Summary
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression by binding to sequences within the 3′UTR of mRNAs. Because miRNAs bind to short sequences with partial complementarity, target identification is challenging. To complement the existing target prediction algorithms, we devised a systematic “reverse approach” screening platform which allows the empirical prediction of miRNA-target interactions. Using Drosophila cells, we screened the 3′UTRs of the Hedgehog pathway genes against a genome-wide miRNA library and identified both predicted and many non-predicted miRNA-target interactions. We demonstrate that miR-14 is essential for maintaining the proper level of Hedgehog signaling activity by regulating its physiological target, hedgehog. Furthermore, elevated levels of miR-14 suppress Hedgehog signaling activity by co-targeting its apparent non-physiological targets, patched and smoothened. Altogether, our systematic screening platform is a powerful approach to identifying both physiological and apparent non-physiological targets of miRNAs, which are relevant in both normal and diseased tissues.
Introduction
MicroRNAs (miRNAs) are endogenously transcribed 19- to 25-nt small non-coding RNAs that post-transcriptionally regulate target mRNAs by pairing to complementary sequences, typically found in their 3′ untranslated regions (3′ UTRs), to repress mRNA translation, promote transcript decay or both (Bartel, 2009; Brodersen and Voinnet, 2009; Ghildiyal and Zamore, 2009; Hendrickson et al., 2009). In normal cells, multiple miRNAs cooperate to maintain a proper balance of various processes, including proliferation, differentiation and cell death. In addition, individual miRNAs can regulate multiple mRNAs further complicating the gene regulatory networks of miRNAs. Therefore, dysregulation of miRNAs can have detrimental cellular consequences and has been associated with several human diseases ranging from metabolic and inflammatory disease to malignancy (Care et al., 2007; Fiore et al., 2008; Krutzfeldt and Stoffel, 2006; Lu et al., 2005; Poy et al., 2004). According to the miR2Disease database (Jiang et al., 2009) that manually curates disease associated miRNAs, both down- and up-regulated miRNA dysregulation are equally prevalent.
Down-regulation of miRNAs in diseased tissue can lead to aberrant expression of their target genes. Such genes are deemed “physiological targets” since their expression is tightly regulated by miRNAs in normal tissues. Conversely, up-regulation of miRNAs in diseased tissue can further down-regulate their physiological targets, preventing normal cell function. In addition, up-regulated miRNAs are able to down-regulate “apparent non-physiological targets,” which correspond to genes bearing miRNA binding sites that in the wild type situation are unaffected. Therefore, identifying both physiological and apparent non-physiological targets of miRNAs is essential for understanding the complex gene regulation by miRNAs in diseased tissues.
Target gene identification of miRNAs is challenging because they bind to their target mRNAs by partial complementarity over a short sequence and the rules of miRNA-mRNA interactions are not fully understood. Unlike the related siRNAs that require a perfect complementary match for cleavage of target mRNAs, miRNAs allow mismatches at positions 1, 9 or 10 relative to their 5′ end, whereas Watson-Crick pairing at positions 2 to 8, referred to as the “seed” region, is the minimal sequence required for silencing of their targets (Brennecke et al., 2005). In addition, despite the large number of target genes predicted to be affected by miRNA loss of function, individual miRNA knockouts lack strong phenotypic consequences. For example, in C. elegans, the majority of individual miRNA mutants display no major phenotype (Miska et al., 2007).
Use of miRNA target prediction algorithms has been valuable in identifying targets of many different miRNAs. However, these prediction algorithms are still not complete. Target prediction algorithms typically predict hundreds to thousands of target genes for an individual miRNA (Betel et al., 2010; Paraskevopoulou et al., 2013; Reczko et al., 2012; Ruby et al., 2007). However, most of these predicted genes may not correspond to true targets (Alexiou et al., 2009) and the algorithms often fail to identify validated miRNA targets (Johnson et al., 2005; Lal et al., 2009). Another complication is that predictions from several different algorithms generate lists of target genes with very little overlap, making decisions about which predictions to investigate difficult. Lastly, the majority of these predictions lack experimental validations.
Here, we devised a general “reverse approach” strategy for rapidly identifying targets of miRNAs whereby, rather than studying specific miRNAs, we screened the 3′ UTR of individual genes against miRNAs. Specifically, we screened the effect of 132 distinct miRNAs on the 3′ UTR of 9 core components of the Hedgehog (Hh) signaling pathway, leading us to identify 59 miRNA-target interactions. The Hh pathway controls multiple developmental processes such as differentiation, pattern formation, and proliferation in various animals ranging from Drosophila to humans (Ingham et al., 2011; Jiang and Hui, 2008; McMahon et al., 2003). In addition, aberrant activation of Hh signaling in humans has been linked to growth and maintenance of various cancers (Barakat et al., 2010; Fan et al., 2004; Jiang and Hui, 2008; Kayed et al., 2004; Ruiz i Altaba, 1999; Taipale and Beachy, 2001; Watkins et al., 2003). We discuss how our experimentally-based miRNA-target interactions approach compare to those obtained with the three most popularly used target prediction algorithms. Further, we focus on one miRNA, miR-14, that we find to regulate the 3′ UTRs of three Hh pathway components. In vivo analysis of miR-14 targets shows how depending on its level of expression, a single miRNA targets different components of the same pathway and highlight the importance of identifying miRNA targets at different miRNA expression levels to fully understand loss or gain of function miRNA phenotypes.
Results
Construction of miRNA screening platform for screening Hh pathway components in tissue culture cells
To facilitate the rapid identification of miRNAs that regulate core components of the Hh pathway, we used a firefly luciferase reporter to assay miRNA activity in Drosophila S2R+ cells. We constructed luciferase reporters for 9 core components of the Hh pathway by cloning their 3′ UTR downstream of the firefly luciferase (Table S1). In addition, for the miRNA overexpression library, we used our previously generated collection of 95 constructs (Bejarano et al., 2012) that we complemented with an additional set of 33 constructs to increase the coverage of the collection (see Experimental Procedures). As some plasmids cover multiple miRNAs, our miRNA overexpression library is composed of 128 overexpression plasmids covering 132 distinct miRNAs (see details in Table S2).
