Nearly all human cancers are preceded by precursor states far more prevalent than clinical malignancy itself. Understanding which premalignant lesions are truly biologically premalignant versus those with impending clinical progression (biologically malignant) is a central question in cancer biology. Tracking progression of human premalignant states (or lack thereof) in vivo is not feasible in most tumors because precursor lesions are typically resected at diagnosis. Multiple myeloma (MM) is characterized by progressive growth of malignant plasma cells in the bone marrow leading to organ dysfunction(1). Nearly all cases of clinical MM (MM) are preceded by asymptomatic monoclonal gammopathies (AMGs) further classified as either monoclonal gammopathy of undetermined significance (MGUS) or asymptomatic myeloma (AMM)(1, 2). MM is a unique model to dissect genetic evolution of early cancer as the precursor state is well defined and not resectable. Several studies have characterized the genetic architecture of malignant plasma cells in both MM and its precursor states (3). MM tumor cells carry several cytogenetic abnormalities (most notably IgH translocations and hyperdiploid karyotypes), as well as copy number abnormalities leading to genomic gains/losses and loss of heterozygosity (LOH). Comparisons of plasma cells (PCs) in cohorts of MM versus MGUS/AMM show greater proportion of cytogenetically abnormal PC in MM, suggesting expansion of preexisting and more proliferative clones during transition to MM(4, 5). However, nearly all of the cytogenetic changes described in MM tumor cells have also been observed in MGUS/AMM, and it has proven difficult to define the malignant phenotype based on genetic changes alone(4, 5). Whole-genome and whole-exome-based sequencing strategies have recently been applied to human MM and shown that there are about 20–40 non-synonymous variants per MM tumor cell, but no single unifying variant has been identified(6–8). The most common targets of recurrent mutation were known cancer genes such as KRAS and TP53, with KRAS mutations in 27% of cases(6). Analysis of serial samples from MM patients undergoing therapy recently demonstrated the presence of subclones, with greater changes in patients with cytogenetically high risk disease(9, 10). Similar findings were made via serial genome sequencing in a patient with t(4:14) myeloma evolving to plasma cell leukemia.(8) In this study, we serially analyzed the exomes of patients with AMGs who progressed to clinical MM and compared the findings with those AMGs that did not progress to clinical MM.
