry on an ABI-433A peptide synthesizer with the inclusion of an N-terminal acetyl cap and a C-terminal amide. Following synthesis, simultaneous side chain deprotection and cleavage from resin were achieved by treating the resin with 95% trifluoroacetic acid, 2.5% 1,2- ethanedithiol, and 2.5% thioanisole for 3 h. Resin was removed by filtration, and the peptide was precipitated by addition of cold diethyl ether. The peptide was pelleted by centrifugation, washed twice with cold diethyl ether, redissolved in water/acetonitrile, and lyophilized. Crude lyophilized peptides were purified by reverse-phase HPLC on a Vydac C18 column (10 μm, 250 × 21.2mm) with water/acetonitrile mobile phases containing 0.1% trifluoroacetic acid. Peptide purity was generally 90–95% as judged by analytical reverse-phase HPLC, and the identity of all peptides was confirmed by MALDI-MS (see Supporting Information). Peptides were dissolved in either 10 mM phosphate (pH 7.1) or 10 mM sodium acetate (pH 4.5), and the concentration was determined by absorbance at 280 nm.
Circular dichroism
Peptide samples were prepared in the appropriate buffer and measurements were performed on a Jasco J-815 spectrometer with a 1 mm quartz cuvette. Peptide concentrations ranged from 35–45 μM as determined by absorbance at 280 nm. CD wavelength scans were obtained with a 0.1 nm step size and a 2 s averaging time. The signal was converted to mean residue ellipticity (θ). Thermal denaturation data were obtained with a 0.5°C step size and 30 s equilibration at each temperature. Nonlinear least squares regression was performed on the ellipticty data using a standard four-parameter logistic equation. The Tm values were derived from the inflection point of the curve.
Analytical Ultracentrifugation
Sedimentation velocity analysis was performed on a Beckman XL-1 analytical ultracentrifuge with a Ti60 rotor. Samples were loaded into double sectors cells at peptide concentrations of 115 μM, blanked against the sample buffer (10 mM sodium acetate, 100 mM NaCl, pH 4.5) in the reference sector. Two hundred scans were acquired at 58,000 rpm and 20 °C with sedimentation boundaries monitored by absorption at a wavelength of 280 nm. The sedimentation boundaries were directly fit as the derivative dc/dt using DCDT+ v2.4.0 to determine the sedimentation and diffusion coefficients, S and D, respectively from which was calculated the apparent molecular weight, Mw,app20,21. Sixty to 80 absorbance scans were globally analyzed for each experiment. The observed values were normalized to standard conditions of 20°C and water (s20,w and D20,w) by correcting for buffer density and viscosity. Buffer density, viscosity, and vbar were calculated using Sednterp22.
X-Ray Crystallography
Diffraction quality crystals were grown by sitting drop vapor diffusion by mixing 1 μL of protein (concentration was 6 mg/mL in 10 mM NaH2PO4, pH 7.5) with 1 μL of reservoir solution and equilibrating the samples against the corresponding reservoir solution. The reservoir solution contained 30% PEG 400, 0.2 M lithium sulfate, 0.1 M sodium cacodylate, pH 6.5. Crystals of HA2-DelEx with dimensions 0.2 × 0.2 × 0.3 mm3, were mounted in cryo-loops directly from the crystallization droplet and flash-cooled in liquid nitrogen. Diffraction data were recorded on a Rayonix 225 HE CCD detector (Ryonyx, L.L.C., Evanston, IL, USA) with 0.979 Å wavelength radiation on the LRL-CAT beamline (Advanced Photon Source, Argone, IL). Intensities were integrated using the HKL2000 program and reduced to amplitudes using the SCALEPACK2MTZ program (see Table I for statistics)23–25. Structures were determined by molecular replacement with PHASER4. Model building and refinement were performed with the programs COOT and REFMAC, respectively26,27. The quality of the final structures was verified with composite omit maps, and stereochemistry was checked with the program MOLPROBITY28. LSQKAB and SSM algorithms were used for structural superpositions29,30. All other calculations were conducted using CCP4 program suite25.
Table I.