Next, to assess the activities of individual miRNAs, we co-transfected the luciferase 3′ UTR reporters with the miRNA overexpression plasmids into S2R+ cells. After 72 hrs, the firefly luciferase levels were measured and normalized to the Renilla luciferase levels (Figure 1A). For each miRNA-target gene pair, we computed a negative log2 median fold-change (LMF) score, where the higher LMF score corresponds to stronger repression of the target gene by the miRNA (Table S2).
Figure 1. Screen for miRNAs that regulate Hh signaling pathway components.
(A) Outline of the primary screen. Genome-wide collection of 128 Drosophila miRNA overexpression plasmids that covers 132 distinct miRNAs were screened against the luciferase 3′ UTR reporters of 9 core genes of the Hh pathway. (B–D) Systematic comparison of the LMF scores from the entire screen to the predicted confidence scores from TargetScan, miRanda, and DIANA, respectively. Grey dashed lines mark the LMF score cutoff value (LMF score ≥ 0.622). Blue lines show the general correlation of the LMF score to the predicted confidence score from target prediction algorithms. Note that the confidence scores for miRanda (mirSVR score) increase as mirSVR scores decrease, thus the inverse in trend line. The significance of correlation is estimated using Pearson’s product moment correlation coefficient (R). (E) Comparison of the LMF scores to the miRNA-target pair predicted by number of tools. Box plots shows that the LMF score distributions of miRNA-target pairs predicted by single tool (1) two tools (2) and three tools (3) compared to the pairs that are not predicted by any of the tools (0). Wilcoxon test was used to test the significance of difference between two distributions. **P < 0.001 and *P < 0.05. (F) Analysis of True Positive Rates (TPR) and False Positive Rates (FPR) at various LMF scores cutoffs. Grey box marks the cutoff value (LMF score ≥ 0.622) at which we achieved 33% TPR and 3% FPR. AUC (area under ROC curve). (G) miRNA-target interaction network. The thickness of interaction lines indicates the range in LMF scores. The thickness in line increases as LMF scores increase. (H) Venn diagram displaying the overlap of miRNA-target interaction predicted by individual prediction algorithms to the screen results.
Using the TargetScan (Ruby et al., 2007), DIANA (Paraskevopoulou et al., 2013; Reczko et al., 2012) and miRanda (Betel et al., 2010) target prediction databases, we compiled a list of miRNAs predicted to regulate the 3′ UTRs of Hh pathway components (Table S3). For each predicted miRNA-target pair, we extracted the confidence score assigned by individual tools. We only considered the miRNAs that are part of our screening library and used the least stringent cutoff values for each tool to compile all possible miRNA-target predictions. Systematic comparison of the LMF score to the predicted confidence score reveals a weak but significant positive correlation (Figure 1B–D). Furthermore, the median LMF scores increase as the number of prediction tools supporting the miRNA-target pair increases (Figure 1E). This suggests that the pairs with high LMF score are likely to be predicted by multiple target prediction tools as high-confidence miRNA-target pairs and demonstrate the reliability of the LMF scores. Next, to systematically evaluate the performance of LMF scores, we created a Positive Reference Set (PRS) and a Random Reference Set (RRS). The PRS includes 24 miRNA-target pairs validated from the literature (6 pairs) and complemented with high-confidence predictions (18 pairs). We constructed 1000 RRS sets sampled from 736 non-specific miRNA-target pairs (Table S4) (see Experimental Procedures). Analysis of True Positive Rates (TPR) and False Positive Rates (FPR) at various cutoff reveals robust performance of the LMF score (area under ROC curve = 0.8). Furthermore, this analysis also facilitated the identification of appropriate cutoff value (LMF score ≥ 0.622) at which we achieved 33% TPR and 3% FPR (Figure 1F). Using this cutoff value, we generated a miRNA-target interaction network composed of 59 interactions connecting 43 miRNAs to 9 Hh pathway members (Figure 1G). Strikingly, all 9 Hh pathway 3′ UTR reporters responded to multiple miRNAs. Consistent with this result, computational analysis of miRNA target sites indicated that most genes bear binding sites of multiple miRNAs (Enright et al., 2003; Grun et al., 2005; Lewis et al., 2003; Maragkakis et al., 2011). Out of our 59 miRNA-target interactions, 9 were supported by at least one of the three target-prediction algorithms, 16 were supported by two of the three target-prediction algorithms, and 11 were supported by all three (Figure 1H). In addition, we identified 23 novel miRNA-target interactions that were not predicted by any of the three target-prediction algorithms highlighting the limitation of the existing algorithms and the need for functional tests.
We identified 43 miRNAs that reduced the activity of the reporters and therefore constitute potential regulators of the Hh pathway. Of these, 30 miRNAs regulated a single Hh pathway component, whereas 13 miRNAs regulated multiple components. We selected miR-14, which was found to regulate both activators (hh and smoothened (smo)) and an inhibitor (patched (ptc)) of the Hh pathway (Figure 1G), for further characterization.
in vitro validations of miR-14 targets
We asked whether miR-14 indeed targets hh, ptc and smo by directly binding to its miRNA responsive elements (MREs) within their 3′ UTRs. To identify possible miR-14 MREs in the 3′ UTR of each target gene, we used RNAhybrid (Rehmsmeier et al., 2004), a bioinformatics tool for finding the minimum free energy (mfe) hybridization sites for miRNAs. For each target gene, we selected the top candidate MREs based on their mfe scores and seed pairing rules, and mutated the sequence complementary to the miR-14 seed (Figure 2A). Mutating the potential MREs within the 3′ UTRs partially (hh and smo) and completely (ptc) relieved the suppression elicited by the addition of miR-14 (Figure 2B). The partial rescue of the luciferase signals observed with the mutated MREs and miR-14 is most likely due to additional, potentially still functional, miR-14 MREs that were not mutated (Figure S1A–C). Collectively, these results demonstrate that direct interactions between miR-14 and the MREs located within the 3′ UTR of the target genes are responsible for suppression of the luciferase signal.