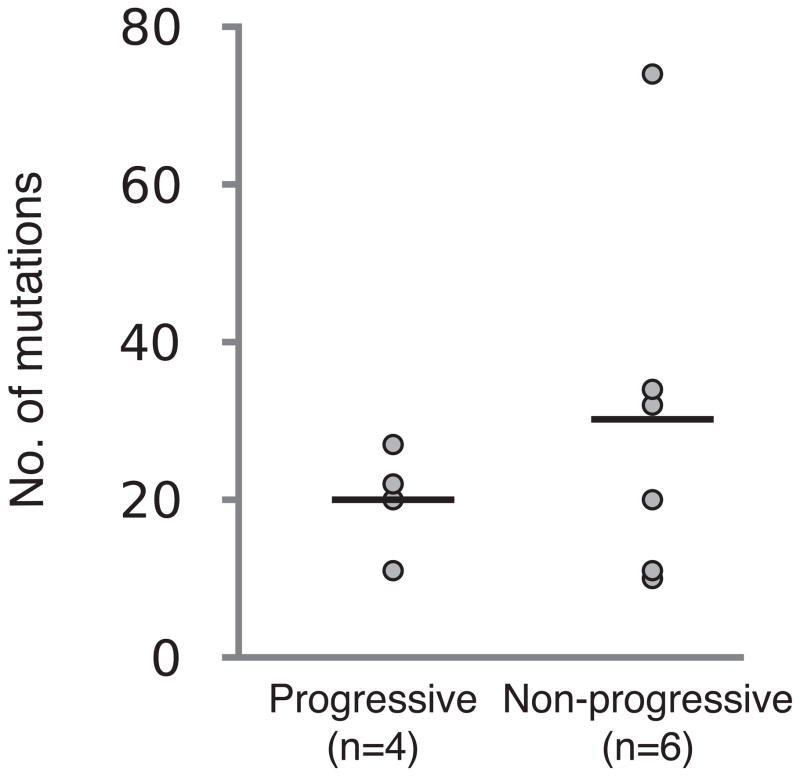
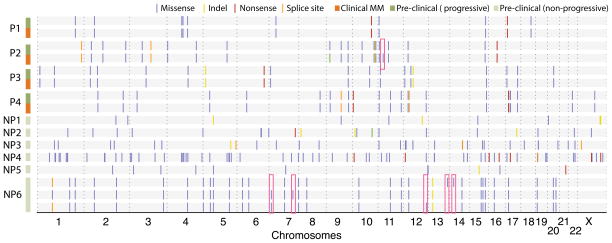
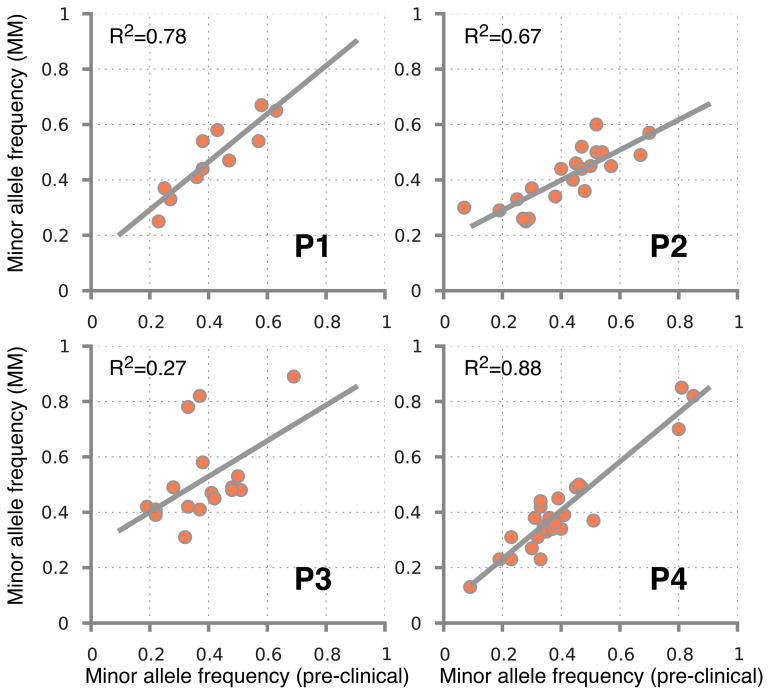
Whole-exome sequencing was performed on tumor and germline DNA from 10 patients with AMGs (exome run summary in supplementary table 1, see supplementary methods online). Clinical characteristics of this cohort are shown in supplementary table 2 and these patients could be classified as MGUS (n= 3) or AMM (n= 7) by current criteria. Four of these patients (P1-P4) progressed to clinical MM requiring therapy, while 6 patients (NP1-NP6) that did not progress with at least 2 years of follow up were studied as controls. Overall 261 somatic non-synonymous variants (SNVs) were seen in these 10 patients, with a mean 26 variants per patient (range 10–74). The mean number of SNVs per sample was similar between patients with progressive or non-progressive disease (Figure 1a). In patients who progressed to clinical MM (P1-P4), the sequencing data from baseline was directly compared to that from the corresponding malignancy. In 3 of the paired samples, there were no new SNVs detected in the progression sample (Figure 1b). In the fourth patient (P2), a new SNV in a single gene (BBOX1) of unclear significance was detected in the progression sample. The lack of significant evolution from baseline to progression was also confirmed through analysis of minor/alternate-allele frequencies in the paired samples (Figure 1c). Variant calls from all progression samples was validated using Sanger sequencing (not shown). Serial samples (at 2 and 4 years of follow up) were also available from a patient (NP6) without disease progression. These samples again demonstrated stable SNV profile, with balanced loss and gain of 2 new detectable SNVs at each time-point (Figure 1b). Together these data indicated that nearly all of the SNVs found at the diagnosis of clinical MM were also present in the precursor state.
Figure 1.
Analysis of somatic variants
1a. Number of somatic variants per sample.
1b. Serial analysis of somatic variants in each patients
1c. Analysis of B allele frequencies in paired samples.