Data Collection and Refinement Statistics for the HA2-Del Crystal Structure
| PDB | 4P67 |
|---|---|
| Data Collection | |
| Wavelength (Å) | 0.979 |
| Space group | P31 |
| Unit cell dimensions (Å) | a = b = 58.44 c = 66.99 a = b = 90°, g = 120° |
| Resolution range (Å) | 20-1.9 |
| Observed reflections | 142,034 |
| Unique reflections | 20,121 |
| Completeness (%) a | 99.5 (100.0)a |
| I/sI | 14.6 (3.0) |
| R-merge (I) b | 0.074 (0.654) |
| Structure Refinement | |
| Rcryst (%) c | 0.185 (0.209) a |
| Rfree (%) c | 0.242 (0.281) a |
| Protein nonhydrogen atoms | 1949 |
| Water molecules | 130 |
| Average B-factor (Å2) | 44.47 |
| RMS Deviations from Ideal Value | |
| Bonds (Å) | 0.013 |
| Angles (°) | 1.47 |
| Torsion angles (°) | 16.8 |
| Overall coordinate error based on R-factor | 0.038 |
|
Ramachandran statistics (%) (for non-Gly/Pro residues) |
|
| most favorable | 99.52 |
| additional allowed | 0.48 |
Values in parentheses indicate statistics for the high resolution bin.
Rmerge = ΣΣ j|Ij(hkl) – <I(hkl)>|/ ΣΣ j|<I(hkl)>|, where Ij is the intensity measurement for reflection j and <I> is the mean intensity over j reflections.
Rcryst/(Rfree) = Σ ||Fo(hkl)| – |Fc(hkl)||/ Σ |Fo(hkl)|, where Fo and Fc are observed and calculated structure factors, respectively. No s-cutoff was applied. 5% of the reflections were excluded from refinement and used to calculate Rfree.
RESULTS
Peptide Design
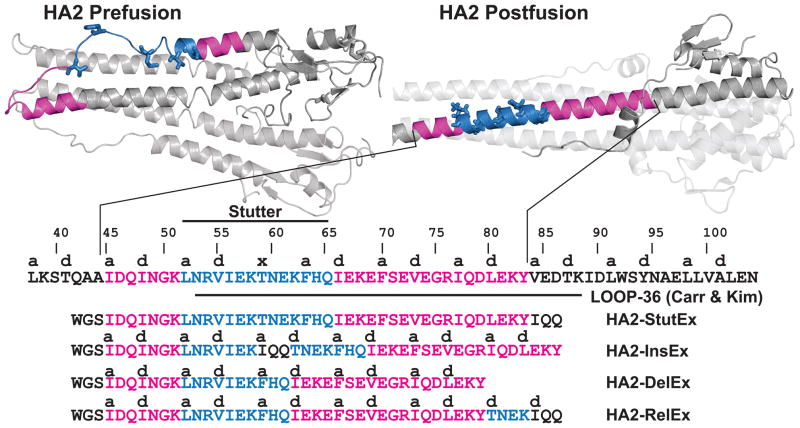
To explore the structural effects of the heptad repeat stutter on the behavior of HA2, we designed a series of peptides based on the central coiled-coil. The HA2 stutter is conserved across subtypes and can be found at position Thr59 in the X-31 strain.14,16,17 Following viral binding and endocytosis, low pH triggers conformational rearrangement of HA2 from an α-helical hairpin structure to an extended trimeric coiled-coil (Figure 1).16–18 The stutter occurs in a region that forms a loop in the prefusion structure, separating two α-helices, but participates in the elongated central trimeric coiled-coil that is the main feature of the postfusion structure. This conformational rearrangement reorients the fusion peptide away from the viral membrane and presumably facilitates its insertion into the host endosomal membrane. Carr and Kim previously characterized the pH-dependent behavior of a peptide corresponding to residues 54–81 (‘LOOP-36’; Figure 1).17 This peptide contains the stutter at its N-terminal end, and forms a trimeric coiled-coil at pH 4 but exists as an unstructured monomer at pH 7.
Figure 1. Peptide Design.
Pre- and postfusion structures of HA2 (PDB entries 2HMG and 1HTM, respectively); location of the heptad repeat stutter (Thr59, indicated with an ‘x’) and designed peptides for this study.
We prepared the peptide HA2-Stut (Figure 1), which overlaps with the first ~4 heptad repeats of LOOP-36 (including the stutter) but is extended toward the HA2 N-terminus by a full heptad repeat. This design places the stutter toward the midsection of the putative HA2-Stut coiled-coil. In addition, a tripeptide ~IQQ~ segment was included at the C-terminus, extending the terminal sequence by an additional half heptad. Incorporation of this segment allows direct comparison to other peptide sequence isomers discussed below. Ile was chosen because it is frequently found at core a positions in trimeric coiled-coils; Gln is highly α-helix promoting but does not impact overall charge or hydrophobicity of the sequence.32,33 An N-terminal Trp residue, separated from the HA2 segment by a ~GS~ linker, was included to facilitate quantification by UV absorbance.
