SUMMARY
Although the oxidative pentose phosphate pathway is important for tumor growth, how 6-phosphogluconate dehydrogenase (6PGD) in this pathway is upregulated in human cancers is unknown. We found that 6PGD is commonly activated in EGF-stimulated cells and human cancer cells by lysine acetylation. Acetylation at K76 and K294 of 6PGD promotes NADP+-binding to 6PGD and formation of active 6PGD dimers, respectively. Moreover, we identified DLAT and ACAT2 as upstream acetyltransferases of K76 and K294, respectively, and HDAC4 as the deacetylase of both sites. Expressing acetyl-deficient mutants of 6PGD in cancer cells significantly attenuated cell proliferation and tumor growth. This is due in part to reduced levels of 6PGD products ribulose-5-phosphate and NADPH, which led to reduced RNA and lipid biosynthesis as well as elevated ROS. Furthermore, 6PGD activity is upregulated with increased lysine acetylation in primary leukemia cells from human patients, providing mechanistic insights into 6PGD upregulation in cancer cells.
INTRODUCTION
Cancer cells appear to coordinate bioenergetics, anabolic biosynthesis and appropriate redox status to provide an overall metabolic advantage to cancer cell proliferation and tumor development (Cairns et al., 2011). The Warburg effect describes a unique metabolic phenomenon in cancer cells, which consists of increased aerobic glycolysis and lactate production. Glycolysis in cancer cells not only generates more ATPs more quickly compared to normal cells that overwhelmingly rely on oxidative phosphorylation (Pfeiffer et al., 2001), but also provides glycolytic intermediates as precursors for anabolic biosynthesis of macromolecules (Vander Heiden et al., 2009). These include nucleotides, amino acids and fatty acids, to produce RNA/DNA, proteins and lipids, respectively, which are necessary for cell proliferation and to fulfill the request of the rapidly growing tumors (Kroemer and Pouyssegur, 2008). For example, glucose-6-phosphate can be diverted into the oxidative pentose phosphate pathways (PPP), which produce ribose-5-phosphate (R-5-P) and/or nicotinamide adenine dinucleotide phosphate (NADPH) (Kroemer and Pouyssegur, 2008). R-5-P is the building block for nucleotide synthesis, while NADPH not only fuels macromolecular biosynthesis such as lipogenesis, but also functions as a crucial antioxidant to quench the reactive oxygen species (ROS) produced during rapid proliferation of cancer cells, which is important for maintenance of cellular redox homeostasis. However, the detailed signaling mechanisms by which cancer cells coordinate bioenergetics (aerobic glycolysis), anabolic biosynthesis and redox homeostasis status to promote cancer cell proliferation and tumor growth remain largely unclear.
6-phosphogluconate dehydrogenase (6PGD) is the third enzyme in the oxidative PPP, which catalyzes the decarboxylating reduction of 6-phosphogluconate (6-PG) to ribulose 5-phosphate (Ru-5-P) and produces NADPH in the presence of NADP+. 6PGD functions as a homodimer in which each monomer acts independently (Bailey-Serres et al., 1992). NADPH is the most crucial metabolite produced in the oxidative PPP by both 6PGD and the first enzyme in the oxidative PPP, glucose-6-phosphate dehydrogenase (G6PD). Increased 6PGD activity has been reported in many cancers, including colorectal cancers (Bravard et al., 1991), cervical intraepithelial neoplasia (Basu et al., 1993; Jonas et al., 1992) and thyroid tumors (Giusti et al., 2008). In addition, 6PGD activity has been documented as a reliable prognostic biomarker in primary breast cancer (Brocklehurst et al., 1986). Yet, how 6PGD is activated in human cancers and whether 6PGD activity is important for cancer pathogenesis and tumor development remain unknown. In this paper, we report that acetylation at K76 and K294 enhances 6PGD activation and is commonly observed in diverse human cancer cells, which is important for coordination of anabolic biosynthesis, redox homeostasis and glycolysis in cells, providing an overall metabolic advantage to cancer cell proliferation and tumor growth.
RESULTS
K76 and K294 acetylation activates 6PGD
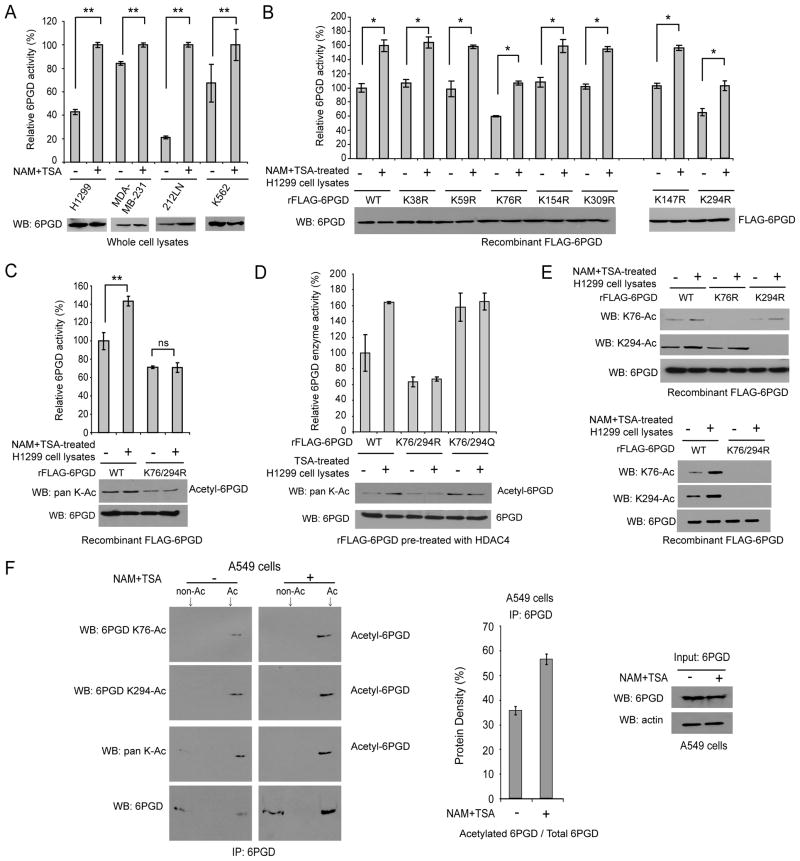
We recently reported that glycolytic enzyme phosphoglycerate mutase 1 (PGAM1) coordinates glycolysis and anabolic biosynthesis in part by regulating 6PGD in the oxidative PPP, suggesting an important role for 6PGD in cell metabolism and tumor growth (Hitosugi et al., 2012). Moreover, proteomics-based studies performed by our collaborators at Cell Signaling Technology (CST) revealed 6PGD as acetylated at a group of lysine residues in human cancer cells (http://www.phosphosite.org/proteinAction.do?id=15053&showAllSites=true). To examine the effect of lysine acetylation on 6PGD activity, we treated diverse human cancer cells including H1299 lung cancer, MDA-MB-231 breast cancer, 212LN head and neck cancer, and K562 leukemia cells with deacetylase inhibitors nicotinamide (NAM) and Trichostatin A (TSA), which led to increased global lysine acetylation in cells. Treatment with NAM+TSA resulted in increased enzyme activity of endogenous 6PGD (Figure 1A). In addition, recombinant FLAG-tagged 6PGD treated with cell lysates of different NAM+TSA-treated cancer cells showed increased enzyme activity and lysine acetylation levels (Figure S1A). These results suggest that lysine acetylation commonly activates 6PGD in human cancer cells.
Figure 1. Lysine acetylation activates 6PGD.
(A) Enzyme activity levels of endogenous 6PGD in diverse human cancer cells treated with NAM+TSA for 16 hours were measured in an in vitro 6PGD enzyme activity assay.