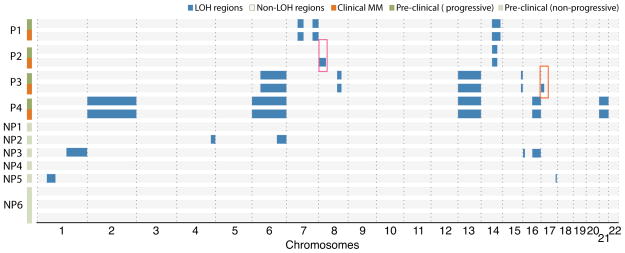
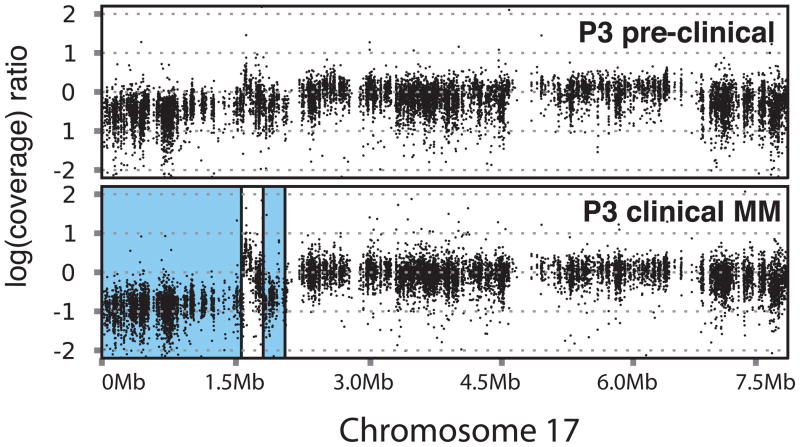
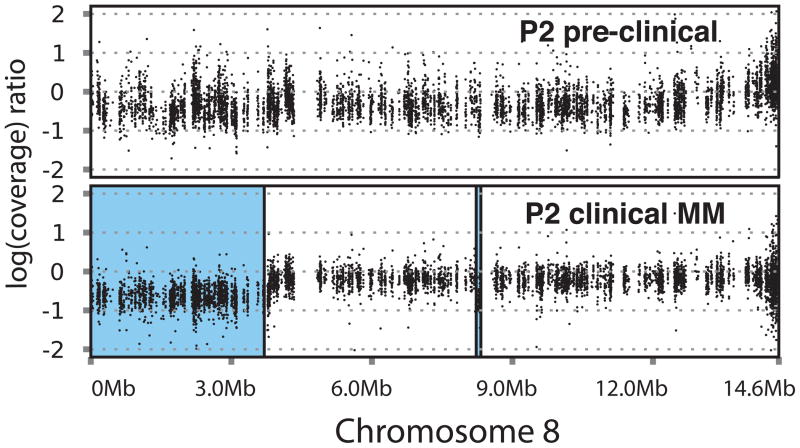
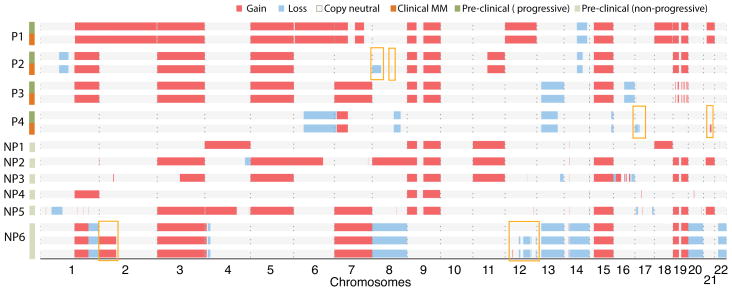
In view of lack of consistent progression-specific SNVs, we turned our attention to regions of somatic copy number alterations (CNAs), intra-clonal variation and pattern of somatic mutations. Analysis of loss-of-heterozygosity (LOH) patterns revealed that degree of LOH at baseline was much greater in patients with Prog-AMG than in NonProg-AMG, although there was no consistent pattern shared by all samples (Figure 2a). In the patients who progressed to MM, the LOH regions at MM stage matched those at baseline with little change. However in two patients (P2 and P3), new regions of MM-specific LOH were identified (Figure 2a–2c). Interestingly, in one of these patients (P3), the region of new LOH correlated with the site of p53 on chromosome 17 (Figure 2b) and possibly involved a dominant subclone as the contribution of the cells carrying the deletion was 10 % lower than the estimated fraction of tumor cells in this sample. Profile of somatic CNAs again revealed several regions of alterations throughout the genome, which were again largely maintained between baseline and progression samples (Figure 2d). Together these data demonstrate that patients with progressive lesions may have greater degree of genomic gains/losses compared to those that remain clinically asymptomatic.