To explore the effect of the heptad repeat stutter, we designed two sequence isomers of HA2-Stut (Figure 1). In peptide HA2-Ins, the auxiliary ~IQQ~ segment is inserted at the stutter position. The outcome of this modification is that the heptad repeat pattern is undisturbed and thus HA2-Ins contains an ideal heptad repeat throughout. In HA2-Rel, the four-residue ~T(59)NEK~ segment that gives rise to the stutter is relocated to the C-terminus of the peptide. This design was intended to allow comparison of the placement of the stutter at the mid-section of the coiled-coil (HA2-Stut) relative to a terminus (HA2-Rel). In a third peptide design, this four-residue segment was deleted (HA2-Del); this modification also restores the ideal heptad repeat pattern but shortens the coiled-coil by seven residues. Thus, HA2-Del is not a sequence isomer of HA2-Stut but nonetheless provides another comparison of effect of including the stutter.
Solution Characterization
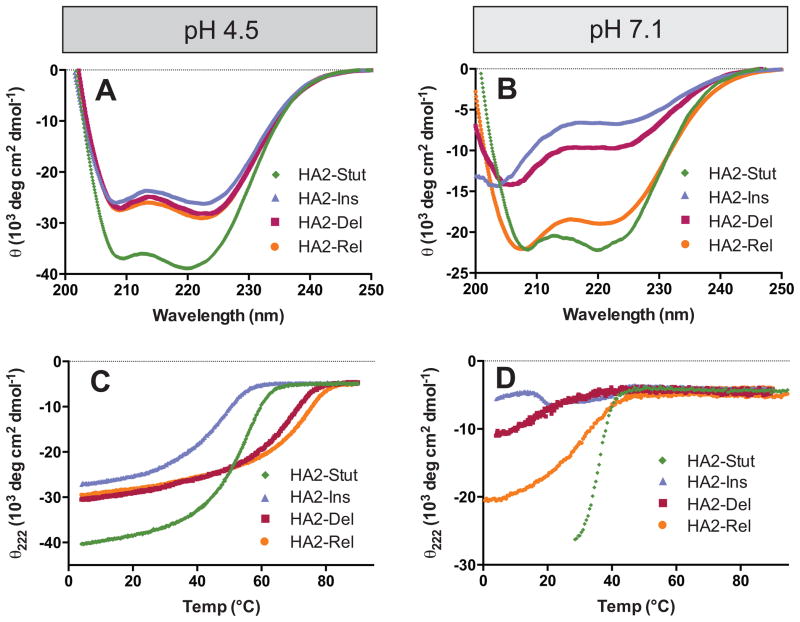
Circular dichroism (CD) spectra indicate that HA2-Stut is strongly α-helical at pH 4.5 and 7.1, with minima at 208 nm and 222 nm (Figures 2A-B). The mean residue ellipticity at 222 nm (θ222) was more intense at pH 4.5 (−38,100 deg cm2 / dmol) than at pH 7.1 (−21,500 mdeg cm2 / dmol), indicating higher α-helical content at low pH. Sedimentation velocity analytical ultracentrifugation analysis at pH 4.5 revealed a single ideal species with apparent molecular weight of 16.1 kDa, consistent with the expected trimer (16.4 kDa, Table II and Supporting Information). The thermal denaturation midpoint (TM) of HA2-Stut was found to be pH-dependent with higher stability at pH 4.5 (TM = 51.4 ± 0.2 °C) than at pH 7.1 (TM = 35.7 ± 0.1 °C) (Figures 2C–D and Table II). We therefore conclude that HA2-Stut forms a trimeric coiled-coil with pH-dependent stability, consistent with studies on LOOP-36 and other peptides from this region of HA2.17
Figure 2. Circular Dichroism.
(A and B) Full wavelength scans at pH 4.5 (A) and pH 7.1 (B). (C and D) Thermal denaturation under similar conditions; TM values are listed in Table II.
Table II.