Figure 2. in vitro validations of miR-14 target genes.
(A) Potential miR-14 binding sites within the 3′ UTRs of the 3 candidate target genes. All predicted binding sites were mutated as shown in red. (B) Secondary luciferase reporter assay. miR-14 was screened against wild-type and mutated 3′ UTRs to determine if the repressive activity of miR-14 requires a direct interaction between miR-14 and the predicted binding sites within the 3′ UTRs of the candidate target genes. Mutating the miR-14 binding sites relieved repression. **P < 0.001 and *P < 0.05.
Overexpression of miR-14 can down-regulate endogenous levels of Hh signaling pathway genes
To determine whether miR-14 affects Hh signaling in vivo, we focused on the Drosophila wing, where Hh regulates both tissue patterning and growth. Cells in the posterior (P) compartment of the developing wing disc express the secreted ligand Hh and induce ptc expression in the anterior (A) compartment to establish the A-P boundary. Expression of Hh and Smo is highest in the P compartment, whereas Ptc is exclusively expressed in the A compartment at the A-P boundary. Since the hh, smo and ptc 3′ UTRs are sensitive to miR-14 expression, we hypothesized that overexpressing miR-14 in the compartments where these target genes are endogenously expressed should phenocopy their loss of function.
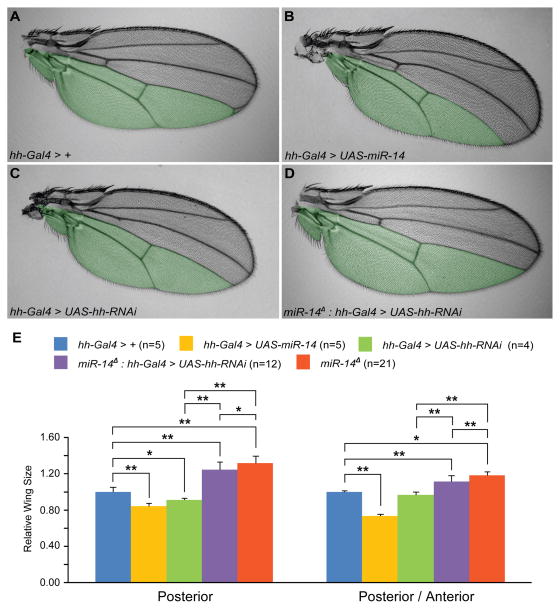
Reduction of Hh expression in the P compartment by RNAi significantly decreased wing size, evident by the slight curving of the wing towards the P compartment and by the quantification of wing size (Figure 3C and 3E). This phenotype is likely due to decreased Decapentaplegic (Dpp) signaling, a downstream target of the Hh signaling that regulates proliferation in both A and P compartments. Consequently, we also observed reduction in the A compartment, resulting in similar P/A compartment ratio to wild-type control (Figure 3E). Interestingly, overexpression of miR-14 in the P compartment caused a significantly greater P compartment wing size defect (Figure 3B and 3E). One possibility is that, in addition to Hh, overexpression of miR-14 in the P compartment represses factors that are required for Hh signaling. Alternatively, factors that are required for responding to Dpp signaling may also be target of miR-14. The wing size defect observed with loss of Hh expression by RNAi was restored when endogenous miR-14 was removed (Figure 3D), demonstrating that endogenous miR-14 likely targets Hh and not Ptc and Smo because Ptc is not expressed in the P compartment, and, while Smo levels are highest in the P compartment, RNAi against smo in the P compartment does not alter the shape of the wing since Smo activity is mainly required at the A-P boundary and A compartment (Figure S2H) (Blair and Ralston, 1997). Interestingly, removal of endogenous miR-14 in the presence of hh-RNAi significantly increased the overall size of the P compartment, further suggesting that additional targets of miR-14 required for Hh and/or Dpp signaling may exist (Figure 3E). However, overall increase in the size of the P compartment was significantly less when compared to miR-14 loss-of-function allele (Figure 3E), suggesting that regulation of Hh signaling by miR-14 is partly to blame for the increase in overall wing size.
Figure 3. miR-14 can modulate wing size by regulating Hh signaling.
Area shaded in green (A–D) marks the region where hh-Gal4 is active. (A) Expression of hh-Gal4 in the wild-type background. (B) Overexpression of miR-14 in the posterior compartment causes curvature of the wing. (C) RNAi against hh in the posterior compartment phenocopies overexpression of miR-14 suggesting that miR-14 modulates Hh activity. (D) Removal of endogenous miR-14 restores the curved wing phenotypes observed in (C). (E) Quantification of P compartment wing size and ratio of P compartment to the A compartment. Reduction of hh expression by RNAi and overexpression of miR-14 significantly reduces the wing size of the P compartment. n=number of wings quantified. **P < 0.001 and *P < 0.05.
Ptc expression is highest at the A-P boundary and RNAi against ptc elevates Hh activity in that region resulting in an increased distance between the L3 and L4 wing veins (Figure S2D–D′). Strikingly, overexpression of miR-14 exhibited the opposite phenotype, decreasing the distance between the L3 and L4 veins, resembling instead decreased Hh signaling (Figure S2E–F′). One possible explanation is that since several rows of cells near the P compartment at the A-P boundary also express Smo, overexpression of miR-14 in this region may inhibit both ptc and smo. Given that Ptc acts upstream of Smo, overexpression of miR-14 will result in an overall decrease in Hh signaling activity. Consistent with this, overexpression of miR-14, as observed with ptc-RNAi, in the A-P region partially reduces the distance between the L3 and L4 veins (Figure S2G–G′), and RNAi against smo in the A-P boundary results in a clear decrease in the L3 and L4 intervein region (Figure S2I). In addition, overexpressing miR-14 in the A compartment also reduced the overall size of the A compartment causing the wing to curve in the anterior direction (Figure S2J). A likely model is that miR-14 down-regulates Smo levels in the A compartment, thus preventing activation of the Hh pathway and resulting in a smaller A compartment. In addition, overexpression of miR-14 in the entire wing decreases the size of the wing (Figure S2C). These results collectively suggest that overexpression of miR-14 in the wing leads to an overall reduction of Hh signaling.