Figure 2.
Analysis of genomic gains/losses with disease progression
2a. Analysis of LOH patterns.
2b. Analysis of CNV pattern in chromosome 17 in patient P3
2c. Analysis of CNV pattern in chromosome 8 in patient P2
2d. Analysis of copy number variations
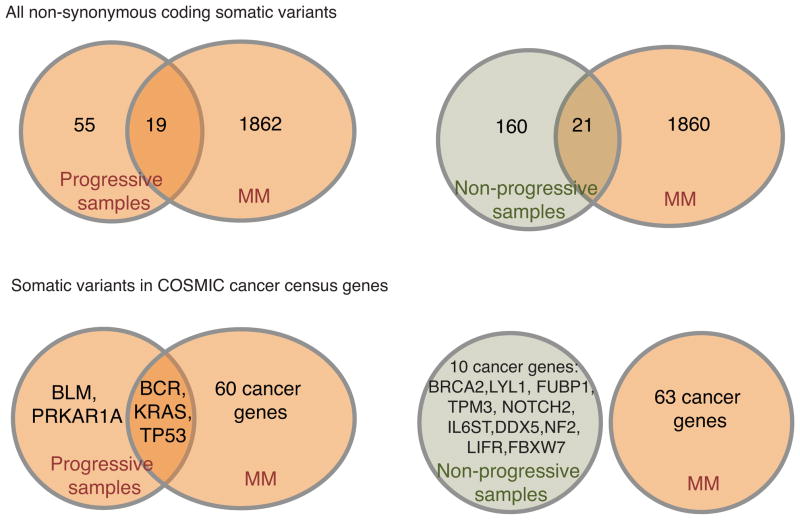
2e. Comparison of mutational profile with published data in patients with clinical myeloma.
An important and unique aspect of this study is the concurrent analysis of samples from patients with AMG that did not progress to clinical MM. Therefore we compared the targets of somatic mutations in this cohort with those published for patients with MM (6–8). Targets of somatic mutations in Prog-AMGs had a greater overlap than NonProg-AMGs with the MM dataset (19 of 74 (26%) in Prog-AMG vs 20 of 181 (11%) in NonProg-AMG, p =0.01) (Figure 2e). This was also true when mutations in known oncogenes in the COSMIC database overlapping between the current AMG cohorts and the MM datasets were compared (3 of 5 in Prog-AMG vs 0 of 10 in NonProg-AMG) (Figure 2e). Notably, this included KRAS and TP53 wherein mutations were only detected in Prog-AMG, but not in patients with NonProg-AMG. Therefore the pattern of mutations at baseline may correlate with the risk of progression to MM.
These data demonstrate that the vast majority of somatic mutations in MM are present at least several months and possibly years before the onset of symptoms indicative of the diagnosis of clinical malignancy. Similar findings were recently reported by Walker et al who compared exomes from 4 patients with AMM and corresponding MM(11). Interestingly, the net mutation load was similar between progressive and non-progressive lesions, demonstrating that long periods of clinical stability are feasible in AMGs in spite of the level of mutational load comparable to that seen in clinical MM. Instead of the number of mutations, the pattern of mutations and genomic changes at baseline may impact the risk of developing clinical malignancy. In such a model, lesions that lack mutations in critical genes are unable to mediate progressive growth needed for malignant phenotype. The list of targets preferentially mutated in patients with Prog-AMGs consisted of well-known cancer genes such as KRAS and TP53 (6). The presence of mutations in KRAS in myeloma has been previously correlated with an aggressive course and disease progression (12). Cytogenetic deletion of chromosome 17p (carrying TP53) has also been well documented as a feature of high risk MM. Mutations in TP53 have also been described in MM and correlate with the presence of del17p(13, 14). Further studies in asymptomatic patients are needed to identify additional mutations that may predict increased risk of malignancy.
Somatic LOH and CNA analyses indicated that the genomes in Prog-AMG also carried greater regions of genomic loss/gains than in patients with NonProg-AMG. These data suggest that the degree of genomic instability may itself be a marker of the risk for malignant transformation. In some instances, it is possible that these secondary changes may contribute more directly to malignant transformation through evolution of a subclone. For example, in patient P3, progression to MM was accompanied with the appearance of new LOH interval in chromosome 17p, including TP53.