Solution Characterization of Peptides
| Peptide | θ222, pH 4.5 (×103 mdeg/cm2 dmol) | θ222, pH 7.1 (×103 mdeg/cm2 dmol) | TM, pH 4.5 (°C) | TM, pH 7.1 (°C) | ΔTM4.5–7.1 (°C) | MWapp (s/D; kDa)A | MWexp (trimer) (kDa) |
|---|---|---|---|---|---|---|---|
| HA2-Stut | −38.1 | −21.5 | 51.4 ± 0.2 | 35.7 ± 0.1 | 15.7 | 14.3 | 16.4 |
| HA2-Ins | −26.1 | −6.7 | 43.3 ± 0.2 | NDB | NDB | 14.8 | 16.4 |
| HA2-Rel | −29.0 | −18.9 | 74.5 ± 1.5 | ~30 | ~44 | 14.4 | 16.4 |
| HA2-Del | −28.2 | −9.7 | 65.5 ± 0.7 | 17.2 ± 0.7 | 48.3 | 16.1 | 13.8 |
| HA2-Stut-E74D | −29.6 | −16.4 | 37.5 ± 0.2 | 32.5 ± 0.2 °C | 5 °C | 15.2 | 16.3 |
| HA2-Stut-E69D | −22.0 | −12.2 | 42.0 ± 0.4 °C | 27.1 ± 0.8 °C | 14.9 °C | 17.4 | 16.3 |
Determined from sedimentation velocity ultracentrifugation at pH 4.5, 20 °C. Full details of the analysis are provided in the Supporting Information. All peptides behaved as a single ideal species.
Accurate TM at pH 7.1 could not be determined due to a broad thermal transition.
HA2-Ins, HA2-Del, and HA2-Rel were α-helical at pH 4.5 but had diminished α-helical character at pH 7.1 (Figures 2A-B). HA2-Rel had the highest degree of α-helical character among these three variants at pH 7.1 (θ222 = −18,900 deg cm2 / dmol); both HA2-Ins and HA2-Del were only partially α-helical as indicated by broad bands at ~205 nm and ~225 nm. This behavior contrasts with that of HA2-Stut, which maintained significant α-helical character at both pHs. Thermal denaturation indicated that HA2-Del and HA2-Rel were more stable than HA2-Stut at pH 4.5 (by 14.1 °C and 23.1 °C, respectively, Figures 2C-D and Table II). However, HA2-Ins was less stable than HA2-Stut by 8.1 °C.
The sensitivity of coiled-coil formation to pH was enhanced for HA2-Ins, HA2-Del, and HA2-Rel in comparison to HA2-Stut (Figures 2C-D and Table II). HA2-Del and HA2-Ins had little α-helical character at pH 7.1 and underwent broad thermal transitions with complete unfolding by ~40 °C. HA2-Rel had higher α-helical content at pH 7.1, but nonetheless underwent broad thermal unfolding with a midpoint of 30.5 ± 0.2 °C. This behavior contrasts with HA2-Stut, which had stronger α-helical character at pH 7.1 and underwent a sharp unfolding transition at 35.7 ± 0.1 °C. The difference in thermal midpoints at pH 4.5 and pH 7.1 (ΔTM4.5–7.1) was 15.7 °C for HA2-Stut but 44.0 °C for HA2-Rel and ~48 °C for HA2-Del. A TM at pH 7.1 was difficult to estimate for HA2-Ins due to the broad unfolding transition; nonetheless, it is evident that coiled-coil formation in HA2-Ins is equally pH-dependent if not more so than HA2-Rel and HA2-Del. Therefore, coiled-coil formation was much less sensitive to pH in HA2-Stut than any of the variants. It is interesting to note that HA2-Del is more stable at pH 4.5 than HA2-Stut despite the fact that it is shorter, but HA2-Del nonetheless shows drastic pH-dependent behavior.
All three peptides were subjected to analytical ultracentrifugation analysis at pH 4.5 and were found to behave as single ideal species with molecular weights consistent with trimers. The observed molecular weights were 16.3 kDa (HA2-Del), 14.8 kDa (HA2-Ins) and 14.3 kDa (HA2-Rel), and the expected trimer molecular weights are 13.9 kDa (HA2-Del), 16.3 kDa (HA2-Ins), and 16.3 kDa (HA2-Rel) (Table II and Supporting Information). Furthermore, size-exclusion gel filtration profiles of HA2-Stut and HA2-Del were similar under these conditions (see Supporting Information).