To further evaluate the effect of miR-14, we examined directly the protein levels of endogenous Hh, Ptc, and Smo when miR-14 was overexpressed. In wild-type wing discs, levels of Hh, Ptc and Smo are identical in the dorsal (D) and ventral (V) compartments (Figure 4A–C). However, when miR-14 is overexpressed in the D compartment using ap-Gal4, the levels of Hh and Smo were significantly reduced (Figure 4A–A‴ and 4C–C‴), indicating that miR-14 can suppress the expression of endogenous Hh and Smo. Interestingly, we observed little or no changes in the levels of Ptc (Figure 4B–B‴). One possible explanation is that since ptc is also a downstream target of Hh signaling, reducing the levels of Ptc by overexpressing miR-14 will result in increased expression of ptc which nullifies the repression by miR-14. Another possibility is that miR-14 may weakly regulate Ptc in contrast to Hh and Smo. In fact, we observed similar results when all three 3′ UTR sensors were treated with miR-14. The ptc sensor was less sensitive to miR-14 treatment compared to both the hh and smo sensors (Figure 2B). We favor the later explanation since the repression of Smo was also weaker when compared to Hh (Figure 4A–A‴ and 4C–C‴) and similar results were also observed with their respective sensors (Figure 2B). Collectively, our results suggest that overexpression of miR-14 can suppress the expression of endogenous Hh, Ptc and Smo at varying levels.
Figure 4. Overexpression of miR-14 reduces the expression of endogenous Hh signaling pathway genes.
(A–D‴) Hh, Ptc, Smo and Dpp-lacZ stainings in Drosophila third instar larva wing discs. Wing discs are oriented dorsal (D) up, ventral (V) down, anterior (A) left, and posterior (P) right. (A–D) Wild–type discs stained for Hh, Ptc, Smo and Dpp-lacZ. (A′–D′) Expression of miR-14 induced in the dorsal region of the wing disc using the ap-Gal4 driver represses the expression of endogenous Hh, Ptc and Smo and the Hh target gene, Dpp. (A″–D″) GFP expression marks the region where ap-Gal4 is active and miR-14 expression is induced. (A‴–D‴) Magnified view of the boxed area in (A′–D′). Red dotted lines mark the ap-Gal4 boundary.
Since overexpression of miR-14 can inhibit both positive (Hh and Smo) and negative (Ptc) regulators of the Hh pathway at varying levels, we analyzed the effect of miR-14 overexpression on Hh pathway signaling output using Dpp expression as readout. Strikingly, Dpp expression was significantly reduced (Figure 4D–D‴), demonstrating that overexpression of miR-14, which can inhibit the endogenous expression of both positive and negative regulators of the Hh pathway, overall exerts a negative effect on Hh signaling activity when expressed at high levels.
Endogenous miR-14 regulates Hh levels
Since overexpression of miR-14 down-regulates protein levels of endogenous Hh, Ptc and Smo, we tested whether endogenous miR-14 also represses these genes in vivo and therefore represent physiological targets. A previous study using a miR-14 sensor has shown that miR-14 is ubiquitously expressed throughout the entire wing disc and that miR-14 homozygous mutants are viable (Varghese et al., 2010). To determine whether miR-14 mutants display any wing phenotypes suggestive of Hh signaling deregulation, we compared the adult wing area of miR-14 mutant flies and found that miR-14 mutant wings are ~18% larger than wild-type (Figure 5A–B). To test whether the increased wing size is due to elevated Hh activity, we reduced Hh level by RNAi in the miR-14 mutant background. Reducing the amount of Hh activity in the mutants partially, but significantly, restored wing size (Figure 5B), suggesting that increased Hh signaling in miR-14 mutants is at least in part responsible for the increased wing size.
Figure 5. Endogenous miR-14 modulates Hh expression.
(A) Wild-type wing (blue) superimposed with miR-14 mutant wing (magenta). (B) Quantification of wing size from wild-type and miR-14 mutants. Wing size defect is partially, but significantly restored when hh levels are reduced by RNAi. **P < 0.001. (C) Severity of hhMrt wing phenotypes displayed in three classes, from mild to strong. Arrowhead marks the region where sensory bristles are missing. L2 and L3 mark the two major veins of the wing. (D) Distribution of hhMrt wing phenotypes. “n” denotes the number of wings counted for each genotype; in parenthesis is the percentage of wings showing the phenotype. Absence of miR-14 enhances both class II and III hhMrt phenotypes (X2=9.27, *P<0.01). Significance calculated using a chi-square test for comparing frequencies.
Next, using a gain-of-function allele of hh called Moonrat (hhMrt), which primarily affects the anterior region of the wing (Felsenfeld and Kennison, 1995), we asked whether loss of miR-14 could enhance the Mrt wing defects. For quantification purposes, we assigned mutant wing phenotypes to three different classes based on their severity. In class I Mrt phenotype, distal anterior region of the wing is slightly expanded with wing vein L2 partially duplicated. In class II, the distal anterior region of the wing is expanded and rounded. The L2 wing vein is frequently duplicated proximally and absent distally while L3 vein is thickened distally. In class III, anterior compartment is almost completely rounded and all phenotypes observed in class II are present, but are more severe; often, both L2 and L3 veins are elaborately broadened. These are also associated with frequent loss of sensory bristles in the anterior region of the wing (Figure 5C). Removing both copies of miR-14 partially, but significantly, enhanced the frequency of class II and III Mrt phenotypes (Figure 5D), suggesting that endogenous miR-14 prevents further enhancement of the Hh activity of hhMrt.
miR-14 ensures correct number of terminal cells in the tracheal system by regulating Hh activity
To determine whether miR-14 can regulate Hh signaling in tissues other than the wing, we examined the Drosophila larval tracheal system where Hh signaling plays an important role in determining terminal cell fates (Glazer and Shilo, 2001). In the dorsal tracheal branch, which typically consists of five or six cells, one cell at the branch tip adopts the terminal cell fate marked by the expression of the Serum Response Factor (SRF) (Guillemin et al., 1996). In addition, a second cell at the branch tip adopts a fusion cell fate and mediates fusion of tracheal branches from the contralateral branches at the dorsal midline (Samakovlis et al., 1996a; Samakovlis et al., 1996b). The two terminal cells from each side then branch extensively to deliver oxygen to neighboring tissues (Samakovlis et al., 1996a; Samakovlis et al., 1996b) (Figure 6A).