To our knowledge, these data provide the first direct comparison of serial exomes in progressive versus non-progressive precursor states in human cancer. In contrast to recent studies with myelodysplasia and acute leukemia, the precursor state in this setting was observed without any specific therapy, therefore the findings are not impacted by possible therapy-induced effects(15). Our findings demonstrate that most somatic mutations and genome alterations predate clinical malignancy and that mutational loads comparable to clinical malignancy could be associated with long periods of dormancy in premalignant states. Instead of net mutational load, the pattern of mutations and the degree of genomic instability may predict malignant fate. Our findings should be considered as preliminary due to the small numbers of AMG exomes sequenced thus far. Analysis of spectrum of mutations in precursor states may identify lesions more likely to progress to clinical cancer.
Supplementary Material
Footnotes
Supplementary information is available at Leukemia website.
Conflict of Interest:
The authors disclose no conflict of interest.
References
- 1.Landgren O, Kyle RA, Rajkumar SV. From myeloma precursor disease to multiple myeloma: new diagnostic concepts and opportunities for early intervention. Clin Cancer Res. 2011;17(6):1243–52. doi: 10.1158/1078-0432.CCR-10-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhodapkar MV, Sexton R, Waheed S, Usmani S, Papanikolaou X, Nair B, et al. Clinical, genomic and imaging predictors of myeloma progression from asymptomatic monoclonal gammopathies (SWOG S0120) Blood. 2013 doi: 10.1182/blood-2013-07-515239. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12(5):335–48. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Corral L, Gutierrez NC, Vidriales MB, Mateos MV, Rasillo A, Garcia-Sanz R, et al. The progression from MGUS to smoldering myeloma and eventually to multiple myeloma involves a clonal expansion of genetically abnormal plasma cells. Clin Cancer Res. 2011;17(7):1692–700. doi: 10.1158/1078-0432.CCR-10-1066. [DOI] [PubMed] [Google Scholar]
- 5.Lopez-Corral L, Sarasquete ME, Bea S, Garcia-Sanz R, Mateos MV, Corchete LA, et al. SNP-based mapping arrays reveal high genomic complexity in monoclonal gammopathies, from MGUS to myeloma status. Leukemia. 2012 doi: 10.1038/leu.2012.128. [DOI] [PubMed] [Google Scholar]
- 6.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker BA, Wardell CP, Melchor L, Hulkki S, Potter NE, Johnson DC, et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood. 2012 doi: 10.1182/blood-2012-03-412981. [DOI] [PubMed] [Google Scholar]
- 8.Egan JB, Shi CX, Tembe W, Christoforides A, Kurdoglu A, Sinari S, et al. Whole genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution and clonal tides. Blood. 2012 doi: 10.1182/blood-2012-01-405977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keats JJ, Chesi M, Egan JB, Garbitt VM, Palmer SE, Braggio E, et al. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012 doi: 10.1182/blood-2012-01-405985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weston-Bell N, Gibson J, John M, Ennis S, Pfeifer S, Cezard T, et al. Exome sequencing in tracking clonal evolution in multiple myeloma following therapy. Leukemia. 2013;27(5):1188–91. doi: 10.1038/leu.2012.287. [DOI] [PubMed] [Google Scholar]
- 11.Walker BA, Wardell CP, Melchor L, Brioli A, Johnson DC, Kaiser MF, et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 2013 doi: 10.1038/leu.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neri A, Murphy JP, Cro L, Ferrero D, Tarella C, Baldini L, et al. Ras oncogene mutation in multiple myeloma. J Exp Med. 1989;170(5):1715–25. doi: 10.1084/jem.170.5.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chng WJ, Price-Troska T, Gonzalez-Paz N, Van Wier S, Jacobus S, Blood E, et al. Clinical significance of TP53 mutation in myeloma. Leukemia. 2007;21(3):582–4. doi: 10.1038/sj.leu.2404524. [DOI] [PubMed] [Google Scholar]
- 14.Xiong W, Wu X, Starnes S, Johnson SK, Haessler J, Wang S, et al. An analysis of the clinical and biologic significance of TP53 loss and the identification of potential novel transcriptional targets of TP53 in multiple myeloma. Blood. 2008;112(10):4235–46. doi: 10.1182/blood-2007-10-119123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366(12):1090–8. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.