Crystal Structure of HA2-Del
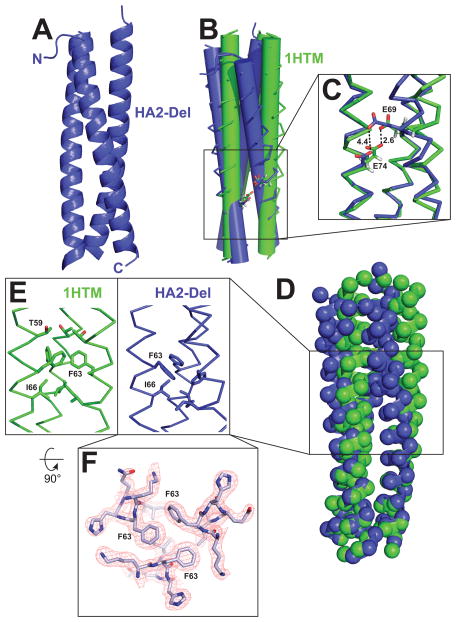
The three-dimensional structure of HA2-Del was determined at 1.9 Å resolution. The diffraction data were consistent with the P31 space group and there were six α-helical chains in the asymmetric unit assembled into two independent and virtually identical trimers. The RMSD values between individual chains ranged from 0.6 – 1.1 Å for Cα atoms, and 1.6–1.8 Å for all atoms. Overall, HA2-Del forms a trimeric coiled-coil with tight packing throughout the core (Figure 3). When compared with the structure from the analogous section the “low” pH structure of WT (pH 5.0; PDB ID 1HTM),16 the HA2-Del trimer has an increased super-helical twist (i.e., decreased pitch, Figure 3B). Overall, the relative packing orientation of the α-helices in HA2-Del has a markedly increased crossing angle between the axes of the individual α-helices and the superhelix. α-Helical net analysis demonstrates that the interhelical a/d and e/g interactions in HA2-Del and HA2-Stut are similar except for the stutter segment that is removed in HA2-Del (see Supporting Information). The basis for the increased superhelical pitch in WT HA2 relative to HA2-Del is the disruption of knobs-into-holes packing at the position of the stutter that distorts interhelical packing arrangement in the WT core trimer throughout the C-terminal end of the molecule. In particular, the packing of the core positions F63, an a position, and I66, a d position are less tightly packed in WT HA2 in comparison to HA2-Del (Figures 3E and 3F). In WT HA2, these residues are located two α-helical layers C-terminal from the stutter.
Figure 3. Crystal structure of the HA2-Del trimer and comparison to WT HA2 (PDB ID 1HTM).
(A) Overall structure of the HA2-coiled-coil. (B) Overlay of α-helical bundles for WT HA2 and HA2-Del; the α-helices are represented as cylinders to illustrate differences in α-helical packing geometry. (C) Potential hydrogen bonding interaction between E69 and E74. In HA2-Del, the residues are separated by 4.5 Å. (D) Locations of Cα carbons by spheres, demonstrating that although there are some differences in overall positioning of residues, there are no gross deviations between the two structures. (E) Differences in packing arrangement of F63 and I66 in the trimer cores. (F) Top-down view of the F63 interaction at the α-helical interface, with sample electron density.
Despite these differences, the relative positioning of many residues within the two trimers are similar (Figure 3D). This analysis suggests that long-range interactions, such as bulk electrostatic interactions, are likely to be similar among HA2-Stut and HA2-Del. Deletion of the stutter did not result in repositioning of any ionizable residues toward the trimer core. In other systems, core acidic residues can result in pH-dependent stability of α-helical bundles proteins derived from viruses.12,34–36
Interactions between E69 and E74
In the WT HA2, there is potential for an interhelical hydrogen bonding interaction between E69 (a g position) and E74 (an e position) with an interatomic Oε-Oε distance of 2.6 Å for all three α-helical interfaces (Figure 3C).16 In HA2-Del the corresponding glutamate residues vary in side chain orientation in the three subunits but in general are spaced further, ranging from 4.4 Å (Figure 3C) to ~9 Å; this increased spacing is due to the increased relative superhelical pitch and local differences in α-helical packing near the stutter site. pH-Dependent conformational behavior in coiled-coil systems is often due to charged residues occupying the e and g positions.6,37 Since a higher degree of pH-dependent structural stability was observed for HA2-Del than was for HA2-Stut (Figure 2), we hypothesized that the loss of a hydrogen bond interaction between E69 and E74 could be playing a role.38,39 Since deprotonation of these carboxylic acids would juxtapose two negatively-charged residues in the context of the trimer, therefore destabilizing it, a hydrogen bond that maintains the protonation state of one or both of these residues might stabilize the coiled-coil across a broader pH range. However, the optimal geometry for this hydrogen bonding pattern is not maintained in HA2-Del.