Figure 6. miR-14 regulates Hh signaling in the larval tracheal system.
(A) Dorsal branches of wild-type third instar larva trachea under bright field illumination. (B) Dorsal branches of miR-14 mutants. In wild-type, the two dorsal branches fuse and give rise to two terminal cells with multiple terminal branches, while miR-14 mutants show excess terminal cells. (A′–B′) Terminal cells are marked using the terminal cell specific Gal4 driver, SRF-Gal4, driving expression of EGFP. (A″–B″) Merged images of dorsal branches and labeled terminal cells. (C) Quantitative analysis of terminal cell numbers in labeled genotypes. miR-14 mutants exhibit more frequent instances of excess terminal cells, as observed when Hh is overexpressed (btl-Gal4 > UAS-hh). Removing one functional allele of hh (miR-14Δ : hhAC / +) or RNAi against hh (miR-14Δ : hh-Gal4 / UAS-hh-RNAi) in the miR-14 mutant background restores the proper number of terminal cells. n= the number of dorsal branch pair examined. *P < 0.05.
Animals with excess levels of Hh signaling exhibit extra SRF-expressing cells that appear to arise from the branch cells located after the fusion cell (Glazer and Shilo, 2001). Since miR-14 regulates the expression levels of Hh in the wing, we tested whether miR-14 mutant larvae have defects in the number of terminal cells in the dorsal tracheal branches. Strikingly, miR-14 mutant larvae possess extra SRF expressing cells (Figure 6B-B″), and expression of UAS-miR-14, using the trachea specific Gal4 driver, btl-Gal4, in miR-14 mutant larvae, rescued the excess terminal cell phenotype (Figure 6C). Next, we tested whether the excess in terminal cells in miR-14 mutant larvae results from an increase in Hh signaling activity. To reduce Hh activity, we either introduced the hh loss of function allele, hhAC, or induced RNAi against hh in the miR-14 mutant background and quantified the number of terminal cells. Strikingly, in both hhAC heterozygotes and hh-RNAi animals, a reduction of excess terminal cells was observed in miR-14 mutant larvae (Figure 6C), indicating that regulation of Hh signaling by miR-14 is critical for determining the correct number of terminal cells. Collectively, our results from both the wing and the tracheal system indicate that miR-14 regulates Hh signaling, a mechanism that may also extend to other tissues.
hh is a physiological target of endogenous miR-14
To further investigate the regulation of target genes by endogenous miR-14, we measured the endogenous protein levels of Hh, Ptc and Smo from whole pupae extracts. Hh levels were significantly elevated in miR-14 mutants, while Ptc and Smo levels remained relatively unchanged (Figure 7A–B), suggesting that endogenous miR-14 primarily functions to dampen Hh signaling by modulating Hh expression, but not Ptc or Smo.
Figure 7. Hh is a physiological target of miR-14.
(A) Western blot analysis of miR-14 mutant pupae. Absence of miR-14 results in elevated Hh protein levels, while Ptc and Smo levels remain relatively unaffected. α-Tubulin was used as loading control. (B) Quantitative analysis of the Western blot from (A). (C) Quantitative PCR analysis from miR-14 mutant pupae. Levels of three Hh pathway genes hh, ptc and smo are up-regulated in miR-14 mutants. Target genes of Hh signaling wg and dpp, and their respective target genes are also up-regulated in miR-14 mutant pupae. Levels of EcR, a validated target of miR-14, and its downstream target genes, E74, E93 and Fbp1, are elevated. **P < 0.001 and *P < 0.05. (E) Model showing regulation of hh signaling pathway by miR-14. Genes labeled in green or red represent predominantly positive or negative regulators of the pathway, respectively. (Left panel) During normal development, miR-14 regulates Hh signaling by buffering Hh levels. Physiological regulation of both Ptc and Smo might also exist, yet no such evidence has been identified in our current work. (Middle Panel) Overexpression of miR-14 results in strong repression of Hh as well as its apparent non-physiological targets, Ptc and Smo, resulting in decreased Hh signaling. (Right panel) In the absence of miR-14, Hh levels increase resulting in increased Hh signaling.
Since miRNAs can also destabilize their target mRNAs, we checked the transcript levels of hh, ptc and smo from total RNA of whole pupae. As expected, hh transcripts were elevated in miR-14 mutants (Figure 7C). Interestingly, ptc mRNAs increased by nearly three-fold in the mutants while smo levels were increased by twofold (Figure 7C). Since ptc is also a direct target of the Hh pathway, one possibility is that increased Hh signaling activity in miR-14 mutants caused an increase in ptc transcript levels. Although it is unclear why an increase in ptc and smo transcript levels do not translate into more protein levels in miR-14 mutants, an alternative mechanism regulating Ptc and Smo proteins production in miR-14 mutants may exist. Alternatively, increase in both ptc and smo transcript levels might be an indirect effect of removing endogenous miR-14. It is possible, given the ability of miRNAs to regulate many different processes, that miR-14 indirectly regulates the expression of ptc and smo.