(B–C) Purified recombinant FLAG-6PGD (rFLAG-6PGD) variants were incubated with cell lysates of H1299 cells treated with or without NAM+TSA for 16 hours, followed by in vitro 6PGD enzyme assay.
(D) Purified rFLAG-6PGD variants were pre-treated with purified upstream deacetylase HDAC4 (identified in Figures 3–4), prior to incubation with cell lysates of H1299 cells treated with or without TSA followed by in vitro 6PGD enzyme assay.
(E) Purified rFLAG-6PGD variants were incubated with cell lysates of H1299 cells treated with or without NAM+TSA. K76 and K294 acetylation levels of 6PGD WT, K76R, K294R (upper) and K76/294R (lower) were determined by Western blot using specific acetyl-6PGD antibodies, K76-Ac and K294-Ac, respectively.
(F) Immunoprecipitates of endogenous 6PGD from A549 cells treated with or without NAM+TSA were analyzed by isoelectric focusing (IEF) experiments to quantify acetylated 6PGD (left). Middle: Quantitative estimate of acetylated 6PGD based on results of density analysis of IEF blots. Right: Western blot results of 6PGD protein input.
The error bars represent mean values +/− SD from three replicates of each sample (*: 0.01<p<0.05; **: 0.001<p<0.01; ns: not significant).
Also see Figures S1–S2.
We next performed mutational analysis and generated diverse acetyl-deficient K→R mutants of 6PGD to replace each of the seven lysine residues identified as acetylated in human cancer cells (Figure 1B). We found that substitution of K76 or K294 with arginine resulted in decreased 6PGD activity of purified rFLAG-6PGD compared to WT control in the presence and absence of lysates from NAM+TSA-treated H1299 cells (Figure 1B). Consistent with this finding, acetyl-deficient mutants K76R and K294R and acetyl-mimetic mutants K76Q and K294Q of FLAG-6PGD that were expressed in H1299 cells via transient transfection demonstrated decreased or similar enzyme activity compared to 6PGD WT, respectively (Figure S1B). However, NAM+TSA treatment still enhanced 6PGD activity of both K76R and K294R mutants (Figure 1B), whereas substitution of both K76 and K294 in the double mutant K76/294R abolished the NAM+TSA treatment-induced activation of 6PGD (Figure 1C), suggesting acetylation at both K76 and K294 is required to achieve 6PGD activation.
We next found that TSA but not NAM treatment resulted in increased 6PGD activity (Figure S1C). This is consistent with our later finding that HDAC4 is the upstream deacetylase of 6PGD (Figures 3–4), because TSA and NAM are considered common inhibitors of HDAC and SIRT deacetylase family members, respectively. In order to determine whether substitution of K76 and/or K294 may affect 6PGD activity independent of lysine acetylation, we compared the responsiveness of double mutants K76/294R and K76/294Q of 6PGD upon treatment with lysates from TSA-treated H1299 cells (Figure 1D). Since recombinant protein purified from bacteria is commonly lysine acetylated (Zhang et al., 2009), the purified rFLAG-6PGD proteins were pre-treated with HDAC4 to remove the background lysine acetylation. We found that treatment with lysates of TSA-treated H1299 cells resulted in enhanced activation of 6PGD WT, whereas the catalytically less active, acetylation-deficient K76/294R mutant and catalytically more active, acetylation-mimetic K76/294Q mutant were resistant to TSA treatment in terms of 6PGD enzyme activity (Figure 1D). These results suggested that TSA-dependent enhanced activation of 6PGD is likely mediated through acetylation at K76 and K294.
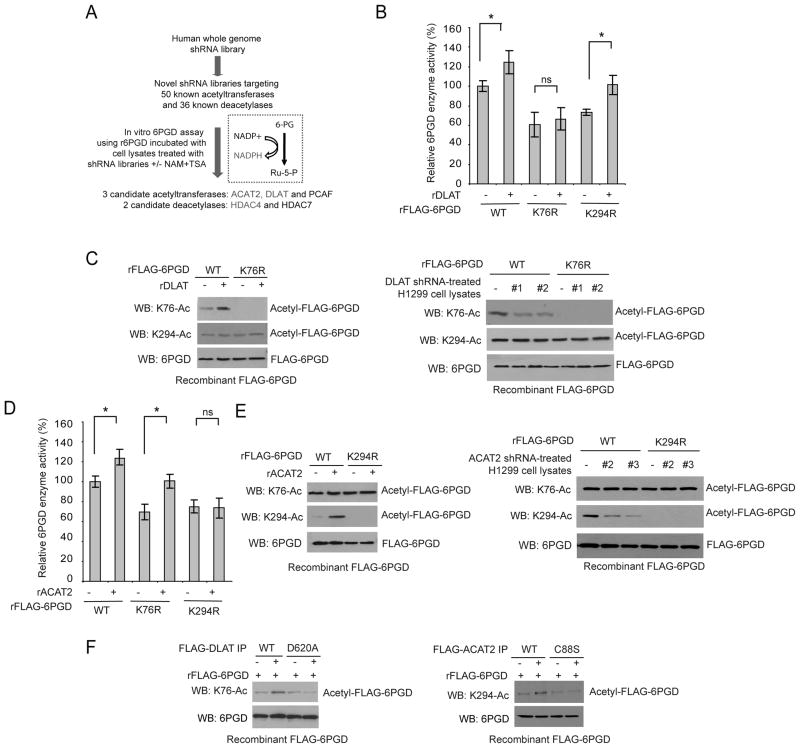
Figure 3. Identification of DLAT and ACAT2 as upstream acetyltransferases for 6PGD K76 and K294, respectively.
(A) Schematic representation of a 6PGD activity assay-based screening strategy to identify lead candidates as upstream acetyltransferases and deacetylases that mediate NAM+TSA treatment-dependent activation of 6PGD. Candidates marked in red color were experimentally confirmed.
(B–E) Purified rFLAG-6PGD WT, K76R and K294R were incubated with purified, recombinant DLAT (rDLAT; B) or ACAT2 (rACAT2; D), followed by 6PGD enzyme activity assay (B and D, respectively). rFLAG-6PGD WT and K76R or K294R proteins were incubated with rDLAT (left; C) or rACAT2 (left; E) or cell lysates of H1299 cells with stable knockdown of DLAT (right; C) or ACAT2 (right; E) by two distinct shRNAs, followed by Western blot to detect K76 and K294 acetylation.
(F) Purified rFLAG-6PGD was incubated with immunoprecipitates of FLAG-DLAT WT or enzyme-deficient D620A mutant (left) or FLAG-ACAT2 WT or enzyme-deficient C88S mutant (right) from 293T cells, followed by Western blot to detect K76 and K294 acetylation, respectively.
The error bars represent mean values +/− SD from three replicates of each sample (*: 0.01<p<0.05; ns: not significant).
Also see Figure S3.