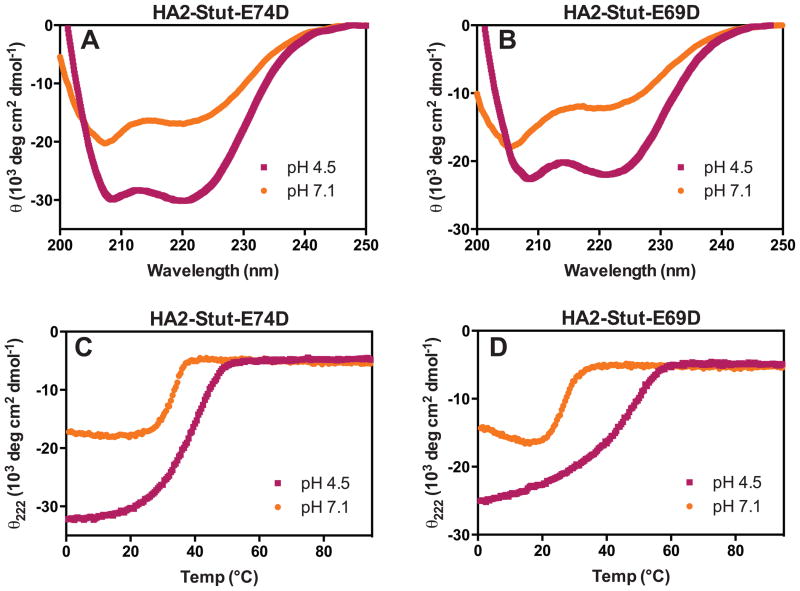
We designed two mutant versions of HA2-Stut, HA2-Stut-E69D and HA2-Stut-E74D, each with a conservative Glu to Asp mutation. By shortening the length of one of the acidic side chains participating in the E69/E74 interaction upon mutation to Asp, the optimal geometry for specific residue-residue interactions is disrupted. However, the Glu → Asp mutation maintains the potential for negative charge, and thus maintains long-range electrostatic interactions. Analytical ultracentrifugation analysis with HA2-Stut-E69D and HA2-Stut-E74D resulted in molecular weight estimates of 15.2 kDa and 17.4 kDa, respectively, both consistent with trimer formation (Table II). Therefore, these mutations did not impact the trimeric state of the coiled-coil. Circular dichroism indicated that both peptides had enhanced α-helicity at pH 4.5 relative to pH 7.1 (Figure 4A-B and Table II), though in general the negative θ222 minima were less intense than for the HA2-Stut peptide. Both peptides underwent cooperative unfolding transitions at pH 4.5 and pH 7.1 (Figure 4C-D). HA2-Stut-E74D was less stable than HA2-Stut under both conditions (by 13.9 °C at pH 4.5 and 3.2 °C at pH 7.1), and the sensitivity of coiled-coil formation to pH was less dramatic than for HA2-Stut (ΔTM4.5–7.1 = 5 °C). HA2-Stut-E69D was also less sable than HA2-Stut at both pHs (by 9.4 °C at pH 4.5 and 8.6 °C at pH 7.1), but the pH-dependence was nearly equivalent to that of HA2-Stut (ΔTM4.5–7.1 = 14.9 °C).
Figure 4. Circular Dichroism of HA2-Stut-E69D and HA2-Stut-E74D.
(A and B) Wavelength scans at pH 4.5 and 7.1. (C and D) Thermal denaturation under similar conditions; TM values are listed in Table 1.
DISCUSSION
Taken together, these results indicate that all peptide variants containing idealized heptad repeats (HA2-Ins, HA2-Rel, and HA-Del) have higher dependency on pH than does HA2-Stut. Therefore, we conclude that the stutter mitigates the pH-dependent conformational behavior of the trimeric coiled-coil from HA2. The structure of HA2-Del, when compared to the low pH WT HA2 structure (PDB ID 1HTM), demonstrates that the stutter results in unwinding of the triple-stranded superhelix and loosening of the core packing. The tight knobs-into-holes packing that occurs without disruption throughout HA2-Del likely explains why this variant is more stable than HA2-Stut at pH 4.5 even though it is seven residues shorter. In the WT HA2 coiled-coil, E69 (a g position) on one α-helix and E74 (e position) on an adjacent α-helix are proximal to one another and have potential for a hydrogen bonding interaction in solution. Such an interaction might contribute to the stability of HA2-Stut at neutral pH; potentially the hydrogen bond could provide a thermodynamic driving force for maintaining protonation of one of these two residues at higher pH. This would prevent unfavorable electrostatic repulsion that would result if both side chains were present as carboxylate species.38,39 In HA2-Del the relative orientation of these two side chains is slightly different than in the WT HA2 coiled-coil, and therefore it is possible that the optimal geometry for hydrogen bonding, which is linked to mitigating pH-dependence, is perturbed in this variant. This conclusion is supported to some extent by the observation that HA2-Stut-E74D coiled-coil formation was less sensitive to pH than HA2-Stut. However, this phenomenon is not completely reciprocal across this pairwise interaction, as HA2-Stut-E69D has essentially similar pH-dependence as did HA2-Stut.