We also measured the expression levels of Hh target genes, wg and dpp, and found that both genes were increased in the miR-14 mutants (Figure 7C). To investigate whether increase in wg and dpp levels leads to hyperactivation of the Wg and Dpp signaling pathways, we examined the expression levels of the Wg target gene, senseless, and Dpp target genes, spalt and omb. As expected, the levels of all three target genes were elevated in the miR-14 mutant (Figure 7C). While it is possible that all three genes are also physiological targets of miR-14, it is most likely an indirect result of increased Hh signaling in the miR-14 mutant since both Wg and Dpp pathways are downstream of Hh signaling. These results collectively show that endogenous miR-14 maintains the proper balance of Hh signaling activity by primarily regulating Hh expression, a physiological target, but not Ptc and Smo, supporting the model that they likely represent apparent non-physiological targets (Figure 7D).
Discussion
Previous miRNA gain-of-function studies using specific phenotypes and pathway sensors as readouts generated many interesting phenotypes and identified several candidate miRNAs in specific signaling pathways (Bejarano et al., 2012; Silver et al., 2007; Szuplewski et al., 2012). However, these approaches faced difficulties in identifying biologically significant targets. Here, we designed a fast and efficient approach to identify miRNA targets, whereby rather than studying the function of individual miRNAs, we screen for all possible targets of all miRNAs in a given signaling pathway. To our knowledge, this is the first study that has used a genome-wide collection of Drosophila miRNAs to screen for potential target genes among specific components of a signaling pathway. Using luciferase as readout, we were able to quickly and easily measure the effect that different miRNAs have on the genes being interrogated. This systematic miRNA screening platform can be used to elucidate miRNA-target relationships for genes in various other processes.
Systematic performance evaluation of the LMF scores by analyzing the TPR and FPR allowed us to identify an appropriate cutoff value at which we achieved 33% TPR and 3% FPR. Our cutoff was stringent enough that previously identified interactions between miR-12 and Costal-2 (Cos2) and miR-283 and Fu were included, but interactions with smo were excluded (Friggi-Grelin et al., 2008). Evaluation of our screen results to three of the popularly used prediction algorithms demonstrated that there is a positive correlation between the LMF scores and the number of miRNA-target predictions made. Furthermore, our screen revealed 23 novel miRNA-target interactions, representing ~39% of the total miRNA-target interactions. Interestingly, the miR-14-hh interaction was predicted by only one prediction algorithm, whereas, miR-14-ptc and miR-14-smo interactions were identified by multiple algorithms. Furthermore, miR-14-hh interaction was only predicted when the least stringent cutoff value was used. The miTG score assigned by DIANA for miR-14-hh is 0.212, which is much lower than the high confidence miTG score of ≥ 0.5. This result further highlights the limitation of the existing algorithms and the need for functional tests.
We identified and characterized in detail miR-14 that can regulate three components of the Hh pathway, hh, ptc and smo. We have shown that miR-14 overexpression can decrease the levels of all three proteins at varying levels and cause an overall decrease in Hh signaling as evidenced by the reduction of Dpp expression in the wing imaginal discs and by the adult wing outgrowth. We have also presented evidence that this regulation is mediated through the direct binding of miR-14 seed sequences to the miR-14 MREs located within the 3′ UTRs of all three genes. We have also presented data demonstrating that hh is a physiological target of miR-14, while ptc and smo appear to be non-physiological targets. However, there are several alternative explanations that might impede us from categorizing both ptc and smo as physiological targets. For example, regulation of Ptc and Smo by miR-14 may be very weak and not detectable by Western blot analysis. In fact, the effect of an individual miRNA on a protein target level tends to be subtle, usually less than 2 folds (Baek et al., 2008). In support of this explanation, the amount of repression elicited by miR-14 on both ptc and smo 3′ UTR sensors and Ptc and Smo levels in the wing discs are far less robust compared to Hh. Alternatively, additional miRNAs may act in a combinatorial manner and down-regulate Ptc and Smo even in the absence of miR-14. In fact, our screen identified several other miRNAs that can also reduce the ptc and smo sensor levels. Finally, since Hh negatively regulates Ptc levels, derepression of Ptc in miR-14 mutants maybe nullified by the increased levels of Hh.
While miR-14 is well conserved in distantly related Drosophila species, it is missing in vertebrates such as Zebrafish and humans. Nevertheless, regulation of Hh signaling by miRNAs seems to be conserved across diverse species. For example, Suppressor of fused (Su(fu)) is targeted by miR-214 in Zebrafish to enable precise specification of muscle cell types by sharpening cellular responses to Hh (Flynt et al., 2007). In non-small cell lung cancer cell line, miR-212 acts as oncogene by targeting PTCH1, human homolog of Drosophila Ptc, to increase cell proliferation, migration and invasion (Li et al., 2012), while in chronic myeloid leukemia (CML), miR-326 targets Smo to decrease cell proliferation and increase apoptosis (Babashah et al., 2013).
Although, ptc and smo seem to be apparent non-physiological targets of miR-14, this information is valuable since dysregulation of miRNAs are common features of many diseases. In addition, given that miRNA replacement therapies are currently being tested as possible treatments for various diseases, information regarding apparent non-physiological targets becomes very important to prevent unnecessary side effects. Therefore, considering the importance of identifying both physiological and apparent non-physiological targets, we propose that our fast and efficient approach of identifying miRNA targets can help understand the complex gene regulatory network of miRNAs.
Experimental Procedures
DNA constructs and cloning
Reporter plasmids were constructed by cloning firefly luciferase into the KpnI/EcoRI site of pAc5.1/V5-His C (Invitrogen). Next, the 3′ UTR of each target gene was cloned into the EcoRI/SacI site, with the exception of Fu that was cloned into the NheI/SacI site. The 3′ UTR for individual genes was cloned based on annotated sequences provided by Flybase (www.flybase.org). A complete list of 3′ UTR primers can be found in Table S1.