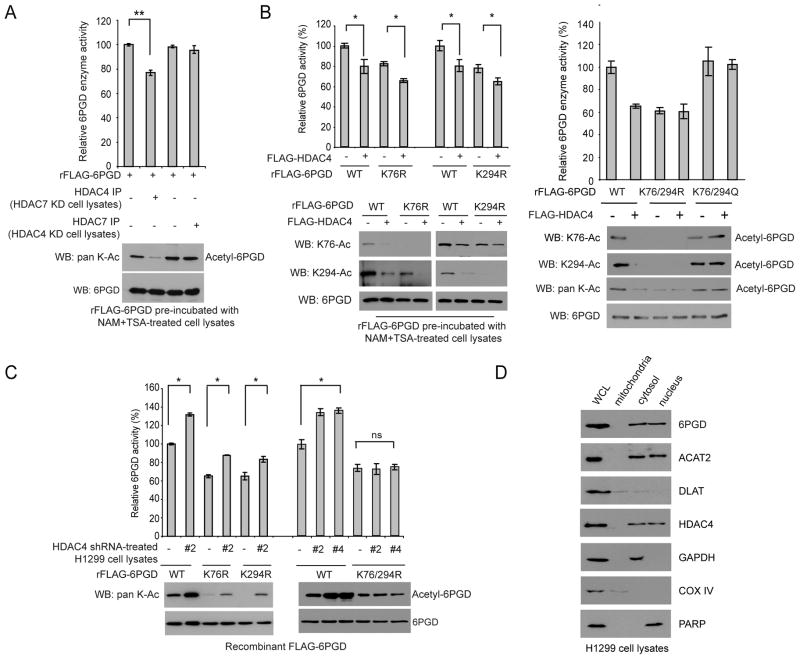
Figure 4. HDAC4 is the upstream deacetylase for both K76 and K294 of 6PGD.
(A) Purified rFLAG-6PGD was pre-incubated with cell lysates of H1299 cells treated with NAM+TSA, followed by incubation with HDAC4 or HDAC7 immunoprecipitated from H1299 cells with stable knockdown of HDAC7 or HDAC4, respectively. The samples were subjected to an in vitro 6PGD enzyme activity assay (upper) and Western blot to detect lysine acetylation levels of 6PGD (lower).
(B) Purified rFLAG-6PGD proteins were incubated with purified FLAG-tagged HDAC4 from 293T cells. The samples were subjected to a 6PGD enzyme activity assay (upper panels) and Western blot (lower panels).
(C) Purified rFLAG-6PGD WT, K76R, K294R and K76/294R were incubated with cell lysates of H1299 cells with stable knockdown of HDAC4, followed by 6PGD enzyme activity assay (upper) and Western blot (lower).
(D) Localization of 6PGD, ACAT2, DLAT and HDAC4 in cytosol of H1299 cells. Cytosolic GAPDH, mitochondrial COX IV and nuclear PARP were included as control markers.
The error bars represent mean values +/− SD from three replicates of each sample (*: 0.01<p<0.05; **: 0.001<p<0.01; ns: not significant).
Also see Figure S4.
Moreover, we generated two specific acetyl-6PGD antibodies that exclusively recognized acetyl-K76 or acetyl-K294 of 6PGD (Figure 1E). Using these antibodies, we found that incubation with lysates of NAM+TSA-treated H1299 cells resulted in increased acetylation at K76 and K294 of 6PGD WT, whereas substitution of K76 and/or K294 abolished acetylation at K76 and/or K294, respectively (Figure 1E). We also performed isoelectric focusing (IEF) experiments using immunoprecipiated endogenous 6PGD and estimated that approximately 36% and 57% of endogenous 6PGD was lysine acetylated in human A549 lung cancer cells in the absence and presence of NAM+TSA treatment, respectively (Figure 1F). Similar results were obtained using human H1299 lung cancer and K562 leukemia cells (Figure S2). Together these data suggested that acetylation at K76 and K294 activates 6PGD in cancer cells.
Acetylation at K76 and K294 promotes NADP+-binding to 6PGD and active 6PGD dimer formation, respectively
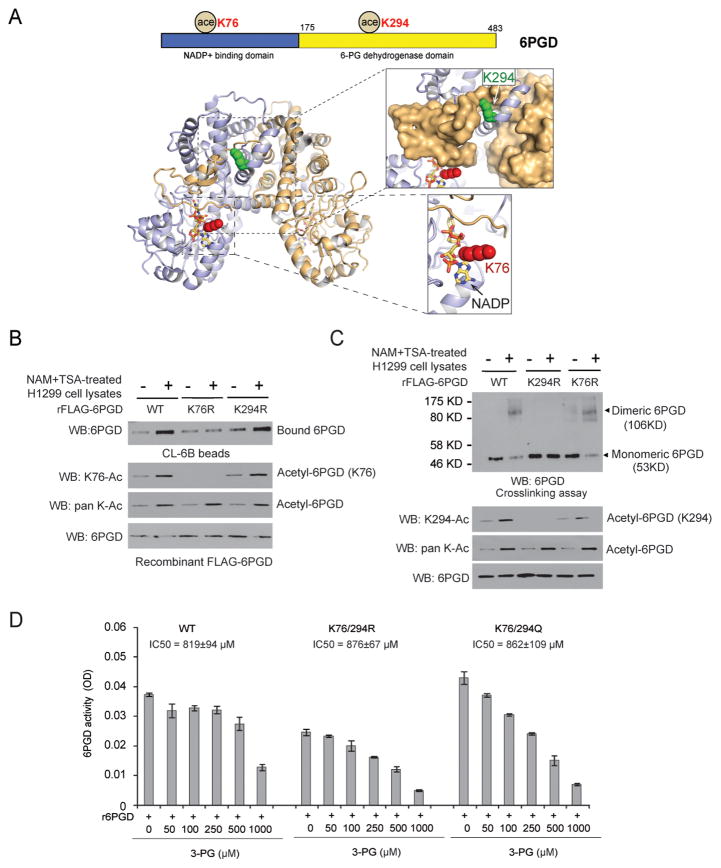
Our structural analysis revealed that K76 is directly proximal (3.7Å) to the cofactor NADP+, while K294 is distal (>20Å) from the catalytic site of 6PGD but forms part of the dimerization interface between the two sister molecules of a 6PGD homodimer (Figure 2A). Monomeric 6PGD was previously suggested to retain activity but is less active compared to dimeric 6PGD (Silverberg and Dalziel, 1973). Thus, we hypothesized that K76 and K294 acetylation may contribute to 6PGD activation by impacting cofactor NADP+ binding and homodimer formation, respectively.
Figure 2. Acetylation at K76 and K294 of 6PGD promotes NADP+-binding to 6PGD and 6PGD dimer formation, respectively.
(A) Upper: Schematic representation of 6PGD with K76 and K294 identified in proteomics studies. Lower: Structural locations of K76 and K294. Crystal structure of human 6-phosphogluconate dehydrogenase in complex with NADP+ (PDB ID: 2JKV). The 6PGD homodimer is shown, with the two chains colored light blue and tan. NADP+ is shown in stick format. K76 is shown as red spheres and K294 as green spheres. Lower right exploded view shows that K76 abuts NADP+ and the C-terminus of the partner molecule. Upper right exploded view shows that K294 forms part of the dimerization interface, with the surface of the partner molecule shown in tan.
(B) Purified FLAG-6PGD variants were incubated with cell lysates of H1299 cells treated with or without NAM+TSA, followed by incubation with CL-6B beads that mimic NADP+ binding.
(C) Purified rFLAG-6PGD variants were incubated with cell lysates of H1299 cells treated with or without NAM+TSA, followed by crosslinking and Western blot experiments to detect dimeric and monomeric 6PGD (upper; 6PGD antibody). 6PGD protein input assessed by Western blot is shown in the lower panel.
(D) Purified rFLAG-6PGD variants were incubated with increasing concentrations of 3-PG, followed by 6PGD enzyme activity assay. IC50 values are shown.
The error bars represent mean values +/− SD.
To test this hypothesis, we performed a “NADP+” binding experiment using Blue Sepharose® CL-6B (Sigma-Aldrich). CL-6B mimics NADP+ and is a pseudo-affinity ligand of many dehydrogenases using NADP+ as a substrate. As shown in Figure 2B, 6PGD WT and K294R incubated with NAM+TSA-treated cell lysates showed a significant increase in the amount of 6PGD protein bound to the CL-6B Agarose beads, indicating increased binding between 6PGD and NADP+. In contrast, substitution at K76 of 6PGD significantly abolished the increased NADP+-binding due to NAM+TSA treatment. To examine whether K294 acetylation promotes homodimer formation, we performed a crosslinking experiment. As shown in Figure 2C, incubation with NAM+TSA treated H1299 cell lysates resulted in increased formation of dimeric 6PGD WT and K76R mutant but not K294R mutant. Together these results suggested that acetylation at K76 and K294 enhances 6PGD activity by different mechanisms, which is consistent with our observations in Figure 1 where 6PGD required acetylation at both K76 and K294 to be fully activated and mutation of each site could not completely abolish acetylation dependent activation of 6PGD.