Another potential explanation for the difference in pH-dependence observed for HA2-Stut in comparison with the three variants is that the local distortion caused by the stutter provides more flexibility and therefore mitigates unfavorable anion-anion (AniAni) interactions at flanking e and g positions. The residues E69, E74, and (to a lesser extent) E67 are proximal to one another in the WT HA2 postfusion structure,16 and are also present at commensurate heptad repeat regions in each of the variants. Since the idealized heptad repeat results in optimal packing geometry at the HA2-Del core, there is less flexibility and therefore these AniAni interactions have an enhanced effect on the pH-dependence than in HA2-Stut, which is less tightly packed at the core. The tighter core packing observed in the HA2-Del structure relative to the low pH structure of WT HA2 support this conclusion.
The presence of the stutter in HA2 is permissive, not restrictive, in that HA2-Stut can adopt the coiled-coil structure at both neutral and low pH, despite the fact that it is more stable at low pH. Why this behavior is important for the complicated process of membrane fusion is not clear. One possibility is that the stutter allows formation of the HA2 postfusion state, at least in this region, across a broad range of pHs. This behavior fits the classical model of metastability, where triggering of the postfusion conformation is kinetically controlled; therefore the post-fusion state is the most globally stable conformation.40,41 The data presented here suggest that the heptad repeat stutter in this region of HA2 is an important architectural feature for allowing coiled-coil formation at neutral pH. Such stutters may be useful design features for environment-dependent coiled-coils.
Supplementary Material
Acknowledgments
FUNDING: Grant sponsor: National Institutes of Allergy and Infectious Diseases of the National Institutes of Health (NIH); Grant number: R01-AI090249; Grant sponsor: National Institutes of General Medical Sciences of the NIH; Grant number: U54-GM094662; Grant sponsor: National Cancer Institute of the NIH; Grant number: P30-CA13330; Grant sponsor: Albert Einstein College of Medicine
We thank Michael Brenowitz and Nina Liu for assistance with analytical ultracentrifugation experiments, Rafael Toro for his assistance with crystallization, and the Lilly Research Laboratory Collaborative Access Team (LRL-CAT) beamtime staff at the Advanced Photon Source, Argonne National Laboratory, for their assistance with diffraction data collection.
Footnotes
CONFLICT OF INTEREST
The authors declare no competing financial interest.
ACCESSION CODE: The X-ray coordinates and structure factors for HA2-Del have been deposited in the Protein Data Bank: 4P67
References
- 1.Liu J, Rost B. Comparing function and structure between entire proteomes. Prot Sci. 2001;10(10):1970–1979. doi: 10.1110/ps.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woolfson DN. The design of coiled-coil structures and assemblies. Adv Prot Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 3.Grigoryan G, Keating E. Structural specificity in coiled-coil interactions. Curr Opin Struct Biol. 2008;18(4):477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lupas AN, Gruber M. The structure of α-helical coiled coils. Adv Prot Chem. 2005;70:73–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 5.Regan L, DeGrado WF. Characterization of a helical protein designed from first principles. Science. 1998;241(4868):976–978. doi: 10.1126/science.3043666. [DOI] [PubMed] [Google Scholar]
- 6.O’Shea EK, Lumb KJ, Kim PS. Peptide ‘Velcro’: Design of a heterodimeric coiled coil. Curr Biol. 1993;3(10):658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 7.Schnarr NA, Kennan AJ. Peptide tic-tac-tie: Heterodimeric coiled-coil specificity from steric matching of multiple hydrophobic side chains. J Am Chem Soc. 2002;124(33):9779–9783. doi: 10.1021/ja0174940. [DOI] [PubMed] [Google Scholar]