Most miRNA overexpression plasmids used to prepare the screening platform have been previously described (UAS-dsRed-miRNA collection described in (Bejarano et al., 2012). To expand coverage, we prepared 33 additional miRNAs overexpression plasmids by amplifying ~400-700 nt fragments encompassing miRNA genes from genomic DNA and cloned them into pAc5.1/V5-His C. Altogether, the resource comprises 128 miRNA overexpression plasmids that covers 132 distinct miRNAs (Table S2). Note that we chose to clone the new miRNAs into a vector under the control of constitutively active Actin promoter rather than UAS to minimize the number of plasmids needed for transfection. Prior to constructing the additional miRNA constructs, we compared the knockdown efficiency of senseless 3′ UTR reporter, which has been previously shown to be regulated by miR-9a (Li et al., 2006), using both pAc and UAS vectors. Both pAc-miR-9a and UAS-dsRed-miR-9a overexpression plasmids were equally effective at reducing the luciferase level of senseless 3′ UTR reporter (Figure S3).
A complete list of miRNAs can be found in Table S2. Primer sequences will be provided upon request. The screening platform will be publicly available at the Drosophila RNAi Screening Center (DRSC).
To mutate the seed sequence of miR-14 and potential MREs in the 3′ UTRs of target genes, we followed the instructions of QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene).
Luciferase reporter assay
Drosophila S2R+ cells were maintained in Schneider’s Drosophila Medium (Gibco) with 10% heat-inactivated FBS (Sigma) and 1% Pen/Strep (Gibco) at 25°C. Experiments were performed in 96 well plates excluding the outer wells. The wells were seeded with 20 ng of plasmid expressing pAc-miRNAs or 20 ng of plasmid expressing UAS-dsRed-miRNAs and 5 ng of Actin-Gal4 plasmid prior to the start of the experiment. Each well was transfected with 5 ng of firefly luciferase reporter plasmid and 5 ng of Renilla luciferase reporter plasmid for transfection control. Transfection was performed using Effectene Transfection Reagent (Qiagen). After 72 hrs, luciferase activities were measured using DualGlo (Promega).
Computing miRNA-target interaction score
We computed the normalized fold change value x for the given miRNA i and 3′ UTR region of gene j as follows:
Where Sij is normalized firefly signal (ratio of firefly/Renilla luciferase levels) from the tested pair of miRNA i and 3′ UTR of gene j, Uj is signal from 3′ UTR control (lacking miRNA) and Mi is signal from miRNA control (lacking 3′ UTR). C is the signal from control without both 3′ UTR and miRNA. Next, we computed the median of fold change values (x̃) from the three replicates. The negative log2 median fold-change score (LMF) is computed as:
Integrative analysis with predicted miRNA-target network
To compare the LMF score with predicted miRNA-target, we collected the potential miRNAs that could regulate the Hh pathway members from TargetScan (http://www.targetscan.org/) (Ruby et al., 2007), miRanda (http://www.microrna.org/microrna/home.do) (Betel et al., 2010) and DIANA (http://diana.cslab.ece.ntua.gr/) (Paraskevopoulou et al., 2013; Reczko et al., 2012) databases. For all three databases, least cutoff values were used to extract all possible miRNA-target relations and each extracted pair was associated with respective scores assigned by the tools (Branch-Length score for TargetScan, mirSVR score for miRanda and miTG score for DIANA). The trend line was fitted using the linear regression model (LMF ~ Prediction score) implemented in R (http://www.r-project.org/). All the integrative analyses were performed using in-house developed perl scripts.
Validation of LMF score performance
In order to choose the LMF cutoff value, we created a positive reference set (PRS) and a random reference set (RRS). The PRS includes 24 miRNA-target gene (member of Hh pathway) interactions curated from literature (6 pairs) and high confidence predictions (18 pairs) that overlap with our screening space. High confidence miRNA-target interactions refer to the pairs that are predicted by 2 or more independent tools as high confidence interactions (TargetScan: Branch-Length score ≥ 0.8; miRanda: mirSVR score ≤ −0.5; DIANA: miTG score ≥ 0.5). To construct RRS, we first compiled a list of 736 potential non-interacting pairs from the screening space (1160 possible pairs) that are not overlapping with PRS and not predicted as miRNA-target by any of the tools even with the least stringency cutoff. From this potential non-interacting pairs, we randomly sampled 1000 RRS sets (size of each RRS is equal to the size of PRS). We analyzed the true positive rate (TPR) and false positive rate (FPR) values for various LMF score cutoff values. For a given LMF score cutoff value i, the TPR and FPR are computed as follows:
TPi, FNi, FPi and TNi correspond to true positive, false negative, false positive and true negative values at given LMF cutoff value i respectively. The Hh miRNA-target network was constructed using at chosen cutoff value (LMF ≥ 0.62). The network is visualized using Cytoscape software (http://www.cytoscape.org/).
Immunostaining, confocal imaging, and analysis
Immunostainings of larval wing imaginal discs were performed as previously described (Belenkaya et al., 2004). Primary antibodies were: mouse anti-Ptc (1:40; DSHB, Apa-1), mouse anti-Smo (1:50; DSHB, 20C6), rabbit anti-Hh (1:50; kindly provided by Dr. Xinhua Lin), and rabbit anti-β-Gal (1:1000; Cappel). Primary antibodies were detected by anti-mouse or anti-rabbit secondary antibodies conjugated to Alexa-Fluor 594 and 647 (1:1000; Invitrogen).
Fluorescent images were acquired with a Leica TCS SP2 AOBS. Images were processed using Adobe Photoshop.
Tracheal terminal branch imaging and analysis
Third instar larvae were heat killed (70°C for 10–15 s), mounted in 50% glycerol and examined under a Zeiss Axioskop 2 compound fluorescence microscope. Average values and their corresponding standard deviations were calculated, and t-test analysis was performed, using Microsoft Excel.
Supplementary Material
Highlights.