We recently reported that metabolite 3-phosphoglycerate (3-PG; substrate of PGAM1) functions as a competitive inhibitor of 6PGD, which binds to 6PGD catalytic site and inhibits 6PGD activity by competing with 6PGD substrate 6-PG for 6PGD binding (Hitosugi et al., 2012). In consonance with these findings, we found that the IC50 values of 3-PG in inhibition of 6PGD WT and acetyl-deficient and –mimetic mutants were comparable (Figure 2D). These results support our findings that lysine acetylation-dependent activation of 6PGD is independent of inhibitory 3-PG that affects 6-PG binding.
Acetyltransferases DLAT and ACAT2 acetylate K76 and K294 of 6PGD, respectively
To identify upstream acetyltransferase(s) and deacetylase(s) of 6PGD, we constructed two novel “targeted” lentiviral shRNA libraries that target 50 out of 71 acetyltransferases and 36 out of 40 deacetylases, respectively (Figure 3A). We chose these targets based on the availability of shRNA clones of each target gene in the purchased shRNA library targeting the whole human genome from Open Biosystems. Using these two libraries, we performed 6PGD enzyme assay-based screening studies using recombinant 6PGD incubated with cell lysates from H1299 cells with knockdown of particular acetyltransferases or deacetylases by lentiviral transduction in the presence or absence of NAM+TSA (Figure 3A). Candidates of 6PGD acetyltransferase or deacetylase were determined by their ability to abolish or mimic NAM+TSA-dependent enhancement of 6PGD activation, respectively, upon lentiviral shRNA-mediated knockdown. We identified acetyl-CoA acetyltransferase 2 (ACAT2), dihydrolipoamide S-acetyltransferase (DLAT) and P300/CBP-associated factor (PCAF) as potential 6PGD acetyltransferases, and histone deacetylase (HDAC) 4 and HDAC7 as potential 6PGD deacetylases.
We next found that knockdown of ACAT2 or DLAT but not PCAF resulted in decreased lysine acetylation of 6PGD (Figure S3A). Treatment with lysates from DLAT stable knockdown H1299 cells (Figure S3B; upper) resulted in decreased enzyme activity and lysine acetylation levels of rFLAG-6PGD WT and K294R mutant, but not K76R mutant (Figure S3C). In contrast, treatment with lysates from ACAT2 stable knockdown H1299 cells (Figure S3B; lower) led to decreased enzyme activity and lysine acetylation levels of rFLAG-6PGD WT and K76R mutant, but not K294R mutant (Figure S3D). In addition, incubation with purified, recombinant DLAT resulted in increased enzyme activity of HDAC4-pretreated, purified rFLAG-6PGD WT and K294R mutant but not K76R mutant (Figure 3B). Such treatment also resulted in increased K76 acetylation of rFLAG-6PGD WT but not K76R mutant, whereas K294 acetylation levels of these 6PGD variants were not affected by FLAG-DLAT treatment (Figure 3C; left). Consistent with these findings, incubation with lysates from H1299 cells with stable knockdown of DLAT resulted in decreased K76 acetylation levels of purified rFLAG-6PGD WT but not K76R mutant, whereas such treatment did not affect K294 acetylation levels of WT or K76R mutant (Figure 3C; right). Similarly, purified, recombinant ACAT2 preferentially increased enzyme activity of HDAC4-pretreated, purified rFLAG-6PGD WT and K76R mutant but not K294R mutant (Figure 3D). Both rACAT2 and lysates of ACAT2 stable knockdown H1299 cells altered K294 acetylation of rFLAG-6PGD WT but not K294R mutant and did not affect K76 acetylation levels (Figure 3E; left and right, respectively).
Moreover, we generated catalytically less active forms of DLAT and ACAT2 (D620A and C88S, respectively; Figure S3E), which demonstrated decreased ability to acetylate 6PGD at K76 and K294, respectively, compared to the corresponding control wild type acetyltransferases in the in vitro acetyltransferase activity assays (Figure 3F). Together these results suggest that DLAT and ACAT2 are upstream acetyltransferases that selectively acetylate K76 and K294 of 6PGD, respectively.
HDAC4 is the upstream 6PGD deacetylase that removes acetylation from both K76 and K294
We previously found that HDAC4 associates with HDAC7 in cells (Figure S4A). In order to exclude the potential influence of HDAC4 and 7 on each other, we performed 6PGD enzyme activity assay using immunoprecipitated HDAC4 or HDAC7 from H1299 cells with stable knockdown of HDAC7 or HDAC4 (Figure S4B), respectively. We used purified rFLAG-6PGD protein with high levels of lysine acetylation due to pre-treatment with lysates from NAM+TSA-treated H1299 cells. As shown in Figure 4A, incubation with immunoprecipitates of HDAC4 but not HDAC7 resulted in decreased enzyme activity and lysine acetylation of 6PGD.
Moreover, incubation with purified FLAG-HDAC4 resulted in decreased enzyme activity as well as decreased K76 and K294 acetylation levels of purified FLAG-6PGD WT (Figure 4B; left and right), as well as decreased enzyme activity of K76R mutant with reduced acetylation at K294 but not K76 (Figure 4B; left) and decreased enzyme activity of K294R mutant with reduced acetylation at K76 but not K294 (Figure 4B; left). In contrast, HDAC4 treatment did not affect enzyme activity or lysine acetylation levels of K76/294R or K76/294Q mutants (Figure 4B; right). In addition, incubation with cell lysates from H1299 cells with stable knockdown of HDAC4 resulted in increased lysine acetylation of purified rFLAG-6PGD WT, K76R and K294R but not the double mutant K76/294R (Figure 4C).
We also found that, despite mitochondrial localization of DLAT and nuclear localization of ACAT2 and HDAC4, 6PGD co-localized with DLAT, ACAT2 and HDAC4 in the cytosol of H1299 cells (Figure 4D). Similar results were obtained using K562 leukemia and Tu212 head and neck cancer cells (Figure S4C). These results suggest that the lysine acetylation and deacetylation of 6PGD regulated by DLAT, ACAT2 and HDAC4 may occur in the cytoplasm.