- 8.Gurnon DG, Whitaker JA, Oakley MG. Design and characterization of a homodimeric antiparallel coiled-coil. J Am Chem Soc. 2003;125(25):7518–7519. doi: 10.1021/ja0357590. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher JM, Harniman RL, Barnes FR, Boyle AL, Collins A, Mantell J, Sharp TH, Antognozzi M, Booth PJ, Linden N, Miles MJ, Sessions RB, Verkade P, Woolfson DN. Self-assembling cages from coiled-coil peptide modules. Science. 2013;340(6132):595–599. doi: 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Igonet S, Vaney MC, Vonrhein C, Bricogne G, Stura EA, Hengartner H, Eschli B, Rey FA. X-ray structure of the arenavirus glycoprotein GP2 in its postfusion hairpin conformation. Proc Natl Acad Sci USA. 2011;108(50):19967–19972. doi: 10.1073/pnas.1108910108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weissenhorn W, Carfí A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2(5):605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 12.Koellhoffer JF, Malashkevich VN, Harrison JS, Toro R, Bhosle RC, Chandran K, Almo SC, Lai JR. Crystal structure of the Marburg virus GP2 core domain in its postfusion conformation. Biochemistry. 2012;51(39):7665–7675. doi: 10.1021/bi300976m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crick FHC. The packing of α-helices: Simple coiled-coils. Acta Crystallographica. 1953;6:689–697. [Google Scholar]
- 14.Brown JH, Coben C, Parry DA. Heptad breaks in α-helical coiled coils: Stutters and stammers. Proteins. 1996;26(2):134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 15.Strelkov SV, Burkhard P. Analysis of alpha-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J Stuct Biol. 2002;137(1–2):54–64. doi: 10.1006/jsbi.2002.4454. [DOI] [PubMed] [Google Scholar]
- 16.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371(6492):37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 17.Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73(4):823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 18.Weis WI, Brünger AT, Skehel JJ, Wiley DC. Refinement of the influenza virus hemagglutinin by simulated annealing. J Mol Biol. 1990;212(4):737–761. doi: 10.1016/0022-2836(90)90234-D. [DOI] [PubMed] [Google Scholar]
- 19.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454(7201):177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Philo JS. A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal Biochem. 2000;279(2):151–63. doi: 10.1006/abio.2000.4480. [DOI] [PubMed] [Google Scholar]
- 21.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354(2):238–46. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 22.Hurton T, Wright A, Deubler G, Bashir B. SEDNTERP. Biomolecular Interactions Technology Center (BITC), University of New Hampshire; Retrieved from http://sednterp.unh.edu/ [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.French S, Wilson K. On the treatment of negative intensity observations. Acta Crystallogr A. 1978;34:517–525. [Google Scholar]
- 25.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr Sect A. 1976;32:922–923. [Google Scholar]
- 31.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–68. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 32.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262(5138):1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 33.O’Neil KT, DeGrado WF. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science. 1990;250(4981):646–651. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- 34.Lau WL, Degrado WF, Roder H. The effects of pK(a) tuning on the thermodynamics and kinetics of folding: design of a solvent-shielded carboxylate pair at the a-position of a coiled-coil. Biophys J. 2010;99(7):2299–2308. doi: 10.1016/j.bpj.2010.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrison JS, Higgins CD, O’Meara MJ, Koellhoffer JF, Kuhlman BA, Lai JR. Role of electrostatic repulsion in controlling pH-dependent conformational changes of viral fusion proteins. Structure. 2013;21(7):1085–1096. doi: 10.1016/j.str.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koellhoffer JF, Dai Z, Malashkevich VN, Stenglein MD, Liu Y, Toro R, S Harrison J, Chandran K, Derisi JL, Almo SC, Lai JR. Structural Characterization of the Glycoprotein GP2 Core Domain from the CAS Virus, a Novel Arenavirus-Like Species. J Mol Biol. doi: 10.1016/j.jmb.2013.12.009. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mason JM, Arndt KM. Coiled coil domains: Stability, specificity, and biological implications. Chembiochem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 38.Northrop DB. Follow the protons: A low-barrier hydrogen bond unifies the mechanisms of the aspartic proteases. Acc Chem Res. 2001;34(10):790–797. doi: 10.1021/ar000184m. [DOI] [PubMed] [Google Scholar]
- 39.Cleland WW. Low-barrier hydrogen bonds and enzymatic catalysis. Arch Biochem Biophys. 2000;382(1):1–5. doi: 10.1006/abbi.2000.2011. [DOI] [PubMed] [Google Scholar]
- 40.Carr CM, Chaudhry C, Kim PS. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci USA. 1997;94(26):14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swalley SE, Baker BM, Calder LJ, Harrison SC, Skehel JJ, Wiley DC. Full-length influenza hemagglutinin HA2 refolds into the trimeric low-pH-induced conformation. Biochemistry. 2004;43(19):5902–5911. doi: 10.1021/bi049807k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.