Construction of a genome-wide Drosophila miRNA screening platform
Systematic method to identify miRNA targets
Identification of physiological and apparent non-physiological targets of miRNAs
miR-14 modulates hedgehog signaling
Acknowledgments
We thank Drs. M. Krasnow, X. Lin and M. Scott for reagents. We thank the Transgenic RNAi Resource Project and the Bloomington Stock Center for flies, the Developmental Studies Hybridoma Bank for monoclonal antibodies, and the Drosophila RNAi Screening Center (Harvard Medical School) for plate-reader equipment. We also thank C. Pitsouli, Y. Kwon, D. Yan, and R. Binari for critical comments on the manuscript and reagents. This work was supported by the National Institute of Health (5P01CA120964 and 5R01DK088718). N.P is a Howard Hughes Medical Institute investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25:3049–3055. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- Babashah S, Sadeghizadeh M, Hajifathali A, Tavirani MR, Zomorod MS, Ghadiani M, Soleimani M. Targeting of the signal transducer Smo links microRNA-326 to the oncogenic Hedgehog pathway in CD34 CML stem/progenitor cells. Int J Cancer. 2013 doi: 10.1002/ijc.28043. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat MT, Humke EW, Scott MP. Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends Mol Med. 2010;16:337–348. doi: 10.1016/j.molmed.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano F, Bortolamiol-Becet D, Dai Q, Sun K, Saj A, Chou YT, Raleigh DR, Kim K, Ni JQ, Duan H, et al. A genome-wide transgenic resource for conditional expression of Drosophila microRNAs. Development. 2012;139:2821–2831. doi: 10.1242/dev.079939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenkaya TY, Han C, Yan D, Opoka RJ, Khodoun M, Liu H, Lin X. Drosophila Dpp morphogen movement is independent of dynamin-mediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell. 2004;119:231–244. doi: 10.1016/j.cell.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair SS, Ralston A. Smoothened-mediated Hedgehog signalling is required for the maintenance of the anterior-posterior lineage restriction in the developing wing of Drosophila. Development. 1997;124:4053–4063. doi: 10.1242/dev.124.20.4053. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Pepicelli CV, Dibble CC, Catbagan W, Zarycki JL, Laciak R, Gipp J, Shaw A, Lamm ML, Munoz A, et al. Hedgehog signaling promotes prostate xenograft tumor growth. Endocrinology. 2004;145:3961–3970. doi: 10.1210/en.2004-0079. [DOI] [PubMed] [Google Scholar]
- Felsenfeld AL, Kennison JA. Positional signaling by hedgehog in Drosophila imaginal disc development. Development. 1995;121:1–10. doi: 10.1242/dev.121.1.1. [DOI] [PubMed] [Google Scholar]
- Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friggi-Grelin F, Lavenant-Staccini L, Therond P. Control of antagonistic components of the hedgehog signaling pathway by microRNAs in Drosophila. Genetics. 2008;179:429–439. doi: 10.1534/genetics.107.083733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer L, Shilo BZ. Hedgehog signaling patterns the tracheal branches. Development. 2001;128:1599–1606. doi: 10.1242/dev.128.9.1599. [DOI] [PubMed] [Google Scholar]
- Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemin K, Groppe J, Ducker K, Treisman R, Hafen E, Affolter M, Krasnow MA. The pruned gene encodes the Drosophila serum response factor and regulates cytoplasmic outgrowth during terminal branching of the tracheal system. Development. 1996;122:1353–1362. doi: 10.1242/dev.122.5.1353. [DOI] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Kayed H, Kleeff J, Keleg S, Guo J, Ketterer K, Berberat PO, Giese N, Esposito I, Giese T, Buchler MW, et al. Indian hedgehog signaling pathway: expression and regulation in pancreatic cancer. Int J Cancer. 2004;110:668–676. doi: 10.1002/ijc.20194. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006;4:9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang D, Chen C, Ruan Z, Huang Y. MicroRNA-212 displays tumor-promoting properties in non-small cell lung cancer cells and targets the hedgehog pathway receptor PTCH1. Mol Biol Cell. 2012;23:1423–1434. doi: 10.1091/mbc.E11-09-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Maragkakis M, Vergoulis T, Alexiou P, Reczko M, Plomaritou K, Gousis M, Kourtis K, Koziris N, Dalamagas T, Hatzigeorgiou AG. DIANA-microT Web server upgrade supports Fly and Worm miRNA target prediction and bibliographic miRNA to disease association. Nucleic Acids Res. 2011;39:W145–148. doi: 10.1093/nar/gkr294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. Functional microRNA targets in protein coding sequences. Bioinformatics. 2012;28:771–776. doi: 10.1093/bioinformatics/bts043. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz i Altaba A. Gli proteins and Hedgehog signaling: development and cancer. Trends Genet. 1999;15:418–425. doi: 10.1016/s0168-9525(99)01840-5. [DOI] [PubMed] [Google Scholar]
- Samakovlis C, Hacohen N, Manning G, Sutherland DC, Guillemin K, Krasnow MA. Development of the Drosophila tracheal system occurs by a series of morphologically distinct but genetically coupled branching events. Development. 1996a;122:1395–1407. doi: 10.1242/dev.122.5.1395. [DOI] [PubMed] [Google Scholar]
- Samakovlis C, Manning G, Steneberg P, Hacohen N, Cantera R, Krasnow MA. Genetic control of epithelial tube fusion during Drosophila tracheal development. Development. 1996b;122:3531–3536. doi: 10.1242/dev.122.11.3531. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Hagen JW, Okamura K, Perrimon N, Lai EC. Functional screening identifies miR-315 as a potent activator of Wingless signaling. Proc Natl Acad Sci U S A. 2007;104:18151–18156. doi: 10.1073/pnas.0706673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuplewski S, Kugler JM, Lim SF, Verma P, Chen YW, Cohen SM. MicroRNA transgene overexpression complements deficiency-based modifier screens in Drosophila. Genetics. 2012;190:617–626. doi: 10.1534/genetics.111.136689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- Varghese J, Lim SF, Cohen SM. Drosophila miR-14 regulates insulin production and metabolism through its target, sugarbabe. Genes Dev. 2010;24:2748–2753. doi: 10.1101/gad.1995910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.