Lysine acetylation of 6PGD is important for cancer cell proliferation and tumor growth
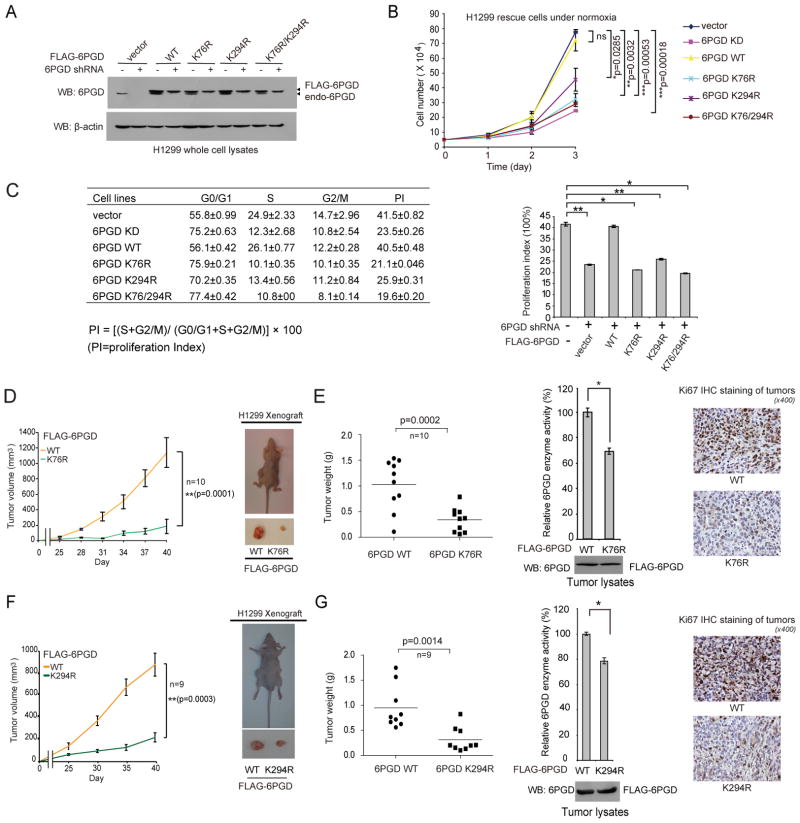
In order to study the importance of lysine acetylation of 6PGD in cancer cells, we generated 6PGD WT, K76R, K294R and K76/294R constructs using a shRNA-resistant FLAG-6PGD form harboring silent mutations in the target region of 6PGD shRNA. Using these shRNA-resistant 6PGD constructs, we generated 6PGD “rescue” H1299 cells with stable knockdown of endogenous 6PGD and rescue expression of different 6PGD variants (Figure 5A). 6PGD knockdown resulted in decreased enzyme activity and cell proliferation, while rescue expression of FLAG-6PGD WT, but not K76R, K294R or K76/294R mutants, rescued not only the decreased 6PGD enzyme activity (Figure S5A) but also cell proliferation under both normoxic and hypoxic conditions (Figures 5B and S5B, respectively) in 6PGD knockdown cells. Consistent with these findings, we found that H1299 cells expressing K→R mutants demonstrated a decreased cell proliferation index (PI) with an increased percentage of cells at G0/G1 phase but decreased cell percentages at S and G2/M phases, compared to 6PGD WT rescue cells (Figure 5C). These results suggested that acetylation at either K76 or K294 of 6PGD itself is necessary and important for the enhanced 6PGD activity that provides a proliferative advantage to cancer cells.
Figure 5. Acetylation of 6PGD K76 and K294 is important for cancer cell proliferation and tumor growth.
(A) Generation of H1299 cells with stable knockdown of endogenous human 6PGD and stable “rescue” expression of an empty vector or WT, K76R, K294R or K76/294R variants of a shRNA-resistant form of human 6PGD.
(B) Cell proliferation rates of 6PGD stable knockdown (KD) and distinct 6PGD rescue H1299 cells under normoxia were determined by cell counting.
(C) Distinct H1299 cell lines were tested for cell proliferation. Left: Percentages of cells in phases of G0/G1, S, G2/M were indicated. Cell proliferation index (PI) was calculated based on the indicated equation (left) and is shown (right).
(D and F) Left panels: Tumor growth was compared between xenograft nude mice injected with K76R (D) or K294R (F) cells compared to mice injected with the control vector cells. Right panels: Dissected tumors in a representative nude mouse injected with H1299 cells expressing 6PGD WT on the left flank and K76R (D) or K294R (F) cells on the right flank.
(E and G) Left panels: Tumor mass in xenograft nude mice injected with K76R (E; n=10) or K294R (G; n=9) cells compared to mice injected with the control H1299 6PGD WT cells are shown. Middle panels show 6PGD enzyme activity (upper) and protein expression (lower) in tumor lysates. Right panels: Representative images of IHC staining of Ki-67 (brown color) in tumor samples. P values were determined by a two-tailed paired Student’s t tests. Center values represent averages.
The error bars represent mean values +/− SD from three replicates of each sample (*: 0.01<p<0.05; **: 0.001<p<0.01; ***: p<0.001; ns: not significant).
Also see Figure S5.
Notably, “rescue” cells expressing K76R or K294R do not have more proliferative potential than cells expressing double mutant K76/294R. This might be due to the comparable 6PGD enzyme activity of these mutants when expressed in cancer cells (Figure S5A). Further discussion is provided in the Discussion section. In contrast, rescue H1299 cells expressing acetyl-mimetic K76Q, K294Q or K76/294Q (Figure S5C) show non-distinguishable 6PGD enzyme activity levels (Figure S5D) and cell proliferation rates under normoxia (Figure S5E), compared to control vector and WT rescue cells. Please see below for detailed discussion.
Furthermore, as shown in Figures S5F–S5G, in a xenograft experiment in which nude mice were injected with control H1299 cells and 6PGD knockdown cells on the left and right flanks, respectively, the growth rate and masses of tumors derived from 6PGD knockdown cells were significantly reduced with decreased Ki67 expression compared to those of tumors formed by control cells over a ~7-week time period. In addition, in xenograft mice injected with FLAG-6PGD WT rescue cells on the left flanks and K76R or K294R rescue cells on the right flanks, the growth rate and masses of tumors derived from K76R (Figures 5D–5E) or K294R (Figures 5F–5G) rescue H1299 cells were significantly reduced with decreased 6PGD enzyme activity and Ki67 expression in tumor cells, compared to those of tumors formed by the control 6PGD WT rescue cells.
We also found that the size of tumors derived from rescue H1299 cells expressing K76/294Q was similar to the size of tumors derived from control cells expressing 6PGD WT with comparable 6PGD enzyme activity (Figure S5H). These results are consistent with our observations that rescue H1299 cells expressing K76/294Q (Figure S5C) show non-distinguishable 6PGD activity (Figure S5D) and cell proliferation rates (Figure S5E), compared to control vector and WT rescue cells. Together these data demonstrate an important role for 6PGD in tumor growth, and that acetylation at either K76 or K294 of 6PGD itself is necessary and important for tumor growth.
Lysine acetylation of 6PGD is important to coordinate biosynthesis, redox homeostasis and glycolysis to confer an overall metabolic advantage to cancer cells
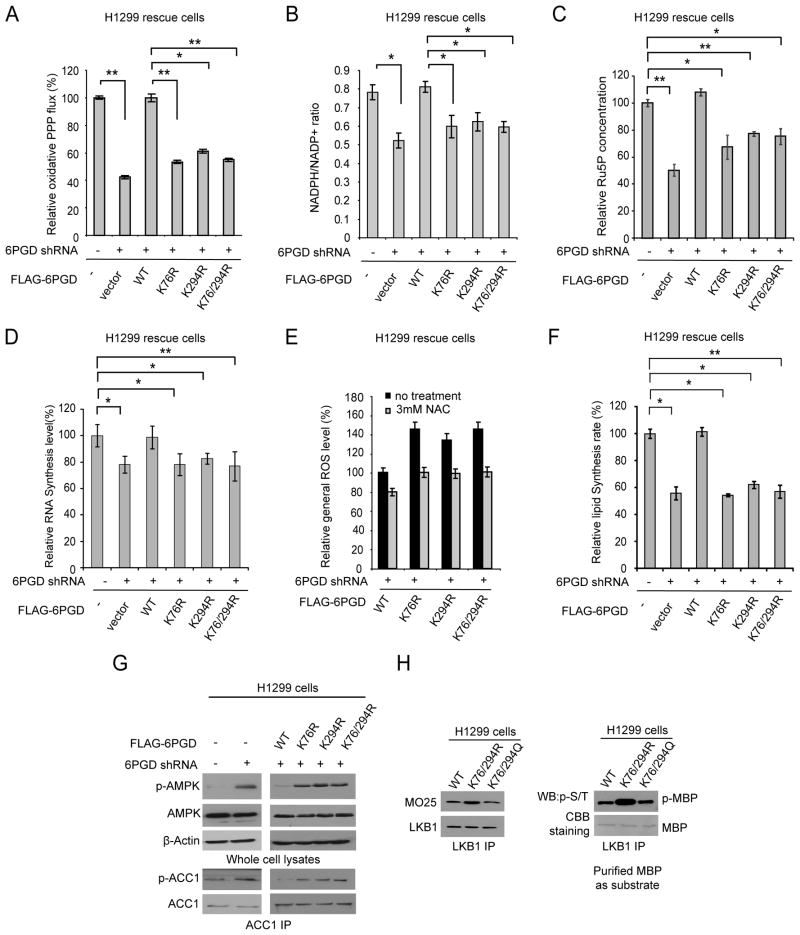
We next found that inhibition of 6PGD by disruption of lysine acetylation in cancer cells resulted in reduced oxidative PPP flux and NADPH/NADP+ ratio, as well as decreased intracellular level of ribulose-5-phosphate (Ru-5-P; 6PGD product) and subsequently RNA biosynthesis, compared to control parental cells (Figures 6A–6D, respectively). We also observed that expressing distinct 6PGD K→R mutants resulted in increased ROS levels, which is consistent with the decreased NADPH/NADP+ ratio, while treatment with an antioxidant agent N-acetylcysteine (NAC) significantly reduced ROS levels (Figure 6E).
Figure 6. Expressing acetyl-deficient mutants of 6PGD results in metabolic defects in cancer cells.
(A–D) Distinct 6PGD rescue H1299 cells and control vector and 6PGD knockdown cells were tested for oxidative PPP flux (A), NADPH/NADP+ ratio (B), intracellular Ru-5-P levels (C), and biosynthesis of RNA (D).
(E) Distinct 6PGD rescue cells were examined for general ROS levels in the absence and presence of NAC (3 mM). The relative general ROS levels were normalized to the WT rescue cells without NAC treatment.
(F) 6PGD rescue cells were tested for lipid biosynthesis.
(G) 6PGD KD and distinct 6PGD rescue H1299 cells as well as control vector cells were examined by Western blot for phosphorylation levels of AMPK (pT172) (upper) and phosphorylation levels of ACC1 (pS79) using ACC1 immunoprecipitates (lower).
(H) Immunoprecipitates of endogenous LKB1 from distinct 6PGD rescue cells were tested for co-immunoprecipitation of MO25 (left) and LKB1 kinase activity using MBP as exogenous substrate (right).
The error bars represent mean values +/− SD from three replicates of each sample (*: 0.01<p<0.05; **: 0.001<p<0.01; ns: not significant).
Also see Figure S6.
In addition, we found that distinct rescue K→R cells showed decreased lipogenesis compared to control WT rescue cells (Figure 6F). Moreover, we found increased levels of activating T172 phosphorylation of adenine monophosphate-activated protein kinase (AMPK) and subsequently increased inhibitory S79 phosphorylation of acetyl-CoA carboxylase 1 (ACC1) in cells with rescue expression of 6PGD K76R, K294R or K76/294R mutants, compared to control WT rescue cells (Figure 6G). ACC1 is a key enzyme in lipogenesis and AMPK inhibits ACC1 by phosphorylating S79, S1200 and S1215 (Mihaylova and Shaw, 2011). Consistently, expressing acetyl-deficient K76/294R mutant resulted in increased LKB1 active complex with increased LKB1 kinase activity, compared to cells expressing 6PGD WT or acetyl-mimetic mutant K76/294Q (Figure 6H). These results are consistent with our recent findings that 6PGD regulates lipogenesis by controlling the intracellular levels of its product Ru-5-P, which inhibits LKB-AMPK signaling cascade by disrupting active LKB1 complex (Shan and Chen, unpublished data).
We also found that inhibition of 6PGD by disrupting lysine acetylation in rescue cells resulted in increased levels of its substrate 6-PG (Figure S6A). It was previously reported that 6-PG activates glycolytic enzyme phosphofructokinase (PFK) (Sommercorn and Freedland, 1982). Consistently, we found that accumulated 6-PG in 6PGD K→R cells resulted in increased PFK activity along with elevated glycolytic rate, lactate production and intracellular ATP levels (Figures S6B–S6C, respectively), while rescue cells expressing distinct acetyl-mimetic K→Q mutants show non-altered oxidative PPP, NADPH/NADP+ ratio, glycolytic rate, ROS level, biosynthesis of RNA and lipids, and intracellular ATP level compared to control cells expressing 6PGD WT (Figure S6D–S6E). Nevertheless, despite the increased glycolysis and ATP levels, rescue cells expressing K→R mutants with attenuated 6PGD activity show reduced cell proliferation and tumor growth (Figure 5), suggesting other metabolic defects including, but not limited to, decreased lipid and RNA biosynthesis and increased ROS level due to disrupted lysine acetylation of 6PGD overshadow the effect of increased bioenergetics on cancer cell proliferation.
Enzyme activity and lysine acetylation levels of 6PGD are upregulated in human leukemia
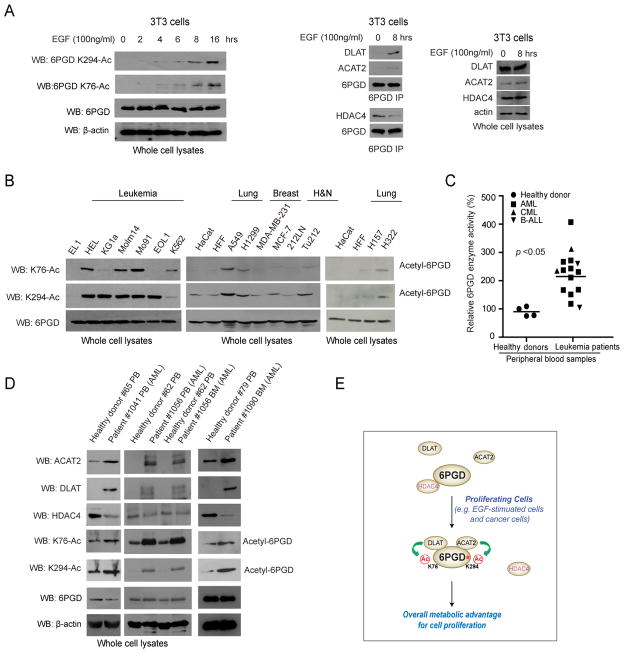
We found that K76 and K294 acetylation of 6PGD was increased in EGF-treated human 3T3 cells (Figure 7A; left), which might be due to increased binding ability of upstream acetyltransferase DLAT and ACAT2 to 6PGD as well as simultaneous dissociation of HDAC4 from 6PGD upon EGF stimulation (Figure 7A; right). Moreover, we found that K76 and K294 acetylation of 6PGD was commonly upregulated in the majority of a spectrum of diverse human leukemia cells, including HEL, KG1a, Molm14, Mo91, EOL1 and K562 cells, as well as a group of solid tumor cells, including A549 and H1299 lung cancer, and Tu212 head and neck cancer cells, compared to normal proliferating human cells including EL1 monocyte/macrophage cells, HaCaT keratinocyte cells and human foreskin fibroblasts (HFF) (Figure 7B). In contrast, MDA-MB-231 and MCF-7 breast cancer and 212LN head and neck cancer cells had similar lysine acetylation levels of 6PGD compared to control HaCaT and HFF cells (Figure 7B).
Figure 7. 6PGD enzyme activity and lysine acetylation levels are commonly upregulated in primary human leukemia cells.
(A) Left: Immunoblotting of lysates of 3T3 cells treated with EGF stimulation for different time points as indicated using specific acetyl-6PGD antibodies. Right: Immunoblotting of 6PGD IP from lysates of 3T3 cells treated with EGF stimulation for 0 or 8 hours to detect co-immunoprecipitated DLAT, ACAT2 and HDAC4.
(B) Western blot results showing acetylation levels of K76 and K294 in diverse human leukemia and tumor cells. Normal proliferating human EL 1 monocyte/macrophage cells, HaCaT keratinocyte cells and human foreskin fibroblasts (HFF) were included as controls.
(C) 6PGD enzyme activity levels were examined using primary leukemia cells from diverse human patients with AML, CML or B-ALL (n=15) and compared to control peripheral blood cells from healthy donors (n=4). The samples were normalized based on total protein levels. Central values represent averages.
(D) K76 and K294 acetylation levels of 6PGD and protein expression levels of 6PGD, ACAT2, DLAT and HDAC4 in primary leukemia cells from three representative AML patients were examined and compared to control peripheral blood cells from healthy donors. PB: peripheral blood; BM: bone marrow.
(E) Proposed model shows that acetyltransferases ACAT2 and DLAT and deacetylase HDAC4 regulates lysine acetylation status of 6PGD, which contributes to 6PGD activation that promotes cell metabolism and proliferation (active 6PGD is marked by asterisk).
Also see Figure S7.
We next found that 6PGD enzyme activity levels were commonly upregulated in primary human leukemia cells from diverse acute myeloid leukemia (AML), chronic myeloid leukemia (CML) and B cell acute lymphoblastic leukemia (B-ALL) patients (n=15), compared to control peripheral blood cells from healthy donors (n=4) (Figure 7C). However, we did not find significantly increased 6PGD expression levels in primary human leukemia cells compared to control peripheral blood (PB) cells from healthy donors (Figure S7A). Instead, compared to peripheral blood cells from healthy donors, primary leukemia cells from representative AML patients showed increased K76 and K294 acetylation levels (Figures 7D and S7B) with increased expression of DLAT and ACAT2 but reduced HDAC4 expression (Figure 7D). These data together suggest that upregulated 6PGD enzyme activity in human leukemias may be due to increased lysine acetylation of 6PGD.
DISCUSSION
Our findings suggest that upregulation of 6PGD by lysine acetylation promotes cell proliferation, which is common and important in cancer cells to provide a metabolic advantage to cell proliferation and tumor growth. Lysine acetylated, active 6PGD may promote oxidative PPP flux to coordinate anabolic biosynthesis and redox homeostasis, at least in part by controlling the intracellular levels of its products Ru-5-P and NADPH, respectively, to promote cell proliferation. EGF stimulation results in increased recruitment of upstream acetyltransferases DLAT and ACAT2 to 6PGD as well as simultaneous dissociation of HDAC4 from 6PGD, leading to elevated lysine acetylation levels of 6PGD. Future studies are warranted to explore the molecular mechanism by which EGF-treatment coordinates the recruitment of DLAT, ACAT2 and HDAC4 to 6PGD to regulate total lysine acetylation levels as well as individual K76 and K294 acetylation levels of 6PGD. Interestingly, in tumor/leukemia cells where such a lysine acetylation-dependent activation mechanism of 6PGD is “hijacked”, 6PGD may be activated due to the upregulated and downregulated protein levels of the 6PGD upstream acetyltransferases (DLAT and ACAT2) and deacetylase (HDAC4), respectively. Together, these findings warrant further studies to decipher the detailed mechanisms and signaling basis by which growth factors such as EGF as well as oncogenic signals may coordinate gene expression levels and/or recruitment of DLAT, ACAT2 and HDAC4 to control lysine acetylation levels of 6PGD to promote cell proliferation (Figure 7E).
Notably, rescue cells expressing 6PGD K76R or K294R mutant do not have more proliferative potential than cells expressing double mutant K76/294R. This might be due to the comparable 6PGD enzyme activity of these mutants when expressed in cancer cells. Since acetylation at K76 and K294 enhances 6PGD activation by different mechanisms through promotion of NADP+ binding and active dimer formation, respectively, these results may suggest that acetylation at both K76 and K294 sites is necessary for 6PGD while abolishing acetylation at either of these two sites is sufficient to prevent 6PGD activation. On the other hand, rescue cells expressing acetyl-mimetic 6PGD K76/294Q mutant show similar 6PGD activity as well as cell proliferation and tumor growth potential compared to cells expressing 6PGD WT. This may suggest that cancer cells might require a certain level of cellular 6PGD activity to function, survive and proliferate. Lysine acetylation is likely important to sustain the 6PGD activity levels above such a threshold; once 6PGD activity reaches the threshold, it cannot be further increased. However, an alternative explanation could be that most of the 6PGD protein is already acetylated in the cells whereas the acetyl-mimetic K76/294Q mutant behaves similarly.
We also identified ACAT2 and DLAT as upstream acetyltransferases and HDAC4 as the deacetylase of 6PGD. Interestingly, DLAT is a mitochondrial protein, which is an enzyme component of the multienzyme pyruvate dehydrogenase complex (PDC) that catalyzes the overall conversion of pyruvate to acetyl-CoA (Hiromasa et al., 2004; Read, 2001). Our findings for the first time describe the cytosolic localization of DLAT and that DLAT functions as a protein acetyltransferase. However, although our data support that DLAT and ACAT2 bind to 6PGD and their catalytic activity is required for acetylation at K76 and K294 of 6PGD, respectively, it remains possible that DLAT and/or ACAT2 may recruit other acetyltransferases to 6PGD that actually acetylate 6PGD. Future studies are warranted to further explore such a possibility.
Although 6PGD was implicated to be a HIF-1α transcription target since 6PGD gene expression is upregulated due to induction of HIF-1α (Guo et al., 2009), we did not observe that the protein expression levels of 6PGD are higher in diverse cancer and leukemia cells compared to normal proliferating cells, or in primary leukemia cells from human patients compared to peripheral blood cells from healthy donors. Instead, we observed that 6PGD is commonly lysine acetylated in diverse cancer and leukemia cells, and the upregulated 6PGD activity in human primary leukemia cells is also likely due to increased lysine acetylation levels of 6PGD rather than increased protein expression. Consistent with these observations, our studies suggest that protein expression and acetylation levels of 6PGD are important for cancer cell proliferation and tumor growth. These results suggest that 6PGD represents a potential therapeutic target in cancer treatment. Indeed, we have developed small molecule inhibitors of 6PGD including Physcion and its derivative S3, which effectively inhibit 6PGD and attenuate cell proliferation and tumor growth potential in cancer cells (Shan and Chen, unpublished data).
EXPERIMENTAL PROCEDURES
Screening for upstream acetyltransferase(s) and deacetylase(s) of 6PGD
The identities of known acetyltransferases and deacetylases in the human genome were provided by the PhosphoSite Plus website of Cell Signaling Technology (http://www.phosphosite.org/psrSearchAction.do). We constructed two “targeted shRNA libraries” that target 50 out of 71 acetyltransferase (Fan et al., 2014) and 36 out of 40 deacetylase genes in the human genome, respectively, which are available in the shRNA library targeting the whole human genome (OpenBioSystems). Each acetyltransferase or deacetylase gene was targeted by a shRNA pool that contains 2–5 different lentiviral-based shRNA constructs that target different regions of the target gene. A 6PGD enzyme assay-based screening strategy was designed to identify upstream acetyltransferase(s) and deacetylase(s) of 6PGD using these shRNA pools. In brief, H1299 lung cancer cells that were infected with lentivirus targeting each acetyltransferase or deacetylase. Four days after lentiviral infection, H1299 cells were treated with NAM+TSA for 16 hours. 6PGD enzyme assay was performed using recombinant 6PGD incubated with 400 μg cell lysate as described above.
Xenograft studies and primary tissue samples from patients with leukemia and healthy donors
Approval of use of mice and designed experiments was given by the Institutional Animal Care and Use Committee of Emory University. Nude mouse xenograft experiments were performed as previously described (Fan et al., 2011; Hitosugi et al., 2011; Hitosugi et al., 2009; Hitosugi et al., 2013) and detailed experimental procedures are provided in Supplemental Information. Approval of use of human specimens was given by the Institutional Review Board of Emory University School of Medicine. All clinical samples were obtained with informed consent with approval by the Emory University Institutional Review Board. Clinical information for the patients was obtained from the pathologic files at Emory University Hospital under the guidelines and with approval from the Institutional Review Board of Emory University School of Medicine and according to the Health Insurance Portability and Accountability Act. Detailed experimental procedures are provided in Supplemental Information.
Supplementary Material
HIGHLIGHTS.
K76 and K294 acetylation activates 6PGD
DLAT and ACAT2 are 6PGD acetyltransferases and HDAC4 is 6PGD deacetylase
Lysine acetylation of 6PGD is important for tumor growth
6PGD is commonly lysine acetylated in EGF-stimulated cells and cancer cells
Acknowledgments
We thank Susan Sunay at the Hematology Division Tissue Bank, Winship Cancer Institute of Emory for providing primary tissue samples from leukemia patients. This work was supported in part by NIH grants CA140515, CA183594, CA174786 (J.C.), CA175316 (S.K.), GM071440 (C.H.) and the Pharmacological Sciences Training Grant T32 GM008602 (S.E.), DoD grant W81XWH-12-1-0217 (J.C.), National Natural Science Funds of China No.20902013 (L.Z.), Charles Harris Run For Leukemia, Inc. (H.J.K.) and the Hematology Tissue Bank of the Emory University School of Medicine and the Georgia Cancer Coalition (H.J.K.). J.X., M.T. and T.-L.G. are employees of Cell Signaling Technology, Inc. H.J.K., F.R.K., S.K. and J.C. are Georgia Cancer Coalition Distinguished Cancer Scholars. S. K. is a Robbins Scholar. S.K. and J.C. are American Cancer Society Basic Research Scholars. J.C. is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Supplemental Information includes detailed experimental procedures and seven figures.
AUTHOR CONTRIBUTIONS
C.S. and S.E. contributed equally to this work. J.X., T.-L.G., K.Y., P.C., D.J.B., M.L.A., S.L., H.J.K. and F.R.K. provided critical reagents. M.T. and T.-L.G. performed mass spectrometry based assays. Q.J., L. Zhou, L. Zhang and C.H. performed biochemical analysis of lysine-acetylated 6PGD and molecular docking studies and analyzed the data. J.S., L.J., M.M., R.J.D., S.W., Y.L. and H.M. performed quantitative mass spectrometry and NMR based assays, and analyzed data. B.H.L. performed the histopathological analyses. T.J.B. performed structural analyses. D.W. and G.Z.C. helped with xenograft experiments. C.S., S.E., H.-B.K., J.-H.S. T.H., and J.F. performed all other experiments. C.S., S.E., H.-B.K., S.K., J.F. and J.C. designed the study and wrote the paper. J.F. and J.C. are senior authors and jointly managed the project. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey-Serres J, Tom J, Freeling M. Expression and distribution of cytosolic 6-phosphogluconate dehydrogenase isozymes in maize. Biochem Genet. 1992;30:233–246. doi: 10.1007/BF02396214. [DOI] [PubMed] [Google Scholar]
- Basu J, Duttagupta C, Vermund SH, Ahn C, Palan PR, Romney SL. Alterations in erythrocyte glutathione metabolism associated with cervical dysplasias and carcinoma in situ. Cancer investigation. 1993;11:652–659. doi: 10.3109/07357909309046937. [DOI] [PubMed] [Google Scholar]
- Bravard A, Luccioni C, Muleris M, Lefrancois D, Dutrillaux B. Relationships between UMPK and PGD activities and deletions of chromosome 1p in colorectal cancers. Cancer genetics and cytogenetics. 1991;56:45–56. doi: 10.1016/0165-4608(91)90361-w. [DOI] [PubMed] [Google Scholar]
- Brocklehurst D, Champion AE, Cheek TR, Dewhurst DG. The value of 6-phosphogluconate dehydrogenase (6-PGDH) activity as a marker of tumour cellularity and prognostic indicator in primary breast cancer. Tumour Biol. 1986;7:99–104. [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature reviews Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Fan J, Hitosugi T, Chung TW, Xie J, Ge Q, Gu TL, Polakiewicz RD, Chen GZ, Boggon TJ, Lonial S, et al. Tyrosine phosphorylation of lactate dehydrogenase A is important for NADH/NAD(+) redox homeostasis in cancer cells. Molecular and cellular biology. 2011;31:4938–4950. doi: 10.1128/MCB.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Shan C, Kang HB, Elf S, Xie J, Tucker M, Gu TL, Aguiar M, Lonning S, Chen H, et al. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Molecular cell. 2014;53:534–548. doi: 10.1016/j.molcel.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti L, Iacconi P, Ciregia F, Giannaccini G, Donatini GL, Basolo F, Miccoli P, Pinchera A, Lucacchini A. Fine-needle aspiration of thyroid nodules: proteomic analysis to identify cancer biomarkers. J Proteome Res. 2008;7:4079–4088. doi: 10.1021/pr8000404. [DOI] [PubMed] [Google Scholar]
- Guo S, Miyake M, Liu KJ, Shi H. Specific inhibition of hypoxia inducible factor 1 exaggerates cell injury induced by in vitro ischemia through deteriorating cellular redox environment. J Neurochem. 2009;108:1309–1321. doi: 10.1111/j.1471-4159.2009.05877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromasa Y, Fujisawa T, Aso Y, Roche TE. Organization of the cores of the mammalian pyruvate dehydrogenase complex formed by E2 and E2 plus the E3-binding protein and their capacities to bind the E1 and E3 components. J Biol Chem. 2004;279:6921–6933. doi: 10.1074/jbc.M308172200. [DOI] [PubMed] [Google Scholar]
- Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, Xie J, Ge Q, Gu TL, Polakiewicz RD, Roesel JL, et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Molecular cell. 2011;44:864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Science signaling. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Zhou L, Elf S, Fan J, Kang HB, Seo JH, Shan C, Dai Q, Zhang L, Xie J, et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22:585–600. doi: 10.1016/j.ccr.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Zhou L, Fan J, Elf S, Zhang L, Xie J, Wang Y, Gu TL, Aleckovic M, LeRoy G, et al. Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation. Nat Commun. 2013;4:1790. doi: 10.1038/ncomms2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas SK, Benedetto C, Flatman A, Hammond RH, Micheletti L, Riley C, Riley PA, Spargo DJ, Zonca M, Slater TF. Increased activity of 6-phosphogluconate dehydrogenase and glucose-6-phosphate dehydrogenase in purified cell suspensions and single cells from the uterine cervix in cervical intraepithelial neoplasia. British journal of cancer. 1992;66:185–191. doi: 10.1038/bjc.1992.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature cell biology. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–507. doi: 10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- Silverberg M, Dalziel K. Crystalline 6-phosphogluconate dehydrogenase from sheep liver. European journal of biochemistry/FEBS. 1973;38:229–238. doi: 10.1111/j.1432-1033.1973.tb03054.x. [DOI] [PubMed] [Google Scholar]
- Sommercorn J, Freedland RA. Regulation of hepatic phosphofructokinase by 6-phosphogluconate. J Biol Chem. 1982;257:9424–9428. [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Molecular & cellular proteomics : MCP. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.